Abstract
Bioengineering advances have made it possible to fundamentally alter the genetic codes of organisms. However, the evolutionary consequences of expanding an organism's genetic code with a non-canonical amino acid are poorly understood. Here we show that bacteriophages evolved on a host that incorporates 3-iodotyrosine at the amber stop codon acquired neutral and beneficial mutations to this new amino acid in their proteins, demonstrating that an expanded genetic code increases evolvability.
The evolution of life on Earth has been dictated by the underlying constancy of the nearly universal twenty amino acid genetic code1,2. However, examples of natural genetic codes that have been functionally expanded with a 21st amino acid3 and the multitude of known post-translational protein modifications suggest that, while aspects of the genetic code have been optimized by evolution4,5, it does not provide the necessary chemical diversity to best perform all functions of potential benefit to organisms. Technologies exist to augment genetic codes with a non-canonical amino acid (ncAA) by introducing an orthogonal aminoacyl-tRNA synthetase (aaRS) and a cognate tRNA recognizing the amber stop codon6,7. We hypothesized that organisms given the ability to encode a 21st amino acid would evolve to utilize this new chemical building block on mutational pathways to higher fitness.
Several challenges can arise when attempting to evolve an organism with a newly expanded genetic code, and the directed evolution of even single proteins with ncAAs has been limited to date8. The organism of interest must first survive the globally disruptive change in how its genomic information is decoded into its proteome2. Next, the reassigned codon must be translated with sufficient efficiency and fidelity for substitutions of the new codon to be beneficial, which is less likely if translating this codon sometimes results in truncated proteins9 or ambiguous amino acid incorporation10. Evolution may also need to proceed for many generations to observe ncAA substitutions because only a small fraction of mutations will result in changes to the reassigned codon. Finally, one must circumvent rejection of the reengineered genetic code in cases where mutations that lead to the ncAA no longer being incorporated into proteins are highly advantageous to the host organism11.
Bacteriophage T7 has been used as a model organism for studies of molecular genetics12,13, experimental evolution14,15, and synthetic biology16. We evolved this bacterial virus in a constant translational environment with an expanded genetic code by passaging it on an RF0 IodoY E. coli host that efficiently incorporates 3-iodotyrosine at amber stop codons due to deletion of protein release factor 1 (RF1) and addition of an engineered aaRS-tRNA pair17,18 (see Supplementary Results, Supplementary Figs. 1–3). Populations of wild-type T7 (WT)12 and a hypermutator variant (Δ2)19 were propagated on cultures of actively growing E. coli hosts by incubating them together until there was visible bacterial lysis due to viral replication and then transferring a fraction of this mixture to a new E. coli culture (Fig. 1a). Lysis time decreased for both WT (from ∼100 to ∼35 minutes) and Δ2 (from ∼120 to ∼45 minutes) populations over 50 serial transfers, indicating that they evolved higher fitness on the RF0 IodoY E. coli host.
Figure 1. Genome evolution of a bacterial virus with a newly expanded genetic code.
a, In the evolution experiment, six populations each of wild-type (WT) and hypermutator (Δ2) bacteriophage T7 were passaged on a Release Factor 1 knockout (RF0) E. coli host strain carrying an orthogonal tRNA and aminoacyl-tRNA synthetase system that recodes the amber stop codon to the non-natural amino acid 3-iodotyrosine. b, E. coli host strains used to test whether evolved phages required amber suppression by 3-iodotyrosine or tyrosine to replicate. c, Relative titer (number of plaques) formed by each evolved bacteriophage population on the host strains in b. Values are the average of three technical replicates with error bars that are 95% confidence limits. P-values for reduced phage number on the alternative hosts compared to RF0 IodoY are for Student's t-tests assuming equal variance and using the Bonferroni correction for multiple testing. d, Locations of point mutations with frequencies ≥5% observed in metagenomic DNA sequencing data for the four bacteriophage populations boxed in c. Red open reading frames encode T7 proteins with known functions13. Mutations of interest in regions highlighted in yellow are discussed in the text and following figures. There were roughly 50 and 400 mutations present with frequencies ≥1% in the evolved WT and Δ2 T7 populations, respectively.
An organism with a newly expanded genetic code is subject to new constraints on codon usage and may have new opportunities for adaptive mutations. The evolutionary response could include: (1) compensatory mutations that restore protein termination, in cases where read-through of a reassigned stop codon is deleterious; (2) genetic drift to reassigned codons at positions where ncAA substitutions are selectively neutral, or (3) mutations to reassigned codons that are beneficial, potentially in ways that would not be possible with a canonical amino acid. Deleterious mutations are highly unlikely to contribute to evolution in our experiment due to the extremely large effective population size (∼107–108 phages transferred each passage).
If amber codons evolve in important or essential genes, these mutations are expected to result in addiction, such that an organism requires an alternative genetic code for viability. To test for this level of dependence, we titered the evolved T7 populations on three E. coli hosts, which either terminate translation (BL21(DE3)), or incorporate 3-iodotyrosine (RF0 IodoY) or tyrosine (RF0 Tyr) at the amber codon (Fig. 1b). All populations produced statistically indistinguishable numbers of phage plaques on RF0 IodoY and RF0 Tyr cells, but one WT and three Δ2 populations produced significantly fewer plaques on the BL21(DE3) host (Fig. 1c), indicating that some phages in these populations had evolved amber codons inside key genes.
To determine exactly what genetic changes that had taken place, we sequenced DNA samples isolated from four evolved T7 populations. Despite a ∼10-fold difference in mutation rates19, the base substitution spectra in Δ2 and WT populations were similar (Supplementary Fig. 4). However, the overall character of genome evolution shifted from directional selection in the wild-type populations (dN/dS = 1.58) to purifying selection in the hypermutator populations (dN/dS = 0.57). Deletions overlapping gene 0.7 that are known to be beneficial13,15 were present, in aggregate, in ∼100% of every sequenced population (Supplementary Fig. 5 and Supplementary Table 1), and there was a deletion overlapping two hypothetical genes in one population (Supplementary Fig. 6).
Among the point mutations present in these populations (Fig. 1d), we observed two substitutions of amber codons that reached high frequencies, one in T7 RNA polymerase (RNAP) and the other in the T7 type II holin. Patching random phage isolates from the respective populations confirmed that each new amber codon was associated with a defect in forming plaques on the BL21(DE3) normal genetic code E. coli host. As anticipated, we also observed compensation for read-through of phage proteins ending in the amber codon. Notably, mutations appeared to restore T7 exonuclease (gene 6) termination in close to 100% of all four sequenced populations (Supplementary Fig. 7).
T7 RNAP is an essential protein that is responsible for transcription of bacteriophage genes13. The Trp-88 to amber mutation in RNAP (Supplementary Fig. 8a) probably has little effect on phage fitness on the RF0 IodoY host. Of 37 base substitutions observed in this gene with frequencies ≥1% across all populations, 21 were synonymous. Thus, the overall molecular signature of selection within RNAP is highly purifying (dN/dS = 0.22). So, even nonsynonymous changes like this amber mutation likely were tolerated only because they did not alter protein function. Furthermore, only one of these 37 mutations was present in a WT population, where fewer neutral mutations are expected. Finally, Trp-88 is located at a surface-exposed site in the structure of T7 RNA polymerase20 where 3-iodotyrosine appears to be accommodated without altering the surrounding molecular structure (Supplementary Fig. 8b).
In contrast, several lines of evidence indicate that the Tyr-39 to amber substitution in the T7 type II holin (gene 17.5) is beneficial to phage fitness (Fig. 2a). Holins regulate lysis timing by forming pores in the bacterial membrane through which lysozyme degrades the cell wall21, and optimizing lysis time is critical to maximizing phage progeny22,23. We found a higher density of point mutations with frequencies ≥1% within gene 17.5 than in the genome at large (Binomial test, P = 1.5 × 10−9). Furthermore, all 22 of these base substitutions in the type II holin were nonsynonymous (Supplementary Table 2), and nearly 100% of phages in each WT and >50% in each Δ2 population have a mutation somewhere in gene 17.5. Together these observations indicate that type II holin activity was under strong directional selection during the evolution experiment.
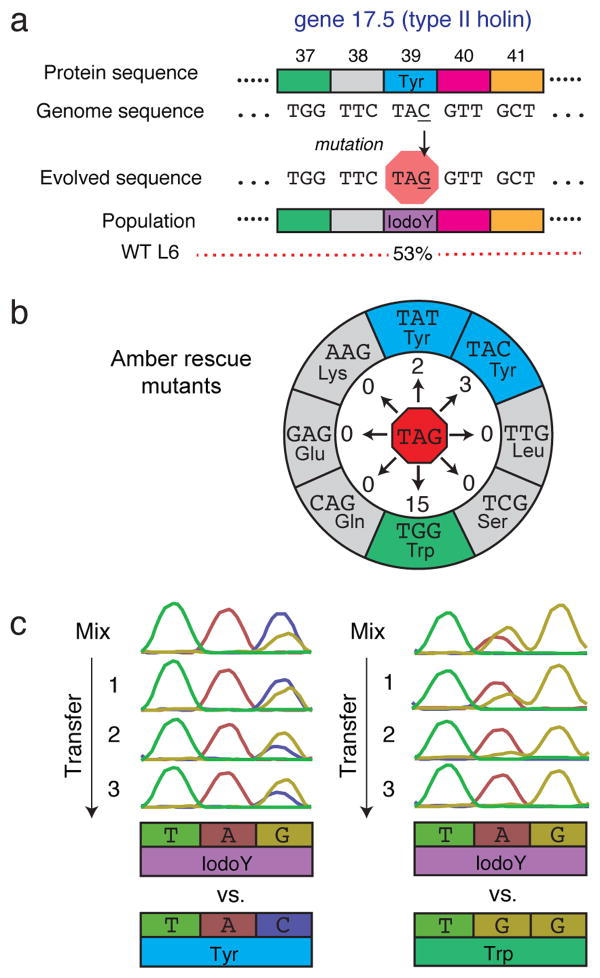
Figure 2. Beneficial amber mutation in the T7 holin II protein.
a, The frequency of an amber mutation resulting in a tyrosine to 3-iodotyrosine substitution in the T7 type II holin (gene 17.5) can explain the reduced titer of population WT L6 on the standard genetic code host (Fig. 1c). Signatures of molecular evolution described in the text strongly suggest that this mutation increases phage fitness on the RF0 IodoY E. coli host. b, Rescue mutants of an evolved WT L6 phage isolate with this amber mutation were obtained by plating on E. coli BL21(DE3) hosts with the standard genetic code and isolating fast-growing plaques. Of the 8 possible neighboring codons accessible by single-base mutations that code for amino acids, only tyrosine and tryptophan mutants were observed. The numbers of rescue mutants found with each codon are indicated at the end of the arrows. c, The evolved phage isolate with the amber codon (TAG) at position 39 of the holin protein was competed against rescue mutants with a tyrosine (TAC) or tryptophan (TGG) at that position. In each competition, lysates of the two phage preconditioned separately were mixed and then passaged on RF0 IodoY cells for three transfers. Sanger sequencing traces at this position show that the amber codon increases in frequency relative to the alternative, indicating that 3-iodotyrosine confers a fitness benefit compared to both the ancestral amino acid and a chemically similar amino acid tolerated at this position.
The amber mutation in the T7 type II holin occurs with the highest frequency (53%) of any coding base substitution in this population and with the highest frequency of any gene 17.5 mutation in any sequenced phage population. This result is even more striking given that it is in a wild-type rather than a hypermutator background. As expected for a highly beneficial mutation, phages with the amber lyse RF0 IodoY cells more completely at earlier times than phages from the same population without it, and this difference is not found on the RF0 Tyr host where tyrosine incorporation at the amber codon restores the normal type II holin sequence (Supplementary Fig. 9).
To directly test whether the 3-iodotyrosine substitution in the type II holin protein was beneficial to phage fitness, we isolated spontaneous rescue mutants of a phage with this amber that could plaque on normal genetic code cells. Only tryptophan and tyrosine mutants were obtained, suggesting that position 39 of the holin is constrained to aromatic amino acids (Fig. 2b). The evolved isolate coding 3-iodotyrosine was significantly more fit than the rescue mutants with tyrosine or tryptophan at this position when they were competed in co-culture under the conditions of the evolution experiment (Fig. 2c). This outcome held for a wide range of transfer volumes that altered the initial multiplicity of infection at each cycle (Supplementary Fig. 10).
We have shown that access to an expanded genetic code increased the evolvability of an organism by characterizing a new beneficial mutation that was only possible with 3-iodotyrosine incorporation into proteins. The readiness with which bacteriophage T7 adapted to and utilized a newly expanded genetic code with a non-canonical amino acid was unexpected. This result establishes that current technologies can effectively thaw Crick's “frozen accident”, opening vast new sequence spaces and novel chemistries for investigation. These capabilities will continue to improve as the genomes of organisms are engineered to have one or more blank codons that can be reassigned to ncAAs without disrupting existing proteins24. Longer evolution experiments are likely to result in codon substitutions that addict organisms to the unique functionality of an introduced ncAA for survival. These may be useful for semantic biocontainment of genetically modified organisms25 and for whole-genome evolutionary approaches for further improving ncAA incorporation. Finally, this general research strategy makes it possible to experimentally address questions about the optimality of the natural genetic code and to discover unexpected utility for genetically encoded ncAAs by the directed evolution of proteins and entire organisms.
Online Methods
E. coli host strains
RF0 IodoY18 was constructed from BL21(DE3) E. coli (Invitrogen), by creating a gentamicin-resistant F plasmid containing several key E. coli genes with re-coded stop codons, knocking out RF1 in the chromosome, and adding a second plasmid containing a modified Methanococcus jannaschii tyrosyl-tRNA synthetase and UAG-decoding tRNA for specifically incorporating 3-iodotyrosine at the amber stop codon17. Cultures of RF0 IodoY were supplemented with 20 μg/mL chloramphenicol and 10 μg/mL gentamycin. RF0 Tyr was generated by replacing the chloramphenicol resistance marker in the RF0 IodoY aaRS-tRNA plasmid with a spectinomycin resistance gene and by replacing the 3-iodotyrosine aaRS with the wild-type M. jannaschii tyrosyl-tRNA synthetase (GenBank: NP_247363.1). RF0 Tyr cultures were supplemented with 60 μg/mL spectinomycin and 10 μg/mL gentamycin. All of these strains contain a copy of T7 RNAP in the E. coli host genome under control of the lac UV5 promoter. This promoter was not induced during any of our experiments, and background expression of this host copy of T7 RNAP was apparently low enough that phages with an amber stop codon in their copy of the T7 RNAP gene could not plaque on BL21(DE3) without amber suppression.
Phage evolution
Wild-type T7 and hyper-mutator Δ2 phage ancestors were obtained from Ian J. Molineux (University of Texas at Austin). A single plaque of each was isolated on BL21(DE3) cells. Frozen aliquots of RF0 IodoY cells were thawed, inoculated into 50 mL Erlenmeyer flasks containing 10 mL Lysogeny Broth (LB), appropriate antibiotics, and 250 μM 3-iodotyrosine (Sigma-Aldrich), and grown to mid-exponential phase (OD600 ∼ 0.5) at 37°C with orbital shaking at 120 rpm. Six lines each of wild-type and mutator phages were initiated by seeding separate E. coli cultures with 1 μL of clonal phage lysate. Phages were allowed to replicate until all flasks showed visible lysis by clearing of their turbidity, a period of time that varied from ∼100-120 minutes at the beginning of the evolution experiment to ∼35-45 minutes at the end. After lysis, 1 μL of each independent culture was transferred to fresh RF0 IodoY cells that had been revived from the same freezer stock and grown to mid-exponential phase. This transfer volume yields a low initial multiplicity of infection (MOI) of ∼0.05 – 0.1. Evolved phage populations were periodically archived at −80°C in LB with 15% (v/v) glycerol added.
Genome sequencing and analysis
Phage DNA from each line was isolated using a CsCl2 gradient and sequenced using standard methods on an Illumina MiSeq instrument at the Genome Sequencing and Analysis Facility (University of Texas at Austin). Adaptor sequences were trimmed from the resulting 250-base paired-end reads using FLEXBAR26 (version 2.32).
We created updated reference sequences for the ancestral T7 wild-type and Δ2 hypermutator phages by comparing the reads from each sample to the published T7 genome sequence12 (GenBank: NC_001604.1) using breseq (version 0.23; http://barricklab.org/breseq) in consensus mode. The wild-type ancestor had only a single difference from the published T7 sequence: an insertion of a single A base after position 1895 within gene 0.6B. The Δ2 ancestor had 112 genetic differences relative to the wild-type ancestor. These differences included 107 base substitutions, two single-nucleotide insertions, two single-nucleotide deletions, and the in-frame 6-bp deletion in the exonuclease domain of T7 DNA polymerase that results in the hypermutator phenotype19. Complete lists of the genetic differences between the Δ2 and WT ancestors and the published T7 genome are available (Supplementary Data Set 1).
Mutations were predicted from the metagenomic sequencing data for the evolved populations using breseq in two stages. The first stage used breseq with both polymorphism and junction prediction enabled to predict large-scale structural variants. From these results, we estimated the fraction of evolved populations containing large deletions by manually examining regions with anomalous read coverage and by counting reads that mapped across new and ancestral junction sequences. The second analysis stage used breseq in polymorphism prediction mode with no prediction of new sequence junctions. This output was used to predict point mutation variants with frequencies ≥1% in each population (Supplementary Data Set 1). Calculated dN/dS ratios accounted for codon abundance and assumed an equal probability of all base substitutions.
Evolved phage population titers
Frozen stocks were first thawed, diluted, and spotted on RF0 IodoY to determine the approximate titer of each stock. Then, 100 μL of an appropriate dilution was added to 3 mL of melted soft agar supplemented with appropriate antibiotics and also 250 μM 3-iodotyrosine for RF0 IodoY. 300 μL of E. coli culture at an optical density of ∼1.0 was immediately added and soft agar was poured across the surface of LB agar plates. Plates were incubated at 37°C for 5 hours, at which time the number of plaques on each host was counted.
Patching assays
Phage dilutions were plated on the permissive RF0 IodoY host at 37°C, and stored at 4°C overnight after clearly discernable plaques were obtained. The next day, E. coli host strains to be tested were inoculated into soft agar containing appropriate supplements and poured over LB plates. Isolated plaques were then picked with a pipette tip and resuspended in 1 mL of LB, and 1 μL of this dilution was spotted on each host strain. Plates were incubated at 37°C and growth was assessed after 5 hours. Lysates of a collection of plaques with and without the amber-suppressor-dependent phenotype were created, and the allelic state of the amber-containing gene in each phage isolate was determined via PCR and Sanger sequencing.
Molecular modeling
PyMOL (Version 1.5.0.4; Schrödinger, LLC) with the SwissSideChain plugin27 was used to model and visualize the 3-iodotyrosine substitution in the structure of RNA polymerase (PDB:1QLN)20.
Lysis assays
We revived RF0 IodoY and RF0 Tyr cells and grew both with 250 μM 3-iodotyrosine and appropriate antibiotics. When cultures reached an OD600 of ∼0.5 we transferred 200 μL aliquots into a clear-bottom 96-well plate and added lysates of individual phage stocks, which had been previously titered on RF0 IodoY cells, to each well at a dilution that gave an MOI of ∼5. These phages had known allelic states of the gene 17.5 amber mutation verified by patching and sequencing. The microplate was incubated at 37°C with OD600 readings taken every minute, preceded by 10 seconds of shaking at intensity 4, in a PowerWave 340 microplate spectrophotometer (BioTek). Lysis curves were corrected for the time it took to aliquot each separate phage and to begin making measurements in the plate reader (<10 min).
Amber rescue mutants
Lysate was created from an evolved WT L6 phage clone that was isolated and confirmed to have the amber mutation in the type II holin gene by patching, PCR, and Sanger sequencing as above. For most rescue mutants, multiple independent cultures of RF0 IodoY were inoculated with ∼104 phage from this stock and again allowed to lyse completely, and 103–105 dilutions were plated on BL21(DE3) cells in soft agar as above. Other rescue mutants were isolated by plating from this stock directly on BL21(DE3) cells. In each case, mutations of the amber to non-termination codons were identified as the fastest-forming plaques, inoculated into RF0 IodoY cells to obtain lysate, and genotyped via PCR and Sanger sequencing.
Competitions
Phages to be competed were revived from frozen stocks through two serial transfers in RF0 IodoY cells under the same conditions as the evolution experiment. After complete lysis of the second cultures, equal volumes of each phage were mixed and inoculated into new RF0 IodoY cultures to initiate competitions at 0.01×, 0.1×, 1×, or 10× the transfer dilution volume used in the evolution experiment to test the effect of different initial phage MOI. Competitions proceeded for three serial transfers at their respective dilutions, and a sample of phage lysate was saved at the end of each infection cycle. After passaging was complete, whole-phage PCR was performed with 1 μL of the mixed lysate as template for the region of interest. Changes in the heights of peaks corresponding to each allele in Sanger sequencing traces for these PCR products over multiple passages were used to infer the more-fit phage genotype.
GFP emission assay
RF0 IodoY cells were transformed with pET21 plasmids containing sfGFP variants under the control of the lac operator and a strong T7 promoter. pET21 sfGFP (WT) contained a version of superfolder GFP (sfGFP) as a positive control. pET21 sfGFP (Y66→TAG) contained a sfGFP variant with an amber codon mutation replacing Y66 in the fluorophore. Individual transformants were isolated and grown overnight with appropriate antibiotics and ncAA supplements. The next day, saturated cultures were diluted 100-fold into the same medium and allowed to reach mid-log phase, at which time cultures were induced with 500 μM IPTG. Samples were taken 150 minutes later and analyzed on a Tecan Safire plate reader. An emission scan with excitation at 480 nm was performed to identify the peak emission wavelength for each sample. RF0 IodoY pET21 GFP(Y66→TAG) was found to emit with a peak at 524 nm, which is 8 nm longer than the WT sfGFP peak of 516 nm, and in accordance with previous studies of GFP with 3-iodotyrosine incorporated at this position28.
Proteomics
RF0 IodoY E. coli cells cultured in LB supplemented with 3-iodotyrosine and appropriate antibiotics were pelleted by centrifugation and resuspended in 50mM Tris (pH 8.0) followed by the addition of 2,2,2-trifluoroethanol (TFE) to a concentration of 50% (v/v) to solubilize the cell membrane and denature proteins. Samples were then reduced with 5 mM dithiothreitol (DTT) for 45 min at 56°C and alkylated with 15 mM iodoacetamide for 30 min. Excess iodoacetamide was quenched by the addition of another 15 mM DTT. Samples were diluted 10-fold in 50mM Tris (pH 8.0) with 4 mM CaCl2 to a final concentration of 5% TFE (v/v) prior to proteolytic digestion with trypsin for 4 h at 37°C.
Alternatively, RF0 IodoY E. coli cells were lysed by a T7 phage isolate from evolved population WT L6 with the amber substitution in the type II holin (gene 17.5), or BL21(DE3) E. coli cells were lysed by wild-type T7. The insoluble pellet from each lysed sample was resuspended in SDS-PAGE loading buffer and proteins were separated on a 4–20% acrylamide gradient gel. Gel slices corresponding to 25–17 kDa, 17–12 kDa, and <12 kDa were cut and subjected to in-gel digestion (8 h at 37°C) as previously described29, but with endoproteinase GluC (New England Biolabs) instead of trypsin. Following digestion, peptides were eluted from the gel, dried by speedvac, resuspended in 50mM Tris (pH 8.0) with 4mM CaCl2 and digested with trypsin as described above.
Peptides resulting from both in-gel and in-solution digestions were desalted on Hypersep C18 spin columns (Thermo Scientific) and analyzed by nanoLC-MS/MS on a Dionex Ultimate 3000 nanoRSLC system coupled to an LTQ-Orbitrap Velos Pro mass spectrometer (Thermo Scientific). Data dependent ion selection was activated with parent ion scans (MS1) collected at 100,000 resolution. Ions with charge > +1 were selected for collision-induced dissociation (CID), with up to 20 fragment spectra (MS2) collected per MS1. Dynamic exclusion was activated, with a 45-second exclusion time for ions selected more than twice in a 30-second window.
Spectra were analyzed by SEQUEST® (Proteome Discoverer 1.4, Thermo Scientific), using a protein sequence database consisting of the E. coli and T7 bacteriophage proteomes, including alternative extended sequences resulting from amber read-through. These protein sequences were generated from GenBank records NC_012971.2 and NC_001604.1, respectively. The amber codon substitution in the type II holin protein was also included for phage T7. Amber sites were designated as residue X in the sequence database, with X alternatively representing 3-iodotyrosine, tyrosine, phenylalanine, or tryptophan in independent searches. The search specified full-proteolytic peptides, allowing up to two missed cleavages. Mass tolerance was set to 10 ppm for precursor and 0.5 for fragment spectra. Carbamidomethylation of cysteine (iodoacetamide) was selected as a static modification and oxidized methionine as a dynamic modification. High confidence peptide-spectrum matches (PSMs) were filtered at <1% FDR (determined by Percolator, Proteome Discoverer v1.3, Thermo Scientific). For the RF0 IodoY E. coli, RF0 IodoY E. coli cells lysed by the T7 phage from population WT L6, and BL21(DE3) E. coli cells lysed by wild-type T7 samples we identified a total of 183,363, 18,527, and 20,639 PSMs, respectively. Full details of predicted PSMs that were informative about the specificity of amber read-through are provided (Supplementary Fig. 2, Supplementary Data Set 2).
Accession codes
National Center for Biotechnology Information Short Read Archive. Genome re-sequencing data for bacteriophage clones and populations have been deposited under accession number SRP021199.
Supplementary Material
Acknowledgments
We thank Jim Bull, Ian Molineux, Randall Hughes, Craig Barnhart, Daniel Deatherage, Razan Alnahhas, Matthew Schmerer, Alex Miklos, Adam Meyer, and Andre Maranhão for plasmids, strains, advice, and technical assistance. The RF0-iodoY strain was provided by RIKEN and by the Targeted Proteins Research Program (TPRP), the Ministry of Education, Culture, Sports, Science and Technology (MEXT), Japan. This research was supported by the U.S. National Institutes of Health (NIH; R00-GM087550 to J.E.B.), the U.S. National Science Foundation (NSF) BEACON Center for the Study of Evolution in Action (DBI-0939454 to J.E.B), the U.S. Army Research Office (W911NF-12-1-0390 to J.E.B and E.M.M.), the U.S. National Security Science and Engineering Faculty (FA9550-10-1-01-69 to A.D.E.), the U.S. Defense Advanced Research Project Agency (HR-0011-12-C-0066 to A.D.E.), and the NSF (MCB-0943383 to A.D.E.). E.M.M. also acknowledges funding from the NIH, NSF, Cancer Prevention Research Institute of Texas, and Welch Foundation (F1515).
Footnotes
Contributions: M.J.H., J.W.E., J.E.B., and A.D.E. conceived the study. M.J.H. performed evolution experiments. J.E.B. and M.J.H. analyzed sequencing data. M.J.H. and J.W.E. created RF0 Tyr and characterized phage lysis times. D.R.B. performed proteomics experiments. J.E.B. performed statistical analyses. J.E.B., M.J.H., J.W.E., and D.R.B. created figures and wrote the manuscript. All authors designed experiments and edited the manuscript.
Competing financial interests: The authors declare no competing financial interests.
References
- 1.Crick FHC. The origin of the genetic code. J Mol Biol. 1968;38:367–379. doi: 10.1016/0022-2836(68)90392-6. [DOI] [PubMed] [Google Scholar]
- 2.Knight RD, Freeland SJ, Landweber LF. Rewiring the keyboard: evolvability of the genetic code. Nat Rev Genet. 2001;2:49–58. doi: 10.1038/35047500. [DOI] [PubMed] [Google Scholar]
- 3.Ambrogelly A, Palioura S, Söll D. Natural expansion of the genetic code. Nat Chem Biol. 2007;3:29–35. doi: 10.1038/nchembio847. [DOI] [PubMed] [Google Scholar]
- 4.Freeland SJ, Hurst LD. The genetic code is one in a million. J Mol Evol. 1998;47:238–248. doi: 10.1007/pl00006381. [DOI] [PubMed] [Google Scholar]
- 5.Itzkovitz S, Alon U. The genetic code is nearly optimal for allowing additional information within protein-coding sequences. Genome Res. 2007;17:405–412. doi: 10.1101/gr.5987307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Liu CC, Schultz PG. Adding new chemistries to the genetic code. Annu Rev Biochem. 2010;79:413–44. doi: 10.1146/annurev.biochem.052308.105824. [DOI] [PubMed] [Google Scholar]
- 7.Davis L, Chin JW. Designer proteins: applications of genetic code expansion in cell biology. Nat Rev Mol Cell Biol. 2012;13:168–82. doi: 10.1038/nrm3286. [DOI] [PubMed] [Google Scholar]
- 8.Brustad EM, Arnold FH. Optimizing non-natural protein function with directed evolution. Curr Opin Chem Biol. 2011;15:201–10. doi: 10.1016/j.cbpa.2010.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang K, Neumann H, Peak-Chew SY, Chin JW. Evolved orthogonal ribosomes enhance the efficiency of synthetic genetic code expansion. Nat Biotechnol. 2007;25:770–777. doi: 10.1038/nbt1314. [DOI] [PubMed] [Google Scholar]
- 10.Bacher JM, Hughes RA, Tze-Fei Wong J, Ellington AD. Evolving new genetic codes. Trends Ecol Evol. 2004;19:69–75. doi: 10.1016/j.tree.2003.11.007. [DOI] [PubMed] [Google Scholar]
- 11.Ohtake K, et al. Efficient decoding of the UAG triplet as a full-fledged sense codon enhances the growth of a prfA-deficient strain of Escherichia coli. J Bacteriol. 2012;194:2606–2613. doi: 10.1128/JB.00195-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dunn JJ, Studier FW. Complete nucleotide sequence of bacteriophage T7 DNA and the locations of T7 genetic elements. J Mol Biol. 1983;166:477–535. doi: 10.1016/s0022-2836(83)80282-4. [DOI] [PubMed] [Google Scholar]
- 13.Molineux IJ. The T7 group The Bacteriophages. Oxford UP: 2006. pp. 277–301. [Google Scholar]
- 14.Bull JJ, et al. Exceptional convergent evolution in a virus. Genetics. 1997;147:1497–1507. doi: 10.1093/genetics/147.4.1497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bull JJ, Badgett MR, Rokyta D, Molineux IJ. Experimental evolution yields hundreds of mutations in a functional viral genome. J Mol Evol. 2003;57:241–248. doi: 10.1007/s00239-003-2470-1. [DOI] [PubMed] [Google Scholar]
- 16.Chan LY, Kosuri S, Endy D. Refactoring bacteriophage T7. Mol Syst Biol. 2005;1:2005.0018. doi: 10.1038/msb4100025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sakamoto K, et al. Genetic encoding of 3-iodo-L-tyrosine in Escherichia coli for single-wavelength anomalous dispersion phasing in protein crystallography. Structure. 2009;17:335–44. doi: 10.1016/j.str.2009.01.008. [DOI] [PubMed] [Google Scholar]
- 18.Mukai T, et al. Genetic-code evolution for protein synthesis with non-natural amino acids. Biochem Biophys Res Commun. 2011;411:757–61. doi: 10.1016/j.bbrc.2011.07.020. [DOI] [PubMed] [Google Scholar]
- 19.Tabor S, Richardson CC. Selective inactivation of the exonuclease activity of bacteriophage T7 DNA polymerase by in vitro mutagenesis. J Biol Chem. 1989;264:6447–6458. [PubMed] [Google Scholar]
- 20.Cheetham GM, Steitz TA. Structure of a transcribing T7 RNA polymerase initiation complex. Science (80- ) 1999;286:2305–9. doi: 10.1126/science.286.5448.2305. [DOI] [PubMed] [Google Scholar]
- 21.Wang IN, Smith DL, Young R. Holins: the protein clocks of bacteriophage infections. Annu Rev Microbiol. 2000;54:799–825. doi: 10.1146/annurev.micro.54.1.799. [DOI] [PubMed] [Google Scholar]
- 22.Wang N, Dykhuizen D, Slobodkin L. The evolution of phage lysis timing. Evol Ecol. 1996;10:545–558. [Google Scholar]
- 23.Heineman RH, Bull JJ, Molineux IJ. Layers of evolvability in a bacteriophage life history trait. Mol Biol Evol. 2009;26:1289–1298. doi: 10.1093/molbev/msp037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lajoie MJ, et al. Genomically recoded organisms expand biological functions. Science (80- ) 2013;342:357–60. doi: 10.1126/science.1241459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Moe-Behrens GHG, Davis R, Haynes KA. Preparing synthetic biology for the world. Front Microbiol. 2013;4:5. doi: 10.3389/fmicb.2013.00005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dodt M, Roehr J, Ahmed R, Dieterich C. FLEXBAR—Flexible barcode and adapter processing for next-generation sequencing platforms. Biology. 2012;1:895–905. doi: 10.3390/biology1030895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gfeller D, Michielin O, Zoete V. SwissSidechain: a molecular and structural database of non-natural sidechains. Nucleic Acids Res. 2013;41:327–332. doi: 10.1093/nar/gks991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Young DD, Jockush S, Turro NJ, Schultz PG. Synthetase polyspecificity as a tool to modulate protein function. Bioorg Med Chem Lett. 2011;21:7502–4. doi: 10.1016/j.bmcl.2011.09.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shevchenko A, Tomas H, Havlis J, Olsen JV, Mann M. In-gel digestion for mass spectrometric characterization of proteins and proteomes. Nat Protoc. 2006;1:2856–60. doi: 10.1038/nprot.2006.468. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.