Abstract
The expression of 4-1BB has been known to be dependent on T cell activation. Recent studies have, however, revealed that 4-1BB expression is not restricted to T cells. We sought to determine the molecular basis for the differential gene expression. Here we report the expression pattern of two mouse 4-1BB transcripts, type I and type II. Whereas the type I transcript was specifically expressed on immune organ as previously reported, the type II transcript was ubiquitously expressed in tissues and various cell lines. However, both type I and type II transcript were highly induced on activated T cells. Primer extension assay of the two 4-1BB transcripts suggested that mouse 4-1BB had more than two transcripts. Using luciferase assay we have identified three promoter regions (PI, PII and PIII), which located on upstream region of second exon 1, first exon 1, and exon 2, respectively. In particular, the type I transcript was preferentially induced when naïve T cells are stimulated by anti-CD3 monoclonal antibody (mAb) since NF-κB specifically binds to the putative NF-κB element of PI. We have also shown that a splice variant, in which the transmembrane domain was deleted, could inhibit 4-1BB signaling. The splicing variant was highly induced by TCR stimulation. Our results reveal 4-1BB also has a negative regulation system through soluble 4-1BB produced from a splice variant induced under activation conditions.
Keywords: 4-1BB, NF-κB, promoter, splice variant
INTRODUCTION
4-1BB (CD137) is a receptor belonging to the tumor necrosis factor receptor (TNFR) superfamily (Goodwin et al., 1993; Kwon et al., 1987; Lotz et al., 1996; Setareh et al., 1995) which is expressed in an activation-dependent manner on T cells (Kwon et al., 1989; Pollok et al., 1993; Schwarz et al., 1996; Shuford et al., 1997) and constitutively on CD4+CD25+ regulatory T cells (McHugh et al., 2002). Although 4-1BB is mainly involved in regulating activated T lymphocyte functions, expression of 4- 1BB has been noted on non-T cells such as B cells, monocytes, macrophages, dendritic cells, eosinophils, and neutrophils (Futagawa et al., 2002; Heinisch et al., 2000; 2001; Schwarz et al., 1995; Watts et al., 2005; Wilcox et al., 2002; Yoon et al., 2005). In addition, the expression of 4-1BB is not restricted to immune cells; its expression has been demonstrated on blood vessel endothelial cells, smooth muscle cells, neurons, astrocytes, and microglia (Boussaud et al., 1998; Broll et al., 2001; Reali et al., 2003; Seko et al., 2004). When T cells are activated by a variety of stimuli such as anti-CD3 mAb, concanavalin A (Con A), phytohaemagglutinin (PHA), interleukin (IL)-2, IL-4, anti- CD28 mAb, phorbol myristyl acetate (P), or ionomycin (I), 4- 1BB is stably upregulated (Garni-Wagner et al., 1996; Kim et al., 2003; Kwon et al., 1989; Pollok et al., 1995). 4-1BB is induced in human peripheral blood mononuclear cells (PBMCs) by DNA-damaging agents such as doxorubicin, bleomycin, and γ- irradiation (Chung et al., 2002).
4-1BB binds to a 4-1BB ligand (4-1BBL) expressed by activated antigen-presenting cells (APCs), including monocytes, macrophages, and B cells (Alderson et al., 1994; DeBenedette et al., 1995; 1997; Langstein et al., 1998; 1999; Pollok et al., 1994). The 4-1BB/4-1BBL interaction provides a costimulatory signal leading to proliferation and differentiation, as well as protection from activation-induced cell death of T cells (Hurtado et al., 1995; 1997; Kwon et al., 2000; Saoulli et al., 1998; Takahashi et al., 1999). The reverse signaling path also exists through 4-1BBL on B cells and dendritic cells resulting in the secretion of immunoglobulins and proinflammatory cytokines, respectively (Langstein et al., 1998; Pollok et al., 1994).
Previous report showed mouse 4-1BB has two different transcripts and their expression may be regulated differently by two putative promoter regions (Kwon et al., 1994). Alternatively spliced mRNA encoding a soluble form of mouse 4-1BB had also been identified (Setareh et al., 1995). Human 4-1BB has two splice variants that produce soluble 4-1BB, which was enhanced in sera of rheumatoid arthritis, leukemia, and lymphoma patients (Furtner et al., 2005; Jung et al., 2004; Michel et al., 1998). Several members of the TNF receptor family, including TNF receptor, NGF receptor, CD27, CD30, GITR, and CD95, have been demonstrated to have soluble forms, which were generated by proteolytic cleavage and alternative splicing (Cascino et al., 1995; 1996; Cheng et al., 1994; Kohno et al., 1990; Nocentini et al., 2000; Pizzolo et al., 1990; Zupan et al., 1989). These soluble receptor forms antagonized the signaling through membrane-bound receptors (Cascino et al., 1996; Cheng et al., 1994; Kohno et al., 1990).
In this study, we investigated the expression of the two mouse 4-1BB transcripts showing different expression patterns on tissues but not T cells and identified three functional promoter regions (PI, PII and PIII). NF-κB binding site in PI region is responsible for activation-dependent 4-1BB expression on T cells. We also investigated the expression of splice variant transcript, which expressed on activated T cells to be comparable for the normal transcript. We determined the splice variant transcript encodes soluble 4-1BB, which has antagonistic effect on 4-1BB signaling.
MATERIALS AND METHODS
Mice, cell lines, and reagents
Six- to eight-week-old Balb/c mice were purchased from Hyo Chang Bioscience (Korea). HEK-293, EL4, 2PK3, A20, and RAW264.7 cells were grown in DMEM medium supple-mented with 10% FBS and 50 mM 2-mercaptoethanol. CTLL-R8, a mouse cytolytic T cell line, was grown in DMEM medium supplemented with 10% FBS and 50 U/ml of rIL2 (R&D systems, USA). Anti 4-1BB mAb (3E1) was provided by Dr. R. Mittler of Emory University. Anti-CD3 mAb, anti-FcγR mAb, and isotype control Ab were purchased from BD biosci-ences (USA). 4- 1BB-Fc protein was purified from supernatants of stable transfected CHO cells by protein G sepharose (Invi-trogen, USA) according to the manufacturer’s instructions.
Cell isolation
A single cell suspension was prepared from balb/c mouse spleen and the red blood cells were lysed. CD4+ T cells or CD8+ T cells were enriched by incubation with anti-CD4 (Miltenyi Biotech, USA) or anti-CD8 microbeads (Miltenyi Biotech), respectively. Cell purification followed using MACS columns (Miltenyi Biotech). Column-bound cells were used as the source of CD4+ T and CD8+ T cells. The purification procedure was performed according to the standard protocol provided by Miltenyi Biotech.
Plasmid construction
For promoter analysis, three ~1.8 kbp upstream regions at positions -6340 to -4541 (PII), -3585 to -1795 (PI), and -1796 to -1 (PIII) of the mouse 4-1BB gene were amplified by PCR with genomic DNA from balb/c mouse spleen, and cloned into the pGL3-basic vector (Promega, USA). Sense and anti-sense primers for PCR amplification are as follows: PI sense, 5′-GGGTACCATCGACTGGAGAGGACAG- 3′; and antisense, 5′-CCTCGAGTCCTTGGCAGCTCGGTGA- 3′; PII sense, 5′-GGGTACCTCTGAATGTGACATTCAG- 3′; and antisense, 5′-CCTCGAGCTGAATTGGTTCCACTCA- 3′; PIII sense, 5′-GGGTACCGAAGCAGAACGCTCCTCG- 3′; and antisense, 5′-CCTCGAGGGCGAAATGTCACATGCA- 3′. Underlines mean flanked KpnI and XhoI restriction enzymes sequences. NF-κB and AP-1 mutant constructs were made by overlap extension PCR with the sense and antisense primers of PI region and the following primers to incorporate a transcription factor binding site mutation: NF-κB sense, 5′-AGCAGCTGGCGATTTCCCAGGAGG- 3′; and antisense, 5′-CCTCCTGGG-AAATCGCCAGCTGCT-3′; AP-1 sense, 5′-GCTCAGTTGA- GTTGCAGGTCCTGT-3′; and antisense, 5′-ACAGGACCTGCAACTCAACTGAGC-3′. The 4- 1BB ligand and 4-1BB splice variant (ΔE8-4-1BB) were amplified by RT-PCR with complementary DNA (cDNA) prepared from purified CD4+ T cells, and then cloned into pCDNA3.1 (Invitrogen, USA). The sense and antisense primers used as follows and HindIII or BamHI restriction enzyme site included: m4-1BB ligand sense, 5′-CAAGCTTATGGACCAG-CAACA-3′; and antisense, 5′-GGGATCCTCATTCCCATGGG-TT-3′; m4- 1BB sense, 5′-CAAGCTTATGGGAAACAACTGT-3′; and antisense, 5′-GGGATCCTCACAGCTCATAGCC-3′. RNase protection assay (RPA) probe fragments for detection of transcripts and splice variants were amplified by PCR and cloned into a pGEM-T Easy vector (Promega). Primer sets of the following sequences were used for amplification of RPA probe fragments. Type I probe sense, 5′-TTTGACCTGTGGTCTTG-TG-3′; type II probe sense, 5′-ATGTCCATGAACTGCTGAG-3′; Type III probe sense, 5′-AGTGTCCTGTGCATGTGACA-3′; type I and II probe antisense, 5′-CGCCATGGGAAACAACT-3′; type III probe antisense, 5′-GAGCTGCCCTCCAAGTACCT-3′; splice variant sense, 5′-AGGGCACTCCTTGC-AGGTC-3′; splice variant antisense, 5′-TCACAGCTCATAGCCTCCT-3′.
Flow cytometry
Cells were harvested at predetermined time points and washed with phosphate-buffered saline containing 2% fetal bovine serum (FBS). To assess 4-1BB expression on HEK 293 cells transfected with pCDNA3.1-4-1BB, pCDNA3.1-4-1BBL, or pCDNA3.1-ΔE8-4-1BB, transfectants were stained with FITCconjugated anti 4-1BB (3E1) mAb for 30 min at 4℃. Stained cells were analyzed on a FACScalibur (BD biosciences).
RT-PCR
Total RNA isolated using TRIzol reagent (Invitrogen) was subjected to cDNA synthesis using Olig dT and AMV reverse transcriptase (Promega). Serial dilution (5- or 10-fold) of complementary DNA (cDNA) reaction mixtures was subjected to PCR amplification using the following primers: type I sense (F1), 5′- GACCTGTGGTCTTGTGGAGCG-3′; type II sense (F2), 5′- TCCATGAACTGCTGAGTGG-3′; type III sense (F3), 5′-GTCCTGTGCATGTGACATT- 3′; 4-1BB antisense (R1), 5′-TCCCGGTCTTAAGCA CAGACCTTCCGT-3′; 4-1BB full antisense (R2) 5′-TACATCACAGCTCATAGCCT-3′; GAPDH sense, 5′- TGAAGGTCGGTGTGAACGGATTTG-3′; GAPDH antisense, 5′-CATGTAGGCCATGAGGTCCACCAC-3′. The conditions for PCR amplification were first denaturation at 95℃ for 5 min, amplification for 30 cycles of 94℃ for 30 s, 53℃ for 30 s, and 72℃ for 30 s, followed by 5 min at 72℃ for type I and type III transcripts, while 35 cycles of the above were required for type II transcript amplification. The GAPDH transcript was amplified by 25 cycles of the above protocol. For PCR amplification of 4- 1BB full transcripts, the sample was denatured at 95℃ for 5 min and amplified for 30 cycles of 94℃ for 1 min, 56℃ for 1 min, and 72℃ for 1 min, followed by 5 min at 72℃ for type I and type III transcripts and 35 cycles of the above for type II transcripts.
RNase protection assay (RPA)
The RNase protection assay was performed according to the Pharmingen standard protocol using the Riboquant in vitro Transcription and RPA kits (BD PharMingen). Briefly, 5 μg of total RNA was hybridized overnight with [α-32P]UTP (Amersham Biosciences, USA) labeled probes at 56℃. Unhybridized ssRNA was digested by RNase treatment, and the dsRNA was purified by phenol/chloroform extraction and ethanol precipitation. The samples were fractionated by electrophoresis on a 6% polyacrylamide/7 M urea gel, dried, and exposed to X-ray film (Agfa, USA) for autoradiographic analysis. The mRNA expression of the corresponding GAPDH was included to normalize for gel loading.
Primer extension analysis
Five micrograms of total RNA from splenocytes, EL4, and CTLL-R8 was annealed with 2 × 104 cpm of the end-labeled oligonucleotide (5′-GGTACTTGGAGGGCAGCTCTTGCAGA- 3′) with [α-32P]dATP (Amersham Biosciences) by T4 polynucleotide kinase (Promega) at 30℃ overnight in a buffer containing 0.4 M NaCl, 40 mM PIPES (pH 7.0), 1.0 mM EDTA (pH 8.0), and 80% formamide. The mixture was ethanol-precipitated and resuspended in a buffer containing 50 mM Tris-HCl (pH 7.6), 60 mM KCl, 10 mM DTT, 1.0 mM each dATP, dGTP, dTTP, and dCTP, 40 U/μl RNAsin (Promega), and 20 U of AMV reverse transcriptase (Promega). The mixture was incubated at 42℃ for 1 h, extracted with phenol-chloroform, and precipitated with ethanol. The precipitate was resuspended in 10 mM Tris-HCl (pH 7.4) and 10 mM EDTA, then incubated with 50 μg/ml RNAse A at 37℃ for 30 min. The reaction product was analyzed by sequencing gel. The resulting gel was dried and exposed to X-ray film (Agfa) for autoradiographic analysis.
Transient transfection and luciferase assay
Cells (5 × 106) were suspended in 0.3 ml of Opti-MEM (Life Technologies, Inc.), transferred to a 4-mm gap cuvette, and then mixed with 10 μg of pCMVβ-gal (Promega) as an internal control and 10 μg of pGL3-basic vector (Promega) as a negative control. In all experiments, 10 μg of the pGL3-basic vector containing different promoter fragments (PI, PII, and PIII) or a control pcDNA3.1 was cotransfected with pCMVβ-gal. Cells were transfected at 960 microfarads and 250 V using a Gene Pulser electroporation apparatus (Bio-Rad, USA). For anti-CD3 stimulation, cells were harvested 12 h after transfection and transferred to wells coated with 10 μg/ml/well of anti-CD3 mAb. In some groups, transfected cells were stimulated with 20 ng/ml of phorbol myristate acetate and 1 ng/ml of ionomycin (P/I) or 10 μg/ml of concanavalin A (Con A). Cells were harvested 24 hr after stimulation, washed with phosphate-buffered saline, and lysed in 100 μl of report lysis buffer (Promega). Luciferase activities were measured using the luciferase assay system (Promega) according to the manufacture’s recommendation, and normalized for transfection efficiency relative to β-galactosidase activity. Reported data were represented as the mean from three independent experiments.
Preparation of nuclear extracts and electrophoretic mobility shift assay (EMSA)
Nuclear extracts were prepared from 2 × 107 of CD4+ T cells stimulated with anti-CD3 for 6 h according to the method described by Dignam et al. (1983) with minor modifications. Briefly, cells were washed with ice-cold phosphate-buffered saline, resuspended in buffer A (20 mM HEPES, pH 7.9, 1.5 mM MgCl2, 10 mM KCl, 0.1 mM EDTA, 0.5 mM dithiothreitol, and 0.5 mM phenylmethylsulfonyl fluoride), and left on ice for 10 min. Nuclei were pelleted by centrifugation at 5,000 rpm for 10 min at 4℃ and resuspended in buffer B (20 mM HEPES, pH 7.9, 1.5 mM MgCl2, 420 mM NaCl, 0.2 mM EDTA, 20% glycerol, and 1 mM dithiothreitol) . After incubation for 30 min at 4℃, the mixture was centrifuged at 13,000 rpm for 15 min at 4℃. The supernatant as nuclear extract was collected and stored at -70℃ until use. Oligonucleotides (-1897/-1874, 5′-AGCAGCTGGGGATTTCCCAGGAGG- 3′ and 5′-CCTCCTGGGAAATCCCCAGCTGCT- 3′) containing putative a NFκB binding site were synthesized (Genomine, Korea). The NFκB control probe was purchased from Santa Cruz. Oligonucleotides were annealed to make double-stranded target DNA, and then endlabeled with [α-32P]dATP (Amersham Biosciences) by T4 polynucleotide kinase (Promega). The nuclear extracts and probe were incubated for 30 min at room temperature in the reaction buffer containing 10 mM HEPES, pH 7.9, 50 mM NaCl, 1 mM EDTA, 1 mM DTT, and 5% glycerol. Each reaction contained 0.02 unit of poly[dI-dC] (Sigma, USA). In some reactions, antip65 (Santa Cruz Biotechnologies, USA), anti-AP-1 (Santa Cruz Biotechnologies), or normal anti-mouse IgG was added prior to incubation with probe. Reactions were electrophoresed on a 4% non-denaturing polyacrylamide gel in 0.5X Tris borate- EDTA buffer at 150 V for 2 h, and then analyzed by autoradiography.
Chromatin immunoprecipitation
Purified of CD4+ T cells (2 × 107) were stimulated with plates coated with 10 μg/ml of anti-CD3 for 6 h, and then fixed by adding formaldehyde (Sigma) to the medium to a final concentration of 1%. After 15 min, cells were washed with ice-cold phosphate- buffered saline containing 1 mM phenylmethylsulfonyl fluoride and then scraped. After centrifugation, cells were lysed in buffer I (50 mM Tris, pH 8.0, 2 mM EDTA, 0.1% Nonidet P-40, 10% glycerol, and protease inhibitors) and centrifuged for 5 min at 3,000 rpm and 4℃. After removal of supernatants, nuclei were resuspended in buffer II (50 mM Tris, pH 8.0, 1% SDS, 5 mM EDTA), and chromatin was sheared by sonication. After removal of nuclear debris by centrifugation for 5 min at 13,000 rpm and 4℃, lysates were diluted ten-fold with dilution buffer (50 mM Tris, pH 8.0, 5 mM EDTA, 200 mM NaCl, 0.5% Nonidet P-40), and then precleared for 2 h using 80 μl of 50% salmon sperm-DNA-saturated protein A-agarose beads. Immunoprecipitation was carried out at 4℃ overnight and immune complexes were collected with salmon sperm-DNA-saturated protein A-agarose beads. Antibodies utilized anti-p65 (Santa Cruz Biotechnologies), anti-AP-1 (Santa Cruz Biotechnologies), or anti-mouse IgG as a control for nonspecific interaction. After washing three times with high salt WB buffer (20 mM Tris, pH 8.0, 0.1% SDS, 1% Nonidet P-40, 2 mM EDTA, and 0.5 M NaCl) and twice with low salt TE buffer (10 mM Tris, pH 8.0, and 1 mM EDTA), immunocomplexes were eluted with TE containing 1% SDS. Protein-DNA cross-links were reversed by incubating at 65℃ overnight. After proteinase K digestion, DNA was extracted with phenol-chloroform and precipitated with ethanol using 15 μg of tRNA as a carrier. PCR was performed (30 cycles, denaturing at 94℃ for 1 min, annealing at 55℃ for 30 s, and extension at 72℃ for 30 s) using primers specific for the PI region (sense, 5′-TGATAAGCGACCAAGTCT-3′; antisense, 5′-CAGGACCTGTGACTCAACTGAGCA-3′), PII region (sense, 5′-TTGGCCACCACACCATGC-3′; antisense, 5′-CACAACTCCATCTGTAGC- 3′), and PIII region (sense, 5′-TGCAGTCGGGGATGGCCTTG- 3′; antisense, 5′-AGCAAAGGAGATTCCGAA- 3′). Intensity of amplified PCR products was quantified by Image J program (available at http://rsb.info.nih.gov/ij/).
ELISA
For detection of soluble 4-1BB, supernatants were collected 24 hr after transfecting EL4 cells with pCDNA3.1-4-1BB or pCDNA3.1-ΔE8-4-1BB using a Gene Pulser electroporation apparatus according to the manufacturer’s procedure. A 96- well ELISA plate (Nunc, USA) was incubated overnight at 4℃ with 100 μl of supernatants and mouse 4-1BB-Fc fusion protein in a standard curve. After blocking with PBS containing 4% BSA, the plate was incubated with biotinylated 3E1 mAb for 2 h at room temperature, washed five times with PBS containing 0.1% tween-20, and then streptavidin-HRP (BD PharMingen) was added to each well for 1 h. The plates were then washed five times with the same solution, developed in 100 μl of 3,5,3′,5′-tetramethylbenzidine (Pierce biotechnology, USA) substrate for 15 min, and stopped by adding 100 μl of 1N HCl. The plates were then read at 450 nm with a Wallac Vector 1420 Multilabel Counter (EG&G Wallac).
RESULTS
Expression pattern of 4-1BB transcripts in various tissues, cell lines, and T cells
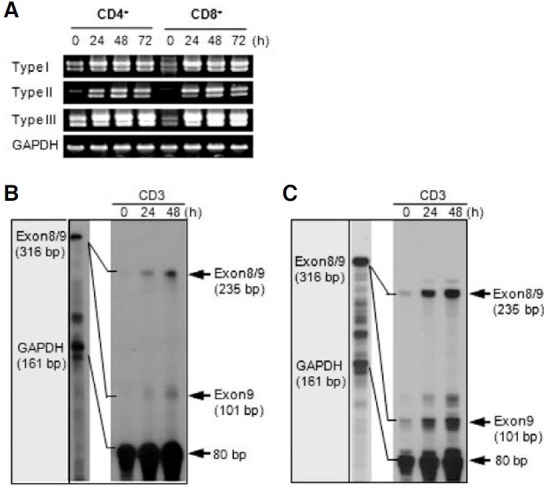
To examine the expression pattern of two 4-1BB transcripts, we used three different sense primers (F1, F2, and F3) and one antisense primer (R1) for RT-PCR analysis (Fig. 1A). PCR products amplified from each sense primer, which were designed in first exon 1, second exon 1, and exon 2 regions, were named as type II, type I, and type III, respectively (Fig. 1A). Type III PCR product represents total 4-1BB transcripts because the type I and type II transcripts include exon 2. In a previous study of the genomic organization of the mouse 4-1BB gene, the first exon 1 and second exon 1 are untranslated regions (UTRs), while exon 2 contains the translational start site. Type II did not contain the second exon 1 as was previously reported (Kwon et al., 1994). Interestingly, type I and type II showed different expression patterns on tissues. While the type I was highly expressed in thymus, spleen, and lymph node, the type II was ubiquitously expressed in tissues and cell lines (Fig. 1B). In particular, CTLL-R8 cells highly expressed two types (Fig. 1B). However, two transcripts were highly induced on T cells stimulated by anti-CD3 (Fig. 1C).
Fig. 1. 4-1BB expression in tissues, cell lines and T cells. (A) Organization of the mouse 4-1BB gene. Exons are shown as boxes; untranslational regions are open and the protein coding regions are grayed and indicated in Roman numerals. Primer regions for RT-PCR are marked by arrows and RPA probe regions are lined. Three distinct putative promoter regions named PI, PII, and PIII. Translational start site is designated +1. (B, C) RT-PCR analysis. Total RNA was extracted from tissues of mice, cell lines, and activated T cells by anti-CD3 (5 μg/ml) and then analyzed for the expression level of the 4-1BB mRNA transcripts. (D, E) RNase Protection Assay (RPA). 5 μg of toal RNA of CD4+ (D) and CD8+ T cells (E) stimulated by anti-CD3 subjected to RPA as described in “Materials and Methods”. The mRNA expression of the corresponding GAPDH was included to normalize for gel loading.
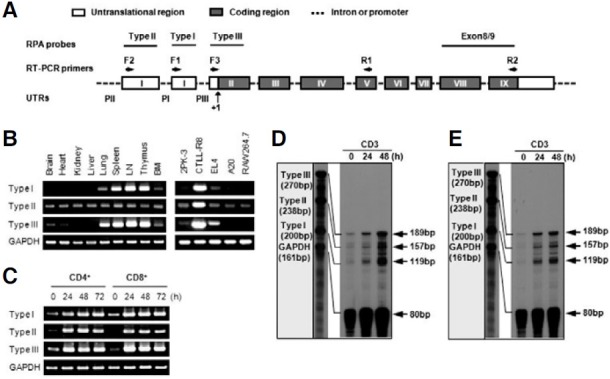
To confirm the result of RT-PCR, RPA analysis was performed. Three RPA probes were designed and the type III RPA probe represents total 4-1BB transcripts because the type I and type II transcripts contain a type III RPA probe region (Fig. 1A). As shown by RT-PCR, Type I and type II transcripts were highly induced on activated T cells (Figs. 1D and 1E). These results suggest that the expression of type I and type II transcripts might be differentially regulated on tissue level but both transcripts are inducible on T cells.
4-1BB has more than two transcripts
A primer extension assay was performed to demonstrate the two different transcripts using primer designed from the exon 2 region. A previous report showed that the transcriptional start site of the type II transcript was only detected by primer extension assay using a specific primer designed in the first exon 1 region, while the type I transcription start site was not detected by a specific primer designed in the second exon 1 region (Kwon et al., 1994). We found more than two transcriptional start sites (Fig. 2). The sizes of the extension products of type I and type II were approximately calculated to be 233 bp, and 297bp products, respectively. In addition to type I and type II matched extension products, 181 bp and 172 bp products were detected. These product sizes suggest the novel transcript without first and second exon I. In particular, a 250 bp product was detected on CTLL-R8 cells. Although it might be another type I transcript that includes second exon I, we concluded that this one is not normal transcript because of its unique expression in CTLL-R8 cells.
Fig. 2. Primer extension analysis for determination of the 4-1BB mRNA start site. 5 μg total RNA of splenocytes (lane 1), CTLL-R8 (lane 2), and EL4 (lane 3) was used for extension analysis. Total mRNA was annealed with 2 × 104 cpm of the end-labeled oligonucleotide using [α-32P]dATP at 30℃, overnight, in a hybridization buffer. Synthesis of cDNA using AMV reverse transcriptase was then performed. The mixture was extracted with phenol-chloroform, and precipitated with ethanol. The precipitate was analyzed by gel sequencing. The nucleotide sequence ladder of the pGEM-T easy vector shown on the left is a size marker for the extension products. The specific extension products for 4-1BB mRNA are indicated by arrows with numbers.
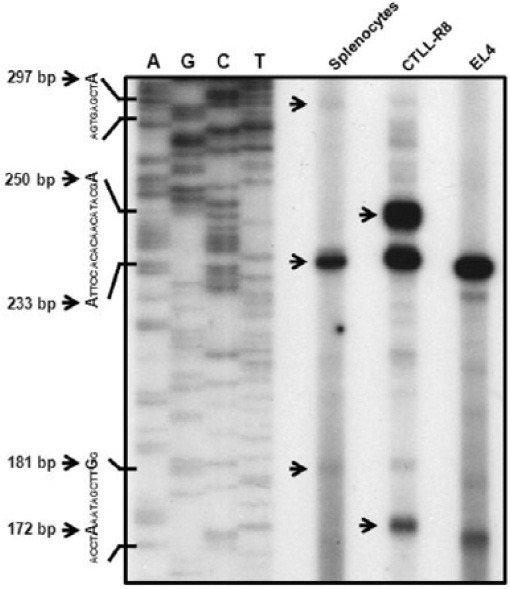
These results support that the mouse 4-1BB gene produce more than two transcripts. It also suggests 4-1BB gene has distinctly different promoters for differential gene expression.
Three distinct upstream regions show promoter activity
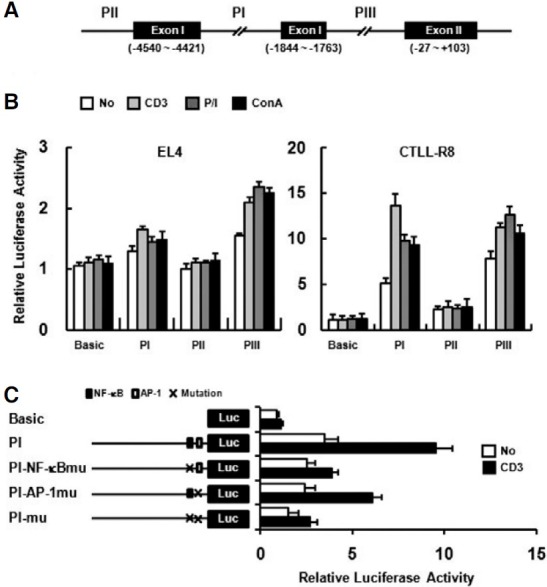
PI, PII, and PIII upstream regions (UTRs) (Fig. 3A) were analyzed for functional activity (Fig. 3B). Promoter activity of each upstream region was examined on EL4 and CTLL-R8 cell lines. While the PIII region showed strong promoter activity on both cell lines, the PI region showed promoter activity in CTLL-R8, but not in EL4 cells. Although the PII region did not show promoter activity on EL4, it showed weak promoter activity on CTLL-R8 cells (Fig. 3B). The promoter activity of PI region was enhanced by stimulation such as anti-CD3 mAb, P/I or Con A treatment on CTLL-R8 cells (Fig. 3B). In a previous report, the NF-κB and AP-1 binding sites found in the PI region (Kwon et al., 1994), suggesting that the PI region might be responsible for activation-dependent expression through TCR stimulation. To address whether the NF-κB and AP-1 binding sites are required for activation-dependent upregulation of the PI region, we mutated the NF-κB (GGGGAC → GGCGAC) and AP-1 binding sites (TGACTCA → TGACTTG) in PI region. Mutated construct of NF-κB site has shown significantly reduced promoter activity upon T cell activation with anti-CD3 (Fig. 3C). AP- 1 site mutation also had defect on promoter response by anti- CD3 stimulation but less than NF-κB site mutation (Fig. 3C). These results suggest that NF-κB and AP-1 binding sites are important for activation-dependent 4-1BB expression and 4- 1BB has at least three distinct functional promoter regions.
Fig. 3. Promoter analysis of the upstream regions (UTRs) of the mouse 4-1BB. (A) Upstream regions used for report analysis are indicated as PI, PII, and PIII. Translational start site is designated +1. Three distinct 1.8 kbp upstream regions of each exon were cloned into the pGL3-basic vector. (B) EL4 and CTLL-R8 cells were transiently transfected with UTR constructs and pCMVβ-gal as an internal control. PGL3-basic (Basic), promoterless luciferase report, was used as negative control. Cells were treated with anti-CD3 mAb (5 μg/ml), P/I (20 ng/ml and 1 ng/ml), or Con A (10 μg/ml), and luciferase activity was determined after 40 h. Anti-CD3 mAb was coated on culture plates for stimulation. (C) CTLL-R8 cells were transiently transfected with NF-κB mutant(PI-NF-κBmu), AP-1 mutant (PI-AP-1mu), and double mutant (PI-mu) constructs. The transfectants were stimulated with plate-bound anti-CD3 mAb for 40 h. Transfection efficiencies were normalized by β-gal activity and data are representative of three independent experiments. Values are presented as means ± s.e.
NF-κB specifically binds to the upstream region of the type I transcript
In a previous report, potential regulatory elements, including NF-κB, AP-1, and AP-2 elements, were identified in upstream regions of the mouse 4-1BB translational start site by comparison with a known consensus sequences (Kwon et al., 1994). In human 4-1BB, it was reported that 4-1BB expression is regulated by NF-κB and AP-1 in T cells (Kim et al., 2003). The putative NFκB element, at positions -1890 to -1880 in the mouse 4- 1BB UTR, is located on the PI region. Three putative AP-1 elements at positions -60 to -50, -1863 to -1855, and -4851 to -4843 were identified by using TFSEARCH V1.3 software. To investigate whether NF-κB cou ld b ind to t he p u tative NF-κB element, nuclear extracts were prepared from CD4+ T cells stimulated by anti-CD3 for 6 h and then subjected to EMSA. As shown in Fig. 4, nuclear extracts binds with a radio-labeled putative NF-κB probe showed a band shift (Fig. 4A, lane 2) that was competed out by a ten-fold excess of unlabeled probe (Fig. 4A, lane 4). Incubation with antibody specific for p65 showed the supershift (Fig. 4A, lane 3). A commercial NF-κB element was used as a positive control (Fig. 4A, lanes 5, 6, and 7).
Fig. 4. NF-κB binds to a putative NF-κB element of the PI region. (A) Nuclear extract prepared from CD4+ T cells was stimulated with anti-CD3 for 6 h, and EMSA was performed with [α-32P] labeled probe. Cold competition assay (lane 4) was performed with a ten-fold excess of unlabelled probe. For antibody mediated supershift analysis, anti-p65 monoclonal antibody was used (lanes 3, 6). A commercial NF-κB probe was used as a positive control (lanes 5-7) (n.s., non-specific binding). (B) CHIP assay performed by using anti-p65, anti-AP-1, or IgG as a negative control. Immunoprecipitated DNA was amplified by PCR with PI, PII, and PIII upstreamspecific primers. (C) Relative intensity of amplified PCR products was quantified by Image J program and graphed. It is normalized by the intensity of input.
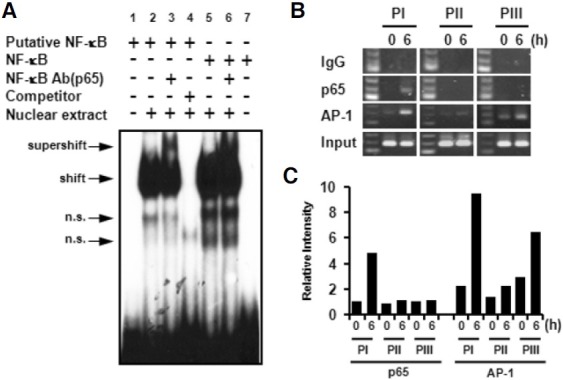
A CHIP assay was performed to confirm the specificity of NF- κB binding to the PI region. After chromatin immunoprecipitation using anti-p65 mAb or anti-AP-1 mAb, the PIII, PI, or PII fragments at positions -217 to -28, -2046 to -1846, -4896 to - 4697, respectively, were amplified by PCR (Fig. 4B). Amplified PCR products were quantified by Image J program (Fig. 4C). In agreement with the EMSA result, NF-κB specifically bound to the PI region but not to PII and PIII regions. In contrast, AP-1 slightly bound to all upstream regions in non-activated condition. While binding activity of AP-1 on the PI and PIII regions was enhanced by anti-CD3 stimulation, no significant change was found in binding activity for the PII region. We demonstrated that anti-CD3 stimulation specifically induced translocation of NF-κB to the PI region for activation-dependent expression.
Alternative 4-1BB splice variant was induced by stimulation
Several splice variants of human 4-1BB and one splice variant of mouse 4-1BB were previously reported (Michel et al., 1998; Setareh et al., 1995). The splice variant may negatively regulate signaling through the 4-1BB receptor by inhibiting receptorligand interaction because it is in soluble form. First, we investigated whether splice variant expression could be induced by stimulation using RT-PCR analysis (Fig. 5A) and RPA analysis (Figs. 5B and 5C) on T cells. For RT-PCR, we utilized specific sense primers of each transcript and an antisense primer (R2) designed in exon 9 (Fig. 1A). In RPA, a probe fragment containing exon 8 and exon 9 was used to discriminate the expression of the splice variant and normal transcript (Fig. 1A). A splice variant was detected from all three transcripts by RT-PCR analysis (Fig. 5A). RPA analysis confirmed that anti-CD3 stimulation induced the expression of splice variants to the extent comparable to the normal transcript containing the transmembrane domain (Figs. 5B and 5C). These results show that the splice variant is also highly induced by increasing normal transcript expression on activated T cells.
Fig. 5. The splice variant without exon 8 was highly induced in activated T cells. (A) The splice variant, which did not contain exon 8, was amplified by PT-PCR using cDNA prepared from activated CD4+ and CD8+ T cells via anti- CD3 stimulation for the indicated time. For RTPCR analysis of full transcripts, each sense primer (F1, F2, and F3) for each transcript, and the identical full reverse primer (R2) were used (Fig. 1A). (B, C) RNase Protection Assay (RPA). Purified CD4+ (B) a nd CD8+ T cells (C) were harvested at the indicated time after stimulation with anti-CD3 and total RNA was extracted. 5 μg of total RNA was hybridized with [α-32P] labeled probes and subjected to RPA analysis as described in the methods section, followed by autoradiography. The mRNA expression of the corresponding GAPDH was included to normalize for gel loading.
Negative regulation of alternative 4-1BB splice variant on 4-1BB signaling
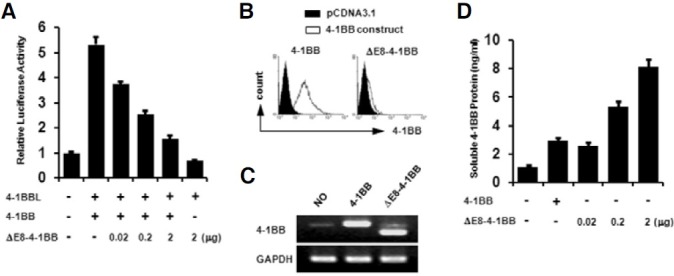
We investigated whether splice variant could inhibit 4-1BB signaling by luciferase assay using a pELAM-luc reporter plasmid containing a NF-κB response element. For transfection experiments, a splice variant was cloned into pCDNA3.1 (pCDNA3.1- ΔE8-4-1BB). Upon NF-κB activation induced on EL4 cells transfected with pCDNA3.1-4-1BB and pCDNA3.1-4-1BBL, it was reduced in a dose-dependent manner by co-transfection with the pCDNA3.1-ΔE8-4-1BB construct (Fig. 6A). After transfection, surface expression of 4-1BB was determined by flow cytometry (Fig. 6B). As we expected, no surface expression was found on EL4 cells transfected with the pCDNA3.1-ΔE8-4- 1BB construct. The expression of the 4-1BB transcript was also determined by RT-PCR (Fig. 6C) and soluble 4-1BB protein was measured by ELISA (Fig. 6D). EL4 cells slightly express endogenous 4-1BB transcript but not detectable on surface without stimulation such as anti-CD3, ConA, and PMA/Inomycin. Soluble 4-1BB protein was increased in a dosedependent manner. Thus, soluble 4-1BB induced under activation conditions is a negative regulator for activated T cells expressing membrane-bound 4-1BB.
Fig. 6. Inhibition effect of a splice variant on 4-1BB signaling. (A) EL4 cells were transiently transfected with the indicated expression plasmids of pCDNA3.1-4-1BB, pCDNA3.1-4-1BBL, or pCDNA3.1-ΔE8-4-1BB. pELAM-luc reporter plasmid and pCMV-β-gal were co-transfected. Luciferase activities were measured 24 h after transfection and normalized on the basis of β-galactosidase activity. Values are presented as means±s.e. Each experiment was performed in triplicate, and the data presented are representative of three different experiments. (B) The surface expression of 4-1BB on EL4 cells transfected with pCDNA3.1, pCDNA3.1-4-1BB, or pCDNA3.1-ΔE8-4-1BB was determined by staining with FITC conjugated 3E1 mAb and an isotype control. (C) RT-PCR performed with identical samples. (D) Soluble 4-1BB protein was measured by ELISA using the supernatant from transfected EL4 cells. A Wallac vector 1420 Multilabel Counter was used for measurement. Data are representative of three independent experiments. Values are presented as means ± s.e.
DISCUSSION
A previous report showed that mouse 4-1BB has two types of transcript and their expression may be regulated differently (Kwon et al., 1994). In this study, we found that mouse 4-1BB has more than two types of mRNA and three distinct promoter regions (PI, PII and PIII) that might be involved in regulating gene expression. Differential expression patterns might be dependent on the differential combination of transcription factors in certain conditions. Previous studies showed that mouse lck is regulated by two independent promoters, called the proximal and distal promoters. The mouse lck proximal promoter was required for tissue-specific expression in the thymus and mtβ was a critical transactivator for the proximal promoter (Reynolds et al., 1990; Yamada et al., 2001). The phenomenon of a differential expression pattern from a single gene by alternative promoters is very common, and is often related to tissuespecific expression of that gene (Christophi et al., 2004; Hai et al., 2001; Jian et al., 2006; Tao et al., 2001; Xiao et al., 2003).
Our result showed that the type I was expressed in tissuespecific manner and was highly induced in activated T cells. Interestingly, the type II transcript was ubiquitously expressed in tissues but inducible on T cells as well. A previous study showed the expression of human 4-1BB is regulated by NF-κB and AP-1 (Kim et al., 2003). In fact, putative AP-1 binding elements were identified in three distinct upstream regions of the mouse 4-1BB gene, and the putative NF-κB binding element was identified at positions -1890 to -1880, which is similarly located to the human 4-1BB gene. In this study, we found that NF-κB specifically binds to the PI region, but not the PII and PIII regions. Although binding activity of AP-1 on the PI region was enhanced by anti-CD3 stimulation, AP-1 bound to PI, PII, and PIII regions in non-activated condition. Therefore, Ap-1 might be responsible for constitutive expression of type II transcript on tissues. The translocation of NF-κB on the PI region is, however, an important event for the induction of 4-1BB transcripts in CD4+ and CD8+ T cells activated by TCR stimulation.
Taken together, each of the two mouse 4-1BB transcripts showed a differential expression pattern, could be regulated by distinct promoters. The PI region is mainly responsible for the activation-dependent expression in T cells through the NF-κB binding.
We have also shown that expression of the 4-1BB splice variant lacking exon 8, which yields a soluble form of 4-1BB, is induced on activated T cells. Previous studies showed that soluble human 4-1BB is enhanced in the sera of patients with rheumatoid arthritis, leukemia, and lymphoma (Furtner et al., 2005; Jung et al., 2004; Michel et al., 1998). The soluble form of 4-1BB seems to be associated with clinical disorders. Whereas two splice variants of human 4-1BB do not have a cytoplasmic domain because of a translational stop codon caused by a frame-shift, the mouse 4-1BB splice variant still has a cytoplasmic domain because it is in-frame. We investigated the expression level of a splice variant in activated T cells. In Fig. 5, the splice variant was highly induced on activated T cells to comparable level to the normal transcript. In addition, the antagonistic activity ofe variant on 4-1BB signaling was determined. These results suggest that the 4-1BB has a negative regulation system which may be required for preventing hyperactivation of T cells by reducing the expression of membrane- bound 4-1BB. In addition, it is possible that soluble 4- 1BB provides the reverse-signal through 4-1BBL on B cells, APCs, and monocytes.
Acknowledgments
This work was supported by grants from the National Cancer Center, Korea (NCC-0810720, NCC-1040830), National Research Foundation (KRF-2005-084-E0001, Stem Cell-2006-2004212), Korea Health 21 R&D (A050260) and Priority Research Center Program through the National Research Foundation of Korea funded by the Ministry of Education, Science and Technology (2009-0094052).
References
- 1.Alderson M.R., Smith C.A., Tough T.W., Davis-Smith T., Armitage R.J., Falk B., Roux E., Baker E., Sutherland G.R., Din W.S. Molecular and biological characterization of human 4-1BB and its ligand. Eur. J. Immunol. (1994);24:2219–2227. doi: 10.1002/eji.1830240943. [DOI] [PubMed] [Google Scholar]
- 2.Boussaud V., Soler P., Moreau J., Goodwin R.G., Hance A.J. Expression of three members of the TNF-R family of receptors (4-1BB, lymphotoxin-beta receptor, and Fas) in human lung. Eur. Respir. J. (1998);12:926–931. doi: 10.1183/09031936.98.12040926. [DOI] [PubMed] [Google Scholar]
- 3.Broll K., Richter G., Pauly S., Hofstaedter F., Schwarz H. CD137 expression in tumor vessel walls. High correlation with malignant tumors. Am. J. Clin. Pathol. (2001);115:543–549. doi: 10.1309/e343-kmyx-w3y2-10ky. [DOI] [PubMed] [Google Scholar]
- 4.Cascino I., Fiucci G., Papoff G., Ruberti G. Three functional soluble forms of the human apoptosis-inducing Fas molecule are produced by alternative splicing. J. Immunol. (1995);154:2706–2713. [PubMed] [Google Scholar]
- 5.Cascino I., Papoff G., De Maria R., Testi R., Ruberti G. Fas/Apo-1 (CD95) receptor lacking the intracytoplasmic signaling domain protects tumor cells from Fas-mediated apoptosis. J. Immunol. (1996);156:13–17. [PubMed] [Google Scholar]
- 6.Cheng J., Zhou T., Liu C., Shapiro J.P., Brauer M.J., Kiefer M.C., Barr P.J., Mountz J.D. Protection from Fas-mediated apoptosis by a soluble form of the Fas molecule. Science. (1994);263:1759–1762. doi: 10.1126/science.7510905. [DOI] [PubMed] [Google Scholar]
- 7.Christophi G.P., Isackson P.J., Blaber S., Blaber M., Rodriguez M., Scarisbrick I.A. Distinct promoters regulate tissue- specific and differential expression of kallikrein 6 in CNS demyelinating disease. J. Neurochem. (2004);91:1439–1449. doi: 10.1111/j.1471-4159.2004.02826.x. [DOI] [PubMed] [Google Scholar]
- 8.DeBenedette M.A., Chu N.R., Pollok K.E., Hurtado J., Wade W.F., Kwon B.S., Watts T.H. Role of 4-1BB ligand in costimulation of T lymphocyte growth and its upregulation on M12 B lymphomas by cAMP. J. Exp. Med. (1995);181:985–992. doi: 10.1084/jem.181.3.985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.DeBenedette M.A., Shahinian A., Mak T.W., Watts T.H. Costimulation of CD28-T lymphocytes by 4-1BB ligand. J. Immunol. (1997);158:551–559. [PubMed] [Google Scholar]
- 10.Dignam J.D., Lebovitz R.M., Roeder R.G. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res. (1983);11:1475–1489. doi: 10.1093/nar/11.5.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Furtner M., Straub R.H., Kruger S., Schwarz H. Levels of soluble CD137 are enhanced in sera of leukemia and lymphoma patients and are strongly associated with chronic lymphocytic leukemia. Leukemia. (2005);19:883–885. doi: 10.1038/sj.leu.2403675. [DOI] [PubMed] [Google Scholar]
- 12.Futagawa T., Akiba H., Kodama T., Takeda K., Hosoda Y., Yagita H., Okumura K. Expression and function of 4-1BB and 4-1BB ligand on murine dendritic cells. Int. Immunol. (2002);14:275–286. doi: 10.1093/intimm/14.3.275. [DOI] [PubMed] [Google Scholar]
- 13.Garni-Wagner B.A., Lee Z.H., Kim Y.J., Wilde C., Kang C.Y., Kwon B.S. 4-1BB is expressed on CD45RAhiROhi transitional T cell in humans. Cell. Immunol. (1996);169:91–98. doi: 10.1006/cimm.1996.0095. [DOI] [PubMed] [Google Scholar]
- 14.Goodwin R.G., Din W.S., Davis-Smith T., Anderson D.M., Gimpel S.D., Sato T.A., Maliszewski C.R., Brannan C.I., Copeland N.G., Jenkins N.A., et al. Molecular cloning of a ligand for the inducible T cell gene 4-1BB: a member of an emerging family of cytokines with homology to tumor necrosis factor. Eur. J. Immunol. (1993);23:2631–2641. doi: 10.1002/eji.1830231037. [DOI] [PubMed] [Google Scholar]
- 15.Hai M., Bidichandani S.I., Patel P.I. Identification of a positive regulatory element in the myelin-specific promoter of the PMP22 gene. J. Neurosci. Res. (2001);65:508–519. doi: 10.1002/jnr.1181. [DOI] [PubMed] [Google Scholar]
- 16.Heinisch I.V., Daigle I., Knopfli B., Simon H.U. CD137 activation abrogates granulocyte-macrophage colony-stimulating factor-mediated anti-apoptosis in neutrophils. Eur. J. Immunol. (2000);30:3441–3446. doi: 10.1002/1521-4141(2000012)30:12<3441::AID-IMMU3441>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- 17.Heinisch I.V., Bizer C., Volgger W., Simon H.U. Functional CD137 receptors are expressed by eosinophils from patients with IgE-mediated allergic responses but not by eosinophils from patients with non-IgE-mediated eosinophilic disorders. J. Allergy Clin. Immunol. (2001);108:21–28. doi: 10.1067/mai.2001.116864. [DOI] [PubMed] [Google Scholar]
- 18.Hurtado J.C., Kim S.H., Pollok K.E., Lee Z.H., Kwon B.S. Potential role of 4-1BB in T cell activation. Comparison with the costimulatory molecule CD28. J. Immunol. (1995);155:3360–3367. [PubMed] [Google Scholar]
- 19.Hurtado J.C., Kim Y.J., Kwon B.S. Signals through 4-1BB are costimulatory to previously activated splenic T cells and inhibit activation-induced cell death. J. Immunol. (1997);158:2600–2609. [PubMed] [Google Scholar]
- 20.Jian J.L., Zhu C.S., Xu Z.W., Ouyang W.M., Ma D.C., Zhang Y., Chen L.J., Yang A.G., Jin B.Q. Identification and characterization of the CD226 gene promoter. J. Biol. Chem. (2006);281:28731–28736. doi: 10.1074/jbc.M601786200. [DOI] [PubMed] [Google Scholar]
- 21.Jung H.W., Choi S.W., Choi J.I., Kwon B.S. Serum concentrations of soluble 4-1BB and 4-1BB ligand correlated with the disease severity in rheumatoid arthritis. Exp. Mol. Med. (2004);36:13–22. doi: 10.1038/emm.2004.2. [DOI] [PubMed] [Google Scholar]
- 22.Kim K.M., Kim H.W., Kim J.O., Baek K.M., Kim J.G., Kang C.Y. Induction of 4-1BB (CD137) expression by DNA damaging agents in human T lymphocytes. Immunology. (2002);107:472–479. doi: 10.1046/j.1365-2567.2002.01538.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kim J.O., Kim H.W., Baek K.M., Kang C.Y. NFkappaB and AP-1 regulate activation-dependent CD137 (4-1BB) expression in T cells. FEBS Lett. (2003);541:163–170. doi: 10.1016/s0014-5793(03)00326-0. [DOI] [PubMed] [Google Scholar]
- 24.Kohno T., Brewer M.T., Baker S.L., Schwartz P.E., King M.W., Hale K.K., Squires C.H., Thompson R.C., Vannice J.L. A second tumor necrosis factor receptor gene product can shed a naturally occurring tumor necrosis factor inhibitor. Proc. Natl. Acad. Sci. USA. (1990);87:8331–8335. doi: 10.1073/pnas.87.21.8331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kwon B.S., Weissman S.M. cDNA sequences of two inducible T-cell genes. Proc. Natl. Acad. Sci. USA. (1989);86:1963–1967. doi: 10.1073/pnas.86.6.1963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kwon B.S., Kim G.S., Prystowsky M.B., Lancki D.W., Sabath D.E., Pan J.L., Weissman S.M. Isolation and initial characterization of multiple species of T-lymphocyte subset cDNA clones. Proc. Natl. Acad. Sci. USA. (1987);84:2896–2900. doi: 10.1073/pnas.84.9.2896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kwon B.S., Kestler D.P., Eshhar Z., Oh K.O., Wakulchik M. Expression characteristics of two potential T cell mediator genes. Cell. Immunol. (1989);121:414–422. doi: 10.1016/0008-8749(89)90040-3. [DOI] [PubMed] [Google Scholar]
- 28.Kwon B.S., Kozak C.A., Kim K.K., Pickard R.T. Genomic organization and chromosomal localization of the T-cell antigen 4-1BB. J. Immunol. (1994);152:2256–2262. [PubMed] [Google Scholar]
- 29.Kwon B., Moon C.H., Kang S., Seo S.K., Kwon B.S. 4-1BB: still in the midst of darkness. Mol. Cells. (2000);10:119–126. doi: 10.1007/s10059-000-0119-0. [DOI] [PubMed] [Google Scholar]
- 30.Langstein J., Schwarz H. Identification of CD137 as a potent monocyte survival factor. J. Leukoc. Biol. (1999);65:829–833. doi: 10.1002/jlb.65.6.829. [DOI] [PubMed] [Google Scholar]
- 31.Langstein J., Michel J., Fritsche J., Kreutz M., Andreesen R., Schwarz H. CD137 (ILA/4-1BB), a member of the TNF receptor family, induces monocyte activation via bidirectional signaling. J. Immunol. (1998);160:2488–2494. [PubMed] [Google Scholar]
- 32.Langstein J., Michel J., Schwarz H. CD137 induces proliferation and endomitosis in monocytes. Blood. (1999);94:3161–3168. [PubMed] [Google Scholar]
- 33.Lee S.C., Ju S.A., Pack H.N., Heo S.K., Suh J.H., Park S.M., Choi B.K., Kwon B.S., Kim B.S. 4-1BB (CD137) is required for rapid clearance of Listeria monocytogenes infection. Infect. Immun. (2005);73:5144–5151. doi: 10.1128/IAI.73.8.5144-5151.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lotz M., Setareh M., von Kempis J., Schwarz H. The nerve growth factor/tumor necrosis factor receptor family. J. Leukoc. Biol. (1996);60:1–7. doi: 10.1002/jlb.60.1.1. [DOI] [PubMed] [Google Scholar]
- 35.McHugh R.S., Whitters M.J., Piccirillo C.A., Young D.A., Shevach E.M., Collins M., Byrne M.C. CD4(+)CD25(+) immunoregulatory T cells: gene expression analysis reveals a functional role for the glucocorticoid-induced TNF receptor. Immunity. (2002);16:311–323. doi: 10.1016/s1074-7613(02)00280-7. [DOI] [PubMed] [Google Scholar]
- 36.Michel J., Langstein J., Hofstadter F., Schwarz H. A soluble form of CD137 (ILA/4-1BB), a member of the TNF receptor family, is released by activated lymphocytes and is detectable in sera of patients with rheumatoid arthritis. Eur. J. Immunol. (1998);28:290–295. doi: 10.1002/(SICI)1521-4141(199801)28:01<290::AID-IMMU290>3.0.CO;2-S. [DOI] [PubMed] [Google Scholar]
- 37.Nocentini G., Ronchetti S., Bartoli A., Spinicelli S., Delfino D., Brunetti L., Migliorati G., Riccardi C. Identification of three novel mRNA splice variants of GITR. Cell Death Differ. (2000);7:408–410. doi: 10.1038/sj.cdd.4400670. [DOI] [PubMed] [Google Scholar]
- 38.Pizzolo G., Vinante F., Chilosi M., Dallenbach F., Josimovic- Alasevic O., Diamantstein T., Stein H. Serum levels of soluble CD30 molecule (Ki-1 antigen) in Hodgkin’s disease: relationship with disease activity and clinical stage. Br. J. Haematol. (1990);75:282–284. doi: 10.1111/j.1365-2141.1990.tb02664.x. [DOI] [PubMed] [Google Scholar]
- 39.Pollok K.E., Kim Y.J., Zhou Z., Hurtado J., Kim K.K., Pickard R.T., Kwon B.S. Inducible T cell antigen 4-1BB. Analysis of expression and function. J. Immunol. (1993);150:771–781. [PubMed] [Google Scholar]
- 40.Pollok K.E., Kim Y.J., Hurtado J., Zhou Z., Kim K.K., Kwon B.S. 4-1BB T-cell antigen binds to mature B cells and macrophages, and costimulates anti-mu-primed splenic B cells. Eur. J. Immunol. (1994);24:367–374. doi: 10.1002/eji.1830240215. [DOI] [PubMed] [Google Scholar]
- 41.Pollok K.E., Kim S.H., Kwon B.S. Regulation of 4-1BB expression by cell-cell interactions and the cytokines, interleukin-2 and interleukin-4. Eur. J. Immunol. (1995);25:488–494. doi: 10.1002/eji.1830250227. [DOI] [PubMed] [Google Scholar]
- 42.Reali C., Curto M., Sogos V., Scintu F., Pauly S., Schwarz H., Gremo F. Expression of CD137 and its ligand in human neurons, astrocytes, and microglia: modulation by FGF-2. J. Neurosci. Res. (2003);74:67–73. doi: 10.1002/jnr.10727. [DOI] [PubMed] [Google Scholar]
- 43.Reynolds P.J., Lesley J., Trotter J., Schulte R., Hyman R., Sefton B.M. Changes in the relative abundance of type I and type II lck mRNA transcripts suggest differential promoter usage during T-cell development. Mol. Cell. Biol. (1990);10:4266–4270. doi: 10.1128/mcb.10.8.4266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Saoulli K., Lee S.Y., Cannons J.L., Yeh W.C., Santana A., Goldstein M.D., Bangia N., DeBenedette M.A., Mak T.W., Choi Y., et al. CD28-independent, TRAF2-dependent costimulation of resting T cells by 4-1BB ligand. J. Exp. Med. (1998);187:1849–1862. doi: 10.1084/jem.187.11.1849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schwarz H., Valbracht J., Tuckwell J., von Kempis J., Lotz M. ILA, the human 4-1BB homologue, is inducible in lymphoid and other cell lineages. Blood. (1995);85:1043–1052. [PubMed] [Google Scholar]
- 46.Schwarz H., Blanco F.J., von Kempis J., Valbracht J., Lotz M. ILA, a member of the human nerve growth factor/ tumor necrosis factor receptor family, regulates T-lymphocyte proliferation and survival. Blood. (1996);87:2839–2845. [PubMed] [Google Scholar]
- 47.Seko Y., Sugishita K., Sato O., Takagi A., Tada Y., Matsuo H., Yagita H., Okumura K., Nagai R. Expression of costimulatory molecules (4-1BBL and Fas) and major histocompatibility class I chain-related A (MICA) in aortic tissue with Takayasu’s arteritis. J. Vasc. Res. (2004);41:84–90. doi: 10.1159/000076437. [DOI] [PubMed] [Google Scholar]
- 48.Setareh M., Schwarz H., Lotz M. A mRNA variant encoding a soluble form of 4-1BB, a member of the murine NGF/TNF receptor family. Gene. (1995);164:311–315. doi: 10.1016/0378-1119(95)00349-b. [DOI] [PubMed] [Google Scholar]
- 49.Shuford W.W., Klussman K., Tritchler D.D., Loo D.T., Chalupny J., Siadak A.W., Brown T.J., Emswiler J., Raecho H., Larsen C.P., et al. 4-1BB costimulatory signals preferentially induce CD8+ T cell proliferation and lead to the amplification in vivo of cytotoxic T cell responses. J. Exp. Med. (1997);186:47–55. doi: 10.1084/jem.186.1.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Takahashi C., Mittler R.S., Vella A.T. Cutting edge: 4-1BB is a bona fide CD8 T cell survival signal. J. Immunol. (1999);162:5037–5040. [PubMed] [Google Scholar]
- 51.Tao L., Dong Z., Zannis-Hadjopoulos M., Price G.B. Immortalization of human WI38 cells is associated with differential activation of the c-myc origins. J. Cell. Biochem. (2001);82:522–534. doi: 10.1002/jcb.1173. [DOI] [PubMed] [Google Scholar]
- 52.Watts T.H. TNF/TNFR family members in costimulation of T cell responses. Annu. Rev. Immunol. (2005);23:23–68. doi: 10.1146/annurev.immunol.23.021704.115839. [DOI] [PubMed] [Google Scholar]
- 53.Wilcox R.A., Chapoval A.I., Gorski K.S., Otsuji M., Shin T., Flies D.B., Tamada K., Mittler R.S., Tsuchiya H., Pardoll D.M., et al. Cutting edge: Expression of functional CD137 receptor by dendritic cells. J. Immunol. (2002);168:4262–4267. doi: 10.4049/jimmunol.168.9.4262. [DOI] [PubMed] [Google Scholar]
- 54.Xiao Z.S., Simpson L.G., Quarles L.D. IRES-dependent translational control of Cbfa1/Runx2 expression. J. Cell. Biochem. (2003);88:493–505. doi: 10.1002/jcb.10375. [DOI] [PubMed] [Google Scholar]
- 55.Yamada A., Takaki S., Hayashi F., Georgopoulos K., Perlmutter R.M., Takatsu K. Identification and characterization of a transcriptional regulator for the lck proximal promoter. J. Biol. Chem. (2001);276:18082–18089. doi: 10.1074/jbc.M008387200. [DOI] [PubMed] [Google Scholar]
- 56.Zupan A.A., Osborne P.A., Smith C.E., Siegel N.R., Leimgruber R.M., Johnson E.M. Jr. Identification, purification, and characterization of truncated forms of the human nerve growth factor receptor. J. Biol. Chem. (1989);264:11714–11720. [PubMed] [Google Scholar]


