Abstract
Cardiac myosin-binding protein C is a key regulator of cardiac contractility and is capable of both activating the thin filament to initiate actomyosin motion generation and governing maximal sliding velocities. While MyBP-C’s C-terminus localizes the molecule within the sarcomere the N-terminus appears to confer regulatory function by binding to the myosin motor domain and/or actin. Literature pertaining to how MyBP-C binding to the myosin motor domain and or actin leads to MyBP-C’s dual modulatory roles that can impact actomyosin interactions are discussed.
Keywords: Cardiac contractility, thick filament, thin filament, myosin, actin, regulation
Introduction
The heart’s ability to pump blood results from the beat-to-beat contraction and relaxation of sarcomeres, the smallest cardiac contractile unit. Sarcomere shortening is mechanically driven by the calcium-regulated sliding of parallel arrays of actin-containing thin filaments past myosin-containing thick filaments (Fig. 1A). Actomyosin force and motion generation is modulated by cardiac myosin-binding protein C (MyBP-C), a 140 kDa immunoglobulin (Ig) protein superfamily member (Fig. 1B). The functional importance of MyBP-C is emphasized by mutations in MyBP-C’s cardiac specific gene (MYBPC3) being the second leading cause of familial hypertrophic cardiomyopathy (for review see [16]). However, the molecular mechanism by which MyBP-C modulates actomyosin function is not well understood.
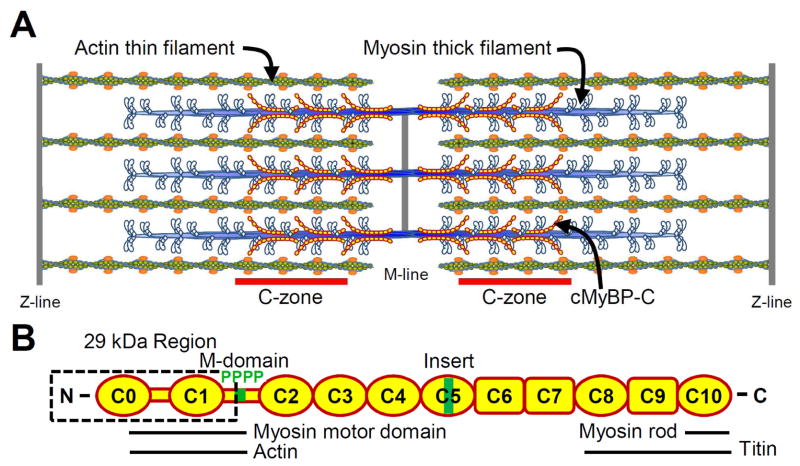
Figure 1.
A) Illustration of cardiac muscle sarcomere with interdigitating myosin-thick and actin-thin filaments. MyBP-C is localized to the C-zones. B) Schematic diagram of MyBP-C’s Ig-like (oval) and fibronectin-like (rectangle) domains structure, functionally significant 29 kDa region, M-domain with 4 phosphorylation sites and cardiac specific C5 insert. Domains involved with sarcomeric protein binding are indicated.
Cardiac MyBP-C consists of a series of eight C2-type immumoglobulin and three type-III fibronectin domains connected by linkers of varying length and flexibility (Fig. 1B). MyBP-C is specifically localized to two regions (C-zones) of the thick filament (Fig. 1A) through high affinity interactions between MyBP-C’s C-terminal domains and the thick filament backbone [11]. MyBP-C’s N-terminal domains extend radially from the thick filament [33,36,56] and are believed to modulate actomyosin contractile function by reversible binding to myosin’s motor domain and/or actin (Fig. 2 B–F). Specifically, MyBP-C’s N terminus can activate actomyosin force generation [18,29] and thin filament sliding at low calcium concentrations [49] while inhibiting the maximal sliding velocity of fully calcium-activated thin filaments [49] as illustrated in Fig 2A. With two potential binding partners (i.e. myosin and actin), are MyBP-C’s functional roles in activation and inhibition of actomyosin contractility the result of two independent mechanisms associated with a given binding partner? Although in vitro experiments confirm cardiac MyBP-C’s N-terminal domains interact with both the myosin motor domain and actin (Fig. 1B), the functional relevance of each binding partner is still vigorously debated [46,50] and will be the focus of this review, with unresolved issues in the literature highlighted in italics.
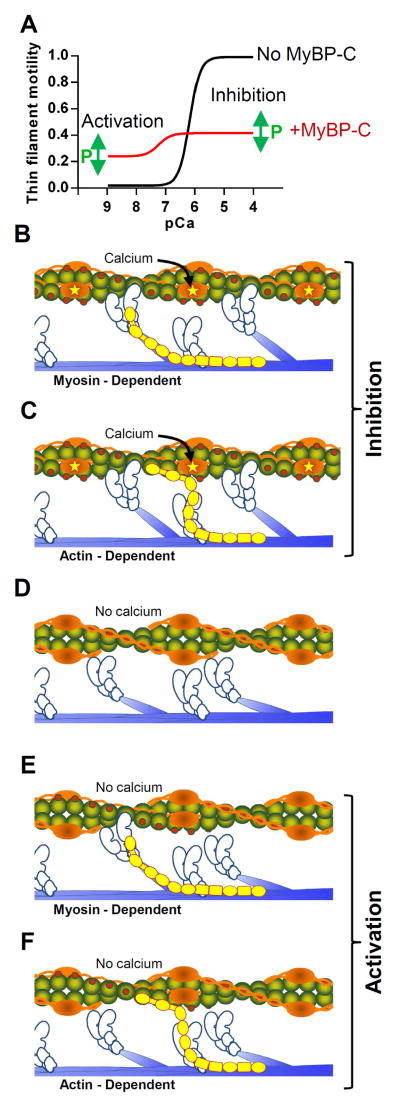
Figure 2.
A) Representative thin filament motility-pCa (−log calcium concentration) plots for regulated thin filaments sliding over myosin in the absence of MyBP-C (black) and presence of N-terminal fragments including domains C1-C2 (red). Phosphorylation within the M-domain presumably has the ability to alter both the activating and inhibitory effects of MyBP-C. B and C) Potential interactions between MyBP-C’s N-terminal domains with the myosin motor domain or actin when troponin is saturated with calcium inhibiting maximal velocity. D) In the absence of calcium or MyBP-C, tropomyosin blocks the myosin binding sites on the thin filament preventing motion. E and F) Potential interactions between MyBP-C’s N-terminal domains with the myosin motor domain or actin in the absence of calcium activating thin filaments.
Cardiac Specific MyBP-C Structure and Phosphorylation
Although MyBP-C is found in both skeletal and cardiac muscles, the cardiac isoform has several unique structural features that may be critical to its molecular function. The first is the presence of C0, an N-terminal Ig domain that is absent in both fast and slow skeletal muscle isoforms. The second is the presence of cardiac specific residues within the ~100 amino acid linker connecting C1 and C2, termed the M-domain or MyBP-C motif [13]. Within the M-domain, four phosphorylatable serines are specific substrates for multiple kinases, such as PKA, PKC, PKD and CaMKII (see review [4]). In human cardiac MyBP-C, a serine within the proline-alanine rich linker between C0 and C1 can also be phosphorylated by GSK3β [30], and additional potential phosphorylation sites have also been located throughout the molecule [26]. The third cardiac specific structure is a 28 amino acid insert in the cardiac C5 Ig domain [21] with as yet unknown function.
Both the proline-alanine rich linker and M-domain are largely unstructured, although the M-domain has three short α-helices near the C-terminus [20]. The M-domain adopts a globular conformation in solution [23] but is freely extensible under low tensile forces applied using atomic force microscopy [24,35]. Phosphorylation of the four serines within the mouse cardiac M-domain stabilizes this globular conformation [35] and may increase α-helical content within the domain [20,35]. Interestingly, these serines are highly phosphorylated in healthy mouse and human hearts but dephosphorylated during cardiac disease [10,22,52]. This suggests that high levels of phosphorylation and its associated stabilized structure are required for normal cardiac function but whether M-domain dephosphorylation leads to contractile dysfunction and heart failure or is a consequence of heart failure has yet to be determined. In addition, with multiple phosphorylation sites within the M-domain, can the M-domain structural stability be graded with the number of phosphorylated sites, thus adding a measure of tunability in response to external stimuli?
MyBP-C Binds both Myosin and Actin
MyBP-C is anchored to the thick filament backbone through C-terminal domain (C8-C10) interactions with myosin rods and titin [11,12,42]. The 7-9 MyBP-C stripes that identify MyBP-C’s location within the C-zone are associated with MyBP-C’s C8-C10 domains interacting with the 11-domain super-repeat region of titin [12]. These interactions position 3 molecules of MyBP-C every 42.9 nm, i.e. the distance between C-zone stripes [32]. Yeast two-hybrid binding assays suggest that these 3 MyBP-C molecules form a trimeric collar around the thick filament through intramolecular contacts between C5 on one MyBP-C molecule and the C8 domain of a neighboring molecule [36]. However, at least 3 MyBP-C domains appear to run axially along both mouse [64] and human [3] native thick filaments, as visualized by electron microscopy, in support of a previously proposed axial arrangement for MyBP-C [56]. Although the arrangement of MyBP-C molecules at each stripe remains unresolved, the presence of MyBP-C increases thick filament flexural rigidity [40] and the stiffness of the myofilament lattice [45,44].
While MyBP-C’s C terminus anchors the molecule to the thick filament backbone and appears to play structural and mechanical roles, what role does MyBP-C’s N terminus play in modulating actomyosin contractile function? After the initial discovery that skeletal MyBP-C was co-purified in myosin preparations [41], its ability to bind myosin’s proteolytic HMM and S2 subfragments [57] as well as actin in vitro were demonstrated [37,38]. However, the specific MyBP-C domains responsible for actin and myosin binding were unknown. Studies using human cardiac MyBP-C N-terminal fragments demonstrate that domains encompassing C1 through C2 can bind to myosin S2 [1,2,14], with binding abolished by phosphorylation within the M-domain [15]. In addition, the C0 domain can bind to the myosin regulatory light chain [48]. These myosin binding data contributed to an early model in which MyBP-C’s N terminus acts as a tether between the myosin motor domain and the thick filament rod to modulate myosin function. In support of this model, x-ray data suggest that MyBP-C maintains the myosin motor domain near the thick filament backbone when the muscle is relaxed [7]. MyBP-C phosphorylation and its resultant diminished myosin S2 binding would release the motor domain and thus allow its center of mass to approach the thin filament [8,31,60]. However, the same MyBP-C N-terminal domains that bind the myosin motor domain can also bind F-actin, suggesting that actin binding may also play a mechanistic role in regulating actomyosin function.
While binding assays show C0 alone can bind actin [28] structural data using small angle X-ray scattering and electron microscopy to visualize F-actin decorated with various expressed C0 through C3 N-terminal fragments show that these N-terminal domains can bind to actin’s subdomain 1 in a stereospecific manner [25,39,43,63]. A direct measure of MyBP-C actin binding was observed at the single molecule level, as expressed mouse C0-C1 fragments were capable of reversibly binding to an F-actin filament in the laser trap assay [61]. Morever, addition of the first 17 amino acids of the M-domain to the C0-C1 fragment (C0-C1f) allowed for stereospecific binding, which was ablated by substituting alanines for the 3 arginines within these 17 amino acids [61]. This finding suggests that at least one binding site exists within C0-C1 and another within the M-domain [53]. Interestingly, M-domain phosphorylation diminishes MyBP-C’s N-terminal binding to F-actin as is the case for myosin binding [5,53,61]. If phosphorylation affects M-domain structure and/or stability, does phosphorylation in effect limit myosin and actin binding site accessibility and can this accessibility be graded by the extent of M-domain phosphorylation?
Are the sites of MyBP-C N-terminal binding to myosin and actin distinct? To complicate matters, a recent yeast two-hybrid assay highlighted the presence of multiple N-terminal binding sites for both myosin S2 and actin within the C1 and M-domains [6]. These sites were associated with clusters of positively charged amino acids, suggesting electrostatic interactions between MyBP-C’s N terminus and both myosin S2 and actin [6] and that these sites may overlap in terms of myosin S2 and actin binding. For example, substitution of alanines for arginines within the first N-terminal 17 amino acids of the mouse M-domain, as mentioned above, ablates stereospecific actin binding in the laser trap assay [61], decreases C1-C2/actin cosedimentation [5], but also reduces myosin S2 binding in the yeast two-hybrid assays [6]. In addition, recent reports challenge the specificity of MyBP-C N-terminal binding to F-actin demonstrating that C-terminal domains can also bind actin [9,50]. These conflicting results raise uncertainty concerning the specific domains and residues involved with myosin motor domain and actin binding.
In vitro model systems of cardiac contractility
Our understanding of MyBP-C’s capacity to modulate cardiac contractility has benefited tremendously from intact heart and muscle fiber studies. However, these multicellular preparations make molecular-level interpretations difficult. Although solution studies in vitro are far simpler; they lack the spatial relations between the thick and thin filaments that exist in vivo. Thus, we recently developed a single fluorescent particle assay to visualize ~250 nm shards of fluorescently-labeled actin filaments sliding along native mouse cardiac thick filaments [47]. These native thick filaments contained MyBP-C in its endogenous stoichiometry and location, i.e. the C-zone. As an actin shard landed near the tip of the thick filament that is devoid of MyBP-C, it was propelled by the myosin heads towards the C-zone (Fig. 1A) at which point the actin’s sliding velocity was reduced ~2-fold [47]. This localized slowing indicates that MyBP-C inhibits actomyosin interactions where it exists within the thick filament rather than propagating its effect through the thick filament backbone. The slowing of actin was attributed to the first 29 kDa of the N terminus (C0-C1f: C0-C1 plus 17 amino acids of the M-domain) as proteolytic removal of this domain prevented MyBP-C’s inhibition of velocity on the thick filament. In parallel experiments, the addition of the expressed 29 kDa fragment (C0-C1f) in the motility assay was sufficient to inhibit the maximal sliding velocity of F-actin sliding over a surface of randomly oriented myosin molecules [47,61]. This finding suggests that the inhibition of actin velocity previously observed in the motility assay using expressed C0-C2, C1-C2, and C0-C3 [49,62,61] must be due to the C1 domain and the first 17 residues of the M-domain, since C0-C1 was incapable of inhibiting actin velocity [49,61]. Through which binding partner, i.e. myosin and/or actin, and how these N-terminal fragments inhibit velocity is still unanswered? For example, MyBP-C’s N-terminal domains binding to myosin’s motor domain may alter myosin’s detachment rate from the actin filament, thus slowing velocity (Fig 2B), independent of a tether between MyBP-C’s C-terminal domains and the thick filament backbone [17,29,49,51,54]. However, actin binding as evidenced by N-terminal interactions with actin in the laser trap assay [61] may also create an internal load slowing actomyosin generated motion (Fig. 2C). Therefore, direct experimental evidence using expressed fragments that are only capable of interacting with a single binding partner may be required to provide unequivocal evidence for MyBP-C N-terminal binding to the myosin motor domain or actin as being the basis for MyBP-C’s mechanical mechanism.
With MyBP-C’s N-terminal binding to either myosin or actin, the potential exists that MyBP-C can modulate thin filament activation, as observed in fiber studies where chemical extraction or genetic mutagenesis of MyBP-C resulted in shifts in the calcium sensitivity of force production [19,27]. Deciphering the molecular mechanism by which MyBP-C can affect thin filament activation by calcium lends itself to the use of the in vitro motility assay. Using thin filaments, i.e. actin filaments decorated with the calcium regulatory proteins, troponin and tropomyosin, one observes sigmoidal calcium-dependant increases in both the sliding velocity and fraction of motile filaments between ~100 nM to 10 uM calcium (Fig 2A). At <100nM calcium, where little to no thin filament motion is expected (Fig. 2A, black line), the presence of whole MyBP-C [51,55] or N-terminal fragments including at least the C1 and M-domains [49] activate the thin filament in a concentration-dependant manner at sub-maximal activating levels of calcium (Fig 2A, red line). As calcium and velocity increase, the inhibitory effect of MyBP-C again limits the sliding velocity (Fig 2A, red line). Therefore, MyBP-C can both activate the thin filament at low calcium while inhibiting actin filament velocity at high calcium, both effects of which should contribute to MyBP-C’s capacity to modulate cardiac contractility.
A widely accepted model of thin filament activation [34,59] suggests that in the absence of calcium, troponin and tropomyosin “block” myosin binding to actin’s subdomain 1 (Fig. 2 D). Upon calcium binding to troponin, tropomyosin moves azimuthally to a “closed” position that partially exposes myosin binding sites on the thin filament so that myosin can weakly bind. Then upon myosin’s transition from the weakly- to the strongly-bound state, myosin physically moves tropomyosin into its “open” position, allowing additional myosin molecules to freely bind within the span of a single regulatory unit. Therefore, thin filament activation by MyBP-C binding to actin could occur if MyBP-C’s N terminus acts as a molecular wedge to pin tropomyosin away from subdomain 1 (Fig. 2F) and expose adjacent myosin binding sites on the thin filament [39]. Thus, MyBP-C would behave functionally like calcium to act as a parallel system for thin filament activation. But once again, thin filament activation could arise from MyBP-C binding to the myosin motor domain and S2 segment (Fig. 2E). For example, the addition of C1-C2 fragments to fibers increased calcium sensitivity of force production [5,17,29]. Since C1-C2 binds the myosin S1-S2 junction, MyBP-C may limit the rotational freedom of the myosin motor domain in turn increasing myosin’s probability of strong-binding to actin and/or limiting myosin’s detachment rate, thus activating the thin filament as do NEM- and rigor-myosin [58].
Summary and Future Perspectives
The importance of cardiac MyBP-C on contractility is clearly evident from the association between mutations in the human MYBPC3 gene and heart disease. Much progress has been made in understanding MyBP-C molecular function within the sarcomere but the ability of its N-terminal domains to bind both the myosin motor domain and actin have generated considerable questions regarding its molecular mechanisms. The fact that MyBP-C plays two functional roles in the heart, i.e. revving the contractile engine during the early phase of contraction and then applying the molecular brakes once the sarcomere begins shortening adds an additional level of mechanistic complexity. However, this also makes MyBP-C an exciting target for therapeutic interventions in disease.
Acknowledgments
This work was supported by National Heart, Lung, Blood Institute grants HL007647, HL007944, and HL059408.
References
- 1.Ababou A, Gautel M, Pfuhl M. Dissecting the N-terminal myosin binding site of human cardiac myosin-binding protein C. Structure and myosin binding of domain C2. The Journal of biological chemistry. 2007;282 (12):9204–9215. doi: 10.1074/jbc.M610899200. [DOI] [PubMed] [Google Scholar]
- 2.Ababou A, Rostkova E, Mistry S, Le Masurier C, Gautel M, Pfuhl M. Myosin binding protein C positioned to play a key role in regulation of muscle contraction: structure and interactions of domain C1. Journal of molecular biology. 2008;384 (3):615–630. doi: 10.1016/j.jmb.2008.09.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Al-Khayat HA, Kensler RW, Squire JM, Marston SB, Morris EP. Atomic model of the human cardiac muscle myosin filament. Proceedings of the National Academy of Sciences of the United States of America. 2013;110 (1):318–323. doi: 10.1073/pnas.1212708110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bardswell SC, Cuello F, Kentish JC, Avkiran M. cMyBP-C as a promiscuous substrate: phosphorylation by non-PKA kinases and its potential significance. Journal of muscle research and cell motility. 2012;33 (1):53–60. doi: 10.1007/s10974-011-9276-3. [DOI] [PubMed] [Google Scholar]
- 5.Bezold KL, Shaffer JF, Khosa JK, Hoye ER, Harris SP. A gain-of-function mutation in the M-domain of cardiac myosin-binding protein-C increases binding to actin. The Journal of biological chemistry. 2013;288 (30):21496–21505. doi: 10.1074/jbc.M113.474346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bhuiyan MS, Gulick J, Osinska H, Gupta M, Robbins J. Determination of the critical residues responsible for cardiac myosin binding protein C’s interactions. Journal of molecular and cellular cardiology. 2012;53 (6):838–847. doi: 10.1016/j.yjmcc.2012.08.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Colson BA, Bekyarova T, Fitzsimons DP, Irving TC, Moss RL. Radial displacement of myosin cross-bridges in mouse myocardium due to ablation of myosin binding protein-C. Journal of molecular biology. 2007;367 (1):36–41. doi: 10.1016/j.jmb.2006.12.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Colson BA, Bekyarova T, Locher MR, Fitzsimons DP, Irving TC, Moss RL. Protein kinase A-mediated phosphorylation of cMyBP-C increases proximity of myosin heads to actin in resting myocardium. Circulation research. 2008;103 (3):244–251. doi: 10.1161/CIRCRESAHA.108.178996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Colson BA, Rybakova IN, Prochniewicz E, Moss RL, Thomas DD. Cardiac myosin binding protein-C restricts intrafilament torsional dynamics of actin in a phosphorylation-dependent manner. Proceedings of the National Academy of Sciences of the United States of America. 2012;109 (50):20437–20442. doi: 10.1073/pnas.1213027109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.El-Armouche A, Pohlmann L, Schlossarek S, Starbatty J, Yeh YH, Nattel S, Dobrev D, Eschenhagen T, Carrier L. Decreased phosphorylation levels of cardiac myosin-binding protein-C in human and experimental heart failure. Journal of molecular and cellular cardiology. 2007;43 (2):223–229. doi: 10.1016/j.yjmcc.2007.05.003. [DOI] [PubMed] [Google Scholar]
- 11.Flashman E, Watkins H, Redwood C. Localization of the binding site of the C-terminal domain of cardiac myosin-binding protein-C on the myosin rod. The Biochemical journal. 2007;401 (1):97–102. doi: 10.1042/BJ20060500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Freiburg A, Gautel M. A molecular map of the interactions between titin and myosin-binding protein C. Implications for sarcomeric assembly in familial hypertrophic cardiomyopathy. European journal of biochemistry/FEBS. 1996;235 (1–2):317–323. doi: 10.1111/j.1432-1033.1996.00317.x. [DOI] [PubMed] [Google Scholar]
- 13.Gautel M, Zuffardi O, Freiburg A, Labeit S. Phosphorylation switches specific for the cardiac isoform of myosin binding protein-C: a modulator of cardiac contraction? The EMBO journal. 1995;14 (9):1952–1960. doi: 10.1002/j.1460-2075.1995.tb07187.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gruen M, Gautel M. Mutations in beta-myosin S2 that cause familial hypertrophic cardiomyopathy (FHC) abolish the interaction with the regulatory domain of myosin-binding protein-C. Journal of molecular biology. 1999;286 (3):933–949. doi: 10.1006/jmbi.1998.2522. [DOI] [PubMed] [Google Scholar]
- 15.Gruen M, Prinz H, Gautel M. cAPK-phosphorylation controls the interaction of the regulatory domain of cardiac myosin binding protein C with myosin-S2 in an on-off fashion. FEBS letters. 1999;453 (3):254–259. doi: 10.1016/s0014-5793(99)00727-9. [DOI] [PubMed] [Google Scholar]
- 16.Harris SP, Lyons RG, Bezold KL. In the thick of it: HCM-causing mutations in myosin binding proteins of the thick filament. Circulation research. 2011;108 (6):751–764. doi: 10.1161/CIRCRESAHA.110.231670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Harris SP, Rostkova E, Gautel M, Moss RL. Binding of myosin binding protein-C to myosin subfragment S2 affects contractility independent of a tether mechanism. Circulation research. 2004;95 (9):930–936. doi: 10.1161/01.RES.0000147312.02673.56. [DOI] [PubMed] [Google Scholar]
- 18.Herron TJ, Rostkova E, Kunst G, Chaturvedi R, Gautel M, Kentish JC. Activation of myocardial contraction by the N-terminal domains of myosin binding protein-C. Circulation research. 2006;98 (10):1290–1298. doi: 10.1161/01.RES.0000222059.54917.ef. [DOI] [PubMed] [Google Scholar]
- 19.Hofmann PA, Hartzell HC, Moss RL. Alterations in Ca2+ sensitive tension due to partial extraction of C-protein from rat skinned cardiac myocytes and rabbit skeletal muscle fibers. The Journal of general physiology. 1991;97 (6):1141–1163. doi: 10.1085/jgp.97.6.1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Howarth JW, Ramisetti S, Nolan K, Sadayappan S, Rosevear PR. Structural insight into unique cardiac myosin-binding protein-C motif: a partially folded domain. The Journal of biological chemistry. 2012;287 (11):8254–8262. doi: 10.1074/jbc.M111.309591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Idowu SM, Gautel M, Perkins SJ, Pfuhl M. Structure, stability and dynamics of the central domain of cardiac myosin binding protein C (MyBP-C): implications for multidomain assembly and causes for cardiomyopathy. Journal of molecular biology. 2003;329 (4):745–761. doi: 10.1016/s0022-2836(03)00425-x. [DOI] [PubMed] [Google Scholar]
- 22.Jacques AM, Copeland O, Messer AE, Gallon CE, King K, McKenna WJ, Tsang VT, Marston SB. Myosin binding protein C phosphorylation in normal, hypertrophic and failing human heart muscle. Journal of molecular and cellular cardiology. 2008;45 (2):209–216. doi: 10.1016/j.yjmcc.2008.05.020. [DOI] [PubMed] [Google Scholar]
- 23.Jeffries CM, Whitten AE, Harris SP, Trewhella J. Small-angle X-ray scattering reveals the N-terminal domain organization of cardiac myosin binding protein C. Journal of molecular biology. 2008;377 (4):1186–1199. doi: 10.1016/j.jmb.2008.01.080. [DOI] [PubMed] [Google Scholar]
- 24.Karsai A, Kellermayer MS, Harris SP. Mechanical unfolding of cardiac myosin binding protein-C by atomic force microscopy. Biophysical journal. 2011;101 (8):1968–1977. doi: 10.1016/j.bpj.2011.08.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kensler RW, Shaffer JF, Harris SP. Binding of the N-terminal fragment C0-C2 of cardiac MyBP-C to cardiac F-actin. Journal of structural biology. 2011;174 (1):44–51. doi: 10.1016/j.jsb.2010.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kooij V, Holewinski RJ, Murphy AM, Van Eyk JE. Characterization of the cardiac myosin binding protein-C phosphoproteome in healthy and failing human hearts. Journal of molecular and cellular cardiology. 2013;60:116–120. doi: 10.1016/j.yjmcc.2013.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Korte FS, McDonald KS, Harris SP, Moss RL. Loaded shortening, power output, and rate of force redevelopment are increased with knockout of cardiac myosin binding protein-C. Circulation research. 2003;93 (8):752–758. doi: 10.1161/01.RES.0000096363.85588.9A. [DOI] [PubMed] [Google Scholar]
- 28.Kulikovskaya I, McClellan G, Flavigny J, Carrier L, Winegrad S. Effect of MyBP-C binding to actin on contractility in heart muscle. The Journal of general physiology. 2003;122 (6):761–774. doi: 10.1085/jgp.200308941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kunst G, Kress KR, Gruen M, Uttenweiler D, Gautel M, Fink RH. Myosin binding protein C, a phosphorylation-dependent force regulator in muscle that controls the attachment of myosin heads by its interaction with myosin S2. Circulation research. 2000;86 (1):51–58. doi: 10.1161/01.res.86.1.51. [DOI] [PubMed] [Google Scholar]
- 30.Kuster DW, Sequeira V, Najafi A, Boontje NM, Wijnker PJ, Witjas-Paalberends ER, Marston SB, Dos Remedios CG, Carrier L, Demmers JA, Redwood C, Sadayappan S, van der Velden J. GSK3beta phosphorylates newly identified site in the proline-alanine-rich region of cardiac myosin-binding protein C and alters cross-bridge cycling kinetics in human: short communication. Circulation research. 2013;112 (4):633–639. doi: 10.1161/CIRCRESAHA.112.275602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Levine R, Weisberg A, Kulikovskaya I, McClellan G, Winegrad S. Multiple structures of thick filaments in resting cardiac muscle and their influence on cross-bridge interactions. Biophysical journal. 2001;81 (2):1070–1082. doi: 10.1016/S0006-3495(01)75764-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Luther PK, Bennett PM, Knupp C, Craig R, Padron R, Harris SP, Patel J, Moss RL. Understanding the organisation and role of myosin binding protein C in normal striated muscle by comparison with MyBP-C knockout cardiac muscle. Journal of molecular biology. 2008;384 (1):60–72. doi: 10.1016/j.jmb.2008.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Luther PK, Winkler H, Taylor K, Zoghbi ME, Craig R, Padron R, Squire JM, Liu J. Direct visualization of myosin-binding protein C bridging myosin and actin filaments in intact muscle. Proceedings of the National Academy of Sciences of the United States of America. 2011;108 (28):11423–11428. doi: 10.1073/pnas.1103216108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.McKillop DF, Geeves MA. Regulation of the interaction between actin and myosin subfragment 1: evidence for three states of the thin filament. Biophysical journal. 1993;65 (2):693–701. doi: 10.1016/S0006-3495(93)81110-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Michalek AJ, Howarth JW, Gulick J, Previs MJ, Robbins J, Rosevear PR, Warshaw DM. Phosphorylation modulates the mechanical stability of the cardiac myosin-binding protein C motif. Biophysical journal. 2013;104 (2):442–452. doi: 10.1016/j.bpj.2012.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Moolman-Smook J, Flashman E, de Lange W, Li Z, Corfield V, Redwood C, Watkins H. Identification of novel interactions between domains of Myosin binding protein-C that are modulated by hypertrophic cardiomyopathy missense mutations. Circulation research. 2002;91 (8):704–711. doi: 10.1161/01.res.0000036750.81083.83. [DOI] [PubMed] [Google Scholar]
- 37.Moos C. Fluorescence microscope study of the binding of added C protein to skeletal muscle myofibrils. The Journal of cell biology. 1981;90 (1):25–31. doi: 10.1083/jcb.90.1.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Moos C, Mason CM, Besterman JM, Feng IN, Dubin JH. The binding of skeletal muscle C-protein to F-actin, and its relation to the interaction of actin with myosin subfragment-1. Journal of molecular biology. 1978;124 (4):571–586. doi: 10.1016/0022-2836(78)90172-9. [DOI] [PubMed] [Google Scholar]
- 39.Mun JY, Gulick J, Robbins J, Woodhead J, Lehman W, Craig R. Electron microscopy and 3D reconstruction of F-actin decorated with cardiac myosin-binding protein C (cMyBP-C) Journal of molecular biology. 2011;410 (2):214–225. doi: 10.1016/j.jmb.2011.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nyland LR, Palmer BM, Chen Z, Maughan DW, Seidman CE, Seidman JG, Kreplak L, Vigoreaux JO. Cardiac myosin binding protein-C is essential for thick-filament stability and flexural rigidity. Biophysical journal. 2009;96 (8):3273–3280. doi: 10.1016/j.bpj.2008.12.3946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Offer G, Moos C, Starr R. A new protein of the thick filaments of vertebrate skeletal myofibrils. Extractions, purification and characterization. Journal of molecular biology. 1973;74 (4):653–676. doi: 10.1016/0022-2836(73)90055-7. [DOI] [PubMed] [Google Scholar]
- 42.Okagaki T, Weber FE, Fischman DA, Vaughan KT, Mikawa T, Reinach FC. The major myosin-binding domain of skeletal muscle MyBP-C (C protein) resides in the COOH-terminal, immunoglobulin C2 motif. The Journal of cell biology. 1993;123 (3):619–626. doi: 10.1083/jcb.123.3.619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Orlova A, Galkin VE, Jeffries CM, Egelman EH, Trewhella J. The N-terminal domains of myosin binding protein C can bind polymorphically to F-actin. Journal of molecular biology. 2011;412 (3):379–386. doi: 10.1016/j.jmb.2011.07.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Palmer BM, McConnell BK, Li GH, Seidman CE, Seidman JG, Irving TC, Alpert NR, Maughan DW. Reduced cross-bridge dependent stiffness of skinned myocardium from mice lacking cardiac myosin binding protein-C. Molecular and cellular biochemistry. 2004;263 (1–2):73–80. doi: 10.1023/B:MCBI.0000041849.60591.45. [DOI] [PubMed] [Google Scholar]
- 45.Palmer BM, Sadayappan S, Wang Y, Weith AE, Previs MJ, Bekyarova T, Irving TC, Robbins J, Maughan DW. Roles for cardiac MyBP-C in maintaining myofilament lattice rigidity and prolonging myosin cross-bridge lifetime. Biophysical journal. 2011;101 (7):1661–1669. doi: 10.1016/j.bpj.2011.08.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pfuhl M, Gautel M. Structure, interactions and function of the N-terminus of cardiac myosin binding protein C (MyBP-C): who does what, with what, and to whom? Journal of muscle research and cell motility. 2012;33 (1):83–94. doi: 10.1007/s10974-012-9291-z. [DOI] [PubMed] [Google Scholar]
- 47.Previs MJ, Beck Previs S, Gulick J, Robbins J, Warshaw DM. Molecular mechanics of cardiac myosin-binding protein C in native thick filaments. Science. 2012;337 (6099):1215–1218. doi: 10.1126/science.1223602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ratti J, Rostkova E, Gautel M, Pfuhl M. Structure and interactions of myosin-binding protein C domain C0: cardiac-specific regulation of myosin at its neck? The Journal of biological chemistry. 2011;286 (14):12650–12658. doi: 10.1074/jbc.M110.156646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Razumova MV, Shaffer JF, Tu AY, Flint GV, Regnier M, Harris SP. Effects of the N-terminal domains of myosin binding protein-C in an in vitro motility assay: Evidence for long-lived cross-bridges. The Journal of biological chemistry. 2006;281 (47):35846–35854. doi: 10.1074/jbc.M606949200. [DOI] [PubMed] [Google Scholar]
- 50.Rybakova IN, Greaser ML, Moss RL. Myosin binding protein C interaction with actin: characterization and mapping of the binding site. The Journal of biological chemistry. 2011;286 (3):2008–2016. doi: 10.1074/jbc.M110.170605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Saber W, Begin KJ, Warshaw DM, VanBuren P. Cardiac myosin binding protein-C modulates actomyosin binding and kinetics in the in vitro motility assay. Journal of molecular and cellular cardiology. 2008;44 (6):1053–1061. doi: 10.1016/j.yjmcc.2008.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sadayappan S, Gulick J, Osinska H, Martin LA, Hahn HS, Dorn GW, 2nd, Klevitsky R, Seidman CE, Seidman JG, Robbins J. Cardiac myosin-binding protein-C phosphorylation and cardiac function. Circulation research. 2005;97 (11):1156–1163. doi: 10.1161/01.RES.0000190605.79013.4d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Shaffer JF, Kensler RW, Harris SP. The myosin-binding protein C motif binds to F-actin in a phosphorylation-sensitive manner. The Journal of biological chemistry. 2009;284 (18):12318–12327. doi: 10.1074/jbc.M808850200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Shaffer JF, Razumova MV, Tu AY, Regnier M, Harris SP. Myosin S2 is not required for effects of myosin binding protein-C on motility. FEBS letters. 2007;581 (7):1501–1504. doi: 10.1016/j.febslet.2007.03.007. [DOI] [PubMed] [Google Scholar]
- 55.Shchepkin DV, Kopylova GV, Nikitina LV, Katsnelson LB, Bershitsky SY. Effects of cardiac myosin binding protein-C on the regulation of interaction of cardiac myosin with thin filament in an in vitro motility assay. Biochemical and biophysical research communications. 2010;401 (1):159–163. doi: 10.1016/j.bbrc.2010.09.040. [DOI] [PubMed] [Google Scholar]
- 56.Squire JM, HAAL-K, Harford JJ, Hudson L, Irving TC, Knupp C, Mok NS, Reedy MK. Myosin filament structure and myosin crossbridge dynamics in fish and insect muscles. Advances in experimental medicine and biology. 2003;538:251–266. doi: 10.1007/978-1-4419-9029-7_24. discussion 266. [DOI] [PubMed] [Google Scholar]
- 57.Starr R, Offer G. The interaction of C-protein with heavy meromyosin and subfragment-2. The Biochemical journal. 1978;171 (3):813–816. doi: 10.1042/bj1710813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Swartz DR, Moss RL. Influence of a strong-binding myosin analogue on calcium-sensitive mechanical properties of skinned skeletal muscle fibers. The Journal of biological chemistry. 1992;267 (28):20497–20506. [PubMed] [Google Scholar]
- 59.Tobacman LS, Butters CA. A new model of cooperative myosin-thin filament binding. The Journal of biological chemistry. 2000;275 (36):27587–27593. doi: 10.1074/jbc.M003648200. [DOI] [PubMed] [Google Scholar]
- 60.Weisberg A, Winegrad S. Alteration of myosin cross bridges by phosphorylation of myosin-binding protein C in cardiac muscle. Proceedings of the National Academy of Sciences of the United States of America. 1996;93 (17):8999–9003. doi: 10.1073/pnas.93.17.8999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Weith A, Sadayappan S, Gulick J, Previs MJ, Vanburen P, Robbins J, Warshaw DM. Unique single molecule binding of cardiac myosin binding protein-C to actin and phosphorylation-dependent inhibition of actomyosin motility requires 17 amino acids of the motif domain. Journal of molecular and cellular cardiology. 2012;52 (1):219–227. doi: 10.1016/j.yjmcc.2011.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Weith AE, Previs MJ, Hoeprich GJ, Previs SB, Gulick J, Robbins J, Warshaw DM. The extent of cardiac myosin binding protein-C phosphorylation modulates actomyosin function in a graded manner. Journal of muscle research and cell motility. 2012;33 (6):449–459. doi: 10.1007/s10974-012-9312-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Whitten AE, Jeffries CM, Harris SP, Trewhella J. Cardiac myosin-binding protein C decorates F-actin: implications for cardiac function. Proceedings of the National Academy of Sciences of the United States of America. 2008;105 (47):18360–18365. doi: 10.1073/pnas.0808903105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zoghbi ME, Woodhead JL, Moss RL, Craig R. Three-dimensional structure of vertebrate cardiac muscle myosin filaments. Proceedings of the National Academy of Sciences of the United States of America. 2008;105 (7):2386–2390. doi: 10.1073/pnas.0708912105. [DOI] [PMC free article] [PubMed] [Google Scholar]