Abstract
We present a case of an interstitial subtelomeric 10q26 deletion in a male child with moderate developmental delay and minor dysmorphic features. Using array comparative genomic hybridization (CGH) and fluorescence in situ hybridization (FISH), we have detected an interstitial deletion at 10q26.2q26.3 encompassing a 5.8 Mb region and spanning 24 genes. Interestingly, losses of this chromosome 10 region have not been previously associated with a phenotype outcome. According to an in silico evaluation, we have suggested that PPP2R2D and BNIP3 losses are likely a cause of developmental delay in the index patient. Our data allow to speculating that haploinsufficiency of these two genes in 10q26.3, which is usually ignored in the context of chromosome 10q deletions, has a phenotypic effect.
1. Introduction
Although subtelomeric chromosomal rearrangements are common in children with intellectual disability, developmental delays and/or dysmorphic features [1–3], deletions affecting subtelomere of chromosome 10 long arm, are rare [4]. This probably explains the small amount of such cases addressed by array comparative genomic hybridization (CGH) [4, 5] in contrast to the number of reports on phenotypic manifestations of chromosome 10q25q26/10q26qter loss analyzed using molecular cytogenetic techniques with a lower resolution [6–8]. Here, we report a case of an interstitial 10q26 deletion in a male child presenting with a phenotypic outcome atypical for subtelomeric deletions at 10qter detected by array CGH and confirmed by fluorescence in situ hybridization (FISH).
2. Case Presentation and Methods
2.1. Clinical Description
A 28-month-old male child with moderate developmental delay and minor dysmorphic features has been referred to (molecular) cytogenetic analysis. The patient was born at 37 weeks of gestation to a 20-year-old mother and 21-year-old father by spontaneous vaginal delivery with a birth weight of 3.16 kg (~25th percentile) and length of 52.6 cm (~75th percentile). Pregnancy history was unremarkable. At 6 months of age, he was able to roll over and sit with support. Physical examination at the age of 2 years and 4 months showed dysmorphic features (flat feet with cutaneous syndactyly of the second and third toes; high forehead and prominent auricles) without any other remarkable congenital malformations and dysmorphic features. A moderate developmental delay was noticed.
2.2. Cytogenetic Analysis
GTG-banding was done according to standard procedures analyzing 30 metaphase plates at a resolution of 550 bands. Karyotype abnormalities were not detected.
2.3. Array CGH
Array CGH was performed using the customized human genomic microarrays (slightly modified Constitutional Chip 4.0) BAC/PAC clones (Human BAC Array-System, Perkin Elmer, USA) achieving a resolution of 0.3 Mb. Technical performance of array CGH (DNA labeling, hybridization, detection, and data analysis) was done according to previously described protocols [9, 10] and to manufacturers' instructions. An interstitial deletion at 10q26.2q26.3 spanning 128,190,760–133,998,503 (confirmed by 14 interchangeable BAC probes (two reverse assays): RP11-16P8, RP11-32H11, RP11-21M8, RP11-42K2, RP11-88B12, RP11-48A2, RP11-168C9, RP11-90B19, RP11-90O13, RP11-113P9, RP11-462G8, RP11-408L20, RP11-142I8, and RP11-140A10) was detected (Figure 1(a)). The deletion size was estimated as 5.8 Mb.
Figure 1.
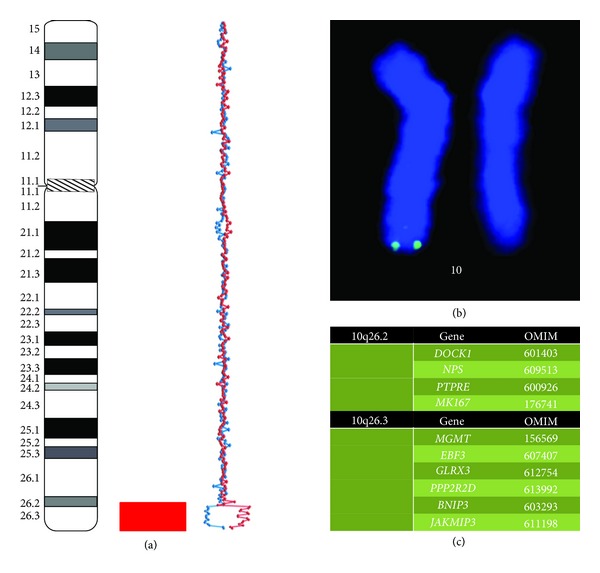
Molecular cytogenetic (array CGH) findings in the index case: (a) array CGH demonstrating arr 10q26.2q26.3(128,190,760–133,998,503) × 1 (two alternative arrays Cy3/Cy5 (red line) and Cy5/Cy3 (blue line) are plotted on the graph); (b) FISH confirmation of subtelomeric 10q deletion; (c) OMIM genes (http://www.omim.org) located at the deleted chromosome 10 region.
2.4. FISH
FISH was performed as described elsewhere [11–13] with a DNA probe located at chromosome 10q26 [12]. Subtelomeric deletion of chromosome 10q was confirmed (Figure 1(b)).
2.5. In Silico Evaluation of the Deleted Chromosomal Region
Bioinformatics analysis of the deletion was made as proposed in our previous studies [9, 10, 13]. The deletion resulted in the loss of 24 genes among which 10 genes are listed in OMIM (http://www.omim.org/) (Figure 1(c)). Using a set of genomic, epigenetic and proteomic databases (for details see [9, 10, 13]), we have evaluated the pathogenic value of 10q26 genes' haploinsufficiency caused by the deletion. We concluded that PPP2R2D and BNIP3 losses are likely to be the cause of intellectual disability in the index patient.
3. Discussion
Subtelomeric chromosome 10q deletions appear to cause a specific phenotype [4–8]. However, wide clinical heterogeneity naturally associated with variability in deletion size has been reported [4, 14]. Interestingly, phenotypic outcomes of all the deletions at 10q26 addressed by array CGH have not been ever associated with haploinsufficiency of PPP2R2D and BNIP3 genes. Molecularly, the detected deletion is similar to a genomic variation, which had resulted from a complex distal 10q rearrangement in a girl with mild intellectual disability reported by Sarri et al. [15]. Unfortunately, the complex chromosome rearrangement has involved other genomic loci impeding correct phenotype correlations. Additionally, Courtens et al. [16] have described subtelomeric 10q deletion spanning the same 10q26.2 genomic loci associated with phenotypically different outcomes. Moreover, bioinformatics analysis has shown that the phenotype is likely to result from simultaneous loss of PPP2R2D and BNIP3. Therefore, we have concluded these two genes to be implicated in the phenotypic outcome of interstitial deletions affecting 10q26.2q26.3. Finally, the present case demonstrates that chromosomal loci ignored in the context of genomic disorders can contribute to the phenotype or cause mild subtypes of the disease.
Acknowledgment
Dr. Ivan Y. Iourov is supported by the Grant of the President of the Russian Federation MD-4401.2013.7.
Disclosure
In the present work, modified Constitutional Chip 4.0 was applied. The latter was purchased from the local Perkin Elmer agency at the time when no other microarray companies were present at the local market. The authors have not ever received any funding or promotion from the Perkin Elmer. None of the authors have been ever involved in the sale and distribution. Therefore, they do not have a direct financial relation with the commercial identity mentioned in their paper.
Conflict of Interests
The authors declare that there is no conflict of interests regarding the publication of this paper.
References
- 1.de Vries BBA, White SM, Knight SJL, et al. Clinical studies on submicroscopic subtelomeric rearrangements: a checklist. Journal of Medical Genetics. 2001;38(3):145–150. doi: 10.1136/jmg.38.3.145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ravnan JB, Tepperberg JH, Papenhausen P, et al. Subtelomere FISH analysis of 11 688 cases: an evaluation of the frequency and pattern of subtelomere rearrangements in individuals with developmental disabilities. Journal of Medical Genetics. 2006;43(6):478–489. doi: 10.1136/jmg.2005.036350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Iourov IY, Vorsanova SG, Yurov YB. Molecular cytogenetics and cytogenomics of brain diseases. Current Genomics. 2008;9(7):452–465. doi: 10.2174/138920208786241216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Plaisancie J, Bouneau L, Cances C, et al. Distal 10q monosomy: new evidence for a neurobehavioral condition? European Journal of Medical Genetics. 2014;57(1):47–53. doi: 10.1016/j.ejmg.2013.11.002. [DOI] [PubMed] [Google Scholar]
- 5.Piccione M, Antona V, Piro E, et al. 10qter deletion: a new case. The American Journal of Medical Genetics A. 2008;146(18):2435–2438. doi: 10.1002/ajmg.a.32467. [DOI] [PubMed] [Google Scholar]
- 6.Lukusa T, Smeets E, Vermeesch JR, Fryns JP. Small terminal 10q26 deletion in a male patient with Noonan-like stigmata: diagnosis by cytogenetic and fish analysis. Genetic Counseling. 2002;13(4):417–425. [PubMed] [Google Scholar]
- 7.Petit P, Devriendt K, Azou M, Gewillig M, Fryns JP. Terminal deletion of chromosome 10q26: delineation of two clinical phenotypes. Genetic Counseling. 1998;9(4):271–275. [PubMed] [Google Scholar]
- 8.Chang YT, Chou IC, Wang CH, et al. Chromosome 10q deletion del (10)(q26.1q26.3) is associated with cataract. Pediatrics and Neonatology. 2013;54(2):132–136. doi: 10.1016/j.pedneo.2012.10.010. [DOI] [PubMed] [Google Scholar]
- 9.Iourov IY, Vorsanova SG, Kurinnaia OS, Zelenova MA, Silvanovich AP, Yurov YB. Molecular karyotyping by array CGH in a Russian cohort of children with intellectual disability, autism, epilepsy and congenital anomalies. Molecular Cytogenetics. 2012;5(1, article 46) doi: 10.1186/1755-8166-5-46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Iourov IY, Vorsanova SG, Voinova VY, et al. Xq28 (MECP2) microdeletions are common in mutation-negative femaleswith Rett syndrome and cause mild subtypes of the disease. Molecular Cytogenetics. 2013;6(1, article 53) doi: 10.1186/1755-8166-6-53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yurov YB, Vorsanova SG, Iourov IY, et al. Unexplained autism is frequently associated with low-level mosaic aneuploidy. Journal of Medical Genetics. 2007;44(8):521–525. doi: 10.1136/jmg.2007.049312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vorsanova SG, Koloti AD, Sharonin VO, Soloviev IV, Yurov YB. FISH analysis of microaberrations at telomeric and subtelomeric regions in chromosomes of children with mental retardation. The American Journal of Human Genetics. 1998;63(supplement 4, article A154) [Google Scholar]
- 13.Iourov IY, Vorsanova SG, Liehr T, Kolotii AD, Yurov YB. Increased chromosome instability dramatically disrupts neural genome integrity and mediates cerebellar degeneration in the ataxia-telangiectasia brain. Human Molecular Genetics. 2009;18(14):2656–2669. doi: 10.1093/hmg/ddp207. [DOI] [PubMed] [Google Scholar]
- 14.Tonk VJ, Wilson GN. Autism spectrum disorder with microdeletion 10q26 by subtelomere FISH. Pediatric Health, Medicine and Therapeutics. 2011;2:49–53. [Google Scholar]
- 15.Sarri C, Douzgou S, Gyftodimou Y, et al. Complex distal 10q rearrangement in a girl with mild intellectual disability: follow up of the patient and review of the literature of non-acrocentric satellited chromosomes. The American Journal of Medical Genetics A. 2011;155(11):2841–2854. doi: 10.1002/ajmg.a.34259. [DOI] [PubMed] [Google Scholar]
- 16.Courtens W, Wuyts W, Rooms L, Pera SB, Wauters J. A subterminal deletion of the long arm of chromosome 10: a clinical report and review. The American Journal of Medical Genetics. 2006;140(4):402–409. doi: 10.1002/ajmg.a.31053. [DOI] [PubMed] [Google Scholar]


