Abstract
Fanconi anemia (FA) is a rare cancer-predisposing genetic disease mostly caused by improper regulation of the monoubiquitination of Fanconi anemia complementation group D2 (FANCD2). Genetic studies have indicated that ubiquitin conjugating enzyme UBE2T and HHR6 could regulate FANCD2 monoubiquitination through distinct mechanisms. However, the exact regulation mechanisms of FANCD2 monoubiquitination in response to different DNA damages remain unclear. Here we report that UBE2W, a new ubiquitin conjugating enzyme, could regulate FANCD2 monoubiquitination by mechanisms different from UBE2T or HHR6. Indeed, UBE2W exhibits ubiquitin conjugating enzyme activity and catalyzes the monoubiquitination of PHD domain of Fanconi anemia complementation group L (FANCL) in vitro. UBE2W binds to FANCL, and the PHD domain is both necessary and sufficient for this interaction in mammalian cells. In addition, overexpression of UBE2W in cells promotes the monoubiquitination of FANCD2 and down-regulated UBE2W markedly reduces the UV irradiation-induced but not MMC-induced FANCD2 monoubiquitination. These results indicate that UBE2W regulates FANCD2 monoubiquitination by mechanisms different from UBE2T and HRR6. It may provide an additional regulatory step in the activation of the FA pathway.
Keywords: DNA repair, FANCD2 monoubiquitination, FA pathway, FANCL, fanconi anemia, UBE2W
INTRODUCTION
Fanconi anemia (FA) is a group of rare human autosomal or X-linked recessive hereditary disorder associated with congenital abnormality, progressive bone marrow failure and cancer sus-ceptibility (Bagby, 2003; D’Andrea and Grompe, 2003; Meetei et al., 2004). Cells from FA patients exhibit spontaneous chro-mosome breakage and cellular hypersensitivity to genotoxic agents (Auerbach, 1988; German et al., 1987; Poll et al., 1985). FA is genetically heterogeneous, and to date at least 13 com-plementation groups (FANCA, B, C, D1, D2, E, F, G, I, J, L, M and N) have been identified to be associated with distinct gene mutations (Grompe and van de Vrugt, 2007; Smogorzewska et al., 2007; Taniguchi and D’Andrea, 2006). These gene products interact functionally with each other in vivo and form a common DNA repair pathway termed as FA pathway to mediate DNA damage repair and maintain the chromosomal integrity.
The monoubiquitination of two proteins in FA pathway, FANCD2 and its new paralog FANCI, are catalyzed by a core complex consisting of eight known FA proteins (FANCA, B, C, E, F, G, L, and M), with FANCL serving as the catalytic ubiquitin E3 ligase (Longerich et al., 2009; Smogorzewska et al., 2007). Subsequently, the monoubiquitinated FANCD2/FANCI heterodi-mers are recruited to the damage-induced nuclear foci on chromatin, along with three other downstream FA members, BRCA2/FANCD1, BRIP1/BACH1/FANCJ and PALB2/FANCN, to facilitate the repair of damaged DNA (Dorsman et al., 2007; Sims et al., 2007; Smogorzewska et al., 2007; Yuan et al., 2009). It is assumed that the monoubiquitination of FANCD2/ FANCI complex is regulated by multi-factors and it plays a cru-cial role in activating the FA pathway. Moreover, the monoubiquitination of paralogs occurs interdependently and a dual activation mechanism is employed to affect the down-stream DNA repair, thus providing an additional regulatory layer in the activation of the FA pathway (Meijer, 2007; Ishiai et al., 2008).
Ubiquitination is a common way of protein modification and it plays important roles in many cellular pathways, including sig-nal transduction and DNA repair (Chen, 2005; Huang and D’Andrea, 2006). Conjugation of ubiquitin to the substrate pro-ceeds via a conserved three-step mechanism, carried out by a complex cascade of enzymes including the ubiquitin-activating enzyme (E1), the ubiquitin-conjugating enzyme (E2), and the ubiquitin-protein ligase (E3) (Hershko et al., 2000). The E2 enzyme typically harbors a so-called conserved UBC domain that catalyzes ubiquitin conjugation (Cook et al., 1992; 1993). The typical E3 enzyme contains RING finger or HECT domains, which work as a scaffold interacting with both the target protein and the UBC, thus allowing transfer of ubiquitin to the target protein (Robinson and Ardley, 2004). E2-E3 interactions are believed to be highly selective, and a specific E2 is expected to interact with a given E3. However, there are a growing number of reports showing that E3 interacts with more than one E2 in different biological processes (Christensen et al., 2007; Dodd et al., 2004; Plans et al., 2006; Zhang et al., 2005).
UBE2T is the newly identified FA E2 enzyme that interacts with FANCL and it is required for the DNA damage-induced FANCD2 monoubiquitination (Machida et al., 2006). It predomi-nantly accumulates on chromatin co-localized with FA core complex and its substrates (Alpi et al., 2007). Moreover, anoth-er E2 enzyme, HRR6, was recently reported to regulate FANCD2/FANCI monoubiquitination in a distinct manner from UBE2T, although HHR6 did not directly interact with FANCL (Zhang et al., 2008a). Thus, the regulation of FANCD2/FANCI modification could be partly, if not solely, determined by the formation of an active E2/E3 holoenzyme involved with different ubiquitin-related enzymes.
UBE2W (also known as CG7220 in D. melanogaster) has a typical UBC domain and was reported to have extensive ex-pression in human and mouse tissues (Yin et al., 2006; Zhang et al., 2008c). In addition, UBE2W is capable of interacting with RING finger domain of BRCA1 and catalyzing the monoubiquitination of RING finger (Christensen et al., 2007). Here, we present data showing that UBE2W, which interacts with the PHD domain of FANCL, could regulate FANCD2 monoubiquitination by mechanisms different from UBE2T or HRR6. We speculate that FANCD2/FANCI monoubiquitination may be one of the nodes in multiple repair pathways regulated by various ubiquitin-related enzymes and plays a more general role in DNA repair or genome maintenance.
MATERIALS AND METHODS
Cell lines and culture conditions
HeLa and NIH3T3 cell lines were purchased from ATCC, and 293FT cell line was purchased from Invitrogen. These cells were maintained in DMEM medium supplemented with 10% fetal calf serum (FCS), and grown in a humidified 5% CO2-containing atmosphere at 37℃.
Plasmid construction
The plasmids expressing mouse UBE2W protein (pCMV-myc/ UBE2W) and FANCL protein (pCMV-HA/FANCL) were gener-ously provided by Dr. Baisong Lu of Wake Forest University Health Sciences. Using these constructs, we have further con-structed plasmids pGADT7/FANCL and pGBKT7/UBE2W for Yeast Two-Hybrid assay. Glutathione-S-transferase (GST) fu-sions with FANCL and UBE2W were generated from pGEX-6P-1 or pGEX-4T-1 vectors (Amersham Biosciences, Sweden), and the fusion proteins were purified with GST affinity chroma-tography. FANCL with EGFP and UBE2W with RFP fusion proteins were constructed, respectively, by insertion of the fancl and ube2w cDNA into pEGFP-C1 (BglII and EcoRI sites) and pDsRed-express-C1 (BglII and HindIII sites, Clontech, USA). The BiFC assay system containing pBIFC-bFosYC155, pBIFC-bFos ΔZIPYC155 and pBIFC-bJunYN155 plasmids were gen-erously provided by Dr. Tom Kerppola of University of Michigan Medical School. The pBIFC-YN173 and pBIFC-YC173 plas-mids were constructed by insertion of the fragment encoding acid residues 1-172, and 173-238 of enhanced YFP (Clontech, USA) into EcoRI and SalI sites of pCMV-HA (Clontech, USA) as described elsewhere (Hu and Kerppola, 2003; Hu et al., 2002). Subsequently, the fancl and ube2w cDNAs were insert-ed into these constructions for protein interaction assay in living cells. Moreover, UBE2W point mutants as controls were gener-ated through overlap-extension PCR. Truncations and deletions of FANCL for further analysis were also generated with stand-ard PCR cloning techniques. All these mutations and truncations were inserted into proper plasmids as the insertion of wild-type genes.
Cell cycle analysis
Cell cycle analysis was performed as described elsewhere with modifications (Taniguchi et al., 2002). Briefly, the cells expressing GFP or GFP-UBE2W were collected and washed with ice cold PBS twice, and fixed in 0.5 ml 1% paraformaldehyde for 20 min on ice in the dark. The fixed cells were further treated with 2 ml cold 75% ethnol/15% PBS (-20℃) overnight and resuspended in 0.5 ml PBST (PBS with 0.1% Triton X-100 and 0.5% BSA, stored at 4℃) with 100 μg/ml RNaseA and 50 μg/ml PI working solution and incubated 1 h at 37℃ in the dark or overnight at 4℃. The cells were analyzed immediately on a Flow Cytometer.
GST pull-down and yeast two-hybrid assay
For GST pull-down assay, GST fusion proteins (10 μg) were incubated with magnetic glutathione particles (Promega) in binding buffer (PBS, pH 7.4) for 1 h at 4℃. After blocked in 10% BSA for 1 h at room temperature, beads were incubated with the extracts of whole cells that expressed myc-UBE2W fusion proteins in the binding buffer for 3 h at 4℃, followed by three washes with the binding buffer. Proteins on beads were eluted by boiling in 1× SDS-PAGE sample buffer and examined by Western blotting. For yeast two-hybrid assay, the construct-ed pGADT7 plasmids containing whole length or truncated FANCL and pGBKT7/UBE2W plasmids were used for the co-transformation of the reporter yeast strain AH109 from the Matchmaker yeast two-hybrid system 3 (Clontech, USA) ac-cording to the manufacturer’s instructions.
Immunoprecipitation
Extracts of whole cells that expressed the myc-UBE2W fusion proteins were prepared in RIPA lysis buffer (50 mM Tris-HCl, pH 7.4, 150 mM NaCl, 1% (vol/vol) Triton X-100) supplemented with protease tablets (Roche). Five hundreds μl lysate was pre-cleared by incubating with 30 μl protein G agarose (Roche) at 4℃ for 1 h. The pre-cleared lysate was incubated with the puri-fied GST fusion protein with FANCL or its truncations and 1 μg anti-myc antibody for 12 h at 4℃. The lysate was further incu-bated with 50 μl protein G agarose (Roche) at 4℃ for 4 h. The beads were spun down, washed several times with ice cold RIPA lysis buffer and re-suspended in 1× SDS loading buffer. The proteins were separated by SDS-PAGE, transferred to PVDF membrane and immunoblotted with anti-GST antibody (Promega).
BiFC assay
Cells transfected with plasmids encoding the indicated combi-nations of fusion proteins were incubated at 37℃ for 24 h and then transferred to 30℃ for 6 h to promote fluorophore matura-tion. The fluorescence emissions of the cells were imaged as described (Hu and Kerppola, 2003; Hu et al., 2002).
Subcellular localization
The fluorescent GFP-FANCL and RFP-UBE2W expressing plasmids were used for the transfection of cells grown on glass coverslip with Lipofectamine 2000 (Invitrogen). Twenty four hours after transfection, the cells were fixed at 4℃ for 30 min with 4% PFA in PBS and then incubated in 300 μl DAPI work-ing solution (1 μg/ml in methanol, Sigma) at 37℃ for 15 min. Following three washes with methanol to remove the DAPI background, the cells were observed with the Bio-Rad Radi-ance 2100 confocal system in conjunction with a Nikon TE300 microscope.
In vitro ubiquitination assay
The reaction was performed essentially as described previously (Garcia-Higuera et al., 2001; Gurtan et al., 2006). Reaction mixtures contained approximately 300 ng E1 (Sigma), 1 μg ubiquitin (Sigma), 300 ng respective E2 (the positive control UbcH2 was purchased from Sigma), and where indicated, 300 ng GST-FANCL and GST-PHD, respectively, in 50 mM Tris-HCl, pH 7.5, 2 mM ATP, 2.5 mM MgCl2, 15 mM KCl, 1 mM dithiothreitol, 1% glycerol, and 0.01% Triton X-100. After incubation at 37℃ for 40 min, the reaction was stopped by adding 10 μl 2× SDS-PAGE sample buffer. The reaction mixture was separated by SDS-PAGE and immunoblotted with anti-GST antibody and anti-Ub antibody.
RNA interference (RNAi) and Western blotting
The H1-U6 dual promoter RNAi plasmids (pGFP-HU) against human UBE2W were constructed as previously described (Zhang et al., 2008b). The functional RNAi targets used were as follows: siControl: ATCTGCCTTCATACTATCTAC, UBE2W- 259: ATGGTCATATCTGTTTATCC. Plasmids described above were used to transfect 293FT or HeLa cells. The FANCD2 mono- ubiquitination was detected by Western blotting as previously reported (Machida et al., 2006), followed by treating cells with Uv (1J) or MMC (120 nM) for 24 h where indicated.
SYBR green quantitative real-time PCR
Total cellular RNA was isolated using a NucleoSpin RNA II kit (MN, Inc.) according to the manufacturer’s instructions. Quantitative PCR (qPCR) was performed using SYBR Green I dye on an IQ™ 5 Real-Time PCR Detection Systems (Bio-Rad). The expression levels of UBE2W are reported as ratios of expression of GAPDH in the same master reaction. The PCR primer pairs (5′ to 3′) used for each gene were: UBE2W, F-AAGGTC CAGTTCTCAGTA and R-GCAACAATAAGGCATTAGTAA; GAPDH: F-TGAACGGGAAGCTCACTGG and R-TCCACCAC CCTGTTGCTGTA.
RESULTS
UBE2W interacts with FANCL in yeast cells
UBE2W is a novel protein, and its homologue in D.melanogaster is predicted to interact with FANCL in a protein-protein interaction database (The BioGrid Database). However, little is known about the interaction of its mammalian homologues with FANCL in the cell. The yeast two-hybrid approach was carried out to identify the interactions of UBE2W and FANCL in yeast. Because FANCL was reported to harbor a PHD domain and exhibit the ubiquitin ligase activity, and because UBE2W was predicted to be a potential ubiquitin conjugating enzyme, we hypothesized that PHD domain of FANCL might mediate the interaction with UBE2W. Therefore, we generated deletion mu-tants of FANCL that lack PHD domain or WD40-repeats and introduced them as well as the wide type FANCL with UBE2W into the AH109 yeast cells. Consistent with the hypothesis, wild-type FANCL, but not the PHD mutated forms (FANCL1-305aa and FANCL 1-275aa), interacted with UBE2W in yeast. More detailed mutation analyses in WD40-repeats demonstrated that FANCL PHD domain (FANCL 306-375aa) is necessary, but not sufficient, to mediate the interaction in yeast cells (Fig. 1A). The sequence upstream the N-terminal of this region (264-375aa) was therefore needed for FANCL to interact with UBE2W.
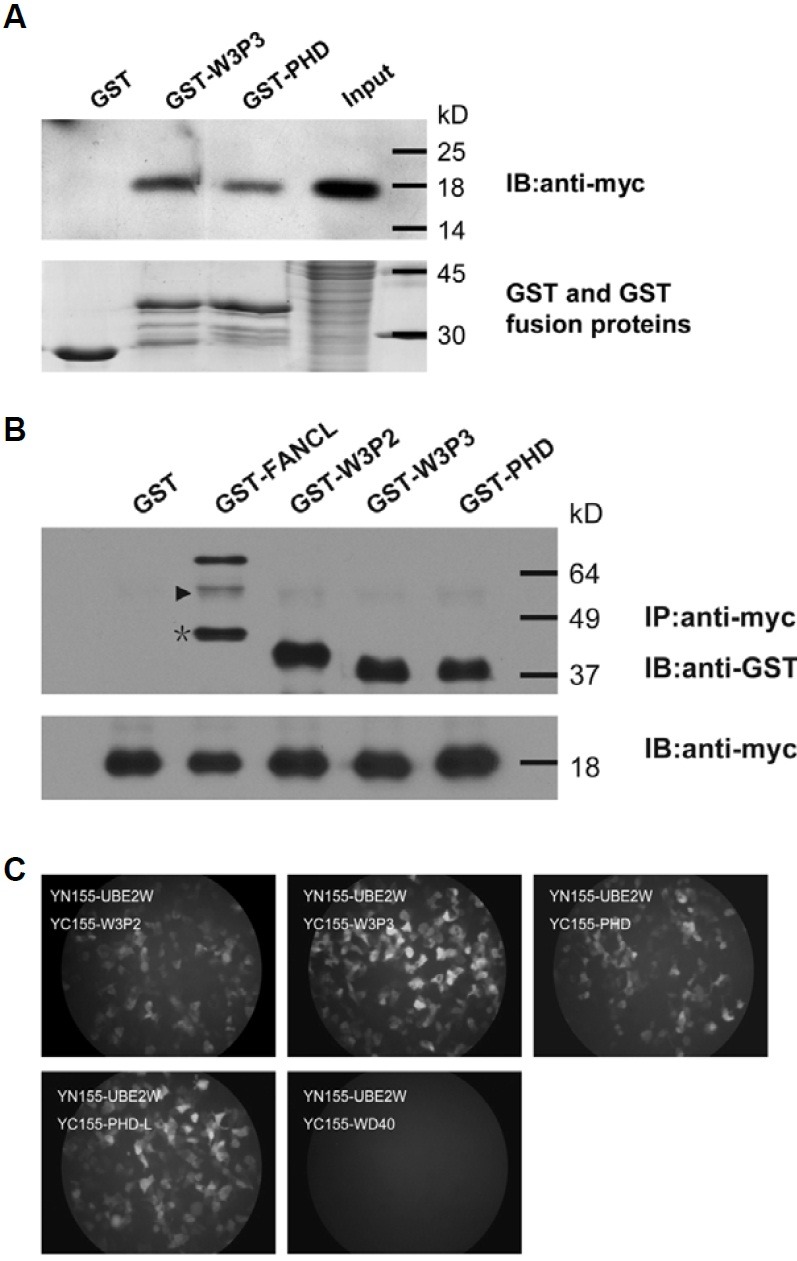
Fig. 1. Interactions of UBE2W with FANCL in vitro and in vivo. (A) Schematic illustration of FANCL WD40-deletion and truncation mutants with a summary of FANCL-UBE2W interactions in yeast cells. Each WD40-repeat, designated as WD40-1, -2, and -3, was deleted as previously reported. The region of WD40-3 was further deleted to 250-, 254-, 264-, and 270-375aa depending on the protein secondary structure. FANCL 254-375aa was suffi-cient for the interaction between FANCL and UBE2W in yeast cells. (B) GST-pulldown experi-ments were used to confirm the interaction be-tween FANCL and UBE2W. The protein complex, precipitated by glutathione particles, was washed four times to demonstrate the interaction between FANCL and UBE2W. The UBE2W could be specif-ically precipitated by full length FANCL followed by washing for four times. (C) DsRed-UBE2W co-localized with EGFP-FANCL in nucleus. The DsRed and DsRed-UBE2W and EGFP-FANCL fusion proteins were transiently expressed in NIH3T3 cells (red and green) and the nuclei were stained with DAPI (blue). The DsRed-UBE2W was found mainly enriched in the nucleus. When the EGFP-FANCL and DsRed-UBE2W were co-expressed, the distribution of EGFP-FANCL changed from a ubiquitous intracellular localization to a subcellular enriched pattern, mainly co-localized with UBE2W in nucleus. (D) UBE2W interacts with FANCL in nucleus visualized in living cells using BiFC analy-sis. Cells transfected with plasmids encoding the indicated combinations of fusion proteins were incubated at 37℃ for 24 h and then transferred to 30℃ for 4-6 h to promote fluorophore maturation. The bFos + bJun group and delta-bFos + bJun group were used as the positive and negative controls. The bar represents 20 μm in all images.

UBE2W binds to FANCL in vitro
GST-Pull down and immunoprecipitation analysis were used to further confirm the interaction between UBE2W and FANCL in vitro. In one direction, purified GST-tagged FANCL, but not GST, co-precipitated with myc-tagged UBE2W when GST-FANCL was pulled down by glutathione-coupled particles (Fig. 1B). In the other direction, myc-tagged UBE2W co- precipitated with GST-FANCL when myc-UBE2W was pulled down by protein G agarose beads linked with myc-tag antibody (Fig. 2B, line 2). These experiments provided further evidence that the two pro-teins physically associated with each other in vitro.
Fig. 2. FANCL PHD domain is necessary and sufficient to interact with UBE2W. (A, B) FANCL PHD domain is sufficient to interact with UBE2W in vitro. (A) The truncated FANCL proteins, GST-W3P3 and GST-PHD, specifically co-precipitated with mammalian expressed UBE2W in vitro. Recombinant GST-W3P3, GST-PHD, and the nega-tive control GST protein were resolved by SDS-PAGE (lower panel). The precipitated protein com-plex was analyzed by western blot with myc tag antibody (upper panel). (B) Immunoprecipitation experiments was performed to confirm the interaction between FANCL PHD domain and UBE2W in the other direction. GST and GST tagged full length or truncated FANCL proteins were purified from E. coli and incubated with the extract of whole cells that expressed myc-UBE2W. The anti-myc tag antibody was used to capture the myc-UBE2W and its interacting proteins. The precipitated proteins were separated by SDS-PAGE and analyzed by Western blot with GST antibody (upper panel) or myc tag antibody (lower panel). The triangle in the upper panel is shown as the non-specific band and the asterisk is shown as the degradation frag-ment of GST-FANCL during the protein purification. (C) FANCL PHD domain is necessary and sufficient to interact with UBE2W in vivo. Visualization of interactions among UBE2W and truncated FANCL proteins in living cells was achieved by BiFC assay. All truncates of FANCL, except the WD40 domain, could complement with UBE2W to emit the visible fluorescence in vivo.
UBE2W interacts with FANCL localized in nucleus in vivo
To confirm the protein-protein interaction between UBE2W and FANCL in vivo, constructs were made for RFP-tagged UBE2W (RFP-UBE2W) and EGFP-tagged FANCL. We first determined the subcellular localization of UBE2W in transfected NIH3T3 cells by tagged RFP or immunostaining with UBE2W antibody. The UBE2W was mainly localized in the nucleus when it was fused to either the C terminal of RFP (Fig. 1C, upper and mid-dle panels). The same localization was also found when the protein was transiently expressed in HeLa or 293FT cells (data not shown). When the two Nuclear Localization Signals (NLS) harbored in the C terminus of UBE2W were deleted, the protein was confined to the cytoplasm, further confirming the nuclear localization of UBE2W (data not shown). FANCL was reported to be an intracellular protein and localized in both the cytoplasm and the nucleus with diffused pattern (Lu and Bishop, 2003). However, when EGFP-FANCL and RFP-UBE2W were co-expressed, the distribution of EGFP-FANCL changed from a ubiquitous intracellular localization to a subcellular enriched pattern, mainly co-localizing with UBE2W in nucleus (Fig. 1C, lower panel).
Since co-localization studies only provide the indirect association of the two proteins and the visualization could be interfered by the fluorescence of each protein, we further used the bimolecular fluorescence complementation (BiFC) approach, which was established by Hu and Kerppola in 2002 (Hu et al., 2002), to visualize the exact subcellular localization of UBE2W-FANCL complex in living cells. The different combinations of YN155-FANCL with YC155-UBE2W and YC155-FANCL with YN155-UBE2W were used to co-transfect the 293FT or NIH3T3 cells. As expected, the complement fluorescence was visible mainly in the nucleus, suggesting that the interaction of UBE2W and FANCL was generally confined in nucleus (Fig. 1D, lower left panel). The similar distribution of complemented fluores-cence was also found in other combinations of YN173-FANCL with YC173-UBE2W, YC173-FANCL with YN155-UBE2W and YN155-FANCL with YN173-UBE2W, excluding the nonspecific fluorescence complementation caused by protein fusion. In addition, we also unexpectedly found that YC155-UBE2W and YN155-UBE2W could complement the fluorescence in nucleus, suggesting that UBE2W was able to form a homodimer with itself in nucleus (Fig. 1D, lower right panel). These data from co-localization and BiFC assay further confirmed the FANCL-UBE2W interaction in vivo.
FANCL PHD-L (FANCL 306-375aa) is sufficient to interact with UBE2W in mammalian cells
To gain insight into the functional role of the UBE2W-FANCL interaction in mammalian cells, the exact region of FANCL that mediates the interaction was further determined by the BiFC approach. The truncated FANCL constructs in pBiFC-YC155 plasmid, named as WD40 (FANCL 1-275aa), W3P2 (FANCL 254-375aa), W3P3 (FANCL 264-375aa), PHD (FANCL 276-375aa) and PHD-L (PHD lacking the linker, FANCL 306-375aa) were respectively co-introduced with YN155-UBE2W into the 293FT cells. Unexpectedly, the YC155-PHD and YC155-PHD-L, as well as the YC155-W3P2 and YC155-W3P3, could comple-ment with YN155-UBE2W to emit the visible fluorescence in mammalian cells, suggesting that FANCL PHD-L (FANCL 306-375aa) is sufficient for the interaction with UBE2W in mammali-an cells (Fig. 2C).
However, these observations were not well in agreement with the previous finding in yeast. Because no homologue of FANCL and UBE2W was found in S. cerevisiae and they probably did not have functional partners in yeast, we hypothesized that the FANCL-UBE2W interaction in mammalian cells might be more complicated and there might be other proteins that stabilize the interaction of PHD with UBE2W. Therefore, we generated simi-lar truncated FANCL constructs fused with GST, and performed the GST-Pull down and immunoprecipitation assay with the mammalian expressed UBE2W in vitro. Consistent with the BiFC assay data, the GST-PHD and myc-UBE2W could co-precipitate with each other as well as with GST-W3P2 or GST-W3P3 (Figs. 2A and 2B). These data further determined the sufficiency of PHD-L (FANCL 306-375aa) for interacting with UBE2W in mammalian cells.
UBE2W but not UBE2WC91S exhibits ubiquitin conjugating activity
The common features of all ubiquitin conjugating enzymes (E2s) characterized to date are that they contain a typical ubiq-uitin conjugating domain (UBC) and they can form a thioester bond between a conserved cysteine residue and ubiquitin in the presence of E1 (Cook et al., 1992). Because the interaction of UBE2W with the ubiquitin ligase FANCL and the PHD of FANCL is necessary for this reaction, and because UBE2W was previously found containing a typical ubiquitin conjugating domain (UBC) (Zhang et al., 2008c), it raises the possibility that UBE2W might have ubiquitin conjugating enzyme (E2) activity. To demonstrate the possibility, the purified GST-UBE2W was incubated with or without E1 and ubiquitin. As expected, in the presence of E1 and ubiquitin, UBE2W could conjugate and transfer ubiquitin in vitro just like the positive control UbcH2 (Fig. 3B, lines 1 and 4).
Fig. 3. C91 in UBC domain is required for in vitro E2 activity of UBE2W. (A) Sequence alignment of UBE2W homologues. The col-ored and shaded region represents con-served residues. C91, marked with bold-face in the box, is completely conserved, despite degeneracy in flanking sequences. Alignment was carried out manually. (B) Catalytic activity of UBE2W and UBE2WC91S in vitro. The GST fused proteins, UBE2W and FANCL, were purified from E. coli, and incubated with E1 activating enzyme, ubiquitin, and ATP. For negative controls, each component (except ATP) was omitted from the reaction, as indi-cated. The E2 (UbcH2), purchased from SIGMA, was used as positive control. The reactions were analyzed by Western blot with ubiquitin antibody (upper panel) or GST anti-body (lower panel). GST-UBE2W and ubiquitinated species are labeled with arrows.

To determine which cysteine residue is involved in the activity of UBE2W, we carried out a sequence alignment study compar-ing the UBE2W homologues from fruit fly to human species and other E2s from S. cerevisiae. Sequence alignment revealed a conserved cysteine residue in UBC domain of UBE2W homo-logues and E2s from S. cerevisiae. In UBE2W, this cysteine, C91, is 100% conserved from fruit fly, mosquito and mouse to human, despite degeneracy in flanking sequences (Fig. 3A). Based on these alignments, we hypothesized that the C91 probably plays a crucial role in UBE2W ubiquitin conjugating activity. To test this hypothesis, we expressed and analyzed the GST-UBE2WC91S ubiquitin conjugating activity. As expected, the mutant GST-UBE2WC91S, lacking the essential cysteine residue in the active site of the UBC domain, failed to conjugate and transfer ubiquitin in vitro, compared to the wild type UBE2W, indicating that C91 residue is crucial to the ubiquitin conjugating activity of UBE2W (Fig. 3B, line 7).
UBE2W is functionally associated with FANCL in vitro
Because the FANCL PHD domain was previously reported to be ubiquitinated in vitro by the UbcH5 family members (a, b and c) and UBE2T (Machida et al., 2006; Meetei et al., 2003), and because UBE2W was demonstrated to exhibit ubiquitin conju-gating activity, we next tested the catalytic ability of UBE2W to mediate PHD auto-ubiquitination in vitro. For these assays, the purified PHD and PHD-L incubated with either UBE2W or posi-tive control UbcH2 were analyzed by anti-Ub antibody as well as silver staining. Interestingly, we could not find any ubiquitination of PHD by UBE2W in our system, comparing to the positive control UbcH2 (Fig. 4A). However, after further deletion of the N-terminal linker, the PHD-L was mono-ubiquitinated by UBE2W as well as by UbcH2, suggesting a more important function of the linker (275-305aa) in ubiquitination mediated by UBE2W (Fig. 4B).
Fig. 4. UBE2W is functionally associated with FANCL in vitro (A, B). The PHD-L of FANCL, but not the PHD, is monoubiquitinated by UBE2W in vitro. The purified GST-PHD and GST-PHD-L were incubated with UBE2W or positive control UbcH2. For negative controls, each component (except ATP) was omitted from the reaction, as indicated. The reactions were analyzed by silver stain (upper panel), or Western blot with GST antibody (middle panel) or ubiquitin anti-body (lower panel). GST-PHD or GST-PHD-L and their ubiquitinated species are labeled with arrows. (A) The GST-PHD could not be ubiquitinated by UBE2W in vitro, comparing with the positive control UbcH2. (B) The GST-PHD-L, lacking the N-terminal linker of PHD, was monoubiquitinated by UBE2W as well as by UbcH2. (C) FANCL stimulates the ubiquitin release from UBE2W. The components of ubiquitination reaction, E1 activating enzyme, UBE2W, and ubiquitin with equal amount, were incu-bated with or without E3 ligase FANCL. The reaction products were separated and analyzed by Western blot with ubiquitin antibody (left panel) or GST anti-body (right panel). Ubiquitinated UBE2W and ubiquitin were labeled with arrows. (D) The N-terminal linker of FANCL PHD shares highly conserved sequence among different species. The colored and shaded regions represent conserved residues.
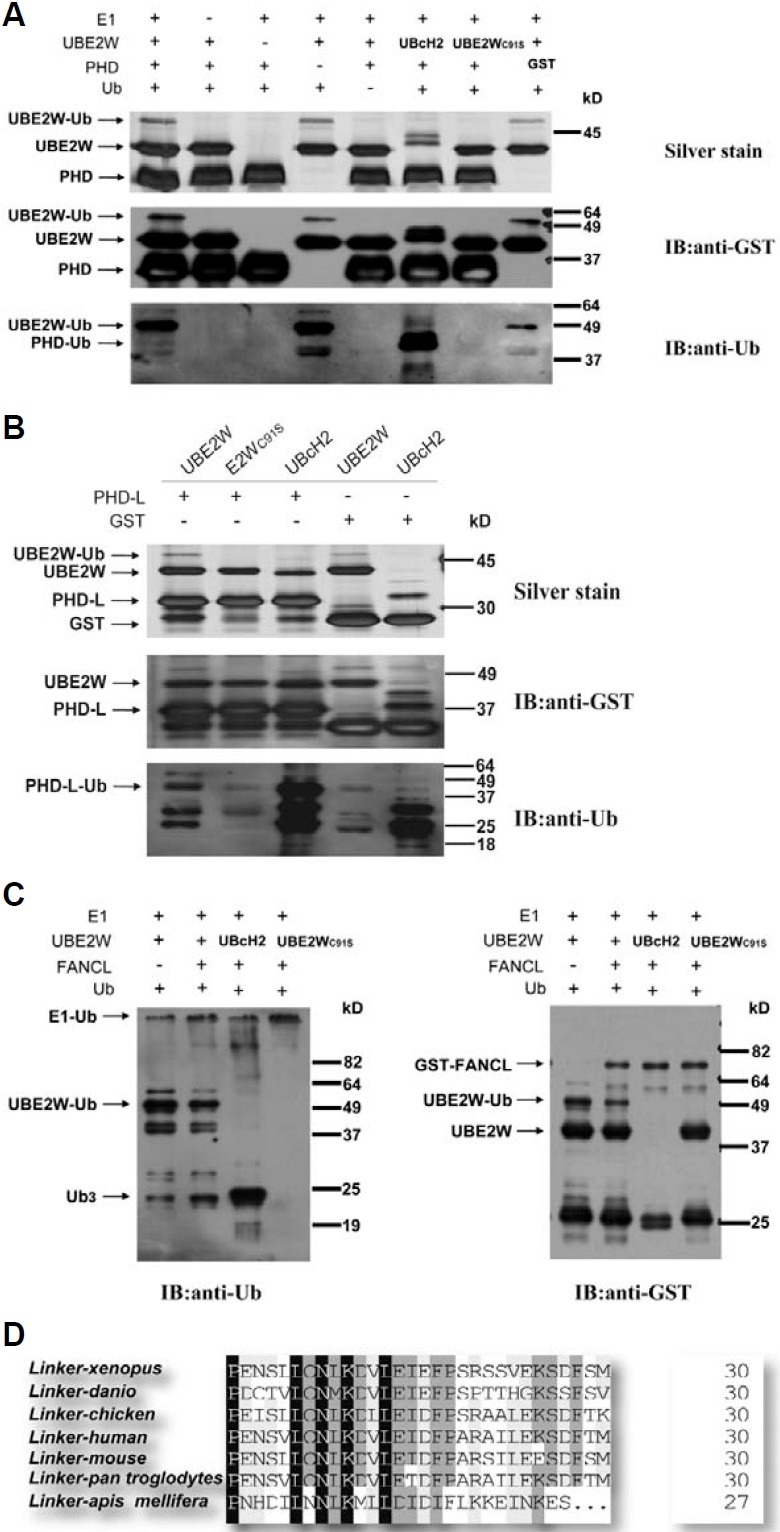
RING containing E3s could act as a scaffold to promote ubiquitin transfer from E2s to substrates, and stimulate the release of ubiquitin from the active-site cysteine residue of E2s in the absence of substrates (Ozkan et al., 2005; Robinson and Ardley, 2004). Moreover, the PHD fingers are usually catego-rized as RING-finger variants (Coscoy and Ganem, 2003; Gurtan et al., 2006). Thus, the PHD finger containing E3 ligase, such as FANCL, might also affect the poly-ubiquitin chain release and formation. Indeed, we found that FANCL could obviously stimulate the ubiquitin release from UBE2W and then form more poly-ubiquitin chain in vitro (Fig. 4C). These data demon-strated that the ubiquitin conjugating activity of UBE2W was functionally associated with FANCL in vitro.
UBE2W regulates UV but not MMC induced FANCD2 monoubiquitination
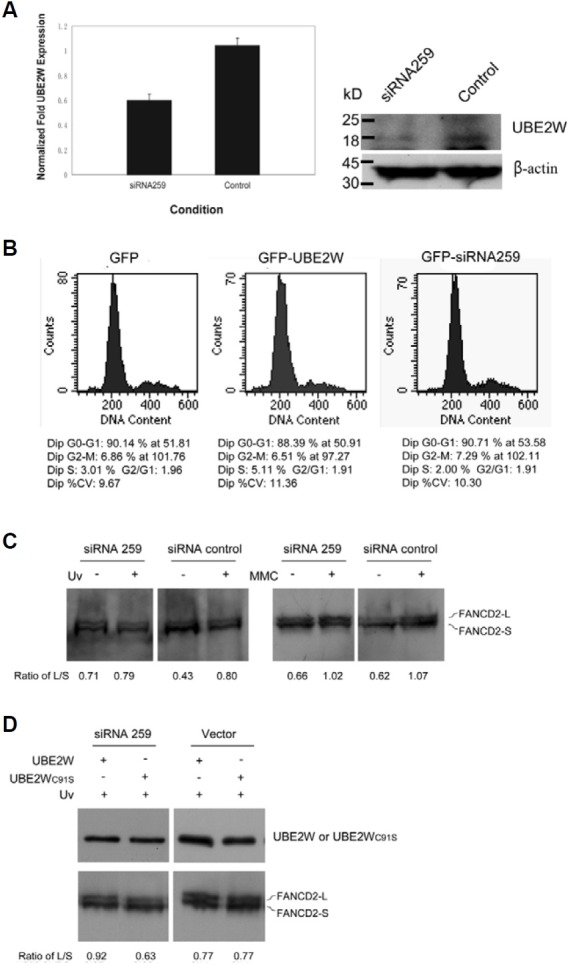
UBE2T had recently been reported to interact with FANCL, and thereby directly regulate DNA damage especially MMC induced FANCD2 monoubiquitination (Alpi et al., 2008; Machida et al., 2006). As evidence presented here showing that UBE2W was an E2 ubiquitin-conjugating enzyme interacting with FANCL, we next determined whether UBE2W might also regulate the DNA damage induced FANCD2 monoubiquitination. We first tested whether decreased UBE2W expression levels might indeed alter the ability of cells to monoubiquitinate FANCD2 in re-sponse to either the DNA cross-linking reagent MMC or UV irradiation. Quantity real-time PCR analysis of human cell total RNA and Western blotting analysis revealed that UBE2W was knocked down efficiently by siRNA256 expressing plasmid targeting UBE2W mRNA (Figs. 5A and 5B), which was con-sistent with our previous report (Zhang et al., 2008b). However, after MMC treatment, the cells expressing UBE2W siRNAshowed no effect on the monoubiquitination of FANCD2, as neither did the control siRNA plasmid (Fig. 5C, right two panels). This phe-notype is completely different from that of cells treated with UBE2T siRNAs. In contrast, after the UV irradiation treatment, down expressed UBE2W had greatly reduced the monoubiquitination of FANCD2 (Fig. 5C, left two panels). More-over, the exogenous murine UBE2W, which was expected to antagonize the effect of hominal UBE2W siRNA, could restore the FANCD2 monoubiquitination induced by UV irradiation. However, the mutant murine UBE2WC91S did not restore the monoubiquitination of FANCD2 (Fig. 5D). Similar results were also obtained in HeLa cells (data not shown). FANCD2 is monoubiquitinated not only following DNA damage, but also during normal S phase (Taniguchi et al., 2002). Thus, any siRNAs or protein expressions that block cell cycle progression have the potential to inhibit FANCD2 monoubiquitination indi-rectly. It is known that down or overexpression of UBE2W does not affect cell cycle progression, especially the S phase population, ruling out this possibility in our study (Fig. 5B, middle and right panels).
Fig. 5. UBE2W is functionally associated with UV induced FANCD2 monoubiquitination in vivo. (A) UBE2W was efficiently knocked down in mammalian cell by RNAi. The siRNA259 targeting UBE2W was generated from H1-U6 dual promoter RNAi plasmid as previously de-scribed. Total RNA and protein were prepared from 293FT cells transfected with indicated siRNA plasmids for 48 h, and the UBE2W ex-pression level was examined by Quantitative real-time PCR and Western blotting respective-ly. (B) Down and up regulation of UBE2W does not affect cell cycle progression or the S phase checkpoint. The cells expressing GPF, GFP-UBE2W and siRNA259 were labeled with Propidium Iodide (PI) and analyzed on a Flow Cytometer. Results indicated similar cell cycle profiles for cells transfected with either GFP-UBE2W or siRNA259 targeting UBE2W. (C) UBE2W is required for UV but not MMC in-duced FANCD2 monoubiquitination in vivo. Cells were transfected with indicated siRNA plasmids (36 h) and treated with 1J UV or MMC (120 nM) for 24 h, and lysates were immuno-blotted for FANCD2. Signal intensity of the monoubiquitinated FANCD2 (FANCD2-L) and the unmodified FANCD2 (FANCD2-S) were quantified with Quantity One 1-D Analysis Soft-ware (BioRad Laboratories). The relative level of monoubiquitinated FANCD2 was normalized to unmodified FANCD2 and represented as the ratio of FANCD2-L/FANCD2-S showing at the bottom. (D) Restoration of FANCD2 monoubiq-uitination in UBE2W-knocked down cells by ex-ogenous murine UBE2W. Overexpression of murine UBE2W or UBE2WC91S was used to antagonize the effect of hominal UBE2W siRNA (upper panel). The empty vector was used as a vector control. The cells were transfected with indicated siRNA plasmids (36 h) and treated with 1J UV where indicated. The FANCD2 were detected by Western blotting (lower panel). The relative level of monoubiquitinated FANCD2 (FANCD2-L) was normalized to unmodified FANCD2 (FANCD2-S) and represented as the ratio of FANCD2-L/FANCD2-S showing at the bottom.
DISCUSSION
It has been well established that monoubiquitination of FANCD2 constitutes a key step in the FA-BRCA DNA repair pathway (Garcia-Higuera et al., 2001). Monoubiquitination of FANCD2 can be induced by various genotoxic stresses including UV light, ionizing radiation, hydroxyurea, and cross-linking agents (MMC, CDDP, DEB). Genetic studies have indicated that ubiquitin conjugating enzyme UBE2T and HHR6 could regulate FANCD2 monoubiquitination through distinct mechanisms (Machida et al., 2006; Zhang et al., 2008a). However, the exact regulation mechanisms of FANCD2 monoubiquitination in response to different DNA damages remain unclear. In this study, we demonstrated that another ubiquitin conjugating enzyme UBE2W also interacts with FANCL and regulates FANCD2 monoubi-quitination in response to UV light exposure.
UBE2W is known to interact with RING finger of BRCA1-BARD1 heterodimer ubiquitin ligase and catalyze BRCA1 monoubiquitination (Christensen et al., 2007). And recently, UBE2W was further described as one of the E2 partners of FANCL and it stimulated FANCD2 monoubiquitination in vitro (Alpi et al., 2008). However, little biological function of UBE2W in vivo is characterized until now. In our in vitro and in vivo pro-tein-protein interaction systems, we observed specific associa-tions of UBE2W and full-length E3 ligase FANCL, consistent with the previous report (Alpi et al., 2008). Interestingly, we also found that FANCL PHD domain was necessary, but not suffi-cient, for the interaction with UBE2W in yeast cells. However, protein interaction assay performed in mammalian cells indicat-ed that FANCL PHD domain was both necessary and sufficient for the interaction with UBE2W. One likely reason could be that there might be additional proteins in mammalian cells stabilizing the interaction of PHD domain and UBE2W, because no homo-logues of FANCL and UBE2W were found in S. cerevisiae and thus they probably did not have functional partners in yeast. Indeed, GST-pull down assay performed in the previous pub-lished study also demonstrated that neither the isolated PHD nor N-terminal WD40 domain expressed in prokaryotic cells was sufficient for the interaction with UBE2W in vitro (Alpi et al., 2008). We found that the interaction of UBE2W with full length FANCL was exclusively confined in the nucleus, and UBE2W formed a homodimer mainly in the nucleus, suggesting that the common biological function of UBE2W and FANCL requires nuclear localization. Moreover, we found that UBE2W could form more stable complex with W3P3 (FANCL 264-375aa) and PHD-L (FANCL 306-375aa) than with W3P2 (FANCL 254-375aa) and PHD (FANCL 276-375aa), whose cellular distribution spread in the whole cell, further indicating the complexity of the FANCL-UBE2W interaction in mammalian cells.
UBE2W has been predicted to be a class I ubiquitin conjugat-ing enzyme because it consists of a ~150aa catalytic UBC-homology domain without additional N- or C-terminal extension (Christensen et al., 2007; Zhang et al., 2008c). We demonstrat-ed that UBE2W indeed had E2 activity and its activity was de-pendent on the conserved cysteine residue C91 in our in vitro ubiquitination assay. Typical class I E2 enzymes such as S. cerevisiae UBC4 and UBC5 were reported to be essential for the degradation of short-lived and abnormal proteins (Seufert and Jentsch, 1990). Interestingly, UBE2W could also directly catalyze multiubiquitination in the absence of an E3 ubiquitin ligase in vitro, similar to the positive control UbcH2. UbcH2 was reported to directly interact with its substrates and conjugate ubiquitin to histone H2A in an E3 independent manner (Kaiser et al., 1995). We do not know whether UBE2W could catalyze such multiubiquitination in vivo and mediate the degradation of proteins in mammalian cells. However, UBE2W could catalyze the monoubiquitination of RING finger of BRAC1 as reported in a previous study (Christensen et al., 2007) and the monoubiquitination of the PHD-L domain of FANCL in this study. We also noted that the PHD protein, which has an additional N-terminal linker (275-305aa) in PHD-L, could not be monoubiquitinated by UBE2W. Moreover, this additional N-terminal linker was highly conserved in various species as shown by sequence alignment (Fig. 4D) and able to attenuate the interaction of UBE2W and PHD (Fig. 2C, upper right panel and lower left panel). Similarly, the linker of E3 ubiquitin ligase c-Cbl, which connected the TBK and RING domain, also played a crucial role in c-Cbl and E2 UbcH7 interaction (Zheng et al., 2000). Alternatively, the linker connecting WD40 and PHD do-main may play important roles in the precise relative arrange-ment of UBE2W and FANCL, and thus mediating the monoubiquitination of substrates.
FANCD2 monoubiquitination reached the highest level 24 h after MMC treatment, but only 8 h after UV irradiation (Garcia-Higuera et al., 2001), suggesting distinct roles of MMC and UV in FANCD2 monoubiquitination. It is well known that UBE2T mediates FANCD2 monoubiquitination in response to MMC treatment (Alpi et al., 2007; 2008; Longerich et al., 2009; Machida et al., 2006). However, our data presented here demonstrated that down-regulation of UBE2W could markedly reduce the UV irradiation but not MMC induced FANCD2 monoubiquitination. MMC is an interstrand DNA cross-linking agent (Talwar et al., 2004), while UV irradiation mainly causes the formation of intrastrand cyclobutane-pyrimidine dimmers (CPDs) (de Lima-Bessa et al., 2008). And the DNA repair path-ways induced by them are also different. For example, cells use homologous DNA repairs (HDR) to eliminate DNA cross-linking damages (Nakanishi et al., 2005) but use excision repair to eliminate CPDs (de Lima-Bessa et al., 2008). It seems that UBE2W and UBE2T interact with FANCL to mediate FANCD2 monoubiquitination in response to different DNA damages. We do not know how UBE2W responds to UV irradiation and stimu-lates FANCD2 monoubiquitination. However, combining our data presented here and previously published data, we con-cluded that FANCD2 monoubiquitination might be regulated by different ubiquitin-related enzymes, which probably include UBE2T and UBE2W, in response to different types of DNA damage agents. FANCD2 monoubiquitination provides an addi-tional regulatory step in the activation of the FA pathway.
Acknowledgments
We thank Dr. Baisong Lu for the plasmids of UBE2W and FANCL (pCMV-myc/UBE2W and pCMV-HA/FANCL) and initiate the project and Dr. Tom Kerppola for the plasmids of BiFC assay system. We also thank members of Laboratory of Protein Engineering for helpful discussion. This work was supported by a grant from the National Natural Sciences Foundation of China (No. 30470379).
References
- 1.Alpi A., Langevin F., Mosedale G., Machida Y.J., Dutta A., Patel K.J. UBE2T, the Fanconi anemia core complex, and FANCD2 are recruited independently to chromatin: a basis for the regulation of FANCD2 monoubiquitination. Mol. Cell. Biol. (2007);27:8421–8430. doi: 10.1128/MCB.00504-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alpi A.F., Pace P.E., Babu M.M., Patel K.J. Mechanistic insight into site-restricted monoubiquitination of FANCD2 by Ube2t, FANCL, and FANCI. Mol. Cell. (2008);32:767–777. doi: 10.1016/j.molcel.2008.12.003. [DOI] [PubMed] [Google Scholar]
- 3.Auerbach A.D. A test for Fanconi’s anemia. Blood. (1988);72:366–367. [PubMed] [Google Scholar]
- 4.Bagby G.C. Jr. Genetic basis of Fanconi anemia. Curr. Opin. Hematol. (2003);10:68–76. doi: 10.1097/00062752-200301000-00011. [DOI] [PubMed] [Google Scholar]
- 5.Chen Z.J. Ubiquitin signalling in the NF-kappaB pathway. Nat. Cell Biol. (2005);7:758–765. doi: 10.1038/ncb0805-758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Christensen D.E., Brzovic P.S., Klevit R.E. E2-BRCA1 RING interactions dictate synthesis of mono- or specific polyubiquitin chain linkages. Nat. Struct. Mol. Biol. (2007);14:941–948. doi: 10.1038/nsmb1295. [DOI] [PubMed] [Google Scholar]
- 7.Cook W.J., Jeffrey L.C., Sullivan M.L., Vierstra R.D. Three-dimensional structure of a ubiquitin-conjugating enzyme (E2). J. Biol. Chem. (1992);267:15116–15121. doi: 10.2210/pdb1aak/pdb. [DOI] [PubMed] [Google Scholar]
- 8.Cook W.J., Jeffrey L.C., Xu Y., Chau V. Tertiary structures of class I ubiquitin-conjugating enzymes are highly conserved: crystal structure of yeast Ubc4. Biochemistry. (1993);32:13809–13817. doi: 10.1021/bi00213a009. [DOI] [PubMed] [Google Scholar]
- 9.Coscoy L., Ganem D. PHD domains and E3 ubiquitin ligases: viruses make the connection. Trends Cell Biol. (2003);13:7–12. doi: 10.1016/s0962-8924(02)00005-3. [DOI] [PubMed] [Google Scholar]
- 10.D’Andrea A.D., Grompe M. The Fanconi anaemia/BRCA pathway. Nat. Rev. Cancer. (2003);3:23–34. doi: 10.1038/nrc970. [DOI] [PubMed] [Google Scholar]
- 11.de Lima-Bessa K.M., Armelini M.G., Chigancas V., Jacysyn J.F., Amarante-Mendes G.P., Sarasin A., Menck C.F. CPDs and 6-4PPs play different roles in UV-induced cell death in nor-mal and NER-deficient human cells. DNA Repair. (2008);7:303–312. doi: 10.1016/j.dnarep.2007.11.003. [DOI] [PubMed] [Google Scholar]
- 12.Dodd R.B., Allen M.D., Brown S.E., Sanderson C.M., Duncan L.M., Lehner P.J., Bycroft M., Read R.J. Solution structure of the Kaposi’s sarcoma-associated herpesvirus K3 N-terminal domain reveals a Novel E2-binding C4HC3-type RING domain. J. Biol. Chem. (2004);279:53840–53847. doi: 10.1074/jbc.M409662200. [DOI] [PubMed] [Google Scholar]
- 13.Dorsman J.C., Levitus M., Rockx D., Rooimans M.A., Oostra A.B., Haitjema A., Bakker S.T., Steltenpool J., Schuler D., Mohan S., et al. Identification of the Fanconi anemia complementation group I gene, FANCI. Cell Oncol. (2007);29:211–218. doi: 10.1155/2007/151968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Garcia-Higuera I., Taniguchi T., Ganesan S., Meyn M.S., Timmers C., Hejna J., Grompe M., D’Andrea A.D. Interaction of the Fanconi anemia proteins and BRCA1 in a common pathway. Mol. Cell. (2001);7:249–262. doi: 10.1016/s1097-2765(01)00173-3. [DOI] [PubMed] [Google Scholar]
- 15.German J., Schonberg S., Caskie S., Warburton D., Falk C., Ray J.H. A test for Fanconi’s anemia. Blood. (1987);69:1637–1641. [PubMed] [Google Scholar]
- 16.Grompe M., van de Vrugt H. The Fanconi family adds a fraternal twin. Dev. Cell. (2007);12:661–662. doi: 10.1016/j.devcel.2007.04.008. [DOI] [PubMed] [Google Scholar]
- 17.Gurtan A.M., Stuckert P., D’Andrea A.D. The WD40 repeats of FANCL are required for Fanconi anemia core complex assembly. J. Biol. Chem. (2006);281:10896–10905. doi: 10.1074/jbc.M511411200. [DOI] [PubMed] [Google Scholar]
- 18.Hershko A., Ciechanover A., Varshavsky A. Basic Medical Research Award. The ubiquitin system. Nat. Med. (2000);6:1073–1081. doi: 10.1038/80384. [DOI] [PubMed] [Google Scholar]
- 19.Hu C.D., Kerppola T.K. Simultaneous visualization of multiple protein interactions in living cells using multicolor fluorescence complementation analysis. Nat. Biotechnol. (2003);21:539–545. doi: 10.1038/nbt816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hu C.D., Chinenov Y., Kerppola T.K. Visualization of interactions among bZIP and Rel family proteins in living cells using bimolecular fluorescence complementation. Mol. Cell. (2002);9:789–798. doi: 10.1016/s1097-2765(02)00496-3. [DOI] [PubMed] [Google Scholar]
- 21.Huang T.T., D’Andrea A.D. Regulation of DNA repair by ubiquitylation. Nat. Rev. (2006);7:323–334. doi: 10.1038/nrm1908. [DOI] [PubMed] [Google Scholar]
- 22.Ishiai M., Kitao H., Smogorzewska A., Tomida J., Kinomura A., Uchida E., Saberi A., Kinoshita E., Kinoshita-Kikuta E., Koike T., et al. FANCI phosphorylation functions as a molecular switch to turn on the Fanconi anemia pathway. Nat. Struct. Mol. Biol. (2008);15:1138–1146. doi: 10.1038/nsmb.1504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kaiser P., Mandl S., Schweiger M., Schneider R. Characterization of functionally independent domains in the hu-man ubiquitin conjugating enzyme UbcH2. FEBS Lett. (1995);377:193–196. doi: 10.1016/0014-5793(95)01323-7. [DOI] [PubMed] [Google Scholar]
- 24.Longerich S., San Filippo J., Liu D., Sung P. Fanci binds branched DNA and is mono-ubiquitinated by UBE2T-FANCL. J. Biol. Chem. (2009);284:23182–23186. doi: 10.1074/jbc.C109.038075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lu B., Bishop C.E. Mouse GGN1 and GGN3, two germ cell-specific proteins from the single gene Ggn, interact with mouse POG and play a role in spermatogenesis. J. Biol. Chem. (2003);278:16289–16296. doi: 10.1074/jbc.M211023200. [DOI] [PubMed] [Google Scholar]
- 26.Machida Y.J., Machida Y., Chen Y., Gurtan A.M., Kupfer G.M., D’Andrea A.D., Dutta A. UBE2T is the E2 in the Fanconi anemia pathway and undergoes negative autoregulation. Mol. Cell. (2006);23:589–596. doi: 10.1016/j.molcel.2006.06.024. [DOI] [PubMed] [Google Scholar]
- 27.Meetei A.R., de Winter J.P., Medhurst A.L., Wallisch M., Waisfisz Q., van de Vrugt H.J., Oostra A.B., Yan Z., Ling C., Bishop C.E., et al. A novel ubiquitin ligase is deficient in Fanconi anemia. Nat. Genet. (2003);35:165–170. doi: 10.1038/ng1241. [DOI] [PubMed] [Google Scholar]
- 28.Meetei A.R., Levitus M., Xue Y., Medhurst A.L., Zwaan M., Ling C., Rooimans M.A., Bier P., Hoatlin M., Pals G., et al. X-linked inheritance of Fanconi anemia complementation group B. Nat. Genet. (2004);36:1219–1224. doi: 10.1038/ng1458. [DOI] [PubMed] [Google Scholar]
- 29.Meijer G.A. The 13th Fanconi anemia gene identified: FANCI-importance of the ‘Fanconi anemia pathway’ for cellular oncology. Cell Oncol. (2007);29:181–182. doi: 10.1155/2007/871608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nakanishi K., Yang Y.G., Pierce A.J., Taniguchi T., Digweed M., D'Andrea A.D., Wang Z.Q., Jasin M. Human Fanconi anemia monoubiquitination pathway promotes homolo-gous DNA repair. Proc. Natl. Acad. Sci. USA. (2005);102:1110–1115. doi: 10.1073/pnas.0407796102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ozkan E., Yu H., Deisenhofer J. Mechanistic insight into the allosteric activation of a ubiquitin-conjugating enzyme by RING-type ubiquitin ligases. Proc. Natl. Acad. Sci. USA. (2005);102:18890–18895. doi: 10.1073/pnas.0509418102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Plans V., Scheper J., Soler M., Loukili N., Okano Y., Thom-son T.M. The RING finger protein RNF8 recruits UBC13 for lysine 63-based self polyubiquitylation. Journal of cellular biochemistry. (2006);97:572–582. doi: 10.1002/jcb.20587. [DOI] [PubMed] [Google Scholar]
- 33.Poll E.H., Arwert F., Joenje H., Wanamarta A.H. Dif-ferential sensitivity of Fanconi anaemia lymphocytes to the clastogenic action of cis-diamminedichloroplatinum (II) and trans-diamminedichloroplatinum (II). Hum. Genet. (1985);71:206–210. doi: 10.1007/BF00284574. [DOI] [PubMed] [Google Scholar]
- 34.Robinson P.A., Ardley H.C. Ubiquitin-protein ligases. J. Cell Sci. (2004);117:5191–5194. doi: 10.1242/jcs.01539. [DOI] [PubMed] [Google Scholar]
- 35.Seufert W., Jentsch S. Ubiquitin-conjugating enzymes UBC4 and UBC5 mediate selective degradation of short-lived and abnormal proteins. EMBO. J. (1990);9:543–550. doi: 10.1002/j.1460-2075.1990.tb08141.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sims A.E., Spiteri E., Sims R.J. 3rd, Arita A.G., Lach F.P., Landers T., Wurm M., Freund M., Neveling K., Hanenberg H., et al. FANCI is a second monoubiquitinated member of the Fanconi anemia pathway. Nat. Struct. Mol. Biol. (2007);14:564–567. doi: 10.1038/nsmb1252. [DOI] [PubMed] [Google Scholar]
- 37.Smogorzewska A., Matsuoka S., Vinciguerra P., McDonald E.R. 3rd, Hurov K.E., Luo J., Ballif B.A., Gygi S.P., Hofmann K., D’Andrea A.D., et al. Identification of the FANCI protein, a monoubiquitinated FANCD2 paralog required for DNA repair. Cell. (2007);129:289–301. doi: 10.1016/j.cell.2007.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Talwar R., Choudhry V.P., Kucheria K. Differentiation of Fanconi anemia from idiopathic aplastic anemia by induced chromosomal breakage study using mitomycin-C (MMC). Indian Pediatr. (2004);41:473–477. [PubMed] [Google Scholar]
- 39.Taniguchi T., D’Andrea A.D. Molecular pathogenesis of Fanconi anemia: recent progress. Blood. (2006);107:4223–4233. doi: 10.1182/blood-2005-10-4240. [DOI] [PubMed] [Google Scholar]
- 40.Taniguchi T., Garcia-Higuera I., Andreassen P.R., Gregory R.C., Grompe M., D’Andrea A.D. S-phase-specific inter-action of the Fanconi anemia protein, FANCD2, with BRCA1 and RAD51. Blood. (2002);100:2414–2420. doi: 10.1182/blood-2002-01-0278. [DOI] [PubMed] [Google Scholar]
- 41.Yin G., Ji C., Wu T., Shen Z., Xu X., Xie Y., Mao Y. Cloning, characterization and subcellular localization of a gene encoding a human ubiquitin-conjugating enzyme (E2) homolo-gous to the Arabidopsis thaliana UBC-16 gene product. Front Biosci. (2006);11:1500–1507. doi: 10.2741/1899. [DOI] [PubMed] [Google Scholar]
- 42.Yuan F., El Hokayem J., Zhou W., Zhang Y. FANCI protein binds to DNA and interacts with FANCD2 to recognize branched structures. J. Biol. Chem. (2009);284:24443–24452. doi: 10.1074/jbc.M109.016006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhang M., Windheim M., Roe S.M., Peggie M., Cohen P., Prodromou C., Pearl L.H. Chaperoned ubiquitylation--crystal structures of the CHIP U box E3 ubiquitin ligase and a CHIP-Ubc13-Uev1a complex. Mol. Cell. (2005);20:525–538. doi: 10.1016/j.molcel.2005.09.023. [DOI] [PubMed] [Google Scholar]
- 44.Zhang J., Zhao D., Wang H., Lin C.J., Fei P. FANCD2 monoubiquitination provides a link between the HHR6 and FA-BRCA pathways. Cell Cycle. (2008a);7:407–413. doi: 10.4161/cc.7.3.5156. [DOI] [PubMed] [Google Scholar]
- 45.Zhang Y., Li C., Yang Z., Xu L., Zhu H., Zhou X., Huang P. Selecting functional siRNA target sites of hUBE2W based on H1-U6 dual promoter RNAi plasmid. Sheng wu gong cheng xue bao = Chin. J. Biotechnol. (2008b);24:1975–1980. [PubMed] [Google Scholar]
- 46.Zhang Y., Zhu H., Zhao L., Zhou X., Huang P. Gene- ration of mouse UBE2W antibody and analysis of UBE2W expression in mouse tissues. Chin. J. Biotechnol. (2008c);24:547–552. [PubMed] [Google Scholar]
- 47.Zheng N., Wang P., Jeffrey P.D., Pavletich N.P. Struc-ture of a c-Cbl-UbcH7 complex: RING domain function in ubiquitin-protein ligases. Cell. (2000);102:533–539. doi: 10.1016/s0092-8674(00)00057-x. [DOI] [PubMed] [Google Scholar]


