Abstract
An industrial microorganism Streptomyces avermitilis, which is a producer of anthelmintic macrocyclic lactones, avermectins, has been constructed as a versatile model host for heterologous expression of genes encoding secondary metabolite biosynthesis. Twenty of the entire biosynthetic gene clusters for secondary metabolites were successively cloned and introduced into a versatile model host S. avermitilis SUKA17 or 22. Almost all S. avermitilis transformants carrying the entire gene cluster produced metabolites as a result of the expression of biosynthetic gene clusters introduced. A few transformants were unable to produce metabolites but their production was restored by the expression of biosynthetic genes using an alternative promoter or the expression of a regulatory gene in the gene cluster that controls the expression of biosynthetic genes in the cluster using an alternative promoter. Production of metabolites in some transformants of the versatile host was higher than that of the original producers and cryptic biosynthetic gene clusters in the original producer were also expressed in a versatile host.
Keywords: secondary metabolism, heterologous expression, biosynthesis, Streptomyces, genome
Introduction
A prominent property of members of the actinomycete microorganisms belonging to the order Actinomycetales is the ability to produce numerous secondary metabolites, including antibiotics and other biologically active compounds of proven value in human and veterinary medicine, agriculture, or useful as biochemical tools. These structurally diverse metabolites possess not only antibacterial, antifungal, antiviral and antitumor activity but also antihypertensive and immunosuppressant properties. Streptomyces are a rich source of pharmaceutical compounds in which common cellular intermediates, including sugars, fatty acids, amino acids and terpenes, are combined to give more complex structures by defined biochemical pathways. In this century, genome analysis of Streptomyces microorganisms, S. avermitilis MA 46804,5, S. bingchenggensis BCW-16, S. cattleya NRRL 80577, S. clavuligerus ATCC 270648, S. coelicolor A3(2)9, S. griseus IFO 1335010, S. scabiei 87.2211, and S. venezuelae ATCC 1071212, has revealed that these microorganisms each have a large linear chromosome that harbors over twenty secondary metabolic gene clusters encoding the biosynthesis of polyketides by polyketide synthases (PKSs), peptides by non-ribosomal peptide synthetases (NRPSs), bacteriocins, terpenoids, shikimate-derived metabolites, aminoglycosides, and other natural products.
The identification and detailed characterization of secondary metabolic gene clusters have proved to be an invaluable tool for elucidation of the biosynthetic mechanism of secondary metabolites. Furthermore, the results show a rich potential source of cryptic metabolites, which appear to be encoded by silent biosynthetic pathways. Genetically engineered biosynthetic gene clusters have in certain cases allowed the production of novel analogs by combinatorial biosynthesis or the production of novel intermediate by specific in-frame deletion(s), respectively. A critical requirement for these applications is the availability of the relevant biosynthetic gene clusters controlling the production of a secondary metabolite of interest as well as appropriate genetic systems for the in vivo manipulation of the corresponding genes in heterologous hosts. Furthermore, successful production of the desired products requires the optimum relationship of the timing and flux between primary and secondary cellular metabolism, since all secondary metabolites are ultimately derived from primary metabolic building blocks and require an adequate source of energy and reducing equivalents derived from primary metabolism such as ATP and NAD(P)H.
In consideration of the above concept, we have constructed a model host for heterologous expression of biosynthetic gene clusters using engineered S. avermitilis13. This pioneer heterologous expression system has been confirmed by the production of streptomycin, cephamycin C and pladienlide in heterologous genetically engineered hosts, S. avermitilis SUKA5 and 17. Since S. avermitilis is used for industrial production of the important anthelmintic agent avermectin14 and the microorganism has already proved to be a highly efficient producing microorganism of secondary metabolites, it seems that the microorganism has been already optimized for the efficient supply of primary metabolite precursors and biochemical energy for the formation of biosynthesis of secondary metabolites. In this paper, we have shown the feasibility of using genetically engineered S. avermitilis as a heterologous host by the effective expression and production of 20 biosynthetic gene clusters for exogenous secondary metabolites.
RESULTS AND DISCUSSION
Properties of S. avermitilis as a versatile host
The genetic instability of Streptomyces strains has been reported15. Not only loss of the development of morphological differentiation but also the marked reduction or loss of secondary metabolite production has often been observed in many Streptomyces strains16,17,18. The genetic instability of producing microorganisms is a serious problem in the industrial production of secondary metabolites. These phenomena have been well documented for years and yet their origins remain unclear; however, the morphology and secondary metabolite production of S. avermitilis is somewhat stable in comparison with other Streptomyces strains. The complete genome sequences of some Streptomyces strains have been revealed and it has been proved that these chromosomes are linear and two telomeres capped by covalently bound terminal proteins are located at both ends of the linear chromosome. Streptomyces chromosomes contain terminal inverted repeats (TIRs), which are generally from tens to hundreds of kilobases. The sequence of TIRs appears to be divergent among different species, and the length of TIRs in S. avermitilis is only 49-bp long5, shorter than other Streptomyces TIRs. Mutants affecting morphological differentiation are sometimes generated in stressful environments (high temperature or hypertonic incubation).
Four genome-sequenced Streptomycetaceae microorganisms, three Streptomyces and one related genus Kitasatospora, were examined for the appearance of bald mutants, which are unable to form aerial hyphae and/or spores, in high temperature incubation (Table 1). S. coelicolor A3(2), which has been used as a model microorganism for studying morphological differentiation in Streptomyces and has relatively long TIRs of 21,653 bp9, generated bald mutants at the frequency of 0.43% under normal culture conditions, which elevated to 7.15% in high temperature incubation (37°C). Both Streptomycetaceae microorganisms, S. griseus IFO 1335010 and K. setae KM-605419, containing more than 100-kb TIRs (132,910 and 127,148 bp, respectively) generated more bald mutants than S. coelicolor A3 (2). Furthermore, the appearance of bald mutants from S. griseus IFO 13350 and K. setae KM-6054 was markedly increased to 44.4 % and 38.4%, respectively, by high temperature incubation. On the other hand, the frequency of bald mutants of S. avermitilis was quite low. The growth characteristics of S. avermitilis were more stable than those of S. coelicolor A3(2), S. griseus IFO 13350 and K. setae KM-6054 and it seemed that genetic stability might depend on the length of TIRs on the chromosome.
Table 1.
Genetic instability of genome-sequenced Streptomycetaceae microorganisms containing linear chromosomes under the high-temperature growth.
| Microorganism | TIR (bp) | Appearance of bld mutants§ | |
|---|---|---|---|
| 28°C (%) | 37°C (%) | ||
| S. avermitilis MA-4680 | 49 | 1/2,675 (0.03) | 4/2567 (0.15) |
| S. coelicolor A3(2) | 21,653 | 9/2,056 (0.43) | 197/2,781 (7.15) |
| S. griseus IFO 13350 | 132,910 | 26/2,138 (1.2) | 893/2,013 (44.4) |
| Kitasatospora setae KM-6054 | 127,148 | 54/2,064 (2.6) | 908/2,367 (38.4) |
About 200 spores of each microorganism were inoculated onto M4 (inorganic salts-starch agar) plate. After each plate had been incubated at 28°C or 37°C for 6 days, bald (bld) mutants were counted.
To evaluate the heterologous expression of genes concerning secondary metabolite biosynthesis, a large DNA fragment containing the entire biosynthetic gene cluster is ligated with an appropriate vector and introduced into the host. In studies of the regulation of the biosynthetic gene cluster or combinatorial biosynthesis to generate hybrid metabolites, more than two vectors carrying genes derived from different microorganisms are convenient for the construction54,55. In general, transformants or exoconjugants generated by transformation or conjugation are selected by antibiotic resistance conferred by a vector marker. S. avermitilis was sensitive to various antibiotics (Table 2). The three resistance genes derived from Escherichia coli, aph(3′), aac(3)IV and aad(3″), were well expressed and transformants carrying these genes responded to resistance to neomycin/ribostamycin, apramycin and streptomycin/spectinomycin, respectively. Ribostamycin was suitable for the selection of transformants carrying aph(3′) because the background growth of non-transformant progenies in the plate containing ribostamycin was extremely low. In these resistance genes, although transformants carrying aac(3)IV were also resistant to ribostamycin, each transformant carrying aph(3′) or aac(3)IV was able to be selected by neomycin or apramycin, respectively, and transformants carrying both aph(3′) and aac(3)IV were selected by neomycin and apramycin. Another four resistance genes derived from actinomycetes, vph (S. vinaceus; viomycin or tuberactinomycin N resistance)20, hph (S. hygroscopicus; hygromycin B resistance)21, tsr (S. azureus; thiostrepton resistance)22 and ermE (Saccharopolyspora erythraea; erythromycin or lincomycin resistance)23, were also used as selective markers for transformation and conjugation. Since some large-deletion derivatives of S. avermitilis (SUKA10 - 17)13 contain ermE, which was replaced with the biosynthetic gene cluster of neopentalenolactone, as a selection marker after interspecific conjugation with other Streptomyces microorganisms or genetic recombination with S. avermitilis derivatives, these SUKA series derivatives were resistant to lincomycin/erythromycin. Interestingly, large-deletion derivatives of S. avermitilis were more sensitive to a polyether compound, lasalocide, than the wild-type strain. Although the resistance mechanism of lasalocide in the wild-type strain has not been examined, the mechanism might be involved in the exclusion of the compound from inside of the cells because helvolic acid and fusidic acid, the targets of which are different from that of lasalocide, also showed a similar response between the wild-type and SUKA mutants.
Table 2.
Antibiotic susceptibility of S. avermitilis wild type, large-deletion mutants and their transformants carrying resistance gene.
| Minimum Inhibitory Concentration (MIC; μg/mL) * | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| neomycin | ribostamycin | apramycin | streptomycin | spectinomycin | hygromycin B | viomycin † | thiostrepton | lincomycin † | lasalocide | |
| Wild type | < 0.1 | 3.13 | 0.39 | 0.1 | 25 | 6.25 | 0.2 | 0.78 | 25 | 12.5 |
| SUKA5 | < 0.1 | 1.56 | 0.39 | 0.1 | 12.5 | 3.13 | 0.2 | 0.39 | 12.5 | 1.56 |
| SUKA17 | < 0.1 | 1.56 | 0.39 | 0.1 | 12.5 | 3.13 | 0.2 | 0.39 | > 200 | 1.56 |
| SUKA22 ‡ | < 0.1 | 1.56 | 0.39 | 0.1 | 12.5 | 3.13 | 0.2 | 0.39 | > 200 | 1.56 |
| aph(3′) § | 25 | > 200 | 0.39 | 0.1 | 12.5 | 6.25 | 0.2 | 0.39 | 12.5 ¶ | 1.56 |
| aac(3)IV § | 0.78 | 200 | > 100 | 0.1 | 12.5 | 6.25 | 0.2 | 0.39 | 12.5 ¶ | 1.56 |
| aad(3″) § | < 0.1 | 1.56 | 0.39 | 25 | > 200 | 3.13 | 0.2 | 0.39 | 12.5 ¶ | 1.56 |
| hph § | 0.1 | 1.56 | 0.39 | 0.1 | 12.5 | > 200 | 0.2 | 0.39 | 12.5 ¶ | 1.56 |
| vph § | 0.1 | 1.56 | 0.39 | 0.1 | 12.5 | 6.25 | > 100 | 0.39 | 12.5 ¶ | 1.56 |
| tsr § | < 0.1 | 1.56 | 0.39 | 0.1 | 12.5 | 3.13 | 0.2 | > 100 | 12.5 ¶ | 1.56 |
| ermE § | < 0.1 | 1.56 | 0.39 | 0.1 | 12.5 | 3.13 | 0.2 | 0.39 | >200 ¶ | 1.56 |
Spores (ca. 104) were spotted onto YMS agar medium containing designated concentration of antibiotic and grown at 30°C for 3 days.
Viomycin and lincomycin replaced tuberactinomycin N and erythromycin, respectively.
S. avermitilis SUKA22 is isogenic to SUKA17. The left side of the deletion region of SUKA22 (corresponding to 8,886,025–8,925,414 nt in wild-type chromosome) was replaced by a mutant-type loxP sequence to prevent recombination between another wild-type loxP sequence on the right side of the deletion region corresponding to 79,455–1,595,563 nt in the wild-type chromosome.
S. avermitilis SUKA22 carrying aph(3′); aminoglycoside 3′-phosphotransferase gene of Tn5, aac(3)IV; aminoglycoside 3-N-acetyltransferase type IV gene of E. coli, aad(3″); streptomycin 3″-adenylyltransferase gene of E. coli, hph; hygromycin B 7″-phosphotransferase gene of S. hygroscopicus, vph; viomycin phosphotransferase gene of S. vinaceus, tsr; rRNA adenosine-2′-O-methyltransferase gene of S. azureus, and ermE; rRNA aminoadenine-6-N-methyltransferase gene of Saccharomyces erythraea).
The MIC values were obtained from S. avermitilis SUKA5 carrying ermE.
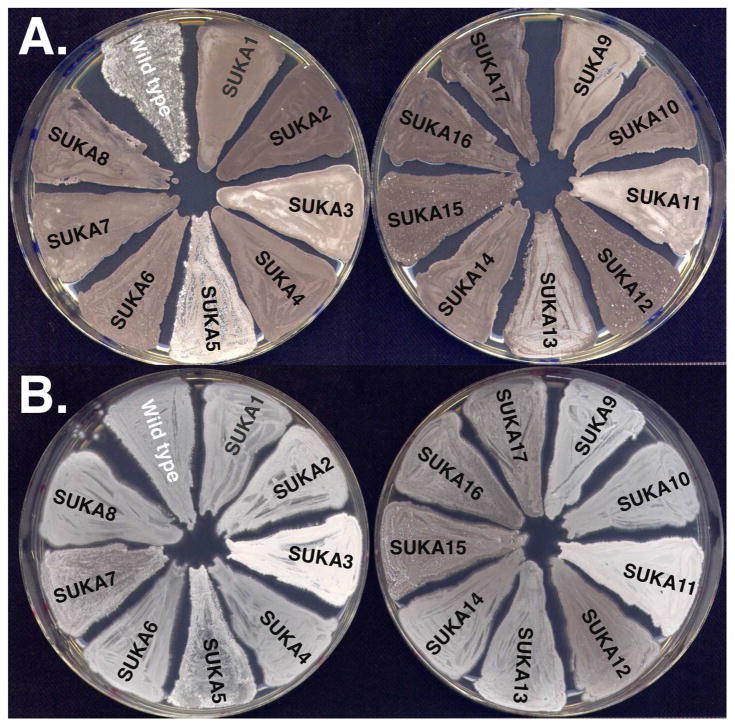
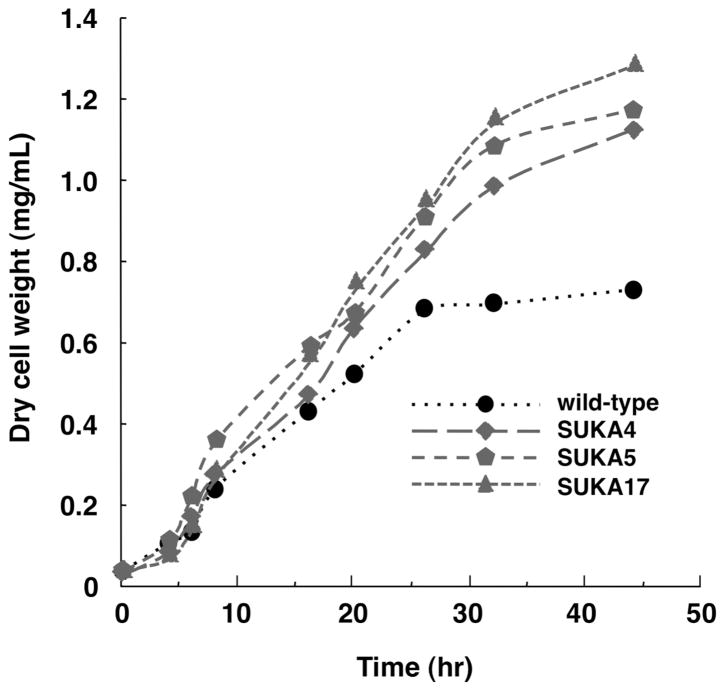
Since all the large-deletion derivatives of S. avermitilis were constructed to remove more than a 1.4-Mb region from the 9.03-Mb linear chromosome, their growth characteristics were examined. All the large-deletion derivatives (SUKA1 to 17) grew well on the sporulation medium and their morphological development on the medium was slightly faster than that of the wild-type strain. As shown in Fig. 1-A, the wild-type strain formed aerial hyphae and started sporulation after 3 days of growth, but all the large-deletion derivatives fully sporulated and formed abundant spores. Large-deletion derivatives could grow on minimum medium without any supplements because the regions deleted from S. avermitilis chromosome included no essential genes (Fig. 1-B). Since the development of morphological differentiation in large-deletion derivatives was faster than that of the wild-type strain, their growth rates were compared with the wild-type strain. The growth rate on liquid complex medium was not clearly different but the growth characteristics of large-deletion derivatives obviously differed from the wild-type strain in liquid minimum medium (Fig. 2). The growth rates of large-deletion derivatives, SUKA4, 5 and 17, until 24 hr incubation were similar and were faster than the wild-type strain. Interestingly, the biomass of the wild-type strain increased until 24 hr but large-deletion derivatives continued to grow until 44 hr and their biomass contents were about 1.7-fold that of the wild-type strain (Fig. 2). Thus, large-deletion derivatives had the ability to grow faster and increase their biomass.
Figure 1.
Growth of S. avermitilis wild type and its large-deletion mutants on (A) YMS medium at 30°C for 3 days and on (B) MM agar medium supplemented with 1% glucose at 30°C for 6 days.
Figure 2.
Growth of S. avermitilis wild type and its large-deletion mutants, SUKA4, SUKA5 and SUKA17, in liquid culture. Spores were cultured on TSB at 30°C for 48 hr. Vegetative culture was diluted to 100-fold by TSB and incubated with shaking for 24 hr at 30°C. A 0.4 mL portion of the culture was inoculated into 20 mL MM liquid medium supplemented with 1% glucose and 0.1% yeast extract, and incubated at 28°C with shaking (180 rpm).
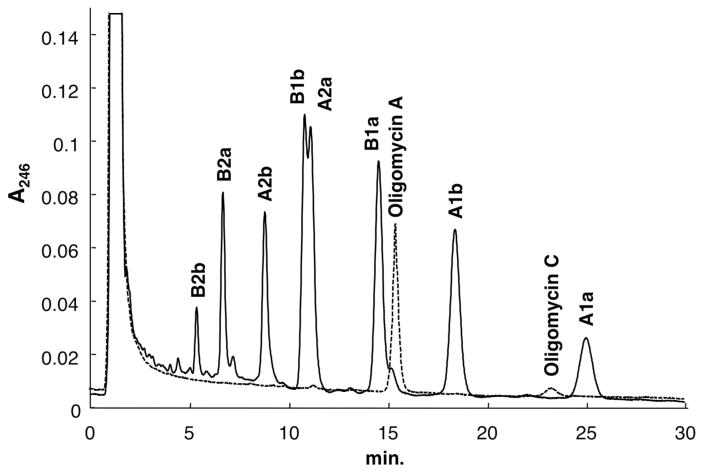
In large-deletion derivatives of S. avermitilis, all biosynthetic gene clusters for endogenous major metabolites, avermectin, oligomycin and filipin, were removed to avoid not only complex analysis and separation of products derived from heterologous expression of the biosynthetic gene cluster and endogenous major products, but also precursor competition between exogenous and endogenous biosynthetic pathways. The latter possibility was confirmed in the production of endogenous products, oligomycin and avermectin. Wild-type S. avermitilis produced a large amount of eight components of avermectins (B2b, B2a, A2b, B1b, A2a, B1a, A1b, and A1a) and a very small amount of oligomcyin A, whereas deletion of the aveR gene, the gene product of which activates the expression of genes involving avermectin biosynthesis, failed to produce avermectins, but oligomcyin productivity was markedly increased more than 50-fold that of the wild-type strain and a minor component, oligomycin C (14-dehydroxyoligomycin A), was also detected in the mutant (Fig. 3). Both avermectin and oligomycin are macrocyclic lactones synthesized from malonyl-CoA and methylmalonyl-CoA by each PKS. Since these two biosynthetic pathways share the same precursors, which are building blocks of these compounds, competition between the precursors would occur in the cell; therefore, almost all malonyl-CoA and methylmalonyl-CoA are consumed by avermectin synthesis and the supply of acyl-CoA for oligomycin synthesis is very low. In the aveR deletion mutant, malonyl-CoA and methylmalonyl-CoA are not consumed by avermectin synthesis because the gene product of aveR acts as an activator of the expression of genes involved in avermectin biosynthesis. These acyl-CoAs will be preferably used for oligomycin synthesis. This suggests that the deletion of biosynthetic gene clusters of endogenous major secondary metabolites is preferred for the synthesis of exogenous secondary metabolites by heterologous expression of the biosynthetic gene cluster.
Figure 3.
HPLC profiles of mycelial extracts of S. avermitilis wild-type strain and its aveR deletion mutant. Solid and dotted lines indicate the production profiles of the wild-type strain and aveR mutant, respectively. Each strain was cultured in avermectin production medium for 168 hr at 28°C. Methanol extract from mycelium was directly applied to ODS-HPLC (HyperSil 3 μm; 4.6 φ × 100 mm) and developed with acetonitrile/methanol/water (65:10:25) at a flow rate of 0.6 mL/min.
Heterologous expression of biosynthetic gene clusters of secondary metabolites
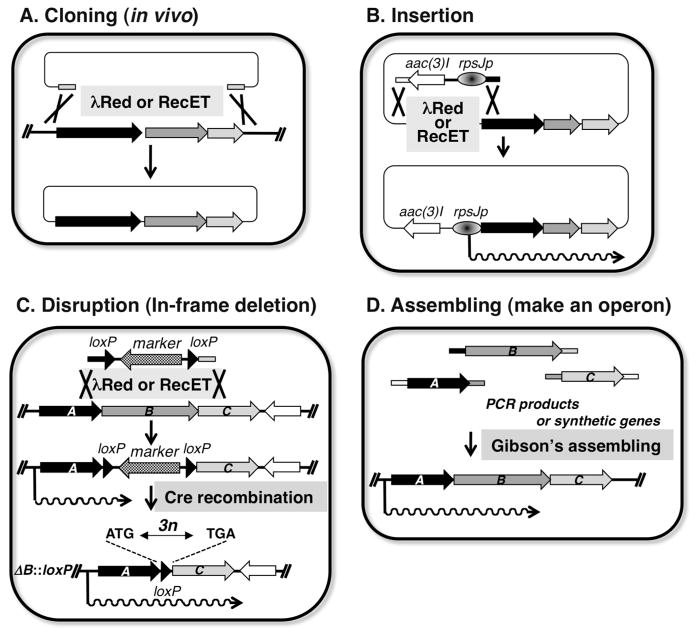
To clone, express and modify biosynthetic gene clusters in heterologous hosts, recent technology will be useful for manipulation of the gene cluster. These technologies are involved in short sequence-driven homologous recombination and site-specific recombination (Fig. 4). Recombination by λ phage gam, bet and exo gene products (λ-Red)24 or E. coli recE/recT gene products25 requires at least 40 homologous nucleotides to recombine. This technology could be applied to clone about up to 15-kb DNA segment from chromosomal DNA preparation and a vector carrying two ~41-bp homologous sequences corresponding to upstream and downstream of the gene cluster (Fig. 5-A). When more than a 15-kb DNA segment was cloned, a vector flanking 125–500-bp homologous regions corresponding to upstream and downstream of the gene cluster was used for λ-Red homologous recombination. This recombination system could be also applied to insert and replace an alternative promoter upstream of the gene operon (Fig. 5-B). In this insertion process, a DNA segment containing a resistant marker gene and alternative promoter sequence was amplified by PCR using a pair of primers consisting of a 24-nt sequence of the 5′ end and ~41-nt homologous sequence corresponding to upstream of the gene cluster, and a 24-nt reversed sequence of the 3′ end and a ~41-nt homologous sequence of the N-terminal region of the first gene in the gene operon, respectively. λ-Red and Cre site-specific recombination26 was combined to construct an in-frame deletion in the gene operon (Fig. 5-C). In vitro assembling of DNA fragments27, called “Gibson’s assembling”, would be a powerful tool for the construction of an artificial gene operon.
Figure 4.
Procedures concerning manipulation of the gene cluster for secondary metabolite biosynthesis. (A) in vivo cloning by λ-Red recombination, (B) construction of in-frame deletion in the gene cluster using λ-Red recombination, (C) introduction of alternative promoter by λ-Red recombination, and (D) construction of synthetic operon by Gibson’s method.
Figure 5.
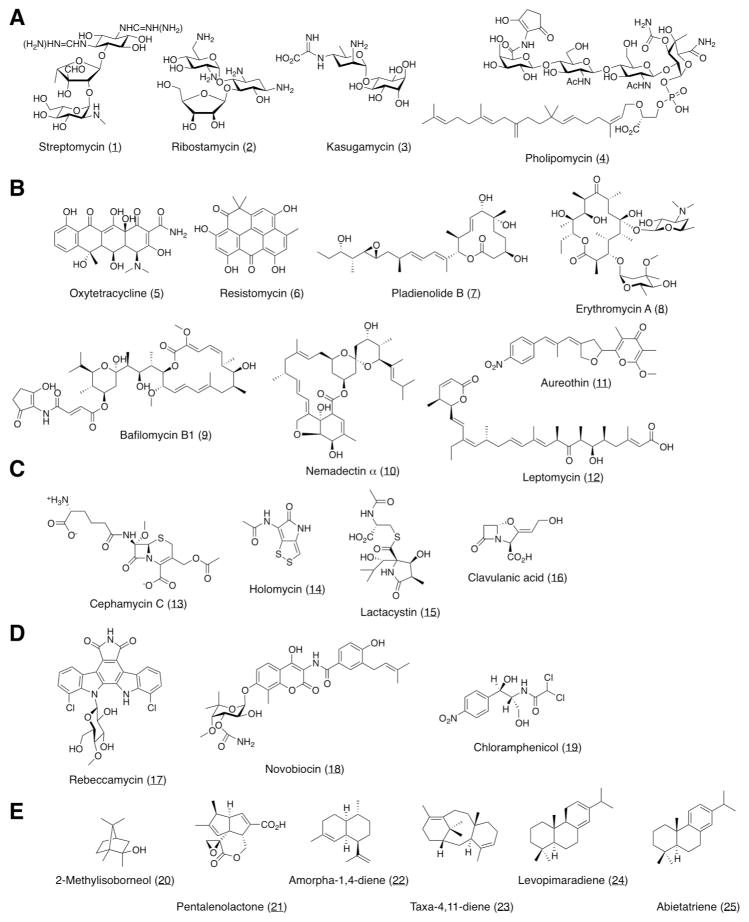
Structures of secondary metabolites (A) derived from sugar pathway, (B) polyketide pathway, (C) amino acid pathway, (D) shikimate pathway, and (E) MVA or MEP pathway.
Heterologous expression of genes involved in secondary metabolite biosynthesis was examined in S. avermitilis large-deletion derivatives, SUKA17 or 22. S. avermitilis SUKA22 was isogenic to SUKA1713. The left side of the deletion region of SUKA17 (corresponding to 8,886,025–8,925,414 nt in wild-type chromosome) was replaced by a wild-type loxP sequence but SUKA22 was used as a mutant-type loxP sequence to prevent recombination between another wild-type loxP sequence on the right side of the deletion region corresponding to 79,455–1,595,563 nt in the wild-type chromosome. The secondary metabolites could be classified into at least five classes on the basis of the pathway for the generation of precursors that are building blocks of the metabolite, sugar, polyketide, peptide (amino acid), shikimate and mevalonate (MVA) or methylerythritol (MEP) pathways. We have been already successful in the expression of biosynthetic gene clusters for streptomycin, cephamycin C and pladienolide13. Large-deletion derivatives should be confirmed to be applicable for use as heterologous hosts for more biosynthetic gene clusters. Genome analysis of S. avermitilis revealed that the microorganism harbored more than thirty biosynthetic gene clusters and many biosynthetic gene clusters for polyketide, non-ribosomal peptide and terpene compounds located in the linear chromosome. No biosynthetic gene clusters for aminoglycoside, polyketide-peptide hybrid and shikimate-derived compounds were found.
(1) Expression of the biosynthetic gene clusters for compounds derived from sugar pathway
Aminoglycoside antibiotics are synthesized from sugar pathway and a streptidine-containing aminoglycoside, streptomycin (1) (Fig. 5-A), has been produced by a S. avermitilis large-deletion derivative carrying the gene cluster for streptomycin biosynthesis (Fig. S. 1-A) of S. griseus IFO 1335013. The biosynthetic gene cluster for deoxystreptamine-containing aminoglycoside, ribostamycin (2) (Fig. 5-A), of S. ribosidificus ATCC 2129428 was cloned by a cosmid vector and about 35-kb segment containing the entire set of the gene cluster (Fig. S. 1-B) was subcloned into the integrating vector pKU465cos13 to generate pRSM1. The resultant transformants produced ribostamycin in the culture broth and their productivity was about 8 mg/L. In general, aminoglycoside antibiotics consist of one molecule of aminocyclitol and two or more molecules of neutral or amino sugars such as streptomycin and ribostamycin. Since kasugamycin (3) (Fig. 5-A) consists of one amino sugar and one neutral cyclitol, myo-inositol, the size of the gene cluster was smaller than biosynthetic gene clustsers for other aminoglycoside antibiotic (about 20-kb). Transformants carrying the entire gene cluster for kasugamycin biosynthesis (Fig. S. 1-C) of S. kasugaensis MB 273 (accession No. AB360380) produced about 5 mg/L kasugamycin in the culture broth. The expression of the kasugamycin biosynthetic genes was controlled by the gene product of regulatory gene kasT. When kasT was expressed in the multi-copy plasmid that gave the desired gene-dosage effect or by the alternative promoter, productivity was improved (7 to 12 mg/L).
A member of phosphoglycolipid compounds, moenomycin, quebemycin, prasinomycin, diumycin, macarbomycin, pholipomycin and nosokomycin, are potent inhibitors for the growth of a broad spectrum of Gram-positive pathogenic bacteria, including multi-drug resistant Staphylococcus aureus, and they inhibit bacterial peptidoglycan synthesis at the transglycosylation stage. Cloning of the gene cluster for moenomcyin biosynthesis in S. ghanaenesis ATCC 14672 was first reported for this member of compounds29. Interestingly, two distinct clusters, in which 30-kb and 5-kb regions contained 20 and 3 protein-coding genes, respectively, are required for the biosynthesis of moenomycin. From bioinformatic analysis of genome data of S. clavuligerus ATCC 27064, the size and organization of genes in about a 26-kb region of mega-plasmid pSCL4 in this microorganism were quite similar to those of a 30-kb region of S. ghanaensis ATCC 146728. The 26-kb region had 17 protein-coding genes (Fig. S. 1-D) that were highly homologous to 17 genes in the 30-kb region of S. ghanaenesis ATCC 14772. The production of phosphoglycolipid compounds from S. clavuligerus ATCC 27064 has not been reported to date. The desired region containing these 17 genes was cloned by λ-Red recombination using a cosmid library of S. clavuligerus ATCC 27064 and pRED vector13 carrying 500-bp homologous regions corresponding to upstream and downstream of sclav_p1274 and sclav_p1290, respectively. About a 28-kb cloned segment was ligated with the integrating vector and the desired plasmid (pPHM1) was introduced into S. avermitilis SUKA22. The resultant transformants produced about 20 mg/L pholipomycin30 (4) (Fig. 5-A), which was mainly accumulated in mycelium and a very small amount of pholipomycin was detected from the culture broth. Interestingly, the original microorganism, S. clavuligerus ATCC 27064, did not produce pholipomycin in any culture conditions.
(2) Expression of the biosynthetic gene clusters for compounds derived from polyketide pathway
Polyketide compounds are synthesized by at least three types of polyketide synthases (PKSs), type I, type II and type III in bacteria31. Macrocyclic lactones are generated by type-I PKSs (modular PKS) and aromatic compounds are produced by type-II or type-III PKSs. Oxytetracycline is widely used in the clinical field as an antibacterial agent. A cosmid clone (pOTC1) contained a putative entire gene cluster for the oxytetracycline biosynthesis (Fig. S. 1-E) of S. rimosus NRRL 223432. A 25 kb region containing 22 genes involved in the biosynthesis, self-resistance and export of oxytetracycline was subcloned into the integrating vector and the desired recombinant plasmid was introduced into S. avermitilis SUKA17. Transformants produced about 20 mg/L oxytetracycline (5) (Fig. 5-B) in the culture broth. In the second example, the entire gene cluster for discoid polyketide, resistomycin, biosynthesis33 was cloned from Streptomyces sp. NA97. A cosmid clone, pKU492resP2-O23-1n, containing at least 18 genes involved in resistomycin biosynthesis (Fig. S. 1-F), was introduced into S. avermitilis SUKA22. Interestingly, the resultant transformants produced a large amount of resistomycin (6) (Fig. 5-B) in the culture broth and the productivity was about 360 mg/L.
As modular PKS contains multi-catalytic domains on a polypeptide, the size of the gene encoding modular PKS is quite large in comparison with genes encoding type-II or type-III PKSs and the entire biosynthetic gene clusters are at least 40-kb. We used an integrating BAC (Bacterial Artificial Chromosome) vector, pKU503, to clone the entire 75-kb gene cluster for macrocyclic lactone, pladienolide B (7) (Fig. 5-B), biosynthesis13 (Fig. S. 1-O). A similar technique was applied to clone the entire biosynthetic gene clusters for erythromycin (8), bafilomycin B1 (9), nemadectin α (10), and linear polyketides, aureothin (11) and leptomycin (12) (Fig. 5-B). A BAC clone, pKU503eryP4-A14, containing the entire 60-kb gene cluster for erythromycin biosynthesis (Fig. S. 1-P) of Sacchropolyspora erythraea NRRL 233834, was isolated and the transformants carrying pKU503eryP4-A14 produced about 4 mg/L erythromycin A in the culture broth. Unfortunately, during several rounds of propagation, productivity was reduced and they accumulated the aglycone moiety of erythromycin, 6-deoxyerythronolide B. The entire 72 kb gene cluster for bafilomycin B1 biosynthesis (Fig. S. 1-Q) of Kitasatospora setae KM-605419 was cloned into BAC vector and the resultant BAC clone, pKU503bafP3-P20 (pBFM1), was introduced into S. avermitilis SUKA22. The transformants produced 16 mg/L bafilomycin B1 (9) and a trace amount of bafilomycin A1. Anthelmintic macrolide, nemadectin α35(10), is structurally related to avermectin and the organization of each gene in the gene cluster was also quite similar to that in the avermectin gene cluster. The gene cluster for nemadectin biosynthesis (Fig. S. 1-R) of S. cyaneogriseus subsp. noncyanogenus NRRL 15774 was cloned into BAC vector. The resultant BAC clone, pKU503nemP11-L5, containing the entire 80-kb gene cluster for nemadectin biosynthesis (accession No. AB363939), was introduced into S. avermitilis SUKA22 and the transformants produced 1.6 mg/L nemadectin α (10) and a trace amount of related components in the mycelium. Two entire biosynthetic gene clusters for linear polyketide compounds, aureothin (11) and leptomycin (12) were cloned into cosmid and BAC vectors, respectively. Transformants carrying a cosmid clone, pKU465aurP2-A6, carrying the entire 35-kb gene cluster for aureothin biosynthesis (Fig. S. 1-S) of Streptomyces sp. MM 2336 produced 116 mg/L aureothin (11) in the culture broth. Since the biosynthetic gene cluster for leptomycin37 was estimated to be larger than 40-kb, the entire biosynthetic gene cluster for leptomycin (Fig. S. 1-T) of Streptomyces sp. EM52 was cloned by BAC vector and a BAC clone, pKU503lepP5-L8, containing the entire 80-kb gene cluster for leptomycin, was isolated. The resultant transformants produced 5 mg/L leptomycin (12) in the mycelium. Thus, large-deletion mutants of S. avermitilis could transform a ~100-kb BAC clone without detectable deletions involved in the highly homologous region of each module in the biosynthetic gene cluster.
(3) Expression of the biosynthetic gene clusters for compounds derived from the amino acid pathway
The biosynthetic process for peptide bonds of peptide compounds derived from amino acids is accomplished by at least three types; peptide bonds are generated by non-ribosomal peptide synthetase (NRPS), ribosomal peptide synthesis and simple modification of the amino residue of amino acid. Cephamycin C (13) (Fig. 5-C), derived from three amino acids, is a typical peptide compound generated by NRPS and we have already shown the heterologous expression in S. avermitilis SUKA17 of the gene cluster for cephamycin C biosynthesis (Fig. S. 1-G) of S. clavuligerus ATCC 2706413. Other two peptide compounds, holomycin (14), synthetized by NRPS, and lactacystin (15), synthesized by NRPS-PKS hybrid, were examined. The entire 18 kb gene cluster for holomycin biosynthesis (Fig. S. 1-H) of S. clavuligerus ATCC 2706438, in which 13 protein-coding genes are located, was cloned from the cosmid library of S. clavuligerus ATCC 27064 by λ-Red recombination using 500-bp homologous regions corresponding to upstream and downstream of sclav_5267 and sclav_5278, respectively. The desired recombinant plasmid (pHLM1), which contained a minimum set of the biosynthetic gene cluster for holomycin, was introduced into S. avermitilis SUKA22 and the transformants produced 8 mg/L holomycin (14) (Fig. 4C) and 2 mg/L deacetylated derivative, holothine, in the culture broth. Lactacystine39 (15) (Fig. 5-C), a specific inhibitor of proteasomes, was thought to be synthesized by the NRPS-PKS hybrid, synthetase. Bioinformatic analysis of the draft sequence data of lactacystin-producing S. lactacystinaeus OM-6519 suggested that the biosynthetic gene cluster for lactacystin was ~12 kb. The entire gene cluster for lactacystin biosynthesis (Fig. S. 1-I) was cloned from the cosmid library of S. lactacystinaeus OM-6519 by λ-Red recombination using 41-bp homologous sequences corresponding to upstream and downstream of the gene cluster. The minimum set of the gene cluster, 12 kb (pLTC1), was introduced into S. avermitilis SUKA22 using an integrating vector, but not all transformants produced lactacystin in both mycelium and culture broth. Many biosynthetic gene clusters for secondary metabolites contain one or more genes encoding transcriptional regulatory proteins that activate the expression of genes involved in biosynthesis in the gene cluster, but no regulatory genes were found in the biosynthetic gene cluster for lactacystin. Since all genes in the gene cluster formed a polycistron, an alternative promoter (a promoter of the gene encoding ribosomal protein S10, rpsJ13) was introduced into the promoter region of the first gene in the operon to generate pLTC1*. Transformants carrying pLTC1* produced 30 mg/L lactacystin (15) in the culture broth.
A strong inhibitor against bacterial β-lactamases, clavulanic acid (16) (Fig. 5-C), belongs to the β-lactam compounds, such as penam (penicillins) and cephem (cephalosporines and cephamycins) compounds. Although these antibacterial penam and cephem compounds are synthesized from three amino acids (α-aminoadipate, cysteine and valine), clavulanic acid was generated from L-arginine and glyceraldehyde-3-phosphate, the biosynthetic process of which does not require activation of the amino acid for the formation of peptide bonds by NRPS40. The gene cluster for clavulanic acid biosynthesis (Fig. S. 1-J) is located adjacent to the gene cluster for cephamycin C biosynthesis in S. clavuligerus ATCC 2706441. A 25 kb region (sclav_4180 - sclav_4197) was cloned from the cosmid library of S. clavuligerus ATCC 27064 by λ-Red recombination using 500-bp homologous regions corresponding to upstream and downstream of sclav_4180 and sclav_4197, respectively, to generate pCLV1. Since it has been demonstrated that the expression of the biosynthetic gene cluster for clavulanic acid was controlled by the gene product of ccaR (sclav_4204) of the gene cluster for cephamycin C biosynthesis42, a ~1.5-kb segment containing ccaR and its promoter region was amplified by PCR and the amplicon was introduced into pCLV1 to generate pCLV1*. The resultant transformants carrying the entire 25-kb biosynthetic gene cluster for clavulanic acid and ccaR gene produced 16 mg/L clavulanic acid (16) in the culture broth, whereas clavulanic acid was not produced by transformants carrying the entire 25-kb biosynthetic gene cluster alone (pCLV1).
(4) Expression of the biosynthetic gene clusters for compounds derived from the shikimate pathway
Since genes including compounds derived from the shikimate pathway were not found in the S. avermitilis genome, the heterologous expression of the biosynthetic gene cluster for compounds derived from shikimate was quite interesting in this microorganism. Aromatic amino acids, phenylalanine, tyrosine and tryptophan, are generated from intermediates derived from the glycolytic and pentose phosphate pathways by the shikimate pathway. Indolocarbazoles are potent inhibitors of protein kinases and the compounds were synthetized from two molecules of L-tryptophan. A cosmid clone, pREB1, carrying the entire 16-kb gene cluster for rebeccamycin biosynthesis (Fig. S. 1-K) of Lechevalieria aerocolonigenes ATCC 3924343, in which 10 protein-coding genes are located, was introduced into S. avermitilis SUKA17 and the transformants produced 7 mg/L rebeccamycin (17) (Fig. 5-D) in the mycelium.
The aromatic ring moieties of antibacterial compounds, novobiocin (18) and chloramphenicol (19), are derived from chorismate, which is an important intermediate in the shikimate pathway. The entire 29-kb gene cluster for novobiocin biosynthesis (Fig. S. 1-L) of S. anulatus 3533-SV4 GM95 was cloned by a cosmid vector and a cosmid clone, pKU492novP-G18-1n, containing a self-resistance gene (gryBr), which was insensitive to novobiocin, and 23 biosynthetic genes (novA-novW). The biosynthetic gene cluster for novobiocin of S. caeruleus (former name; spheroides) NCBI 11891 was cloned previously44 and the size, organization and transcriptional direction of each gene of S. caeruleus NCBI 11891 was the same as that of S. anulatus 3533-SV4 GM95. S. avermitilis SUKA22 transformants carrying pKU492novP-G18-1n, produced 1 mg/L novobiocin (18) (Fig. 5-D) in the culture broth. The entire gene cluster for chloramphenicol (19) (Fig. 5-D) biosynthesis (Fig. S. 1-M) of S. venezuelae ATCC 1071245 was isolated and expressed in S. coelicolor A3(2) mutants previously46. We subcloned the minimum set of the gene cluster for chloramphenicol biosynthesis from pAH91, which is a cosmid clone of S. venezuelae ATCC 10712. The entire 24-kb gene cluster for chloramphenicol biosynthesis was cloned from pAH91 by λ-Red recombination using 500-bp homologous regions corresponding to upstream and downstream of sven_0913 and sven_0931, respectively. The desired recombinant plasmid, pCML1, carrying 19 protein-coding genes, was introduced into S. avermitilis SUKA22 and the resultant transformants produced abundant chloramphenicol (250 mg/L) in the culture broth.
(5) Expression of the biosynthetic gene clusters for compounds derived from MVA or MEP pathway
A degraded sesquiterpenoid alcohol, geosmin, which is responsible for the characteristic odor of moist soil, is produced not only by many actinomycete microorganisms but also by Cyanobacteria and Myxobacteria. Another common metabolite with a characteristic musty, camphor-like odor is the monoterpenoid alcohol 2-methylisoborneol. The genes involved in 2-methylisoborneol biosynthesis of S. avermitilis have been deleted naturally during evolution. Genes encoding 2-methylisoborneol synthase and GPP 2-methyltransferase (Fig. S. 1-N) in Saccharopolyspora erythraea NRRL 2338 and S. ambofaciens ATCC 15154 were expressed in S. avermitilis SUKA17 and transformants produced abundant 2-methylisoborneol47 (20) (Fig. 5-E).
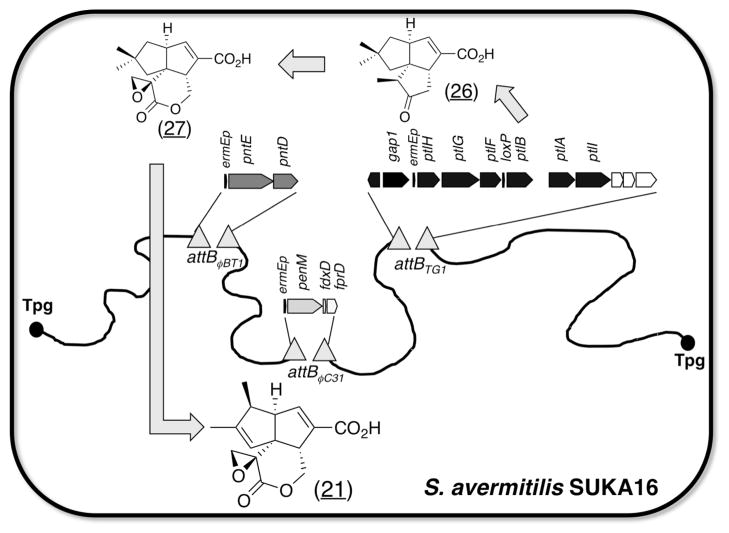
Pentalenolactone (21) (Fig. 5-E) is an anti-bacterial, -fungal and -viral sesquiterpenoid antibiotic produced by many Streptomyces microorganisms48,49. Pentalenene synthase, which catalyzes the cyclization of farnesyl diphosphate to the parent sesquiterpenoid hydrocarbon pentalenene, was firstly isolated from S. exfoliatus U5319 and subsequently cloned and expressed in E. coli50. The sav2998 gene of S. avermitilis encodes a 336-aa protein (PtlA) with 76% identity to the well-characterized pentalenene synthase of S. exfoliatus U531951. The ptlA gene is itself located within a 13.4-kb ptl gene cluster encoding 13 predicted protein coding sequences that we initially thought might represent the set of biosynthetic genes for pentalenolactone itself. Ultimately, S. avermitilis mutants, in which the expression of ptl cluster was controlled by an alternative promoter, did not produce pentalenolactone but produced a novel sesquiterpenoid compound, neopentalenoketolactone, an isomer of the expected product pentalenolactone F52. Thereafter, pentalenolactone biosynthetic gene clusters were identified from established producers of the antibiotic, S. exfoliatus UC5319 (pen gene cluster) and S. arenae TÜ469 (pnt gene cluster). Each of the two gene clusters contained a homologous set of 10 unidirectionally transcribed protein-coding genes, including close orthologues of each of the previously characterized gene products in the S. avermitilis ptl gene cluster. The biosynthetic process from the formation of pentalenene by PtlA, PenA or PntA to the formation of a key intermediate, 11-oxo-1-deoxypentalenic acid (26), by PtlF53, PenF or PntF was identical in ptl, pen and pnt gene clusters. The following stepwise process was introduced into S. avermitilis SUKA17 carrying ptl cluster (Fig. 6). (i) The double deletion mutant, S. avermitilis ΔptlE ΔptlD, which produces a key intermediate, 11-oxopentalenic acid (26), was transformed by pntE, the gene product of which catalyzed region-specific Baeyer-Villiger oxidation exclusively to pentalenolactone D, and pntD-encoded dioxygenase catalyzed the α-ketoglutarate- and non-heme iron-dependent, two step desaturation/epoxidation of pentalenolactone D to pentalenolactones E and F (27). (ii) The resultant recombinants were further introduced into penM, the gene product of which was cytochrome P450, which catalyzed the unusual oxidative rearrangement of pentalenolactone F (27) to pentalenolactone54 (21). The final recombinant strain, S. avermitilis SUKA17 carrying ptl cluster (ΔptlE ΔptlD), processed pntD-pntE55 and penM produced pentalenolactone but not neopentalenoketolactone, which is the natural product of S. avermitilis.
Figure 6.
Construction of engineered pentalenolactone biosynthesis in S. avermitilis. Construction steps were as follows: (i) an intermediate of pentalenolactone-family metabolites, 11-oxo-1-deoxypentalenic acid (26) producer (S. avermitilis SUKA16 ΔptlE ΔptlD) was constructed by introducing pKU462::ptlH-ptlG-ptlF-ptlB-ptlA-ptlI into S. avermitilis SUKA16. (ii) Pentalenolactone (21) producer was constructed by stepwise introduction of pKU464aac(3)IV::pntE-pntD and pKU460hph::penM-fdxD-fprD into S. avermitilis SUKA16 ΔptlE ΔptlD recombinant. All gene cassettes were controlled by ermE promoter and penM was expressed as an operon with fdxD (sav3129) and fprD (sav5675) encoding ferredoxin and ferredoxin reductase, respectively. pKU460aac(3)IV, pKU462 and pKU464hph each contained an attachment site and integrase gene of bacteriophages, φC31, TG1 or φBT1, respectively.
We have recently examined the heterologous expression of the gene encoding sesquiterpene synthase, amorpha-4,11-diene synthase (ads), of plant origin, Artemisia annua, in which codon usage was optimized for expression in S. avermitilis13. A synthetic gene was expressed in S. avermitilis under control of the rpsJ promoter that was expressed constitutively. The requisite farnesyl diphosphate precursor was provided by introducing a copy of the native of the S. avermitilis ptlB gene (sav2997) encoding farnesyl diphosphate synthase downstream of the ads gene, and transformants produced abundant amorpha-1,4-diene (22) (Fig. 5-E) in the mycelium. We have now extended this observation to the expression of plant diterpene synthase genes in S. avermitilis. Two synthetic genes encoding taxa-4,11-diene (tds) and levopimaradine synthases (lps) of taxol producer Taxus brevifolia (accession No. Q41594) and ginkgolide A producer Ginkgo biloba (accession No. Q947C4), respectively, in which codon usage was optimized for expression in S. avermitilis, were prepared and joined with the native of S. avermitilis crtE gene (sav1022) encoding geranylgeranyl diphosphate synthase upstream of the synthetic diterpene synthase gene, respectively56. Each synthetic operon was introduced into S. avermitilis SUKA17 by transformation. A single major product with mass [M++1] m/z 273 corresponding to C20H32 was detected in the n-hexane extract of transformants carrying synthetic tds and the product was identical to taxa-1,4-diene (23) (Fig. 5-E) in several spectrum data. Similarly, two products with mass [M+] m/z 272 (C20H32) and [M+] m/z 270 (C20H30) were observed from the extract of transformants carrying synthetic lpd, but their productivity was lower than that of transformants carrying tds. These two products were thought to be levopimaradine (24) and abietatriene (25) (Fig. 5-E), respectively. Production of the parent intermediates of plant sesqui- and diterpenoid compounds by S. avermitilis SUKA17 transformants was observed by growth in a simple synthetic medium without the addition of precursors and each terpene synthase gene was efficiently expressed as a synthetic operon with the gene encoding farnesyl diphosphate synthase or geranylgeranyl diphosphate synthase.
Productivity of secondary metabolites in heterologous host
More than twenty biosynthetic gene clusters were introduced into engineered S. avermitilis and many of these biosynthetic gene clusters showed efficient expression. Although a few gene clusters were inefficiently expressed and the transformants did not produce metabolites, production was successfully restored by controlling the expression of the regulatory gene or directly expressing the biosynthetic gene operon using an alternative promoter. As mentioned above, S. avermitilis harbors many biosynthetic gene clusters for secondary metabolites derived from polyketide, amino acid pathways, the backbones of which were synthesized by PKS and NRPS, but no biosynthetic gene clusters for secondary metabolites derived from sugar and shikimate pathways. Furthermore, although at least eight biosynthetic gene clusters harboring NRPS genes are located in the S. avermitilis genome, no peptide compounds were detected under any culture conditions. The production of these secondary metabolites in S. avermitilis is interesting. As described above, transformants of large-deletion derivatives of S. avermitilis produced aminoglycoside and phosphoglycolipid antibiotics, shikimate-derived compounds and peptide compounds.
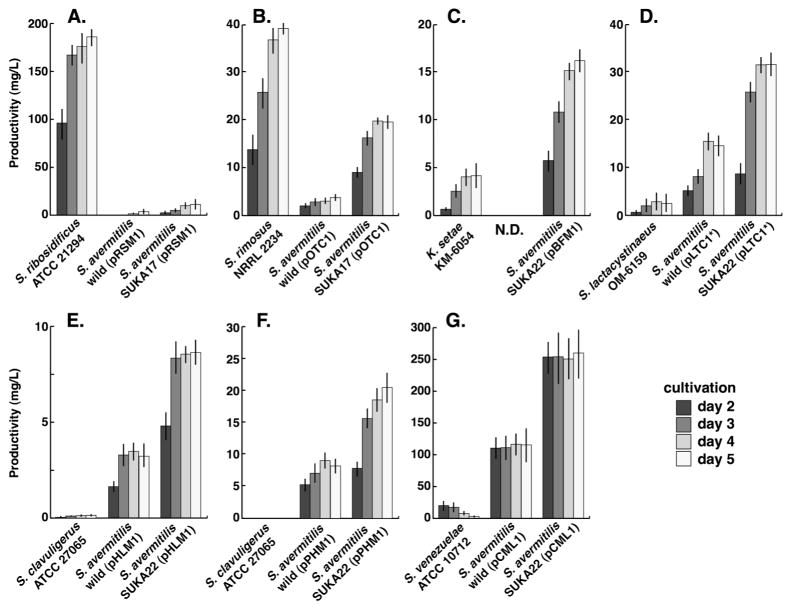
Transformants of S. avermitilis wild-type strain carrying the biosynthetic gene cluster for streptomycin (1) or cephamycin C (13) produced about half of the amount of each metabolite as the original producer S. griseus IFO 13350 or S. clavuligerus ATCC 27065. The productivity of these metabolites in S. avermitilis large-deletion mutant SUKA17 transformants was improved, being 4.5 to 7-fold higher than that of wild-type transformants and 2 to 2.5-fold higher than that of the original producers as described previously13. Both transformants of S. avermitilis wild-type and its large deletion-mutant SUKA17 carrying biosynthetic gene cluster for deoxystreptamine-containing aminoglycoside, ribostamycin (2), of S. ribosidificus ATCC 21294 produced small amount of ribostamycin (Fig. 7-A; about 1.5 – 4% productivity of that of S. ribosidificus ATCC 21294). Similarly, transformants of S. avermitilis SUKA17 carrying the biosynthetic gene cluster for oxytetracycline produced about half of the amount of metabolite as the original producer S. rimosus NRRL 2234 and the productivity of S. avermitilis wild type transformants was inefficient (Fig. 7-B). These two biosynthetic gene clusters might be inefficiently expressed in S. avermitilis.
Figure 7.
Production profiles of (A) ribostamycin (2), (B) oxytetracycline (5), (C) bafilomycin B1 (9), (D) lactacystin (15), (E) holomycin (14), (F) pholipomycin (4) and (G) chloramphenicol (19) in original producers S. ribosidificus ATCC 21294, S. rimosus NRRL 2234, K. setae KM-6054, S. lactacystinaeus OM-6159, S. clavuligerus ATCC 27065 and S. venezuelae ATCC 10712, respectively, S. avermitilis wild type and SUKA17 or SUKA22 carrying each biosynthetic gene cluster (pRSM1, pOTC1, pBFM1, pLTC1*, pHLM1, pPHM1 or pCML1). Each productivity was calculated by duplicated experiments and vertical lines indicate standard deviation. Transformants of S. avermitilis wild type strain by pBFM1 were not obtained.
Since S. avermitilis wild-type produced principal endogenous polyketide compounds synthesized by type-I PKSs (avermectin, oligomycin and filipin), it is quite interested in heterologous expression of biosynthetic gene cluster for polyketide compounds. Transformation or conjugation efficiency by BAC clone (ca. 100-kb) in Streptomyces strains was extremely low in comparison to that by cosmid clone (ca. 45-kb). A few transformants of SUKA22 were obtained, but no transformants of wild type appeared. Bafilomycin B1 production in the SUKA22 transformants carrying the entire gene cluster for bafilomycin biosynthesis was 4-fold higher than that of the original producer K. setae KM-6054 (Fig. 7-C). No all transformants carrying the entire gene cluster for lactacystin biosynthesis of S. lactacystinaeus OM-6159 produced lactacystin. But lactacystin production was extremely improved by expression of the gene cluster using an alternative promoter (promoter of rpsJ). Lactacystin production in transformants of both the wild type and SUKA22 was 7 to 12-fold higher than that of the original producer S. lactacystinaeus OM-6159 (Fig. 7-D). Holomycin production in the wild type S. clavuligerus ATCC 27065 was extremely low level and variable38. Holomycin production in wild-type S. clavuligerus ATCC 27065 was less than 0.2 mg/L. The production of holomycin and its deacetylated derivative holothin in both S. avermitilis wild type and SUKA22 was relatively stable (Fig. 7-E). The productivity of holomycin in SUKA22 transformants was improved at least 40-fold higher than that of the original producer S. clavuligerus ATCC 27065 (Fig. 7-E). Since pholipomycin production has been never detected from the culture of S. clavuligerus ATCC 27065 in any culture conditions, the biosynthetic cluster would be cryptic in S. clavuligerus ATCC 27065. On the other hand, transformants of both the wild type and SUKA22 carrying the entire biosynthetic gene cluster for pholipomycin produced 9 mg/L and 20 mg/L pholipomycin, respectively (Fig. 7-F). These results indicate that S. avermitilis will possess the upper regulatory system for the control of the expression of the gene cluster for pholipomycin biosynthesis. Chloramphenicol, derived from shikimate pathway, production in transformants of both the wild type and SUKA22 was 6 to 12-fold higher than that of the original producer S. venezuelae ATCC 10712 (Fig. 7-G). It seems that the biosynthetic gene cluster for chloramphenicol of S. venezuelae ATCC 10712 is efficiently expressed and precursors of the shikimate pathway are also efficiently supplied to chloramphenicol synthesis in S. avermitilis.
Among of them, production of bafilomycin B1, lactacystin, holomycin, pholipomycin and chloramphenicol was extremely efficient. The productivity of lactacystin, holomycin, pholipomycin and chloramphenicol in S. avermitilis wild-type transformants was higher than that of original producer K. setae KM-6054, S. lactacystinaeus OM-6159, S. clavuligerus ATCC 27065 and S. venezuelae ATCC 10712. The productivity of these metabolites in S. avermitilis SUKA22 transformants was improved, being 2 to 2.5-fold higher than that of wild-type transformants. Because S. avermitilis SUKA22 lacks the biosynthetic gene clusters for the principal endogenous metabolites of S. avermitilis, the natural precursors and biochemical energy of the host are apparently efficiently used in these metabolites. In lactacystin, holomycin, pholipomycin and chloramphenicol production of S. avermitilis transformants, since each precursor for biosynthesis is different from that of the principal endogenous polyketide metabolites, the productivity of these metabolites in the wild-type strain was not too low.
CONCLUSION
Genetic instability has been observed in many Streptomyces microorganisms and could be an important driving force in evolution. This phenomenon disagrees with the industrial production of secondary metabolites. Among the Streptomyces microorganisms genome-sequenced to date, an industrial anthelmintic macrocyclic lactone, avermectin, a producer S. avermitilis, has a stable phenotype in comparison with other Streptomycetaceae microorganisms. Comparative analysis of these genome data suggests that the size of TIRs found at the end of the linear chromosome ends correlated with genetic instability and the genetically stable characteristics of S. avermitilis are due to the shortest TIRs in the linear chromosome. The intrinsic genetically stable characteristics of S. avermitilis make it especially suitable as a source of endogenous secondary metabolites and as a host for the heterologous production of secondary metabolites derived from exogenous biosynthetic gene clusters.
Using large-deletion derivatives of S. avermitilis, from which biosynthetic gene clusters for endogenous major products were removed, it has been planned to construct the heterologous expression of biosynthetic gene clusters for secondary metabolites; therefore, large-deletion derivatives did not produce endogenous main metabolites. The characteristics of large-deletion derivatives of S. avermitilis are fundamentally similar to those of the wild-type strains. Not only morphological differentiation, which is observed in many Streptomyces, of large-deletion derivatives was more developed than that of the wild type, but also the growth rate and biomass in the stationary phase was enhanced.
The feasibility of using large-deletion derivatives of S. avermitilis as a heterologous host has been shown by the effective expression of 18 intact biosynthetic gene clusters, two biosynthetic gene clusters controlled by an alternative promoter and three codon-optimized genes encoding plant terpenoid intermediates. Furthermore, because large-deletion derivatives of S. avermitilis no longer produced major endogenous metabolites, primary metabolism seemed to be efficiently exploited to generate precursors of exogenous biosynthetic gene clusters.
METHODS
Bacterial strains, and growth conditions
Streptomyces avermitilis K13947 (isogenic to MA-4680 but lacking a large linear plasmid SAP2) was used as the wild-type strain and its large-deletion derivatives, SUKA17 and SUKA22, were used for heterologous expression of biosynthetic gene clusters for secondary metabolites. Each spore preparation for the production and storage was obtained by growth on YMS medium57 at 30°C for 3 to 5 days. Two minimum media (MM), agar medium containing 1.0 g (NH4)2SO4, 0.5 g K2HPO4, 0.2 g MgSO4•7H2O, 0.003 g FeSO4•7H2O and 15.0 g agar per liter (pH 7.0), and liquid medium containing 2.0 g (NH4) 2SO4, 0.6 g MgSO4•7H2O, 1.0 mL of trace elements solution (40 mg ZnCl2•7H2O, 200 mg FeCl3, 10 mg CuCl2•2H2O, 10 mg MnCl2•4H2O, 10 mg Na2B4O7•10H2O and 10 mg (NH4)2MoO24•4H2O per liter), and 20 mL of 0.1 M phosphate buffer (pH7.0) were used to assess the growth characteristics of S. avermitilis. Actinomycete microorganisms were used as the source of chromosomal DNA preparation, S. ribosidificus ATCC 21294 (JCM 4923), S. clavuligerus ATCC 27064 (JCM 4710), and S. venezuelae ATCC 10712 (JCM 4526) were obtained from the culture collection of the RIKEN Bioresource Center (Wako, Japan) and Saccharopolyspora erythraea NRRL 2338, S. cyaneogriseus subsp. noncyanogenus NRRL 15774 and S. rimosus NRRL 2234 were from the Agricultural Research Service Culture Collection (National Center for Agricultural Utilization Research, US Department of Agriculture, Peoria, IL, USA). Other microorganisms were from our culture collection, isolated from soil samples. The construction of the genomic library using a cosmid or BAC vector and in vivo cloning of the entire biosynthetic gene cluster by λ-RED recombination are described in “SI methods”.
Cultivation for secondary metabolite production
Conditions for vegetative culture and fermentation culture were described previously13. In almost all cases, semi-synthetic medium was used for the production and another medium containing 4.0 g yeast extract, 10.0 g malt extract and 10.0 g glucose (pH 7.0) per liter was used when the productivity was low in the semi-synthetic medium13. Secondary metabolites caused by introduction of the exogenous entire biosynthetic gene cluster for secondary metabolites into large-deletion derivatives of S. avermitilis were detected by HPLC-MS analysis and/or biological activity with comparison with each authentic sample. The metabolite was purified and the structure was confirmed by several spectrum data (1H and 13C NMR, MS analyses and so on) when authentic samples could not be purchased or obtained. In each case, the productivity of the metabolite(s) was measured from 4 to 6 transformants.
Supplementary Material
Acknowledgments
We thank I. Kojima of Akita Prefecture University, H. Onaka of Toyama Prefecture University and M. J. Bibb for kindly providing gene clusters for kasugamycin biosynthesis, rebeccamycin biosynthesis and chloramphenicol biosynthesis (pAH91), respectively. We also thank M.C.M. Smith of the University of Aberdeen, UK for provision of attP and integrase genes of bacteriophage φBT1. This work was supported by a research Grant-in-Aid for Scientific Research on Innovative Areas from the Ministry of Education, Culture, Sports, Science and Technology of Japan (to H.I.), a research grant from the Institute for Fermentation, Osaka, Japan (to H.I.) and a research Grant-in-Aid for Scientific Research from the New Energy and Industrial Technology Development Organization (NEDO; to K.S. and H.I.).
References
- 1.Waksman SA. Streptomycin: Background, isolation, properties, and utilization. Science. 1953;118:259–266. doi: 10.1126/science.118.3062.259. [DOI] [PubMed] [Google Scholar]
- 2.Stackebrandt E, Sproer C, Rainey FA, Burghardt J, Pauker O, Hippe H. Int J Syst Bacteriol. 1997;47:1134–1139. doi: 10.1099/00207713-47-4-1134. [DOI] [PubMed] [Google Scholar]
- 3.Berdy J. Bioactive microbial metabolites - A personal view - J Antibiot. 2005;58:1–26. doi: 10.1038/ja.2005.1. [DOI] [PubMed] [Google Scholar]
- 4.Omura S, Ikeda H, Ishikawa J, Hanamoto A, Takahashi C, Shinose M, Takahashi Y, Horikawa H, Nakazawa H, Osonoe T, Kikuchi H, Shiba T, Sakaki Y, Hattori M. Genome sequence of an industrial microorganism Streptomyces avermitilis: Deducing the ability of producing secondary metabolites. Proc Natl Acad Sci USA. 2001;98:12215–12220. doi: 10.1073/pnas.211433198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ikeda H, Ishikawa J, Hanamoto A, Shinose M, Kikuchi H, Shiba T, Sakaki Y, Hattori M, Omura S. Complete genome sequence and comparative analysis of the industrial microorganism Streptomyces avermitilis. Nat Biotechnol. 2003;21:526–531. doi: 10.1038/nbt820. [DOI] [PubMed] [Google Scholar]
- 6.Wang XJ, Yan YJ, Zhang B, An J, Wang JJ, Tian J, Jiang L, Chen YH, Huang SX, Yin M, Zhang J, Gao AL, Liu CX, Zhu ZX, Xiang WS. Genome sequence of the milbemycin-producing bacterium Streptomyces bingchenggensis. J Bacteriol. 2010;192:4526–4527. doi: 10.1128/JB.00596-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Barbe V, Bouzon M, Mangenot S, Badet B, Poulain J, Segurens B, Vallenet D, Marlière P, Weissenbach J. Complete genome sequence of Streptomyces cattleya NRRL 8057, a producer of antibiotics and fluorometabolites. J Bacteriol. 2011;193:5055–5056. doi: 10.1128/JB.05583-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Medema MH, Trefzer A, Kovalchuk A, Berg M, Müller U, Heijne W, Wu L, Alam MT, Ronning CM, Nierman WC, Bovenberg RAL, Breitling R, Takano E. The sequence of a 1.8-Mb bacterial linear plasmid reveals a rich evolutionary reservoir of secondary metabolic pathways. Genome Biol Evol. 2010;2:212–224. doi: 10.1093/gbe/evq013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bentley SD, Chater KF, Cerdeno-Tarraga AM, Challis GL, Thomson NR, James KD, Harris DE, Quail MA, Kieser H, Harper D, Bateman A, Brown S, Chandra G, Chen CW, Collins M, Cronin A, Fraser A, Goble A, Hidalgo J, Hornsby T, Howarth S, Huang CH, Kieser T, Larke L, Murphy L, Oliver K, O’Neil S, Rabbinowitsch E, Rajandream MA, Rutherford K, Rutter S, Seeger K, Saunders D, Sharp S, Squares R, Squares S, Taylor K, Warren T, Wietzorrek A, Woodward J, Barrell BG, Parkhill J, Hopwood DA. Complete genome sequence of the model actinomycete Streptomyces coelicolor A3(2) Nature. 2002;417:141–147. doi: 10.1038/417141a. [DOI] [PubMed] [Google Scholar]
- 10.Ohnishi Y, Ishikawa J, Hara H, Suzuki H, Ikenoya M, Ikeda H, Yamashita A, Hattori M, Horinouchi S. Genome sequence of the streptomycin-producing microorganism Streptomyces griseus IFO 13350. J Bacteriol. 2008;190:4050–4060. doi: 10.1128/JB.00204-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bignell DRD, Seipke RF, Huguet-Tapia JC, Chambers AH, Parry RJ, Loria R. Mol Plant Microbe Interact. 2010;23:161–175. doi: 10.1094/MPMI-23-2-0161. [DOI] [PubMed] [Google Scholar]
- 12.Pullan ST, Chandra G, Bibb MJ, Merrick M. Genome-wide analysis of the role of GlnR in Streptomyces venezuelae provides new insights into global nitrogen regulation in actinomycetes. BMC Genomics. 2011;12:175. doi: 10.1186/1471-2164-12-175. (2011) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Komatsu M, Uchiyama T, Omura S, Cane DE, Ikeda H. Genome-minimized Streptomyces host for the heterologous expression of secondary metabolism. Proc Natl Acad Sci USA. 2010;107:2646–2651. doi: 10.1073/pnas.0914833107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Burg RW, Miller BM, Baker EE, Birnbaum J, Currie SA, Hartman R, Kong YL, Monaghan RL, Olson G, Putter I, Tunac JB, Wallick H, Stapley EO, Oiwa R, Omura S. Avermectins, new family of potent anthelmintic agents: producing organism and fermentation. Antimicrob Agents Chemother. 1979;15:361–367. doi: 10.1128/aac.15.3.361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen CW, Huang CH, Lee HH, Tsai HH, Kirby R. Once the circle has been broken: dynamics and evolution of Streptomyces chromosomes. Trends in Genet. 2002;18:522–529. doi: 10.1016/s0168-9525(02)02752-x. [DOI] [PubMed] [Google Scholar]
- 16.Redshaw PA, McCann PA, Pentella MA, Pogell BM. Simultaneous loss of multiple differentiated functions in aerial mycelium-negative isolates of streptomycetes. J Bacteriol. 1979;137:891–899. doi: 10.1128/jb.137.2.891-899.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fishman SE, Hershberger CL. Amplified DNA in Streptomyces fradiae. J Bacteriol. 1983;155:459–466. doi: 10.1128/jb.155.2.459-466.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dyson P, Schrempf H. Genetic instability and DNA amplification in Streptomyces lividans 66. J Bacteriol. 1987;169:4796–4803. doi: 10.1128/jb.169.10.4796-4803.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ichikawa N, Oguchi A, Ikeda H, Ishikawa J, Kitani S, Watanabe Y, Nakamura S, Katano Y, Kishi E, Sasagawa M, Ankai A, Fukui S, Hashimoto Y, Kamata S, Otoguro M, Tanikawa S, Nihira T, Horinouchi S, Ohnishi Y, Hayakawa M, Kuzuyama T, Arisawa A, Nomoto F, Miura H, Takahashi Y, Fujita N. Genome sequence of Kitasatospora setae NBRC 14216T: an evolutionary snapshot of the family Streptomycetaceae. DNA Res. 2010;17:393–406. doi: 10.1093/dnares/dsq026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Thomas MG, Chan YA, Ozanick SG. Deciphering tuberactinomycin biosynthesis: isolation, sequencing, and annotation of the viomycin biosynthetic gene cluster. Antimicrob Agents Chemother. 2003;47:2823–2830. doi: 10.1128/AAC.47.9.2823-2830.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zalacain M, Gonzalez A, Guerrero MC, Mattaliano RJ, Malpartida F, Jimenez A. Nucleotide sequence of the hygromycin B phosphotransferase gene from Streptomyces hygroscopicus. Nucleic Acids Res. 1986;14:1565–1581. doi: 10.1093/nar/14.4.1565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bibb MJ, Bibb MJ, Ward JM, Cohen SN. Nucleotide sequences encoding and promoting expression of three antibiotic resistance genes indigenous to Streptomyces. Mol Gen Genet. 1985;199:26–36. doi: 10.1007/BF00327505. [DOI] [PubMed] [Google Scholar]
- 23.Uchiyama H, Weisblum B. N-Methyl transferase of Streptomyces erythraeus that confers resistance to the macrolide-lincosamide-streptogramin B antibiotics: amino acid sequence and its homology to cognate R-factor enzymes from pathogenic bacilli and cocci. Gene. 1985;38:103–110. doi: 10.1016/0378-1119(85)90208-2. [DOI] [PubMed] [Google Scholar]
- 24.Datsenko KA, Wanner BL. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci USA. 2000;97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang Y, Muyrers JPP, Testa G, Stewart AF. DNA cloning by homologous recombination in Escherichia coli. Nat Biotechnol. 2000;18:1314–1317. doi: 10.1038/82449. [DOI] [PubMed] [Google Scholar]
- 26.Suzuki N, Tsuge Y, Inui M, Yukawa H. Cre/loxP-mediated deletion system for large genome rearrangements in Corynebacterium glutamicum. Appl Microbiol Biotechnol. 2005;67:225–233. doi: 10.1007/s00253-004-1772-6. [DOI] [PubMed] [Google Scholar]
- 27.Gibson DG, Young L, Chuang RY, Venter JC, Hutchison CA, III, Smith HO. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat Methods. 2009;6:343–345. doi: 10.1038/nmeth.1318. [DOI] [PubMed] [Google Scholar]
- 28.Subba B, Kharel MK, Lee HC, Liou K, Kim BG, Sohng JK. The ribostamycin biosynthetic gene cluster in Streptomyces ribosidificus: comparison with butirosin biosynthesis. Mol Cells. 2005;20:90–96. [PubMed] [Google Scholar]
- 29.Ostash B, Saghatelian A, Walker S. A streamlined metabolic pathway for the biosynthesis of moenomycin A. Chem & Biol. 2007;14:257–267. doi: 10.1016/j.chembiol.2007.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Arai M, Nakayama R, Yoshida K, Takeuchi M, Teramoto S, Torikata A. Pholipomycin, a new member of phosphoglycolipid antibiotics. II Physico-chemical properties and comparison with other members of this family of antibiotics. J Antibiot. 1977;30:1055–1059. doi: 10.7164/antibiotics.30.1055. [DOI] [PubMed] [Google Scholar]
- 31.Shen B. Polyketide biosynthesis beyond the type I, II and III polyketide synthase paradigms. Current Opinion Chem Biol. 2003;7:285–295. doi: 10.1016/s1367-5931(03)00020-6. [DOI] [PubMed] [Google Scholar]
- 32.Zhang W, Ames BD, Tsai SC, Tang Y. Engineered biosynthesis of a novel amidated polyketide, using the malonamyl-specific initiation module from the oxytetracycline polyketide synthase. Appl Environ Microbiol. 2006;72:2573–2580. doi: 10.1128/AEM.72.4.2573-2580.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jakobi K, Hertweck C. A gene cluster encoding resistomycin biosynthesis in Streptomyces resistomycificus; exploring polyketide cyclization beyond linear and angucyclic patterns. J Am Chem Soc. 2004;126:2298–2299. doi: 10.1021/ja0390698. [DOI] [PubMed] [Google Scholar]
- 34.Oliynyk M, Samborskyy M, Lester JB, Mironenko T, Scott N, Dickens S, Haydock SF, Leadlay PF. Complete genome sequence of the erythromycin-producing bacterium Saccharopolyspora erythraea. NRRL 23338. Nat Biotechnol. 2007;25:447–453. doi: 10.1038/nbt1297. [DOI] [PubMed] [Google Scholar]
- 35.Carter GT, Nietsche JA, Hertz MR, Williams DR, Siegel MH, Morton GO, James JC, Borders DB. LL-F28249 antibiotic complex: A new family of antiparasitic macrocyclic lactones. Isolation, characterization and structures of LL-28249 α, β, γ, λ. J Antibiot. 1988;41:519–529. doi: 10.7164/antibiotics.41.519. [DOI] [PubMed] [Google Scholar]
- 36.Ueda J, Hashimoto J, Nagai A, Nakashima T, Komaki H, Anzai K, Harayama S, Doi T, Takahashi T, Nagasawa K, Natsume T, Takagi M, Shin-ya K. New aureothin derivative, alloaureothin, from Streptomyces sp. MM23. J Antibiot. 2007;60:321–324. doi: 10.1038/ja.2007.41. [DOI] [PubMed] [Google Scholar]
- 37.Hu Z, Reid R, Gramajo H. The leptomycin gene cluster and its heterologous expression in Streptomyces lividans. J Antibiot. 2005;58:625–633. doi: 10.1038/ja.2005.86. [DOI] [PubMed] [Google Scholar]
- 38.Li B, Walsh CT. Identification of the gene cluster for the dithiolopyrrolone antibiotic holomycin in Streptomyces clavuligerus. Proc Natl Acad Sci USA. 2010;107:19731–19735. doi: 10.1073/pnas.1014140107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Omura S, Fujimoto T, Otoguro K, Matsuzaki K, Moriguchi R, Tanaka H, Sasaki Y. Lactacystin, a novel microbial metabolite, induces neurogenesis of neuroblastoma cells. J Antibiot. 1991;44:113–116. doi: 10.7164/antibiotics.44.113. [DOI] [PubMed] [Google Scholar]
- 40.Jensen SE, elder KJ, Aidoo KA, Paradkar AS. Enzymes catalyzing the early steps of clavulanic acid biosynthesis are encoded by two sets of paralogous genes in Streptomyces clavuligerus. Antimicrob Agents Chemother. 2000;44:720–726. doi: 10.1128/aac.44.3.720-726.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Paradkar AS, Aidoo KA, Wong A, Jensen SE. Molecular analysis of a β-lactam resistance gene encoded within the cephamycin gene cluster of Streptomyces clavuligerus. J Bacteriol. 1996;178:6266–6274. doi: 10.1128/jb.178.21.6266-6274.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bignell DRD, Tahlan K, Colvin KR, Jensen SE, Leskiw BK. Expression of ccaR, encoding the positive activator of cephamycin C and clavulanic acid production in Streptomyces clavuligerus, is dependent on bldG. Antimicrob Agents Chemother. 2005;49:1529–1541. doi: 10.1128/AAC.49.4.1529-1541.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Onaka H, Taniguchi S, Igarashi Y, Furumai T. Characterization of the biosynthetic gene cluster rebeccamycin from Lechevalieria aerocolonigenes ATCC 39243. Biosci Biotechnol Biochem. 2003;67:127–138. doi: 10.1271/bbb.67.127. [DOI] [PubMed] [Google Scholar]
- 44.Steffensky M, Muhlenweg A, Wang ZX, Li SM, Heide L. Identification of the novobiocin biosynthetic gene cluster of Streptomyces spheroides NCIB 11891. Antimicrob Agents Chemother. 2000;44:1214–1222. doi: 10.1128/aac.44.5.1214-1222.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.He J, Magarvey N, Piraee M, Vining LC. The gene cluster for chloramphenicol biosynthesis in Streptomyces venezuelae ISP5230 includes novel shikimate pathway homologues and a monomodular non-ribosomal peptide synthetase gene. Microbiology. 2001;147:2817–2829. doi: 10.1099/00221287-147-10-2817. [DOI] [PubMed] [Google Scholar]
- 46.Gomez-Escribano JP, Bibb MJ. Engineering Streptomyces coelicolor for heterologous expression of secondary metabolite gene clusters. Microb Biotechnol. 2010;4:207–215. doi: 10.1111/j.1751-7915.2010.00219.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Komatsu M, Tsuda M, Omura S, Oikawa H, Ikeda H. Identification and functional analysis of genes controlling biosynthesis of 2-methylisoborneol. Proc Natl Acad Sci USA. 2008;105:7422–7427. doi: 10.1073/pnas.0802312105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Koe BK, Sobin BA, Celmer WD. PA 132, a New Antibiotic. I. Isolation and Chemical Properties. Antibiot Annu. 1957:672–675. [PubMed] [Google Scholar]
- 49.Takeuchi S, Ogawa Y, Yonehara H. The Structure of pentalenolactone (PA-132) Tetrahedron Lett. 1969:2737–2740. doi: 10.1016/s0040-4039(01)88256-3. [DOI] [PubMed] [Google Scholar]
- 50.Cane DE, Tillman AM. Pentalenene biosynthesis and the enzymatic cyclization of farnesyl pyrophosphate. J Am Chem Soc. 1983;105:122–124. [Google Scholar]
- 51.Tetzlaff CN, You Z, Cane DE, Takamatsu S, Omura S, Ikeda H. A gene cluster for biosynthesis of the sesqui-terpenoid antibiotic pentalenolactone in Streptomyces avermitilis. Biochemistry. 2006;45:6179–6186. doi: 10.1021/bi060419n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Jiang J, Tetzlaff CN, Takamatsu S, Iwatsuki M, Komatsu M, Ikeda H, Cane DE. Genome mining in Streptomyces avermitilis: A biochemical Baeyer-Villiger reaction and discovery of a new branch of the pentalenolactone family tree. Biochemistry. 2009;48:6431–6440. doi: 10.1021/bi900766w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Cane DE, You Z, Omura S, Ikeda H. Pentalenolactone biosynthesis. Molecular cloning and assignment of biochemical function to PtlF, a short-chain dehydrogenase from Streptomyces avermitilis, and identification of a new biosynthetic intermediate. Archi Biochem Biophys. 2007;459:233–240. doi: 10.1016/j.abb.2006.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhu D, Seo MJ, Ikeda H, Cane DE. Genome mining in Streptomyces. Discovery of an unprecedented P450-catalyzed oxidative rearrangement that is the final step in the biosynthesis of pentalenolactone. J Am Chem Soc. 2011;133:2128–2131. doi: 10.1021/ja111279h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Seo MJ, Zhu D, Endo S, Ikeda H, Cane DE. Genome mining in Streptomyces Elucidation of the role of Baeyer-Villiger monooxygenases and non-heme iron-dependent dehydrogenase/oxygenases in the final steps of the biosynthesis of pentalenolactone and neopentalenolactone. Biochemistry. 2011;50:1739–1754. doi: 10.1021/bi1019786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yamada Y, Cane DE, Ikeda H. Diversity and Analysis of Bacterial Terpene Synthases. In: Hopwood DA, editor. In Chapter seven, Natural Product Biosynthesis by Microorganisms and Plants, Part A, Methods in Enzymology. Vol. 515. Elsevier Inc. Academic Press; New York: 2012. pp. 123–166. [DOI] [PubMed] [Google Scholar]
- 57.Ikeda H, Kotaki H, Omura S. Genetic studies of avermectin biosynthesis in Streptomyces avermitilis. J Bacteriol. 1987;169:5615–5621. doi: 10.1128/jb.169.12.5615-5621.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.