Abstract
Despite similar structures and DNA binding profiles, two recently synthesized dinuclear platinum compounds are shown to elicit highly divergent effects on cell cycle progression. In colorectal HCT116 cells, BBR3610 shows a classical G2/M arrest with initial accumulation in S phase, but the derivative compound BBR3610-DACH, formed by introduction of the 1,2-diaminocyclohexane (DACH) as carrier ligand, results in severe G1/S as well as G2/M phase arrest, with nearly complete S phase depletion. The origin of this unique effect was studied. Cellular interstrand crosslinking as assayed by comet analysis was similar for both compounds, confirming previous in vitro results obtained on plasmid DNA. Immunoblotting revealed a stabilization of p53 and concomitant transient increases in p21 and p27 proteins after treatment with BBR3610-DACH. Cell viability assays and cytometric analysis of p53 and p21 null cells indicated that BBR3610-DACH-induced cell cycle arrest was p21-dependent and partially p53-dependent. However, an increase in the levels of cyclin E was observed with steady state levels of CDK2 and Cdc25A, suggesting that the G1 block occurs downstream of CDK/cyclin complex formation. The G2/M block was corroborated with decreased levels of cyclin A and cyclin B1. Surprisingly, BBR3610-DACH-induced G1 block was independent of ATM and ATR. Finally, both compounds induced apoptosis, with BBR3610-DACH showing a robust PARP-1 cleavage that was not associated with caspase-3/7 cleavage. In summary, BBR3610-DACH is a DNA binding platinum agent with unique inhibitory effects on cell cycle progression that could be further developed as a chemotherapeutic agent complementary to cisplatin and oxaliplatin.
Keywords: Dinuclear platinum compounds, interstrand crosslinks, cell cycle, p53, DNA damage response, apoptosis
1. INTRODUCTION
Platinum drugs are a mainstay of cancer therapeutics especially for the treatment of head and neck, testicular, ovarian, lung, and colorectal cancers [1,2]. To date, all clinically useful platinum drugs contain a single Pt (II) center with two exchangeable ligands in cis geometry. Interaction of these drugs with cellular biomolecules such as sulfur-containing glutathione and metallothionein can render them inactive before reaching their pharmacological target, DNA [3,4]. Subsequently, polynuclear platinum complexes (PPCs) that are structurally different from cisplatin, and exhibit a different mode of DNA binding, were developed to circumvent the cellular resistance arising toward the mononuclear compounds, primarily decreased uptake, increased efflux, and increased DNA repair. Especially, structural conformational changes induced by long-range inter and intrastrand crosslinks are distinctly different from those induced by the mononuclear cisplatin and oxaliplatin [5]. The prototype of this class, BBR3464 [{trans-PtCl (NH3)2}2{µ-trans-Pt (NH3)2 (H2N(CH2)6NH2)}]4+ (Fig. 1A), is the only platinum compound without two exchangeable ligands in cis to have reached human Phase II clinical trials [2]. It is cytotoxic in cisplatin-resistant cell lines and shows high efficacy in p53 mutant tumor cells [6]. However, despite promising responses in Phase II clinical trials, BBR3464 had an unacceptably low therapeutic index, possibly due to increased metabolism in human plasma [7,8].
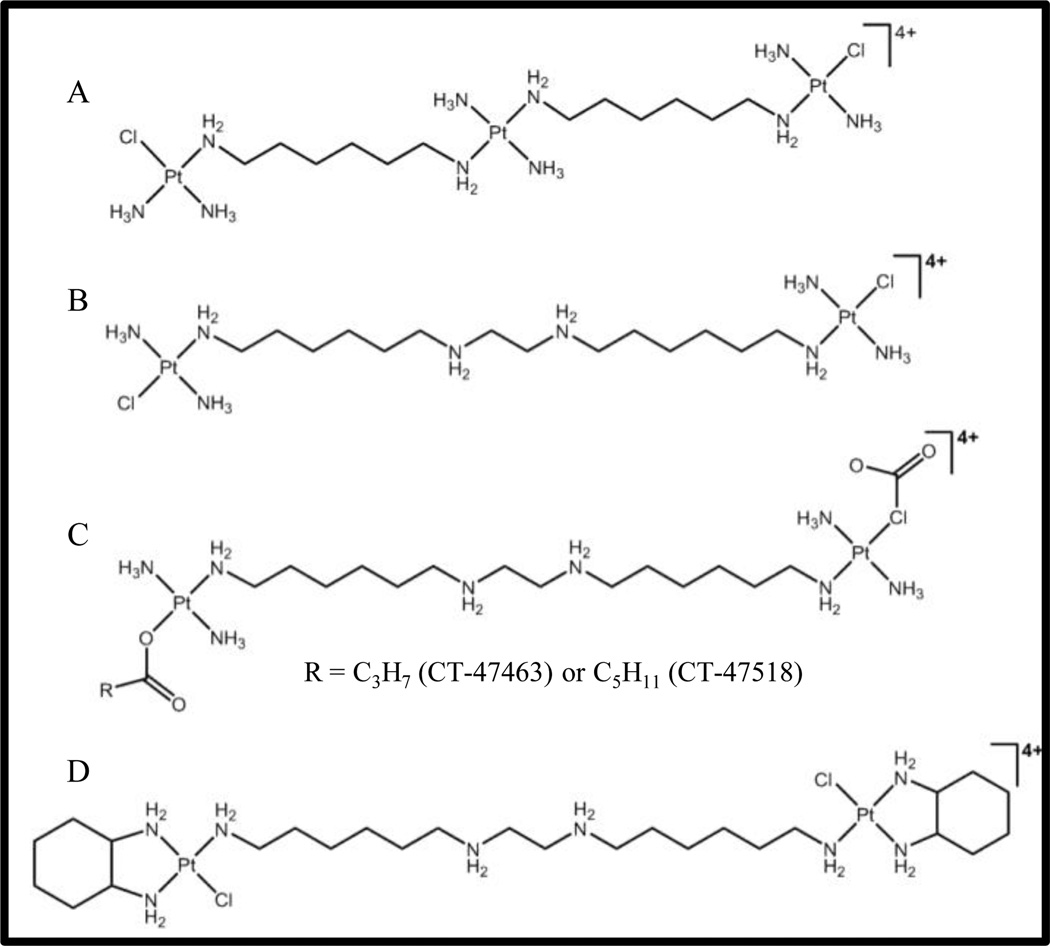
Figure 1. Structures of the polynuclear platinum compounds (PPC) mentioned in the present study.
(A) BBR3464; (B) BBR3610; (C) CT-47463 and CT-47518; (D) BBR3610-DACH
In analyzing the structure-activity relationship of the trinuclear complexes, it was seen that the charged central platinum atom provided H-bonding and an electrostatic pre-association with duplex DNA in the minor groove. Replacement of this central platinum with linear polyamines has yielded second-generation analogues of BBR3464 that retain its biological activity [9]. BBR3610 (Fig. 1B), which was developed by replacing the central platinum with a spermine-like linker, is one of the most cytotoxic platinum compounds, with nanomolar toxicity in gliomas and colon cancer cells [10,11]. However, like its predecessor, BBR3610 was also found to undergo serum biotransformation leading to its degradation[12,13]. This was attributed to the substitution of the Pt-Cl bond by thiol compounds, resulting in bridge cleavage.
To improve metabolic stability and circumvent irreversible plasma protein binding, different analogues of BBR3610 have been developed by modifying either the leaving groups or the carrier ligands. Replacement of the chloride leaving groups in BBR3610 with butyrate or capronate alkylcarboxylates (CT-47518 and CT-47463 respectively, Fig. 1C), improved pharmacokinetic and pharmacodynamic profiles of the parent drug [14]. These compounds were found to overcome resistance due to defects in DNA mismatch repair and were highly effective in cisplatin- and oxaliplatin-resistant cell lines. The use of the 1,2-diaminocyclohexane (DACH) as the carrier ligand gave BBR3610-DACH (Fig. 1D), where the chelating effect of the DACH ring contributed to the increased metabolic stability of BBR3610-DACH in the presence of sulfur-containing compounds at physiological pH [15]. In vitro studies showed this compound formed DNA adducts that persisted longer, escaped DNA repair, and inhibited transcription [16].
Deregulation of cell cycle progression is one of the key contributors toward cancer development [17]. Exposure to DNA damaging agents induces G1/S or G2/M cell cycle arrest by activating cell cycle checkpoint proteins that inhibit cyclin-dependent kinases (CDKs) [18]. The resulting cell cycle arrest can afford an opportunity for DNA repair and have negative effects on apoptosis [19]. Like cisplatin and most of its analogues, BBR3464 and BBR3610 disrupt DNA synthesis, resulting in transient S phase accumulation, followed by arrest in G2. However, as shown below, BBR3610-DACH unexpectedly induced a dramatically different cellular response, consisting of a biphasic cell cycle arrest in G1 and G2, with a complete depletion of cells in S phase. These unusual cellular effects indicate a marked divergence from other PPCs and raise the possibility that BBR3610-DACH may have a unique spectrum and/or mechanism of antitumor activity.
2. MATERIALS AND METHODS
2.1. Chemicals
Cisplatin was obtained from Sigma-Aldrich (St. Louis, MO, USA; Cat# 479306) and dissolved in water. BBR3610 and BBR3610-DACH were synthesized as discussed earlier [12,15]. The stock solutions of platinum compounds were prepared at the concentration of 1 mM in water, stored at −20°C and diluted in complete medium. The ATM inhibitor, KU-60019, was obtained from Selleck Chemicals (Houston, TX, USA). BrdU was obtained from Sigma-Aldrich (Cat# 9285).
2.2. Cell Culture and PPC treatments
The human colorectal cancer cell line, HCT116, and its p53 and p21 knockout derivatives (denoted as p53−/− and p21−/−; kind gift from Dr.Bert Vogelstein) were cultured in RPMI 1640 (Invitrogen, Grand Island, NY, USA; Cat# 11875-093) supplemented with 10% fetal bovine serum (Quality Biologicals, Gaithersburg, MD, USA; Cat# 110-001-101US) and 100 U/mL penicillin and 100 µg/mL streptomycin (Invitrogen, Grand Island, NY, USA; Cat# 15140). The mismatch repair-proficient HCT116+chr3 cell line (a kind gift from Dr. C.Richard Boland, Baylor Health Care System, Dallas, Texas) was cultured in IMDM (Invitrogen, Grand Island, NY, USA; Cat# 12440-053) supplemented with 10% fetal bovine serum and 100 U/mL penicillin and 100 µg/mL streptomycin. Cells were maintained in logarithmic growth as a monolayer in T75 tissue culture flask at 37°C in a 5% CO2 incubator. For the treatment, cells were seeded at a density of 5×105 cells/10 mL medium and treated with 20 µM compound, unless otherwise specified.
2.3. Growth Inhibition Assay
Five thousand cells/well in 100 µL were seeded in a 96-well plate and allowed 24 hr to attach. Cells were then treated with the PPCs for 72 hr. Following PPC removal, the cells were incubated in the presence of 0.5 mg/mL MTT reagent [(3, 4, 5-dimethylthiazol-2-yl)-2, 5-diphenyl tetrazolium bromide; Sigma-Aldrich, St.Loius, MO, USA; Cat# M2128] in RPMI medium for 3 hr at 37°C. The MTT reagent was removed and 100 µL DMSO was added to each well. The plate was then incubated on a shaker at room temperature in the dark. The spectrophotometric reading was taken at 570 nm using a microplate reader.
2.4. Analysis of cell cycle distribution
HCT116, HCT116 p53−/−, HCT116 p21−/−, and HCT116 +chr3 cells were seeded in 100-mm tissue culture dishes at a density of 5×105 cells per dish. After 24 hr, cells were treated with an equimolar dose of the PPCs (20 µM) for 6, 24, and 48 hr. Both attached and floating cells were harvested at the different timepoints. One million cells were then suspended in 1 mL of propidium iodide solution (3.8 mM sodium citrate; 0.05 mg/mL propidium iodide; 0.1% Triton X-100) with added RNaseB at a final concentration of 7 Kunitz units/mL and kept in the dark at 4°C. Cells were then analyzed using flow cytometry.
2.5. Flow Cytometry
A Becton Dickinson (San Jose, CA, USA) FACSCanto II flow cytometer was used for these studies. The argon ion laser set at 488 nm was used as an excitation source. Cells having DNA content of 2N and 4N were designated as being in G1 and G2 phases respectively, with those in between as being in S. Twenty thousand events were acquired for each sample and the data obtained were analyzed using the Modfit software.
2.6. G2/M checkpoint assay
HCT116 cells were incubated with 20 µM BBR3610 or 20 µM BBR3610-DACH for 6, 24, and 48 hr. Cells were then harvested, fixed with 1 % paraformaldehyde followed by 80 % ethanol, and permeabilized with 0.25 % Triton X-100 in PBS for 5 min. Cells were stained with anti phospho-histone H3 antibody (Cell Signaling, Danvers, MA, USA; Cat#3377P), followed by anti-IgG Alexa 647. DNA was stained with propidium iodide in the presence of RNAse B. Phospho-histone H3 positive cells were determined using a FACSCanto II flow cytometer (Becton Dickinson, San Jose, CA, USA). Ten thousand events were acquired for each sample.
2.6. Cell Synchronization studies
To obtain G0 synchronization, HCT116 cells were first seeded at a density of 5×105 cells/10 cm dish and allowed 24 hr to attach. The cells were then serum-starved in medium containing 0.5% FBS for 96 hr. Flow cytometry analysis of synchronized cells showed approximately 80% cells arrested in G0/G1 phase.
2.7. Antibodies
The primary antibodies used are listed in Table 1 and were used at the dilutions recommended by the manufacturer.
TABLE 1.
List of antibodies used in this study
| Antibody | Description | Source |
|---|---|---|
| p53 | Rabbit Polyclonal | Cell Signaling Cat# 9282 |
| p-p53 | Rabbit Polyclonal | Cell Signaling Cat# 9284 |
| p21 | Rabbit Monoclonal | Cell Signaling Clone 12D1, Cat# 2947 |
| p27 | Mouse Monoclonal | Cell Signaling Clone SX53G8, Cat# 3698 |
| ATR | Mouse Monoclonal | Abcam Clone 2B5, Cat# 4471 |
| DNA-PK | Mouse IgG1, κ | BD Pharmingen Clone 4F10C5, Cat# 2348 |
| Phospho-Chk1 (Ser 345) | Rabbit Monoclonal | Cell Signaling Clone 133D3, Cat# 2348 |
| Cdc25A | Rabbit Polyclonal | Santa Cruz Clone 144, Cat# 97 |
| Phospho-Rb (Ser 780) | Rabbit Polyclonal | Cell Signaling, Cat# 9307 |
| CDK2 | Rabbit Polyclonal | Santa Cruz Clone M2, Cat# 163 |
| Cyclin E | Rabbit Polyclonal | Santa Cruz Clone C-19, Cat# 198 |
| Cyclin D1 | Rabbit Monoclonal | Cell Signaling Clone 92G2, Cat# 2978 |
| Cyclin D3 | Mouse Monoclonal | Cell Signaling Clone DCS22, Cat# 2936 |
| Cyclin A | Rabbit Polyclonal | Santa Cruz Clone H-432, Cat# 751 |
| Cyclin B1 | Rabbit Polyclonal | Cell Signaling Cat# 4138 |
| Caspase-8 | Mouse Monoclonal | Cell Signaling Clone 1C12, Cat# 9746 |
| Caspase-9 | Rabbit Polyclonal | Cell Signaling Cat# 9502 |
| Caspase-9 | Rabbit Polyclonal | Cell Signaling Cat# 9502 |
| Caspase-3 | Rabbit Polyclonal | Cell Signaling Cat# 9662 |
| Cleaved Caspase-3 | Rabbit Polyclonal | Cell Signaling Clone 5AE1, Cat# 9664 |
| Caspase-7 | Rabbit Polyclonal | Cell Signaling Cat# 9492 |
| Cleaved PARP | Rabbit Polyclonal | Cell Signaling Cat# 9541 |
| Phospho-histone H3 (S10) | Rabbit Monoclonal | Cell Signaling Clone D2C8, Cat# 3377P |
| β-actin | Rabbit Monoclonal | Cell Signaling Clone 13E5, Cat# 4970 |
2.8. Immunoblotting
Following PPC treatments, both floating and adherent cells were harvested, washed once with ice-cold PBS and pelleted (10,000 rpm, 5 min, 4°C). The pellets were then lysed in 50–200 µL SDS lysis buffer [62.5 mM Tris-HCl, pH 7.5, 5% glycerol, 4% SDS, 4% complete protease inhibitor (Roche, Indianapolis, IN, USA; Cat# 11873580001), 5% BME], passed through 21-gauge needle 10–15 times on ice, and centrifuged (12,000 rpm, 20 min, 4°C). Protein concentrations were determined by the Bradford Assay, resolved on 10%, 12%, or 4–20% gradient polyacrylamide gels, and transferred to PVDF membrane (90 V, 75 min, 4°C). For the detection of ATR in the siRNA knockdown experiment, the proteins were resolved on 5% gel and transferred to PVDF membrane (0.1 A, 7 hr, 4°C). The membrane was blocked in 5% non-fat dry milk in 1X Tris buffered saline containing 0.1% Tween-20 for 60–90 min. The membranes were then probed with the primary antibodies in blocking buffer overnight at 4°C, followed by secondary antibody (anti-rabbit or anti-mouse) conjugated to horseradish peroxidase (Cell Signaling, Danvers, MA, USA or Thermo Scientific, Pittsburgh, PA, USA) for 1 hr at room temperature. Chemiluminescent protein bands were visualized on X-ray films.
2.9. siRNA Transfection
ATR expression was down-regulated using the ON-Target plus Smart Pool, L-003202-00 and Non-targeting siGENOME control siRNA, D-001210-01–05 (Dharmacon, Thermo Scientific, Pittsburgh, PA, USA). 50 nM siRNAs were transfected 24 hr after seeding of HCT116 cells using Dharmafect 1 according to the manufacturer’s instructions.
2.10. BrdU Incorporation Assay
To determine the percentage of cells in the S phase, the cells were treated with PPCs for various times, pulse-labeled with 10 µM BrdU for 3 hr and then fixed in 70% ethanol overnight. Following fixation, the cells were incubated in 2.5 M HCl for 30 min and neutralized in 0.1 M sodium borate for 2 min at room temperature. The cells were then blocked in PBS, 2% BSA, 0.1% Tween-20, RT, 1 hr, incubated with mouse anti-BrdU antibody (BD Biosciences, San Jose, California, ISA) for 1 hr and then probed with the secondary Alexa Fluor 488 rabbit anti-mouse antibody (Molecular Probes, Invitrogen, Grand Island, NY, USA) for 1 hr. Finally, the cells were stained with propidium iodide and analyzed by flow cytometry.
2.11. Alkaline Comet Assay
Single-cell gel electrophoresis or comet assay was carried out using a Trevigen kit to quantitatively assess PPC-induced DNA damage [20]. Briefly, HCT116 cells (1×105cells/mL) were plated in 35 mm or 60 mm dishes and allowed to attach for 24 hr. The cells were then treated with different concentrations of each PPC for 1 hr. The medium was removed, the cells washed once, and further incubated in drug-free medium for 6 hr. The cells were then irradiated with 15 Gy γ-radiation (MDS Nordion Gammacell 40 research irradiator (ON, Canada), with a 137Cs source. Cells were harvested by scraping and resuspended in 1 mL of ice-cold phosphate buffered saline (PBS, pH 7.4). Then, 50 µL of the cell suspension was mixed with 500 µL of low melting agarose at 37°C, and 50 µL of this mixture was spread on comet slides, and solidified in the dark for 30 min at 4°C. The slides were then treated with ice-cold lysis buffer in the dark for 1 hr at 4°C and then incubated in an alkaline solution (pH>13) for 1 hr at room temperature to allow for alkaline unwinding. Electrophoresis was carried out under alkaline conditions at 21 V, 300 mA, and 30 min at 4°C. Slides were washed twice with distilled water, once with 70% ethanol, and then allowed to dry for 10 min at around 37–45°C. The slides were then stained with SYBR green for 5 min at 4°C. Comet images were obtained using a fluorescence microscope (Olympus IX70). The comet analysis was performed using the Comet Score software from TriTek Corporation.
The extent of crosslinking was expressed as the percentage decrease in tail moment and was calculated using the formula:
% Decrease in tail moment = [1 − (TMdi − TMcu)/(TMci − TMcu)] × 100
where, TMdi is the tail moment of the PPC treated irradiated samples, TMci is the tail moment of the untreated irradiated samples, and TMcu is the tail moment of the untreated, unirradiated samples [21]. At least 40 comets were scored for each concentration and the reduction in comet tail length and intensity was apparent within each sample.
2.12 Statistical Analysis
Data are shown as a mean and standard deviation or standard error of the mean from two or three independent experiments. Student’s t-test was used to analyze the significance of difference between different treatments wherever applicable. *p-value < 0.05. **p-value < 0.01, ***p-value < 0.001.
3. RESULTS
3.1. BBR3610-DACH induces interstrand crosslinks in HCT116 cells
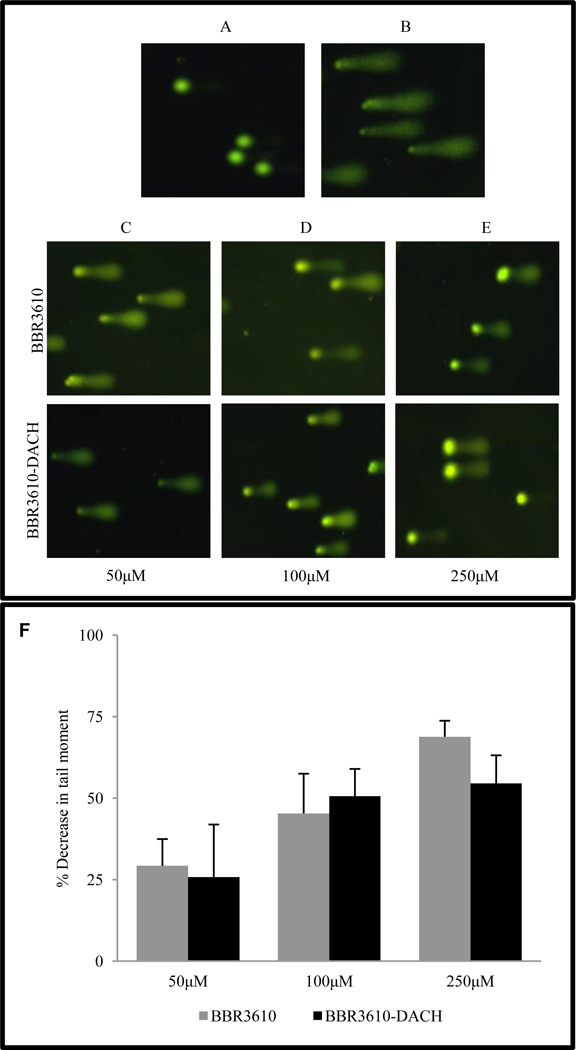
PPCs are known to cause long-range intrastrand and interstrand crosslinks in DNA [5]. We utilized the alkaline comet assay to assess interstrand crosslinking by BBR3610-DACH in cells. (Fig. 2). In unirradiated untreated HCT116 cells, no DNA damage was detected (Fig. 2A), nor were any strand breaks detectable following treatment with BBR3610-DACH (250 µM) alone (data not shown). Following irradiation with 15 Gy to introduce a fixed level of DNA strand breaks, shorter DNA fragments migrated from the bulk of DNA during alkaline electrophoresis to produce comet tails (Fig. 2B). When the cells were treated with 50–250 µM BBR3610-DACH prior to irradiation, a decrease in the comet tail length and intensity was observed, in comparison to irradiated controls (Fig. 2C–E), consistent with the retention of more DNA in the head due to the PPC-induced interstrand crosslinks. By quantifying the decrease in the moment of the comet tails, BBR3610-DACH-induced interstrand crosslinks were compared to those formed by BBR3610 at the same concentrations, and while both analogues showed concentration-dependent decreases in tail moments, there were no statistically significant differences between the effects of the two PPCs (Fig. 2F).
Figure 2. Comet assays of interstrand crosslinks in HCT116 cells treated with BBR3610 or BBR3610-DACH.
(A) Untreated, unirradiated cells. (B) Cells receiving 15 Gy irradiation only. (C–E) Cells were treated with 50, 100, or 250 µM BBR3610 or BBR3610-DACH for 1 hr, incubated for 6 hr in PPC-free medium to allow interstrand crosslink formation, and then irradiated. Interstrand crosslink formation was represented as mean percentage decrease in the tail moment ± SE from three independent experiments (F).
3.2. BBR3610-DACH exhibits an anti-proliferative effect
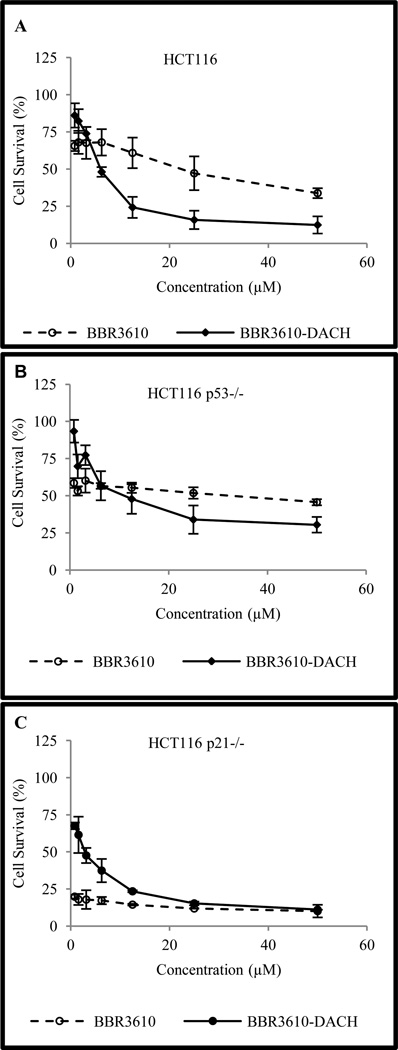
BBR3610-DACH was evaluated in HCT116 and in isogenic cell lines deficient in p53 or p21. Cells were exposed to the platinum complex (0.78 – 50 µM) and the cytotoxicity was assessed using the MTT assay. In the case of BBR3610, the cells showed an approximately 50% reduction in cell viability at 1–10 nM (data not shown), but were highly resistant to any further reduction (Fig. 3A). The basis of this biphasic response in uncertain, but it could reflect either a resistant subpopulation of cells or a high-affinity transport mode that saturates at low concentrations of BBR3610. No such biphasic response was seen in cells treated with BBR3610-DACH (Fig. 3A) and as a result the IC50 (5.9 ± 0.87µM, Table 2) was lower than that for BBR3610. However, the IC50 was higher in the p53-deficient cells (17.14 ± 5.06) than in parental cells (Fig. 3B) indicating a pivotal role of p53 in the antiproliferative response. However, in p21-null HCT116 cells, BBR3610-DACH produced a greater anti-proliferative effect on growth than in the wild type cells (Fig. 3C and Table 2). This could indicate that p21-mediated cell cycle arrest in G1 led to enhanced DNA repair and a reduction in cell death that outweighed the initial arrest.
Figure 3. Cytotoxicity of PPCs in HCT116 and isogenic cell lines.
Sensitivity of (A) HCT116; (B) HCT116 p53−/−; or (C) HCT116 p21−/− cells to BBR3610 and BBR3610-DACH was determined by MTT assay following exposure to micromolar concentrations of the polynuclear platinum complexes for 72 hr. Error bars indicate mean ± SD from three independent experiments.
TABLE 2.
Cytotoxicity of BBR3610-DACH in HCT116 and isogenic cell lines.
| Cell Line | IC50 (µM)* |
|---|---|
| HCT116 | 5.9 ± 0.87 |
| HCT116 p53−/− | 17.14 ± 5.06 |
| HCT116 p21−/− | 2.7 ± 0.75 |
IC50 values are means ± SD from 2 or 3 independent experiments, each conducted in triplicate
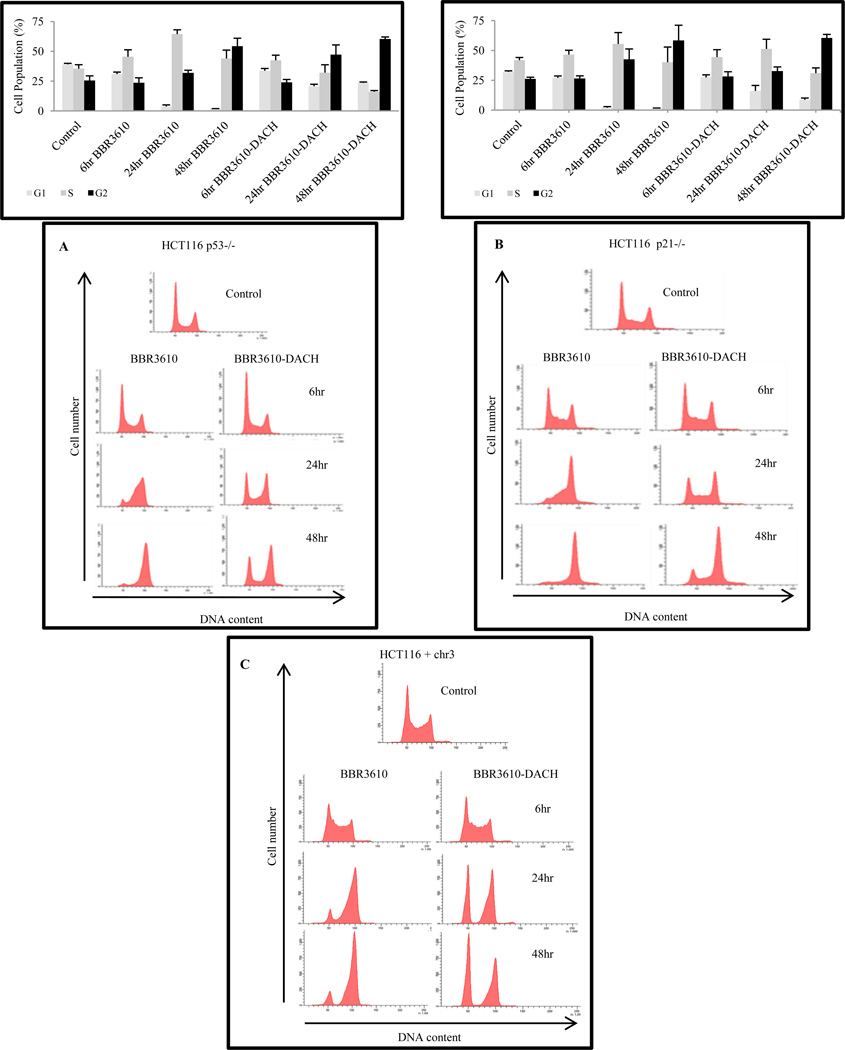
3.3. BBR3610-DACH induces G1 and G2 arrest
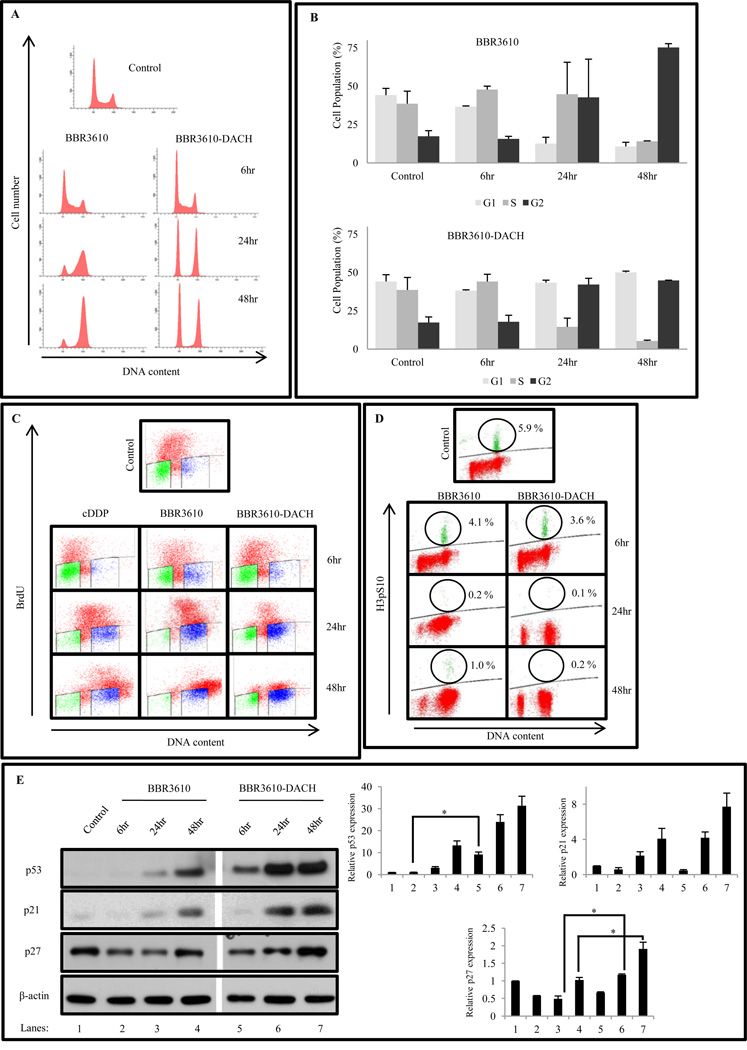
Following genotoxic stress induced by antitumor agents, p53 levels are stabilized resulting in induction of p21, which mediates transient cell cycle arrest or apoptosis, depending on the damage incurred. As expected from previous work [22,23], treatment of HCT116 cells with cisplatin caused accumulation of S phase cells within 24 hr, followed by a G2 arrest at 48 hr (data not shown). However, treatment with BBR3610-DACH induced an early robust G1 and G2 phase arrest with a significant reduction in S phase, compared to BBR3610 which like cisplatin showed only G2 arrest (Fig. 4A, B). To assess the possibility that the high proportion of BBR3610-DACH-treated cells having 2N DNA content might represent an arrest in very early S rather than in G1, BrdU incorporation and DNA content were measured simultaneously. The results gave no evidence of an early S block, which would have been manifested as an accumulation of BrdU-positive cells with near-G1 DNA content. Instead, the results confirmed the depletion of BrdU-positive S-phase cells following BBR3610-DACH treatment, as well as the transient accumulation of late S cells following BBR3610 treatment (Fig. 4C). Also, the distinct cell cycle effects of the two analogues were observed over a range of concentrations, with the specific G2 block detectable with as low as 0.5 µM BBR3610 and G1 arrest with S phase depletion being apparent with as low as 5 µM BBR3610-DACH treatment (data not shown), suggesting that the qualitative differences between these compounds in cellular responses cannot be ascribed to differences in uptake, overall potency, or stability. Finally, cytometry of the mitotic marker phospho-histone H3 indicated that both PPCs induced complete depletion of mitotic cells, implying a block in G2 rather than M phase (Fig. 4D).
Figure 4. BBR3610-DACH induces G1/S and G2/M cell cycle arrest in HCT116 cells with robust S phase depletion.
(A) Cells were treated with 20 µM BBR3610 or 20 µM BBR3610-DACH and analyzed for cell cycle distribution using propidium iodide. Modfit analysis of G1, S, and G2 populations is also shown. (The significance of differences between BBR3610-DACH-and BBR3610-treated cells were p<0.02 for G1 fraction at 24 hr, p<0.03 for G1 fraction, p<0.003 for S fraction, and p<0.005 for G2 fraction at 48 hr) (C) BrdU staining assay following treatment with 20 µM BBR3610-DACH as compared to BBR3610 or 20 µM cDDP. The data shown are a representative of 2 or 3 independent experiments.(D) G2/M checkpoint assay. HCT116 cells were exposed to 20 µM BBR3610 or 20 µM BBR3610-DACH for 6, 24, and 48 hr. Cells were fixed and stained with phospho-histone H3 (Ser10), followed by anti-rabbit Alexa 647 secondary antibody and PI staining. Ten thousand events were analyzed by flow cytometry. The data shown are a representative of two independent experiments. (E) HCT116 cells were treated with 20 µM BBR3610 or 20 µM BBR3610-DACH for 6, 24, and 48 hr. Representative immunoblots are shown from two or three independent experiments. The gap between BBR3610 and BBR3610-DACH lanes was a lane for the Mr standards. Relative band intensities of the proteins were normalized with β-actin as an internal control. Error bars indicate ± SEM from two or three independent experiments. *p-value < 0.05
Concurrently, a stabilization of p53 protein resulting in an increase in its steady-state level was seen in BBR3610-DACH-treated cells, as early as 6 hr and continuing to 24 and 48 hr, accompanied by an increase in p21 and p27 expression levels (Fig. 4E). This result indicates that the classical p53 pathway is being triggered for the G1/S phase arrest and possibly the G2/M arrest. However, p21 induction was seen in HCT116 p53−/− cells (data not shown), indicating that expression of p21 did not require p53 stabilization. Although BBR3610 also induced both p53 and p21, the response was slower and less robust (Fig 4E). Similar studies carried out in the HCT116 isogenic cell line lacking p53 or p21, showed that BBR3610-DACH-induced G1 arrest was largely p21-dependent (Fig. 5B) but only partially p53-dependent (Fig. 5A). However, the G2/M arrest was found to be independent of p53 and p21.
Figure 5. Effect of BBR3610-DACH on isogenic HCT116 cell lines.
Following treatment with 20 µM BBR3610 or 20 µM BBR3610-DACH, cell cycle distribution was analyzed using propidium iodide. BBR3610 treatment showed a similar cell cycle profile in HCT116 p53−/− (A) and p21−/− cells (B) as observed in the parental cells. Data shown are a representative of 2 or 3 independent experiments. Modfit analysis is shown above each histogram for HCT116 p53−/− and p21−/− null cells. (C) Wild type HCT116 and hMLH1 complemented cells, HCT116+chr3, were treated with 20 µM BBR3610 or BBR3610-DACH for 6, 24, and 48 hr. They were then analyzed for cell cycle distribution using propidium iodide.
DNA mismatch repair is a post-replication DNA repair mechanism which removes the mismatch from newly synthesized daughter strand [24,25]. Defects in mismatch repair confer resistance to some platinum drugs, especially cisplatin [26], but not to analogues with bulky DACH ligands such as oxaliplatin [27]. In this context, we investigated whether the disparate cell cycle effects elicited by BBR3610 and BBR3610-DACH were dependent on the mismatch repair status of the cells. We used HCT116+chr3 cells wherein the absence of the mismatch repair hMLH1 gene is complemented by the transfer of an additional copy of chromosome 3, which carries the MLH1 gene [28]. Treatment of HCT116+chr3 cells with BBR3610 or BBR3610-DACH resulted in cell cycle profiles similar to those of parental HCT116 cells (Fig. 5C). Treatment with cisplatin elicited a classical G2/M arrest 24–48 hrs after PPC treatment (data not shown) that was similar for the two cell lines [29]. Thus, the disparate cellular effects of BBR3610 and BBR3610-DACH do not depend on either the presence or the absence of mismatch repair.
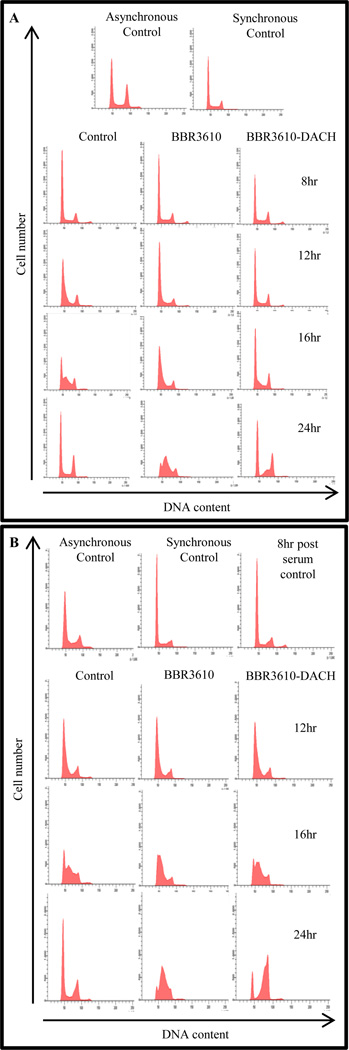
3.4. BBR3610-DACH specifically targets the G1 phase population of cells
To define precisely the phase of the cell cycle where BBR3610-DACH exerts its anti-proliferative effect, we carried out a “cell mapping” experiment with cells blocked in G0/G1 phase by serum starvation. As shown in Fig. 6A, progression into S phase was seen at 12 hr after release from the block. Immediately following release of the block, PPC treatment was initiated and cells were harvested 8, 12, 16, or 24 hr later. The cells that were synchronized in G0/G1 continued to be arrested in G0/G1 following treatment with BBR3610-DACH (Fig. 6A). However, with BBR3610 treatment, they progressed normally out of G1 and halted in the S phase. These results show that the cells are specifically halted in the G1 phase by BBR3610-DACH. To further confirm this finding, the cells were released from the block and allowed to progress through G1 phase into S. A 4-hr treatment with each PPC in the early S phase, ~8 hr after release of block, resulted in a blockade of cells at the G2/M phases in the case of BBR3610-DACH and a delayed accumulation in S phase in the case of BBR3610 (Fig. 6B). These results corroborate the observation with the asynchronous population of cells wherein a prominent G1/S and G2/M arrest is seen following treatment with BBR3610-DACH, and further show that G1 accumulation in BBR3610-DACH-treated cells is due almost exclusively to a persistent and complete G1 arrest, and not to cells that successfully traversed mitosis to re-enter G1.
Figure 6. BBR3610-DACH specifically inhibits the progression of G1 phase cells.
HCT116 cells were synchronized in G0/G1 by serum starvation using 0.5% serum for 96 hr. (A) The cells were then released immediately into medium containing BBR3610 or BBR3610-DACH. FACS was performed at different timepoints to determine the progression of cell cycle after PPC treatment. (B) Cells were released into medium and allowed to progress into S phase. They were then treated with BBR3610 or BBR3610-DACH and FACS was performed at different timepoints to determine the progression of cell cycle following PPC treatment‥
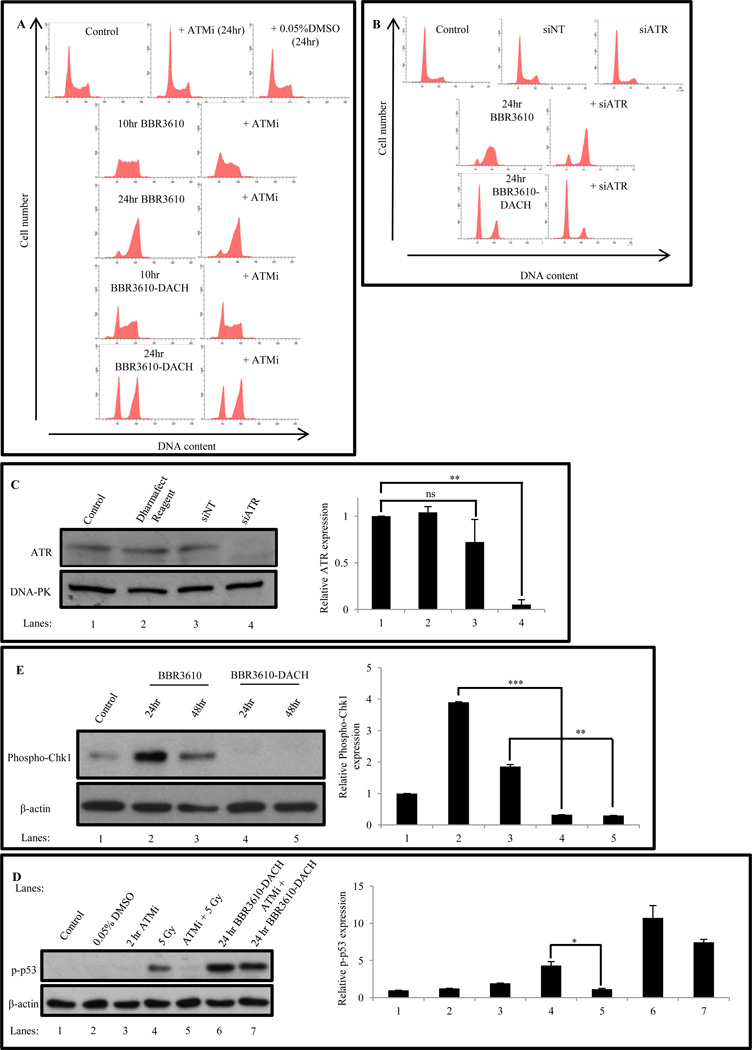
3.5. BBR3610-DACH-induced S phase depletion is independent of ATM and ATR activation
ATM and ATR are DNA damage sensor protein kinases that activate a signal transduction cascade involving Chk1 and Chk2 and culminating in cell cycle arrest [30]. To test whether the cell cycle perturbations elicited by BBR3610-DACH were ATM/ATR dependent, we inhibited ATM/ATR activation by, (i) KU-60019, a small molecule inhibitor of ATM (3 µM), and (ii) knockdown of ATR by 50 nM ATR-targeting siRNA.
Inhibition of ATM using KU-60019 did not abrogate the cell cycle arrest elicited either by BBR3610 or BBR3610-DACH (Fig. 7A), indicating that ATM activation was not required for arrest by these agents. The effectiveness of KU-60019 in inhibiting ATM was confirmed by its suppression of radiation-induced p53 phosphorylation on Ser15 (Fig. 7D). Intriguingly, the robust Ser15 phosphorylation induced by BBR3610-DACH was not affected by KU-60019, confirming that BBR3610-DACH activates the p53 pathway by an ATM-independent route.
Figure 7. BBR3610-DACH induced cell cycle arrest is independent of ATM or ATR activity.
(A) Effect of ATM inhibition on BBR3610 or BBR3610-DACH-induced cell cycle arrest. Cells were pretreated with 3 µM KU-60019 (ATMi) for 1 hr and then treated with 20 µM BBR3610 or BBR3610-DACH for 10 hr and 24 hr respectively in the continued presence of inhibitor. As a control, cells were also treated with 0.05% DMSO (solvent for KU-60019). (B) Effect of ATR knockdown on BBR3610 or BBR3610-DACH induced cell cycle arrest. Cells were transfected with 50 nM siRNA against ATR for 3 days. Each PPC was then added and the cells incubated for 24 hr. (C) Immunoblot showing ATR knockdown in HCT116 cells by 50 nM siRNA in comparison to non-targeting siRNA control and Dharmafect Reagent Control. Relative band intensities of the protein was normalized with DNA-PK as an internal control. Error bars indicate ± SEM from two or three independent experiments. *p-value < 0.05. **p-value<0.01. (D) Immunoblot analysis of p-p53 levels in HCT116 cells following treatment with 5 Gy radiation in the presence and absence of 3 µM KU-60019 and 20 µM BBR3610-DACH for 24 hr in the presence and absence of 3 µM KU-60019. Relative band intensities of the protein was normalized with β-actin as an internal control. Error bars indicate ± SEM from two or three independent experiments. **p-value<0.01, ***p-value<0.001 (E) Immunoblot analysis of Phospho-Chk1 levels in HCT116 cells following treatment with 20 µM BBR3610 or BBR3610-DACH. Relative band intensities of the protein was normalized with β-actin as an internal control. Error bars indicate ± SEM from two or three independent experiments. *p-value<0.05.
Similarly, while ATR knockdown by siRNA (Fig. 7C) allowed faster progression of BBR3610-treated cells through S phase to G2/M arrest, it did not relieve the G1 block in BBR3610-DACH-treated cells at all (Fig. 7B). The absence of ATR activation by BBR3610-DACH was further confirmed by examining Phospho-Chk1 levels [31], which were increased dramatically after 24 hr in the cells treated with BBR3610 (Fig. 7E) or cisplatin (data not shown). These results suggest a significant S phase arrest following treatment with BBR3610, which is then resolved, allowing cells to progress into the G2 phase. However, BBR3610-DACH treatment did not induce any Phospho-Chk1, indicating the absence of ATR activation, which is consistent with the lack of delay in progression through S phase. In summary, ATM/ATR activation is not required for BBR3610-DACH-induced S phase depletion, but BBR3610-induced S phase accumulation is abrogated by knockdown of ATR (Fig. 7B).
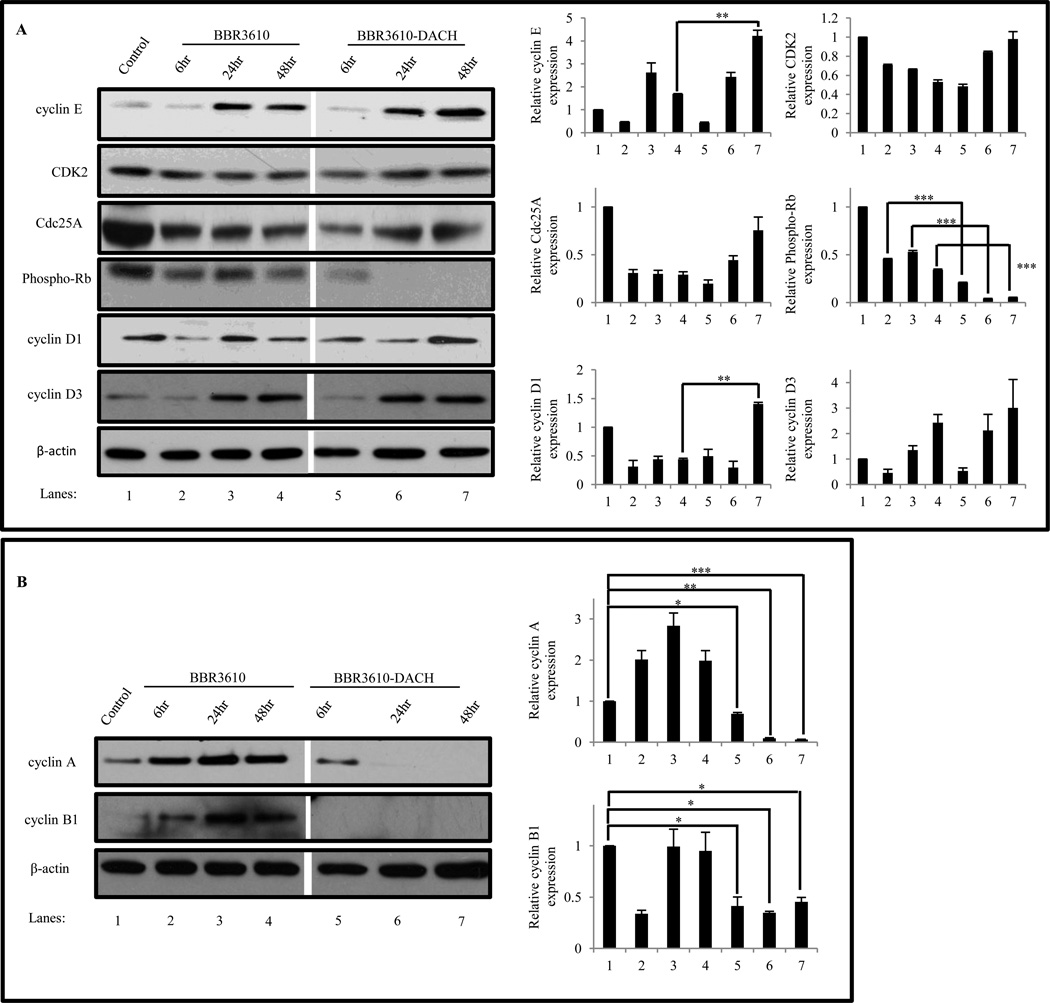
3.6. Effect of BBR3610-DACH on G1/S and G2/M phase cell cycle regulators
The interaction of CDKs with their corresponding cyclins mediates the normal cell cycle progression from G1 to S, S to G2, and G2 to M phases [32]. To assess their roles in cell cycle perturbations, we investigated the effect of BBR3610-DACH treatment on the protein levels of G1 CDKs and cyclins. Immunoblot analysis revealed that treatment with BBR3610-DACH, which elicits a G1 arrest, was accompanied by an increase in cyclin E (Fig. 8A), which normally accumulates in G1 and is degraded in S. CDK2 and Cdc25A were maintained at steady state levels while phospho-Rb decreased markedly after 24 and 48 hr of treatment. Thus, in BBR3610-DACH-treated cells CDK2/cyclin E complexes presumably were formed, but kinase activity was apparently suppressed by p21, resulting in Rb dephosphorylation and G1 arrest. An increase in cyclins D1 and D3 was observed, which could be due to cells exiting the G0 phase and getting arrested in G1. Also, the levels of cyclins A and B fell dramatically after 24 and 48 hr of BBR3610-DACH treatment (Fig. 8B), which may result from p53-mediated suppression of their synthesis [33,34] and may contribute to G2 arrest.
Figure 8. Effect of BBR3610-DACH on G1/S and G2/M regulators.
Cells were treated with 20 µM BBR3610 or BBR3610-DACH for 6, 24, or 48 hr. Protein lysates were prepared and 30 µg protein was resolved on SDS-PAGE and detected by probing the immunoblot with antibodies against (A) pRb, CDK2, Cdc25A, cyclin E, cyclin D1 and cyclin D3 and (B) cyclin A and cyclin B1. β-actin was used as a loading control. Relative band intensities of the proteins were normalized with β-actin as an internal control. Error bars indicate ± SEM from two or three independent experiments. *p-value<0.05, **p-value<0.01, ***p-value<0.001.
3.7. BBR3610-DACH induces early PARP cleavage independent of caspase-3 and caspase-7 activation
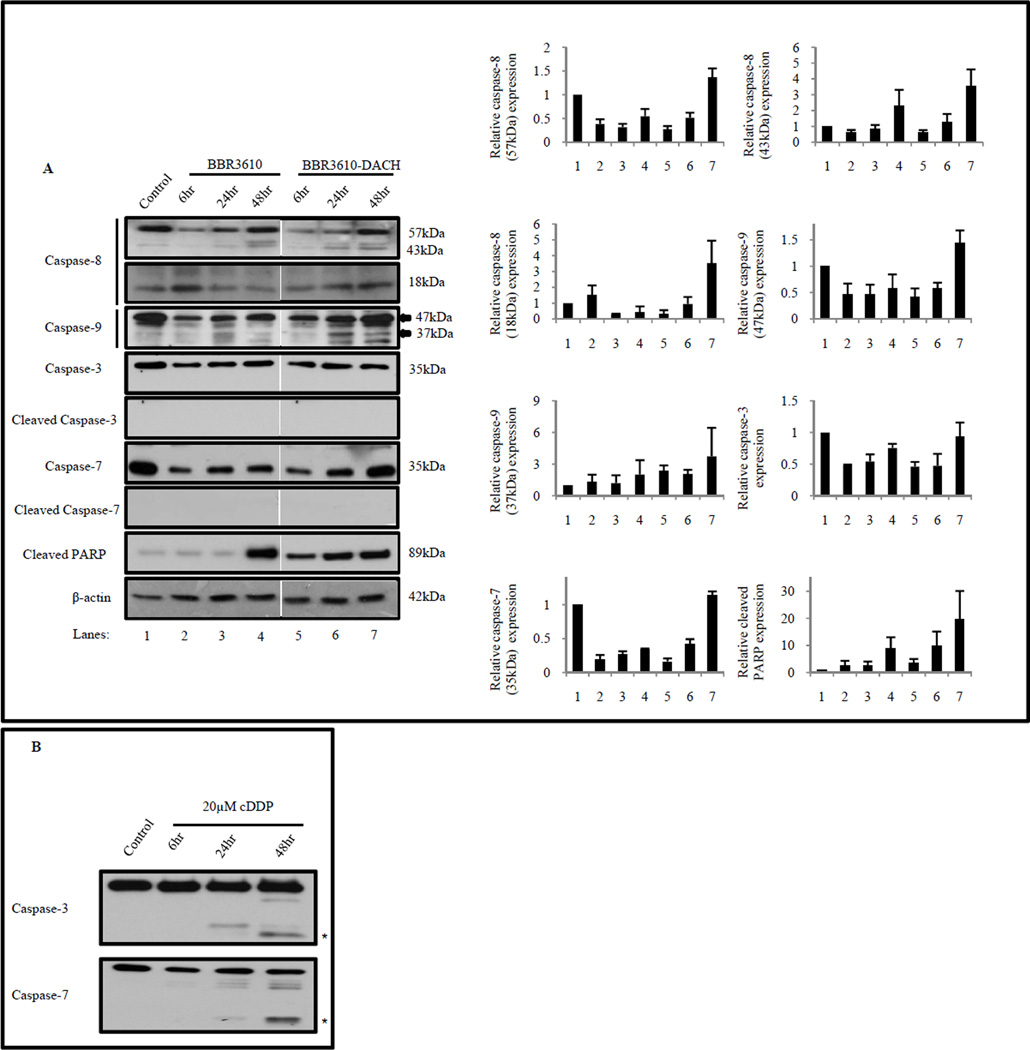
Following prolonged cell cycle arrest, damaged cells eventually either recover or undergo programmed cell death or apoptosis. During this event, activated cysteine aspartate proteases (caspases), primarily caspase-3 and caspase-7, cleave downstream substrates such as the DNA repair protein PARP [35]. Treatment with BBR3610-DACH elicited robust PARP cleavage between 6–24 hr (Fig. 9A), along with a weak but reproducible increase in cleavage of caspase-8 and caspase-9, which are markers for the extrinsic and intrinsic apoptotic pathways respectively. However, unlike cisplatin (Fig. 9B), neither of the PPCs induced detectable caspase-3 or caspase-7 cleavage, suggesting a caspase-independent PARP cleavage (Fig. 9A). There was no internucleosomal DNA fragmentation in BBR3610-DACH-treated cells, as analyzed by agarose gel electrophoresis (data not shown). Overall, our results suggest that BBR3610-DACH induces a non-classical apoptotic pathway.
Figure 9. BBR3610-DACH induces an early apoptotic response.
HCT116 cells were treated with 20 µM BBR3610 or 20 µM BBR3610-DACH for 6, 24, or 48 hr and then, (A) western blots of lysates were probed with antibodies against caspase-8, caspase-9, full length and cleaved caspase-3, full length and cleaved caspase-7, and cleaved PARP. β-actin was used as a loading control. Arrow indicates the 37 kDa fragment of cleaved caspase-9. (B) Time course analysis of cleaved caspase-3 and cleaved caspase-7 in HCT116 cells treated with 20 µM cisplatin for 6, 24, and 48 hr. β-actin was used as a loading control. Relative band intensities of the proteins were normalized with β-actin as an internal control. Error bars indicate ± SEM from two or three independent experiments. Asterisks indicate cleaved fragments of caspase-3 (17 and 19 kDa) and caspase-7 (20 kDa) respectively.
4. DISCUSSION
Platinum-based drugs constitute one of the major classes in the armamentarium of cancer therapies. However, their use is often compromised by intrinsic or acquired drug resistance in cells, including increased scavenging by thiol-containing compounds such as glutathione and metallothionein [3,4]. The dinuclear platinum compound, BBR3610-DACH, used in this study, was previously shown to be resistant to metabolic decomposition by sulfur nucleophiles including glutathione [15,16]. Subsequent in vitro studies showed that the DNA adducts formed by BBR3610-DACH persisted longer, escaped DNA repair, and inhibited RNA pol II transcription much more efficiently than cisplatin adducts [16]. Considering these promising early findings and the potential for clinical combinations of platinum drugs with DNA-repair modifying agents such as PARP inhibitors [36] it was important to examine whether the cell signaling responses elicited by BBR3610-DACH were similar to those of other platinum-based compounds, both mononuclear and multinuclear.
As reported previously for glioma cells, BBR3610 induced a classical G2/M arrest in HCT116 cells [10], which is a typical response to PPCs. However, BBR3610-DACH unexpectedly showed a completely different response wherein a biphasic G1 and G2 cell cycle arrest was accompanied by near total depletion of S phase (Fig. 4). Assuming that these effects represent responses to DNA damage, it is likely that at least some of the crosslinks formed by BBR3610-DACH had structural features not present in BBR3610 adducts, resulting in recruitment of specific proteins that trigger these unique responses. In this context it is of interest that the main effects of oxaliplatin in a panel of human tumor cell lines were also G2/M cell cycle arrest and a transient delay in entry into S phase [37,38].
Typical of DNA damaging agents, treatment with both BBR3610 and BBR3610-DACH induced an increase in the level of p53, which can mediate arrest at both G1 and G2 phases of the cell cycle. However, BBR3610-DACH elicited a more rapid and robust p53 stabilization, accompanied by increased levels of its transcriptional target, p21. Binding of p21 to the proliferating cell nuclear antigen (PCNA), even in p53-null cells, causes both G1 and G2 arrest [39]. Experiments with isogenic HCT116 p21-null and p53-null cells revealed that BBR3610-DACH-induced G1 arrest was strictly p21-dependent but only partially p53-dependent, indicating a p53-independent mechanism for induction of p21. However, p21 was not required for the G2 arrest by either of these PPCs. Compared to the sensitivity of wild-type HCT116 cells to platinum agents, there is a greater resistance of HCT116 p53-null cells to oxaliplatin than observed here for BBR3610-DACH [40]
Ataxia Telangiectasia Mutated (ATM) and Ataxia Telangiectasia and Rad3-related (ATR) are members of the PI3K–like family of serine/threonine protein kinases, playing critical roles as DNA damage sensors upstream of the p53/p21 pathway and thereby promoting cell cycle arrest, DNA repair, and apoptosis [30]. Typically, ATR senses stalled replication forks such as those resulting from DNA interstrand crosslinks, while ATM senses double-strand breaks. Experiments with pharmacological inhibitors of ATM and/or ATR, along with ATR knockdown, showed that BBR3610-induced G2 arrest was ATM-independent but ATR-dependent, consistent with interstrand crosslinks as the initiating lesion (Fig. 7A–E). The observed S phase accumulation could be due to replication fork arrest that is subsequently recognized by ATR, which then activates Phospho-Chk1 (Fig. 7F). However, after 24 hr treatment, BBR3610-DACH induced an ATR-dependent G2 arrest, as well as a p21-dependent G1 arrest that was independent of both ATM and ATR. The exact mechanism behind the dual phase arrest is not known. While interstrand crosslinks could be converted to double-strand breaks in the process of crosslink repair, the lack of ATM dependence argues against such conversion as an intermediate in checkpoint activation. Although an increase in Phospho-Chk2 levels is seen after BBR3610-DACH treatment (data not shown), the absence of S phase accumulation (Fig. 4) and lack of ATR activation (Fig. 7) appear to exclude stalled replication forks as the signal for cell cycle arrest as well, raising the possibility of a novel mechanism which senses DNA adducts directly and causes the activation of DNA damage checkpoints [41]. In this context, it has been reported earlier that adducts formed by the mononuclear platinum compound oxaliplatin, containing a DACH ligand, likewise does not activate ATR, which points out the possibility that compared to most platinum adducts, DACH adducts may trigger unique recognition mechanisms and thereby elicit unique responses [42].
The normal progression through the different phases of the cell cycle depends on the pivotal interaction of CDKs with the cyclins and thus disruption of these interactions could in principle mediate the effects of PPCs. In our study, the steady state levels of CDK2 and Cdc25A remained relatively constant following PPC treatment (Fig. 8A). Perturbations of the G1/S cyclins E, D1 and D3 were similar for BBR3610 and BBR3610-DACH, although cyclin E reached much higher levels after treatment with BBR3610-DACH, perhaps reflecting the large number of cells that accumulate in late G1 but fail to enter S phase, during which cyclin E is normally degraded. Overall, however, these data suggest that the unique effect of BBR3610-DACH on G1/S progression reflects events downstream of CDK/cyclin complex formation, such as CDK inhibition by p21.
The G2-M transition is primarily governed by the activities of cyclin A and cyclin B. Following treatment with BBR3610-DACH, a robust decrease in levels of both of these cyclins (Fig. 8B) was observed, which may result from their suppression by p53, and could contribute to G2 arrest. Treatment of HepG2 cells with 5-fluorouracil or methotrexate reduced cyclin B2 mRNA levels accompanied with increased p53 levels [34]. Likewise, DLD-1 colorectal cancer cells harboring a p53 mutation showed decreased cyclin B1 and B2 mRNA levels after expression of a wild-type p53 [34]. Similarly, stabilization of p53 following BBR3610-DACH treatment could be interfering with the transcription of cyclin B or cyclin A since the latter contains a p53 binding site in the 5′ untranslated region [33].
Based on weak caspase-8/caspase-9 cleavage, lack of caspase-3/caspase-7 cleavage, and absence of internucleosomal cleavage, BBR3610-DACH is not a potent inducer of classical apoptosis, compared to cisplatin. An apparently caspase-independent BBR3610-DACH-induced PARP-1 cleavage (Fig. 9A) may instead be mediated by cathepsins or TGF-β activation [43]. This effect is mechanistically different from other platinum-based compounds, and it will be of interest to determine the structural reasons behind such differences.
One of the key features that distinguish PPCs from their mononuclear counterparts is the formation of long-range DNA intrastrand and interstrand crosslinks. PPC-induced crosslinks cause conformational changes in DNA that are not recognized by many of the DNA binding proteins and repair proteins that bind to the adducts formed by cisplatin or its congeners [44,45]. Earlier work using plasmid DNA showed that both BBR3610 and BBR3610-DACH form a high proportion of highly toxic interstrand crosslinks (26% and 23% of total adducts respectively) [16]. These interstrand crosslinks irreversibly block major DNA transactions such as replication, transcription, and recombination [46,47]. Consistent with the in vitro studies, BBR3610 and BBR3610-DACH formed similar levels of interstrand crosslinks in cells, as analyzed by the comet assay (Fig. 2). Thus, while it is likely that the intracellular effects observed after treatment with these compounds are a consequence of the crosslinks formed, there are no obvious qualitative or quantitative differences in adducts formed in cells that would explain the dramatic differences between the cellular effects of BBR3610 and BBR3610-DACH, other than the presence of the DACH ligand. The results show that despite retaining superficially similar DNA-binding profiles, relatively minor modifications in PPCs can dramatically alter the effects of these compounds on cell signaling pathways, suggesting new avenues for rational combination drug development involving platinum cytotoxics.
In summary, BBR3610-DACH induces DNA interstrand crosslinks, eventually eliciting a unique dual mode of cell cycle arrest. Since this compound exhibits significant anti-proliferative activity and also has been shown previously to be metabolically stable to nucleophilic attack, it would be an ideal candidate to be developed further and tested in clinical trials. Also, it shows a promising paradigm shift for the structure-activity relationship of novel platinum based compounds, wherein a ligand modification in the original compound, BBR3610 yields a structurally similar compound but with distinctly different cellular effects.
ACKNOWLEDGMENTS
We thank John Ryan for a critical reading of the manuscript. This work was supported by grants supported by R01CA078754-14 (N.P.F.) and R01CA040615 (L.F.P.) from the National Cancer Institute, US Public Health Service, DHHS. The VCU Flow Cytometry Core Facility is supported by NIH-P30CA16059.
ABBREVIATIONS
- ATM
Ataxia Telangiectasia Mutated
- ATR
Ataxia Telangiectasia Mutated and Rad3 related
- BrdU
Bromodeoxyuridine
- CDK
Cyclin Dependent Kinase
- Chk1
Checkpoint Kinase 1
- DACH
1, 2 diaminocyclohexane
- Gy
Gray
- MTT
(3, 4, 5-dimethylthiazol-2-yl)-2, 5-diphenyl tetrazolium bromide
- PAGE
Polyacrylamide Gel Electrophoresis
- PARP
Poly (ADP-Ribose) Polymerase
- PBS
Phosphate buffered saline PCNA - Proliferating Cell Nuclear Antigen
- PPC
Polynuclear Platinum Complex
- PVDF
Polyvinylidene difluoride
- SDS
Sodium Dodecyl Sulfate
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
CHEMICAL COMPOUNDS STUDIED IN THE ARTICLE
Cisplatin (PubChem CID: 441203); BrdU (PubChem CID: 6035); Propidium Iodide (PubChem CID: 104981); MTT (PubChem CID: 64965); SYBR Green (PubChem CID: 56841760)
REFERENCES
- 1.O’Dwyer PJ, Stevenson JP, Johnson SW. Clinical pharmacokinetics and administration of established platinum drugs. Drugs. 2000;59(Suppl 4):19–27. doi: 10.2165/00003495-200059004-00003. [DOI] [PubMed] [Google Scholar]
- 2.Farrell NP. Progress in Platinum-Derived Drug Development. Drugs of the future. 2012;37(11):795–806. [Google Scholar]
- 3.Wang And X, Guo Z. The role of sulfur in platinum anticancer chemotherapy. Anticancer Agents Med.Chem. 2007;7:19–34. doi: 10.2174/187152007779314062. [DOI] [PubMed] [Google Scholar]
- 4.Meijer C, Mulder NH, Timmer-Bosscha H, Sluiter WJ, Meersma GJ, de Vries EG. Relationship of cellular glutathione to the cytotoxicity and resistance of seven platinum compounds. Cancer Res. 1992;52:6885–6889. [PubMed] [Google Scholar]
- 5.Mangrum JB, Farrell NP. Excursions in polynuclear platinum DNA binding. Chem.Commun.(Camb) 2010;46:6640–6650. doi: 10.1039/c0cc01254h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Manzotti C, Pratesi G, Menta E, Di Domenico R, Cavalletti E, Fiebig HH, et al. BBR3464: a novel triplatinum complex, exhibiting a preclinical profile of antitumor efficacy different from cisplatin. Clin.Cancer Res. 2000;6:2626–2634. [PubMed] [Google Scholar]
- 7.Oehlsen ME, Qu Y, Farrell N. Reaction of polynuclear platinum antitumor compounds with reduced glutathione studied by multinuclear (1H, 1H-15N gradient heteronuclear single-quantum coherence, and 195Pt) NMR spectroscopy. Inorg.Chem. 2003;42:5498–5506. doi: 10.1021/ic030045b. [DOI] [PubMed] [Google Scholar]
- 8.Vacchina V, Torti L, Allievi C, Lobinski R. Sensitive species-specific monitoring of a new triplatinum anti-cancer drug and its potential related compounds in spiked human plasma by cation-exchange HPLC-ICP-MS -J. Anal. At. Spectrom. 2003;18:884–890. [Google Scholar]
- 9.McGregor TD, Hegmans A, Kasparkova J, Neplechova K, Novakova O, Penazova H, et al. A comparison of DNA binding profiles of dinuclear platinum compounds with polyamine linkers and the trinuclear platinum phase II clinical agent BBR3464. J.Biol.Inorg.Chem. 2002;7:397–404. doi: 10.1007/s00775-001-0312-4. [DOI] [PubMed] [Google Scholar]
- 10.Shingu T, Chumbalkar VC, Gwak HS, Fujiwara K, Kondo S, Farrell NP, et al. The polynuclear platinum BBR3610 induces G2/M arrest and autophagy early and apoptosis late in glioma cells. Neuro Oncol. 2010;12:1269–1277. doi: 10.1093/neuonc/noq095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mitchell C, Kabolizadeh P, Ryan J, Roberts JD, Yacoub A, Curiel DT, et al. Low-dose BBR3610 toxicity in colon cancer cells is p53-independent and enhanced by inhibition of epidermal growth factor receptor (ERBB1)-phosphatidyl inositol 3 kinase signaling. Mol.Pharmacol. 2007;72:704–714. doi: 10.1124/mol.107.038406. [DOI] [PubMed] [Google Scholar]
- 12.Ruhayel RA, Zgani I, Berners-Price SJ, Farrell NP. Solution studies of dinuclear polyamine-linked platinum-based antitumour complexes. Dalton Trans. 2011;40:4147–4154. doi: 10.1039/c1dt00001b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Summa N, Maigut J, Puchta R, van Eldik R. Possible biotransformation reactions of polynuclear Pt(II) complexes. Inorg.Chem. 2007;46:2094–2104. doi: 10.1021/ic061990d. [DOI] [PubMed] [Google Scholar]
- 14.Gatti L, Perego P, Leone R, Apostoli P, Carenini N, Corna E, et al. Novel bis-platinum complexes endowed with an improved pharmacological profile. Mol.Pharm. 2010;7:207–216. doi: 10.1021/mp900211j. [DOI] [PubMed] [Google Scholar]
- 15.Williams JW, Qu Y, Bulluss GH, Alvorado E, Farrell NP. Dinuclear platinum complexes with biological relevance based on the 1,2-diaminocyclohexane carrier ligand. Inorg.Chem. 2007;46:5820–5822. doi: 10.1021/ic700410y. [DOI] [PubMed] [Google Scholar]
- 16.Zerzankova L, Suchankova T, Vrana O, Farrell NP, Brabec V, Kasparkova J. Conformation and recognition of DNA modified by a new antitumor dinuclear PtII complex resistant to decomposition by sulfur nucleophiles. Biochem.Pharmacol. 2010;79:112–121. doi: 10.1016/j.bcp.2009.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Deep G, Agarwal R. New combination therapies with cell-cycle agents. Curr.Opin.Investig Drugs. 2008;9:591–604. [PMC free article] [PubMed] [Google Scholar]
- 18.O’Connor PM, Fan S. DNA damage checkpoints: implications for cancer therapy. Prog.Cell Cycle Res. 1996;2:165–173. doi: 10.1007/978-1-4615-5873-6_16. [DOI] [PubMed] [Google Scholar]
- 19.Shapiro GI, Harper JW. Anticancer drug targets: cell cycle and checkpoint control. J.Clin.Invest. 1999;104:1645–1653. doi: 10.1172/JCI9054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kim EH, Deng CX, Sporn MB, Liby KT. CDDO-imidazolide induces DNA damage, G2/M arrest and apoptosis in BRCA1-mutated breast cancer cells. Cancer.Prev.Res.(Phila) 2011;4:425–434. doi: 10.1158/1940-6207.CAPR-10-0153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Olive PL, Banáth JP, Durand RE. Heterogeneity in Radiation-Induced DNA Damage and Repair in Tumor and Normal Cells Measured Using the “Comet” Assay. Radiat.Res. 1990;122:86–94. [PubMed] [Google Scholar]
- 22.Zamble DB, Jacks T, Lippard SJ. p53-Dependent and -independent responses to cisplatin in mouse testicular teratocarcinoma cells. Proc.Natl.Acad.Sci.U.S.A. 1998;95:6163–6168. doi: 10.1073/pnas.95.11.6163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pani E, Stojic L, El-Shemerly M, Jiricny J, Ferrari S. Mismatch repair status and the response of human cells to cisplatin. Cell.Cycle. 2007;6:1796–1802. doi: 10.4161/cc.6.14.4472. [DOI] [PubMed] [Google Scholar]
- 24.Li GM. Mechanisms and functions of DNA mismatch repair. Cell Res. 2008;18:85–98. doi: 10.1038/cr.2007.115. [DOI] [PubMed] [Google Scholar]
- 25.Jiricny J. The multifaceted mismatch-repair system. Nat.Rev.Mol.Cell Biol. 2006;7:335–346. doi: 10.1038/nrm1907. [DOI] [PubMed] [Google Scholar]
- 26.Aebi S, Kurdi-Haidar B, Gordon R, Cenni B, Zheng H, Fink D, et al. Loss of DNA mismatch repair in acquired resistance to cisplatin. Cancer Res. 1996;56:3087–3090. [PubMed] [Google Scholar]
- 27.Mu D, Tursun M, Duckett DR, Drummond JT, Modrich P, Sancar A. Recognition and repair of compound DNA lesions (base damage and mismatch) by human mismatch repair and excision repair systems. Mol.Cell.Biol. 1997;17:760–769. doi: 10.1128/mcb.17.2.760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fang Y, Tsao CC, Goodman BK, Furumai R, Tirado CA, Abraham RT, et al. ATR functions as a gene dosage-dependent tumor suppressor on a mismatch repair-deficient background. EMBO J. 2004;23:3164–3174. doi: 10.1038/sj.emboj.7600315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pani E, Stojic L, El-Shemerly M, Jiricny J, Ferrari S. Mismatch repair status and the response of human cells to cisplatin. Cell.Cycle. 2007;6:1796–1802. doi: 10.4161/cc.6.14.4472. [DOI] [PubMed] [Google Scholar]
- 30.Abraham RT. Cell cycle checkpoint signaling through the ATM and ATR kinases. Genes Dev. 2001;15:2177–2196. doi: 10.1101/gad.914401. [DOI] [PubMed] [Google Scholar]
- 31.Zhao H, Piwnica-Worms H. ATR-mediated checkpoint pathways regulate phosphorylation and activation of human Chk1. Mol.Cell.Biol. 2001;21:4129–4139. doi: 10.1128/MCB.21.13.4129-4139.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sherr CJ, Roberts JM. Living with or without cyclins and cyclin-dependent kinases. Genes Dev. 2004;18:2699–2711. doi: 10.1101/gad.1256504. [DOI] [PubMed] [Google Scholar]
- 33.Desdouets C, Ory C, Matesic G, Soussi T, Brechot C, Sobczak-Thepot J. ATF/CREB site mediated transcriptional activation and p53 dependent repression of the cyclin A promoter. FEBS Lett. 1996;385:34–38. doi: 10.1016/0014-5793(96)00330-4. [DOI] [PubMed] [Google Scholar]
- 34.Krause K, Wasner M, Reinhard W, Haugwitz U, Dohna CL, Mossner J, et al. The tumour suppressor protein p53 can repress transcription of cyclin B. Nucleic Acids Res. 2000;28:4410–4418. doi: 10.1093/nar/28.22.4410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chang HY, Yang X. Proteases for cell suicide: functions and regulation of caspases. Microbiol.Mol.Biol.Rev. 2000;64:821–846. doi: 10.1128/mmbr.64.4.821-846.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cheng H, Zhang Z, Borczuk A, Powell CA, Balajee AS, Lieberman HB, et al. PARP inhibition selectively increases sensitivity to cisplatin in ERCC1-low non-small cell lung cancer cells. Carcinogenesis. 2013;34:739–749. doi: 10.1093/carcin/bgs393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.William-Faltaos S, Rouillard D, Lechat P, Bastian G. Cell cycle arrest and apoptosis induced by oxaliplatin (L-OHP) on four human cancer cell lines. Anticancer Res. 2006;26:2093–2099. [PubMed] [Google Scholar]
- 38.William-Faltaos S, Rouillard D, Lechat P, Bastian G. Cell cycle arrest by oxaliplatin on cancer cells. Fundam.Clin.Pharmacol. 2007;21:165–172. doi: 10.1111/j.1472-8206.2007.00462.x. [DOI] [PubMed] [Google Scholar]
- 39.Cayrol C, Knibiehler M, Ducommun B. p21 binding to PCNA causes G1 and G2 cell cycle arrest in p53-deficient cells. Oncogene. 1998;16:311–320. doi: 10.1038/sj.onc.1201543. [DOI] [PubMed] [Google Scholar]
- 40.Hata T, Yamamoto H, Ngan CY, Koi M, Takagi A, Damdinsuren B, et al. Role of p21waf1/cip1 in effects of oxaliplatin in colorectal cancer cells. Mol.Cancer.Ther. 2005;4:1585–1594. doi: 10.1158/1535-7163.MCT-05-0011. [DOI] [PubMed] [Google Scholar]
- 41.Jiang G, Sancar A. Recruitment of DNA damage checkpoint proteins to damage in transcribed and nontranscribed sequences. Mol.Cell.Biol. 2006;26:39–49. doi: 10.1128/MCB.26.1.39-49.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lewis KA, Lilly KK, Reynolds EA, Sullivan WP, Kaufmann SH, Cliby WA. Ataxia telangiectasia and rad3-related kinase contributes to cell cycle arrest and survival after cisplatin but not oxaliplatin. Mol.Cancer.Ther. 2009;8:855–863. doi: 10.1158/1535-7163.MCT-08-1135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yang Y, Zhao S, Song J. Caspase-dependent apoptosis and -independent poly(ADP-ribose) polymerase cleavage induced by transforming growth factor beta1. Int.J.Biochem.Cell Biol. 2004;36:223–234. doi: 10.1016/s1357-2725(03)00215-2. [DOI] [PubMed] [Google Scholar]
- 44.Kasparkova J, Zehnulova J, Farrell N, Brabec V. DNA interstrand cross-links of the novel antitumor trinuclear platinum complex BBR3464. Conformation, recognition by high mobility group domain proteins, and nucleotide excision repair. J.Biol.Chem. 2002;277:48076–48086. doi: 10.1074/jbc.M208016200. [DOI] [PubMed] [Google Scholar]
- 45.Zehnulova J, Kasparkova J, Farrell N, Brabec V. Conformation, recognition by high mobility group domain proteins, and nucleotide excision repair of DNA intrastrand cross-links of novel antitumor trinuclear platinum complex BBR3464. J.Biol.Chem. 2001;276:22191–22199. doi: 10.1074/jbc.M103118200. [DOI] [PubMed] [Google Scholar]
- 46.Deans AJ, West SC. DNA interstrand crosslink repair and cancer. Nat.Rev.Cancer. 2011;11:467–480. doi: 10.1038/nrc3088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Vare D, Groth P, Carlsson R, Johansson F, Erixon K, Jenssen D. DNA interstrand crosslinks induce a potent replication block followed by formation and repair of double strand breaks in intact mammalian cells. DNA Repair (Amst) 2012;11:976–985. doi: 10.1016/j.dnarep.2012.09.010. [DOI] [PubMed] [Google Scholar]