Abstract
Ubiquitination is a unique protein degradation system utilized by eukaryotes to efficiently degrade detrimental cellular proteins and control the entire pool of regulatory components. In plants, adaptation in response to various abiotic stresses can be achieved through ubiquitination and the resulting degradation of components specific to these stress signalings. Arabidopsis has more than 1,400 E3 enzymes, indicating E3 ligase acts as a main determinant of substrate specificity. However, as only a minority of E3 ligases related to abiotic stress signaling have been studied in Arabidopsis, the further elucidation of the biological roles and related substrates of newly identified E3 ligases is essential in order to clarify the functional relationship between abiotic stress and E3 ligases. Here, we review the current knowledge and future prospects of the regulatory mechanism and role of several E3 ligases involved in abiotic stress signal transduction in Arabidopsis. As another potential approach to understand how ubiquitination is involved in such signaling, we also briefly introduce factors that regulate the activity of cullin in multi-subunit E3 ligase complexes.
Keywords: 26S proteasome, abiotic stress signal transduction, Arabidopsis, E3 ubiquitin ligase, ubiquitination
UBIQUITINATION AND E3 LIGASES
Ubiquitination is a post-translational modification of proteins that plays an important role in several processes in plants such as biotic and abiotic stress responses, cell cycle progression and hormone response. Ubiquitin, so named because of its ubiquitous presence in all eukaryotic species, is a highly conserved 8kDa protein. This small protein attaches to lysine residues on the target protein by three successive enzymes; Ub-activating enzyme (E1), Ub-conjugating enzyme (E2), and Ub-ligase (E3) (Kraft et al., 2005; Mukhopadhyay and Riezman, 2007; Smalle and Viestra, 2004; Stone et al., 2005) (Fig. 1A). The target protein can be tagged with a single ubiquitin molecule (monoubiquitination), multiple ubiquitin molecules (multiubiquitination), or a ubiquitin chain (polyubiquitination). In general, monoubiqui-tination and multiubiquitination function in the regulation of protein activity, protein-protein interaction, and subcellular localization. In contrast, polyubiquitination, the most frequently found type of ubiquitination, mainly triggers the transfer of the ubiquityinated target protein to the 26S proteasome complex, resulting in further degradation (Mukhopadhyay and Riezman, 2007; Pickart and Eddins, 2004).
Fig. 1. Ubiquitination process and E3 ligases. (A) Overview of the polyubiquitination process. Ubiquitination requires the sequential action of three enzymes; ubiquitin-activating enzyme (E1), ubiquitin-conjugating enzyme (E2) and ubiquitin ligase (E3). Ubiquitinated substrates are targeted to the 26S proteasome for subsequent degradation. (B) Single subunit and multi-subunit E3 ligases. Based on the E2-interacting region, single subunit E3 ligases can be largely categorized into two types; HECT-type and RING/U-box E3 ligases. Cullin-based E3 ligases are composed of several components; cullin, RBX1, adaptor, and substrate receptor. The resulting complex acts as an E3 ligase in the ubiquitination of substrates. (C) Several types of cullin-based E3 ligases in plants. Cullin 1 assembles into the SCF complex with ASK as an adaptor and F-box protein as a substrate receptor. As cullin 2 was reported to interact with F-box proteins, cullin 2 may be another subunit of the SCF complex (Risseeuw et al., 2003). In the case of cullin 4 complex, DDB1 and DWD proteins are utilized as adaptors and substrate receptors, respectively. Unlike cullin 1 and cullin 4, cullin 3 only requires BTB-containing proteins as substrate receptors without additional adaptor proteins.
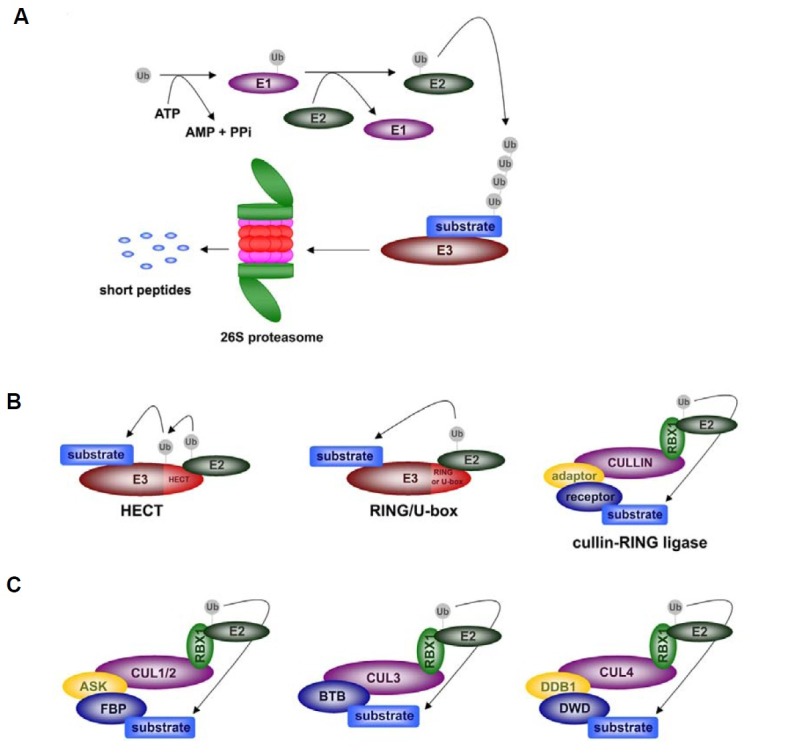
Arabidopsis has 2 E1, at least 37 E2, and more than 1,400 E3 enzymes (Downes et al., 2003; Dreher and Callis, 2007; Gagne et al., 2002; Gingerich et al., 2005; Hatfield et al., 1997; Kraft et al., 2005; Vierstra, 2009). The large diversity of E3 ligases suggests that E3 is important in determining substrate specificity, that is, identifying which protein will be ubiquitinated. E3 ligases can be largely divided into two groups based on their structure: single-subunit E3 ligases and multi-subunit E3 ligases.
Single-subunit E3 ligases
Single-subunit E3 ligases are further categorized into distinct families based on the presence of specific domains, such as homology to E6AP C terminus (HECT), U-box, and really interesting new gene (RING) domains (Fig. 1B).
HECT E3 ligases contain a conserved C-terminal HECT domain that was first detected in human E6AP (Huibregtse et al., 1995). While other single-subunit E3 ligases are indirectly involved in the transfer of ubiquitin from E2 to the target protein, HECT E3 ligases generate an intermediate form of Ub-E3 through a cysteine residue that pre-exists in the HECT domain. This intermediate form acts as a proximal ubiquitin donor for the ligation of ubiquitin to the substrate protein. Although approxi-mately 7 HECT E3 ligases have been identified in Arabidopsis (UPL1-7), there are only a few reports on their roles in plant signalings so far. For instance, UPL3 has been suggested to be involved in trichome development and UPL5 was shown to play a negative role in leaf senescence through degradation of the transcription factor WRKY53 (Downes et al., 2003; Miao and Zentgraf, 2010; Vierstra, 2003).
RING finger-containing proteins are the most abundant group among the single-subunit ligases. It is estimated that more than 460 RING proteins exist in the Arabidopsis genome. This group is defined by the presence of a RING finger motif, a zinc-binding motif composed of approximately 70 amino acids. Most RING motifs coordinate two zinc ions so that it forms a platform that can bind E2 (Freemont et al., 1991). RING finger-contain-ing proteins also function as a component of the cullin-based multi-subunit E3 ligase complex (Santner and Estelle, 2010).
U-box domain is composed of 75 amino acids (Hatakeyama et al., 2001) and is structurally related to RING domain. But unlike RING, zinc is not required for the stabilization of the U-box structure (Aravind and Koonin, 2000). Similar to RING domain-containing proteins, U-box proteins participate in several events in the life of a plant such as hormone and biotic/abiotic stress signalings (Yee and Goring, 2009). There are 64 U-box genes in the Arabidopsis genome, a significantly larger number than that in human or yeast (21 and 2, respectively). As other single subunit E3 ligases share a similar number of gene families among eukaryotes, the diversity of plant U-box genes suggests that the U-box domain is important in determining substrate specificity for various cellular processes in plants (Aravind and Koonin, 2000; Koegl et al., 1999; Ohi et al., 2003; Wiborg et al., 2008).
Multi-subunit E3 ligases: cullin-based E3 ligases
Among the multi-subunit E3 ligases, cullin-based E3 ligases have been best characterized to date. In Arabidopsis, five cullins, namely, cullin 1, cullin 2, cullin 3a, cullin 3b, and cullin 4 have been identified as components of the cullin-based E3 ligases. Cullin proteins serve as a scaffold that provides a framework for assembling the ubiquitination machinery. In addition to cullin proteins, two other modules are required for the complex: the RING finger protein, RBX1, which recruits E2, and the substrate-recognition module that contains the adaptor and substrate receptor. After all proteins are assembled, the complex acts as an E3 ligase for subsequent target modification (Petroski and Deshaies, 2005; Schwechheimer and Calderon Villalobos, 2004; Thomann et al., 2005) (Fig. 1B).
At least three types of cullin-based E3 ligases exist in plants (Fig. 1C). In the well-known Skp1-Cullin-F-box (SCF) complex, cullin 1 directly interacts with Arabidopsis-SKP1-like (ASK), the counterpart of SKP1 in animals, and ASK serves as an adaptor protein that connects cullin 1 and the F-box protein. There are more than 700 different F-box proteins in Arabidopsis. As F-box proteins act as substrate receptors in the SCF complex, such diversity ensures that a specific target protein is recruited to the complex (Cardozo and Pagano, 2004; Cope and Deshaies, 2006; Schulman et al., 2000; Zheng et al., 2002). In Arabidopsis, it has been reported that cullin 2 also interacts with F-box proteins, suggesting cullin 2 could also be a subunit of the SCF complex (Risseeuw et al., 2003).
The function and expression pattern of cullin 3a and cullin 3b largely overlap with each other and both proteins are essential for embryo development in Arabidopsis (Figueroa et al., 2005). Cullin 3a and cullin 3b can assemble into E3 ligase complexes in the absence of adaptor proteins because BTB/POZ-domain proteins directly associate with CUL3 and also act as receptors for substrate recognition (Gingerich et al., 2005; Pintard et al., 2004). It appears that Arabidopsis possesses at least 37 BTB/POZ-domain proteins (from Protein Families Database of Alignments, PFAM) (Bateman et al., 2004).
Cullin 4 utilizes damaged DNA binding 1 (DDB1) protein as an adaptor to assemble the E3 ligase complex. Arabidopsis has two closely related forms of DDB1; DDB1a and DDB1b. The expression pattern of DDB1a and DDB1b overlap and the protein sequences of the two proteins are highly similar (91% amino acid identity) (Schroeder et al., 2002). Nevertheless, the fact that ddb1b mutants are lethal while ddb1a does not show any distinct phenotype indicates each protein also has a unique function during plant development (Bernhardt et al., 2010). As a substrate receptor for the cullin 4 E3 ligase complex, DWD domain-containing proteins have been identified (Angers et al., 2006; He et al., 2006; Higa et al., 2006; Jin et al., 2006; Lee et al., 2008). DWD proteins contain WD40 repeats that, in turn, share a conserved motif, the DWD box, that is usually com-posed of 16 amino acids. In general, the 16th arginine (or lysine) of the DWD domain acts as the major determinant for the direct interaction with CUL4-DDB1 (Angers et al., 2006; He et al., 2006; Jin et al., 2006). There have been 85 and 78 DWD proteins reported in Arabidopsis and rice, respectively (Lee et al., 2008). In some cases, the WDxR motif in the WD40 region rather than the entire DWD box is sufficient to bind DDB1. This suggests that an additional 34 Arabidopsis proteins and 32 rice proteins that contain the WDxR motif may also be potential substrate receptors for the E3 complex (Zhang et al., 2008a). Interestingly, besides the typical CUL4-DDB1-DWD (or WDxR) complex, Arabidopsis CUL4 is able to associate with a different complex named CDD (COP10-DET1- (COP10-DET1-DDB1) as well, and the resulting CUL4-CDD complex mediates the repression of photomorphogenesis (Bernhardt et al., 2006; Chen et al., 2006).
FUNCTIONAL RELATIONSHIP BETWEEN ABIOTIC STRESS SIGNALING AND E3 LIGASES
Water-deficit stress signaling and the related E3 ligases
Water-deficit stress such as drought and high salinity results in a marked reduction of crop productivity on as much as half of the world's irrigated land (Boyer, 1982; Cushman and Bohnert, 2000). This stress brings about a variety of cellular processes such as stomatal closure and changes in cell wall/membrane composition which, in turn, enable plants to overcome the unfavorable conditions (Bohnert et al., 2006). As shown in Fig. 2A, signal transduction pathways triggered by drought and high salt content share a number of signaling components that transduce the related signalings into downstream processes that endow resistance to such stresses.
Fig. 2. Abiotic stress signalings and the related E3 ligases. (A, B) Schematic representation of water-deficit stress signaling in the presence or absence of ABA (A) and the involvement of E3 ligases in ABA-mediated drought signaling via AREB/ABF/ ABI5/DPBF bZIP subfamily (B). ‘Substrate modification’ indicates either negative regulation by polyubiquitination or positive regulation by monou-biquitination. (C, D) Simplified scheme of cold signaling pathway (C) and the negative regulation of ICE1 by HOS1 E3 ligase (D). The protein stability of ICE1 is negatively regulated by ubiquitination, whereas its activity is positively regulated by phosphorylation and sumoylation. The activated ICE1 binds to MYCRS in the promoter region of CBF3 and then positively regulates the expression of CBF3 and its downstream genes. ‘P’ and ‘S’ indicates phosphate and small ubiquitin-related modifier (SUMO), respectively.
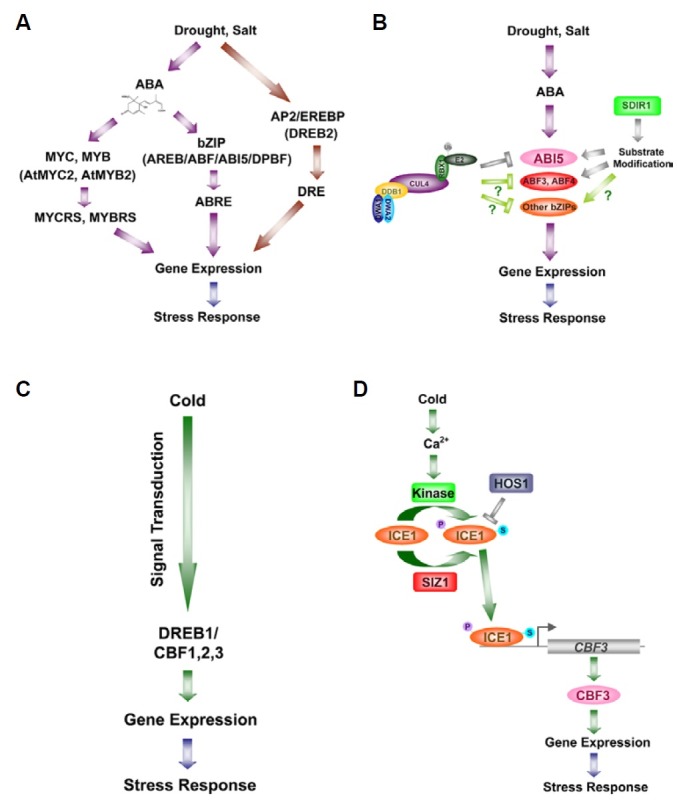
Drought/salt stress signaling can be divided into two types of pathways based on the dependence on the plant hormone, abscisic acid (ABA). A large number of ABA-inducible genes are up-regulated by drought and high-salinity, indicating ABA acts as a positive regulator in such stress signal transduction pathways (Takahashi et al., 2004). The ABA-dependent pathway is mediated by two different systems (Agarwal et al., 2006). First, in the basic leucine zipper (bZIP)/ABA-responsive element (ABRE) system, leucine zipper transcription factors such as ABRE binding protein (AREB), ABRE BINDING FACTOR (ABF), ABA INSENSITIVE 5 (ABI5) and Dc3 promoter binding factor (DPBF) bind to ABRE, a cis-acting element for ABA-responsive genes such as RD29B, AIL1 and RAB18, and transactivate the responding genes (Fujita et al., 2005; Nakashima et al., 2009). Nine types of proteins have been identified as members of the AREB/ABF/ABI5/DPBF bZIP subfamily in Arabidopsis. Among them, AREB1/ABF2, AREB2/ABF4, and ABF3/DPBF5 are inducible in response to dehydration, high-salt treatments, and ABA in vegetative tissues (Fujita et al., 2005). The second system is controlled by MYC/MYB transcription factors. These proteins bind to cis-acting elements such as MYCRS and MYBRS (MYC- and MYB-recognition sequence, respectively) and activate transcription of the drought-respon-sive gene, RD22 (Abe et al., 1997).
The first insight into ABA-independent dehydration stress signaling came from findings that various dehydration stress-responsive genes were upregulated in ABA-deficient (aba) and ABA-insensitive (abi) mutants under drought stress (Shinozaki and Yamaguchi-Shinozaki, 2000). This also led to the discovery of dehydration-responsive element (DRE), a cis-acting element responsible for dehydration signal transduction (Yamaguchi-Shinozaki and Shinozaki, 1994). Although DRE binding 1 (DREB1) and DREB2, transcription factors that belong to the APETALA2 (AP2)/ethylene response factor (ERF) family, both interact with DRE, DREB2 functions as a transcription factor only under drought/salt stresses and DREB1 only under low temperature (Liu et al., 1998).
Several reports have demonstrated that single E3 ligases are involved in ABA-dependent or -independent pathways in drought/ salt stress responses. In Arabidopsis, the RING finger E3 ligase, SALT-AND DROUGHT-INDUCED RING FINGER1 (SDIR1) was identified as a positive regulator in ABA response, as loss of SDIR1 resulted in ABA-insensitive phenotypes. Although SDIR1 is induced by drought and salt stress, and not by ABA, transgenic plants that overexpress SDIR1 display ABA-hyper-sensitive phenotypes such as enhanced ABA-induced stomatal closure and drought tolerance. The heterologous overexpression of Arabidopsis SDIR1 in tobacco and rice also confers enhanced drought tolerance, suggesting the role of SDIR1 in drought-tolerance is conserved across dicotyledons and mono-cotyledons (Zhang et al., 2008b). Since SDIR1 fails to rescue abi5-1, whereas ABI5, ABF3 and ABF4 rescue the ABA-insen-sitive phenotype of sdir1-1, SDIR1 may function upstream of the bZIP proteins in ABA signaling (Fig. 2B) (Zhang et al., 2007).
The U-box containing E3 ligase, AtCHIP, is also involved in ABA-mediated stress response. AtCHIP, named because of its structural resemblance to animal CHIP proteins, was originally reported to play a crucial role in tolerance against low temperature stress (Yan et al., 2003). Interestingly, AtCHIP monoubiq-uitylates PP2A in vitro and the activity of PP2A increases in AtCHIP-overexpressing plants under low-temperature conditions. Moreover, these overexpressing plants are more sensitive to ABA treatment in terms of stomatal aperture and germination rate (Luo et al., 2006). Since monoubiquitination can regulate substrate activity (Pickart, 2001; Wojcik, 2001), this suggests that the activity of PP2A is increased by AtCHIP and that the increase in PP2A activity leads to enhanced ABA sensitivity.
Recently, Ryu et al. (2010) acquired 100 different T-DNA insertion mutants for RING E3 ligase genes in Arabidopsis and screened for mutants that exhibited altered ABA-sensitivity phenotypes. Here, the loss-of-function mutant of AtAIRP1 (Arabidopsis thaliana ABA Insensitive Ring Protein 1) displayed ABA-insensitive phenotypes, while AtAIRP1-over-expressors were significantly more resistant to drought stress. These results implicate that AtAIRP1 acts as a positive regulator in drought-stress responses mediated by ABA (Ryu et al., 2010).
PUB22 and PUB23 were first identified as Arabidopsis proteins most closely related to the hot pepper (Capsicum annuum) CaPUB1, a U-box-containing E3 ligase induced by drought stress (Cho et al., 2006; 2008). In a previous report, transgenic plants harboring 35S::PUB22 and 35S::PUB23 were hypersensitive to drought, while pub22 and pub23 mutants displayed drought-tolerance, and pub22 pub23 double mutants showed even stronger tolerance to drought. From these results, PUB22 and PUB23 are thought to act as negative regulators in response to water-deficit stress (Cho et al., 2008). Interestingly, PUB22 and PUB23 interact with and ubiquitinate RPN12a, a subunit of the 19S regulatory particle (RP) in the 26S proteasome. Moreover, RPN12a dissociates from the 19S RP in wildtype and PUB-overexpressing plants under water stress. Thus, PUB22 and PUB23 have been proposed to function in the finetuning of drought response by ubiquitinating cytosolic RPN12a and then altering the characteristics of the 19S RP in response to water-deficit stress. Similar to PUB22 and PUB23 overexpressors and mutants, the overexpression of CaPUB1 in Arabidopsis does not alter ABA-sensitivity (Cho et al., 2006). Thus, it is most likely that PUB22 and PUB23 function is independent of ABA.
The hot pepper Rma1H1 is a homolog of human RING membrane-anchor 1 E3 ligase and possesses E3 ligase activity in vitro. Rma1H1 overexpression in plants inhibits PIP2;1 trafficking from the ER to the plasma membrane and increases tolerance to drought stress. The reduction of PIP2;1 in Rma1H1-overexpressing plants is inhibited by MG132, a 26S proteasome inhibitor. In addition, Rma1H1 interacts with an aquaporin isoform PIP2;1 in vitro and ubiquitinates it in vivo. Rma1, an Arabidopsis homolog of Rma1H1, also inhibits PIP2;1 trafficking and the reduced expression of Rma1 leads to an increase in the level of PIP2;1. Taken together, it has been suggested that Rma1H1 and Rma1 play a positive role in drought tolerance by repressing the trafficking of aquaporin and decreasing its protein stability (Lee et al., 2009). On the other hand, an Arabidopsis homolog AtRma2 interacts auxin binding protein 1, suggesting that Rma1 homologs are associated not only with abiotic stress responses but also with hormone responses (Son et al., 2009; 2010).
Cullin-RING ubiquitin E3 ligases (CRLs) are also involved in ABA-mediated water-deficit stress in Arabidopsis. Drought tolerance repressor (DOR) is a F-box protein that interacts with CUL1, and is, therefore, a substrate receptor for Cullin1-RING ubiquitin E3 ligases (CRL1). Loss of DOR results in ABA-hypersensitive stomatal closing and increased drought tolerance, and overexpression of DOR leads to higher susceptibility to drought. Thus, DOR is thought to function as a negative regulator in ABA-mediated guard cell signaling (Zhang et al., 2008c).
DWD hypersensitive to ABA1 (DWA1) and DWA2 have also been reported as negative regulators in ABA signal transduction pathways (Lee et al., 2010). Both proteins possess DWD domains, the conserved domain found in most substrate receptors for Cullin4-RING ubiquitin E3 ligases (CRL4), and interact with components of the CRL4 complex such as DDB1 and CUL4 in vivo, indicating they function as substrate receptors for CRL4. DWA1 and DWA2 directly interact with each other and both associate with and mediate the degradation of ABI5, a key component for ABA responses such as inhibition of seed germination and post-germinational growth arrest (Fig. 2B). However, dwa1 dwa2 mutants exhibit drought tolerance in adult plants as well as ABA-hypersensitivity in seed germination. Thus, DWA1 and DWA2 have been speculated to be involved in the destruction of other positive regulators of ABA response, such as additional AREB/ABF/DPBF bZIP proteins that are highly expressed in adult plants (Nakashima et al., 2009). In a recent report, DWA3 was also reported as a negative regulator in ABA response (Lee et al., 2011). Loss of DWA3 resulted in increased ABA sensitivity and, in particular, dwa3 leaves lost less water than those of wild-type after detachment. Unlike DWA1 and DWA2, DWA3 does not appear to be directly involved in the destruction of ABI5 even though ABI5 level is higher in dwa3 than in wild-type, indicating DWA3 may not act together with other DWA proteins in ABA signaling.
Cold stress signaling and HOS1
In response to low temperature, C-repeat (CRT)-binding factors (CBFs), namely CBF1 (DREB1b), CBF2 (DREB1c) and CBF3 (DREB1a), bind to the cis-acting element, CRT/DRE, and function as key regulators of cold-responsive (COR) genes (Liu et al., 1998; Stockinger et al., 1997; Yamaguchi-Shinozaki and Shinozaki, 1994) (Fig. 2C). It is thought that CBFs are differentially regulated during cold acclimation as CBF2 is induced later than CBF1 and CBF3 (Novillo et al., 2004). There have been reports on upstream regulators of CBF3 that are involved in low temperature signaling. Inducer of CBF expression 1 (ICE1) is a MYC-like basic helix-loop-helix (bHLH) transcription activator that binds to MYCRS within the promoter of CBF3. ice1 mutants display impaired coldinduced transcription of CBF3 and its downstream genes and exhibit decreased chilling and freezing tolerance (Chinnusamy et al., 2003). Phosphorylation and sumoylation of ICE1 by an unknown kinase and SIZ1, respectively, are required for its activity in cold signaling (Chinnusamy et al., 2007). In contrast, ICE1 activity is inhibited through ubiquitination. The RING finger E3 ligase, HOS1, interacts with and ubiquitinates ICE1 (Fig. 2D). Mutations in HOS1 lead to the hyper-induction of both the CBFs and their downstream cold-responsive genes in response to cold treatment (Lee et al., 2001). The overex-pression of HOS1 lowers the level of ICE1 protein and reduces the expression of CBFs and their downstream target genes involved in freezing tolerance (Dong et al., 2006). Thus, HOS1 functions as a negative regulator in low temperature signaling by mediating the degradation of ICE1 and inhibiting the expression of ICE1 target genes.
The role of COP1 and CRL4 in UV-B signaling
In the past years, there has been an astonishing increase in the amount of UV-B (280 to 320 nm) that penetrates the ozone layer and reaches the surface of the Earth. Due to this phenomenon, UV-B has become a major source of stress to plants. While high fluence UV-B generates reactive oxygen species (ROS) that damage macromolecules such as DNA, low fluence UV-B acts as a signal that initiates regulatory responses involved in repairing UV damage. In addition, low fluence UV-B also triggers photomorphogenic processes such as anthocya-nin accumulation, inhibition of hypocotyls, and cotyledon opening (Jenkins, 2009). Although little is known about the UV-B signaling pathway, there have been several reports on UV RESISTANCE LOCUS 8 (UVR8), a key component that positively regulates UV-B response. UVR8 rapidly accumulates in the nucleus upon UV-B irradiation and associates with chromatin regions that contain UV-B-responsive genes such as HY5 (ELONGATED HYPOCOTYL5) and HY5 HOMOLOG (HYH). This association may function in recruiting transcription factors that positively modulate UV-B signaling (Brown et al., 2005; Cloix and Jenkins, 2008). In the presence of UV-B, UVR8 also interacts with the RING finger E3 ligase, CONSTITUTIVELY PHOTOMORPHOGENIC 1 (COP1) (Favory et al., 2009). In white light conditions, COP1 nega-tively regulates the level of HY5 protein. However, under UV-B, the COP1-mediated protein degradation of HY5 is inhibited and, instead, COP1 contributes in the increase of HY5 expression. Therefore, it has been proposed that UVR8 is involved in the inhibition of HY5 degradation under UV-B and that UVR8-COP1 interaction is important to properly tranduce UV-B signaling into downstream pathways (Favory et al., 2009; Oravecz et al., 2006). Although the mechanism of action of COP1 under UV-B is still poorly understood, it is thought that the E3 ligase activity of COP1 contributes to the positive regulation of UV-inducible gene expression.
Recently, two DWD genes, an ortholog of the mammalian Cockayne Syndrome type-A (AtCSA-1) and damage-specific DNA binding protein 2 (DDB2), were reported as crucial components in UV-B response (Biedermann and Hellmann, 2010; Zhang et al., 2010). As AtCSA-1 and DDB2 form CRL4 complexes with DDB1 and CUL4, it is possible that CRL4 E3 ligase acts as an important regulator in UV-B response (Biedermann and Hellmann, 2010; Zhang et al., 2010).
CONCLUSION AND FUTURE PERSPECTIVES
Higher plants are frequently affected by diverse stresses from the environment throughout their life cycles. This stress can have a huge impact on the growth of plants. In particular, abiotic stresses such as drought, cold and salt greatly impair plant growth and development (Boyer, 1982). Although many genes involved in such stress signalings have been studied by biochemical and genetic methods, the physiological role of these genes in relation to stress tolerance or sensitivity is largely unknown in higher plants (Bohnert et al., 2006; Oono et al., 2003; Vij and Tyagi, 2007; Zhu, 2002). Therefore, a more detailed understanding of stress signal transduction pathways would serve as a platform to increase the quality and productivity of agronomically useful plants.
Although only a minor part of E3 genes in the entire Arabidopsis genome have been studied, the rapidly growing amount of novel findings on E3 ligases has significantly advanced our knowledge in this field over the last decade. However, in spite of several reports on E3 ligases in abiotic stress signaling in this study, there is only a limited amount of information on the associated target proteins. Further studies on E3 ligase substrates will offer a broader insight into how plants specifically modulate the activity and/or stability of substrates via E3 ligases and utilize the ubiquitination system to adapt to unfavorable conditions. Comparative genomics and proteomics with mutants or overexpressors of E3 ligases of interest would also help elucidate the related substrates.
In some cases, the target proteins destined for ubiquitination are also under the control of additional post-translation modifications. For example, phosphorylation is required for the increased activity of ICE1 (as previously mentioned) and ABI5, which are negatively modulated via ubiquitination (Chinnusamy et al., 2007; Lopez-Molina et al., 2001). The proper balance between these modifications would most likely act as a determining factor in how plants transduce upstream signals into downstream pathways. Thus, it would be interesting to further investigate whether ubiquitination substrates found by com-parative genomics and proteomics are dependent on other modification processes, and if so, how the different modifications are regulated during abiotic stress signaling.
In CRL E3 ligases, cullin is another factor that determines the activity of the complex. The ubiquitin-like protein, RUB (Related to Ubiquitin)/NEDD8 (Neural precursor cell-Expressed Developmentally Downregulated 8) can bind to cullin and this binding (named rubylation) seems to stimulate the activity of CRL in vitro by promoting E2 recruitment (Kawakami et al., 2001; Read et al., 2000; Wu et al., 2000). Moreover, it has been reported that COP9 signalosome (CSN) modulates the activity of CRL via the derubylation/deneddylation of cullin (Lyapina et al., 2001; Schwechheimer et al., 2001). Therefore, future studies on the role of CSN and rubylation in the aspect of cullin in abiotic stress signaling would help us understand how ubiquitination mediated by multi-subunit E3 complexes contribute to abiotic stress tolerance.
Here, we described the recent advances and future prospects in abiotic stress signaling mediated by E3-dependent ubiquitination in Arabidopsis. Because an enormous amount of E3 ligases still remain to be identified, the further pursuit of E3 enzymes involved in abiotic stress transduction could provide a functional connection between this signaling and the ubiquitination process, and furthermore, aid the generation of stress-resistant plants.
Acknowledgments
The authors acknowledge Sunglan Chung (Yale University) for critical reading of the manuscript. This work was supported by grants from the National Research Foundation (Project No. 2009-0078317 funded by the Ministry of Education, Science, and Technology, Republic of Korea), the Technology Develop-ment Program for Agriculture and Forestry (Project No. 309017-5 funded by the Ministry for Agriculture, Forestry and Fisheries, Republic of Korea), and the BioGreen 21 Program (funded by the Rural Development Administration, Republic of Korea) to W.T.K.
References
- 1.Abe H., Yamaguchi-Shinozaki K., Urao T., Iwasaki T., Hosokawa D., Shinozaki K. Role of Arabidopsis MYC and MYB homologs in drought- and abscisic-acid-regulated gene expression. Plant Cell. (1997);9:1859–1868. doi: 10.1105/tpc.9.10.1859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Agarwal P.K., Agarwal P., Reddy M.K., Sopory S.K. Role of DREB transcription factors in abiotic and biotic stress tolerance in plants. Plant Cell Rep. (2006);25:1263–1274. doi: 10.1007/s00299-006-0204-8. [DOI] [PubMed] [Google Scholar]
- 3.Angers S., Li T., Yi X., MacCoss M.J., Moon R.T., Zheng N. Molecular architecture and assembly of the DDB1-CUL4A ubiquitin ligase machinery. Nature. (2006);443:590–593. doi: 10.1038/nature05175. [DOI] [PubMed] [Google Scholar]
- 4.Aravind L., Koonin E.V. The U box is a modified RING finger: a common domain in ubiquitination. Curr. Biol. (2000);10:R132–R134. doi: 10.1016/s0960-9822(00)00398-5. [DOI] [PubMed] [Google Scholar]
- 5.Bateman A., Coin L., Durbin R., Finn R.D., Hollich V., Griffiths-Jones S., Khanna A., Marshall M., Moxon S., Sonnhammer E.L., et al. The Pfam protein families database. Nucleic Acids Res. (2004);32:D138–141. doi: 10.1093/nar/gkh121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bernhardt A., Lechner E., Hano P., Schade V., Dieterle M., Anders M., Dubin M.J., Benvenuto G., Bowler C., Genschik P., et al. CUL4 associates with DDB1 and DET1 and its downregulation affects diverse aspects of development in Ara-bidopsis thaliana. Plant J. (2006);47:591–603. doi: 10.1111/j.1365-313X.2006.02810.x. [DOI] [PubMed] [Google Scholar]
- 7.Bernhardt A., Mooney S., Hellmann H. Arabidopsis DDB1a and DDB1b are critical for embryo development. Planta. (2010);232:555–566. doi: 10.1007/s00425-010-1195-9. [DOI] [PubMed] [Google Scholar]
- 8.Biedermann S., Hellmann H. The DDB1a interacting proteins ATCSA-1 and DDB2 are critical factors for UV-B toler-ance and genomic integrity in Arabidopsis thaliana. Plant J. (2010);62:404–415. doi: 10.1111/j.1365-313X.2010.04157.x. [DOI] [PubMed] [Google Scholar]
- 9.Bohnert H.J., Gong Q., Li P., Ma S. Unraveling abio-tic stress tolerance mechanisms - Getting genomics going. Curr. Opin. Plant Biol. (2006);9:180–188. doi: 10.1016/j.pbi.2006.01.003. [DOI] [PubMed] [Google Scholar]
- 10.Boyer J.S. Plant productivity and environment. Science. (1982);218:443–448. doi: 10.1126/science.218.4571.443. [DOI] [PubMed] [Google Scholar]
- 11.Brown B.A., Cloix C., Jiang G.H., Kaiserli E., Herzyk P., Kliebenstein D.J., Jenkins G.I. A UV-B-specific signaling component orchestrates plant UV protection. Proc. Natl. Acad. Sci. USA. (2005);102:18225–18230. doi: 10.1073/pnas.0507187102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cardozo T., Pagano M. The SCF ubiquitin ligase: insights into a molecular machine. Nat. Rev. Mol. Cell Biol. (2004);5:739–751. doi: 10.1038/nrm1471. [DOI] [PubMed] [Google Scholar]
- 13.Chen H., Shen Y., Tang X., Yu L., Wang J., Guo L., Zhang Y., Zhang H., Feng S., Strickland E., et al. Arabidopsis CULLIN4 forms an E3 ubiquitin ligase with RBX1 and the CDD complex in mediating light control of development. Plant Cell. (2006);18:1991–2004. doi: 10.1105/tpc.106.043224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chinnusamy V., Ohta M., Kanrar S., Lee B.H., Hong X., Agarwal M., Zhu J.K. ICE1: a regulator of cold-induced transcriptome and freezing tolerance in Arabidopsis. Genes Dev. (2003);17:1043–1054. doi: 10.1101/gad.1077503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chinnusamy V., Zhu J., Zhu J.K. Cold stress regulation of gene expression in plants. Trends Plant Sci. (2007);12:444–451. doi: 10.1016/j.tplants.2007.07.002. [DOI] [PubMed] [Google Scholar]
- 16.Cho S.K., Chung H.S., Ryu M.Y., Park M.J., Lee M.M., Bahk Y.Y., Kim J., Pai H.S., Kim W.T. Heterologous expression and molecular and cellular characterization of CaPUB1 encoding a hot pepper U-Box E3 ubiquitin ligase homolog. Plant Physiol. (2006);142:1664–1682. doi: 10.1104/pp.106.087965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cho S.K., Ryu M.Y., Song C., Kwak J.M., Kim W.T. Arabidopsis PUB22 and PUB23 are homologous U-Box E3 ubiquitin ligases that play combinatory roles in response to drought stress. Plant Cell. (2008);20:1899–1914. doi: 10.1105/tpc.108.060699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cloix C., Jenkins G.I. Interaction of the Arabidopsis UV-B-specific signaling component UVR8 with chromatin. Mol. Plant. (2008);1:118–128. doi: 10.1093/mp/ssm012. [DOI] [PubMed] [Google Scholar]
- 19.Cope G.A., Deshaies R.J. Targeted silencing of Jab1/Csn5 in human cells downregulates SCF activity through reduction of F-box protein levels. BMC Biochem. (2006);7:1. doi: 10.1186/1471-2091-7-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cushman J.C., Bohnert H.J. Genomic approaches to plant stress tolerance. Curr. Opin. Plant Biol. (2000);3:117–124. doi: 10.1016/s1369-5266(99)00052-7. [DOI] [PubMed] [Google Scholar]
- 21.Dong C.H., Agarwal M., Zhang Y., Xie Q., Zhu J.K. The negative regulator of plant cold responses, HOS1, is a RING E3 ligase that mediates the ubiquitination and degradation of ICE1. Proc. Natl. Acad. Sci. USA. (2006);103:8281–8286. doi: 10.1073/pnas.0602874103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Downes B.P., Stupar R.M., Gingerich D.J., Vierstra R.D. The HECT ubiquitin-protein ligase (UPL) family in Ara-bidopsis: UPL3 has a specific role in trichome development. Plant J. (2003);35:729–742. doi: 10.1046/j.1365-313x.2003.01844.x. [DOI] [PubMed] [Google Scholar]
- 23.Dreher K., Callis J. Ubiquitin, hormones and biotic stress in plants. Ann. Bot. (2007);99:787–822. doi: 10.1093/aob/mcl255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Favory J.J., Stec A., Gruber H., Rizzini L., Oravecz A., Funk M., Albert A., Cloix C., Jenkins G.I., Oakeley E.J., et al. In-teraction of COP1 and UVR8 regulates UV-B-induced photomorphogenesis and stress acclimation in Arabidopsis. EMBO J. (2009);28:591–601. doi: 10.1038/emboj.2009.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Figueroa P., Gusmaroli G., Serino G., Habashi J., Ma L., Shen Y., Feng S., Bostick M., Callis J., Hellmann H., et al. Ara-bidopsis has two redundant Cullin3 proteins that are essential for embryo development and that interact with RBX1 and BTB proteins to form multisubunit E3 ubiquitin ligase complexes in vivo. Plant Cell. (2005);17:1180–1195. doi: 10.1105/tpc.105.031989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Freemont P.S., Hanson I.M., Trowsdale J. A novel cysteine-rich sequence motif. Cell. (1991);64:483–484. doi: 10.1016/0092-8674(91)90229-r. [DOI] [PubMed] [Google Scholar]
- 27.Fujita Y., Fujita M., Satoh R., Maruyama K., Parvez M.M., Seki M., Hiratsu K., Ohme-Takagi M., Shinozaki K., Yamagu-chi-Shinozaki K. AREB1 is a transcription activator of novel ABRE-dependent ABA signaling that enhances drought stress tolerance in Arabidopsis. Plant Cell. (2005);17:3470–3488. doi: 10.1105/tpc.105.035659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gagne J.M., Downes B.P., Shiu S.H., Durski A.M., Vierstra R.D. The F-box subunit of the SCF E3 complex is encoded by a diverse superfamily of genes in Arabidopsis. Proc. Natl. Acad. Sci. USA. (2002);99:11519–11524. doi: 10.1073/pnas.162339999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gingerich D.J., Gagne J.M., Salter D.W., Hellmann H., Estelle M., Ma L., Vierstra R.D. Cullins 3a and 3b assemble with members of the broad complex/tramtrack/bric-a-brac (BTB) protein family to form essential ubiquitin-protein ligases (E3s) in Arabidopsis. J. Biol. Chem. (2005);280:18810–18821. doi: 10.1074/jbc.M413247200. [DOI] [PubMed] [Google Scholar]
- 30.Hatakeyama S., Yada M., Matsumoto M., Ishida N., Naka-yama K.I. U box proteins as a new family of ubiquitin-protein ligases. J. Biol. Chem. (2001);276:33111–33120. doi: 10.1074/jbc.M102755200. [DOI] [PubMed] [Google Scholar]
- 31.Hatfield P.M., Gosink M.M., Carpenter T.B., Vierstra R.D. The ubiquitin-activating enzyme (E1) gene family in Ara-bidopsis thaliana. Plant J. (1997);11:213–226. doi: 10.1046/j.1365-313x.1997.11020213.x. [DOI] [PubMed] [Google Scholar]
- 32.He Y.J., McCall C.M., Hu J., Zeng Y., Xiong Y. DDB1 functions as a linker to recruit receptor WD40 proteins to CUL4-ROC1 ubiquitin ligases. Genes Dev. (2006);20:2949–2954. doi: 10.1101/gad.1483206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Higa L.A., Wu M., Ye T., Kobayashi R., Sun H., Zhang H. CUL4-DDB1 ubiquitin ligase interacts with multiple WD40-repeat proteins and regulates histone methylation. Nat. Cell Biol. (2006);8:1277–1283. doi: 10.1038/ncb1490. [DOI] [PubMed] [Google Scholar]
- 34.Huibregtse J.M., Scheffner M., Beaudenon S., Howley P.M. A family of proteins structurally and functionally related to the E6-AP ubiquitin-protein ligase. Proc. Natl. Acad. Sci. USA. (1995);92:2563–2567. doi: 10.1073/pnas.92.7.2563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jenkins G.I. Signal transduction in responses to UV-B radiation. Annu. Rev. Plant Biol. (2009);60:407–431. doi: 10.1146/annurev.arplant.59.032607.092953. [DOI] [PubMed] [Google Scholar]
- 36.Jin J., Arias E.E., Chen J., Harper J.W., Walter J.C. A family of diverse Cul4-Ddb1-interacting proteins includes Cdt2, which is required for S phase destruction of the replication factor Cdt1. Mol. Cell. (2006);23:709–721. doi: 10.1016/j.molcel.2006.08.010. [DOI] [PubMed] [Google Scholar]
- 37.Kawakami T., Chiba T., Suzuki T., Iwai K., Yamanaka K., Minato N., Suzuki H., Shimbara N., Hidaka Y., Osaka F., et al. NEDD8 recruits E2-ubiquitin to SCF E3 ligase. EMBO J. (2001);20:4003–4012. doi: 10.1093/emboj/20.15.4003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Koegl M., Hoppe T., Schlenker S., Ulrich H.D., Mayer T.U., Jentsch S. A novel ubiquitination factor, E4, is involved in multiubiquitin chain assembly. Cell. (1999);96:635–644. doi: 10.1016/s0092-8674(00)80574-7. [DOI] [PubMed] [Google Scholar]
- 39.Kraft E., Stone S.L., Ma L., Su N., Gao Y., Lau O.S., Deng X. W., Callis J. Genome analysis and functional characterization of the E2 and RING-type E3 ligase ubiquitination enzymes of Arabidopsis. Plant Physiol. (2005);139:1597–1611. doi: 10.1104/pp.105.067983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lee H., Xiong L., Gong Z., Ishitani M., Stevenson B., Zhu J.K. The Arabidopsis HOS1 gene negatively regulates cold signal transduction and encodes a RING finger protein that displays cold-regulated nucleocytoplasmic partitioning. Genes Dev. (2001);15:912–924. doi: 10.1101/gad.866801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lee J.H., Terzaghi W., Gusmaroli G., Charron J.B., Yoon H.J., Chen H., He Y.J., Xiong Y., Deng X.W. Characterization of Arabidopsis and rice DWD proteins and their roles as substrate receptors for CUL4-RING E3 ubiquitin ligases. Plant Cell. (2008);20:152–167. doi: 10.1105/tpc.107.055418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lee H.K., Cho S.K., Son O., Xu Z., Hwang I., Kim W.T. Drought stress-induced Rma1H1, a RING membrane-anchor E3 ubiquitin ligase homolog, regulates aquaporin levels via ubiquitination in transgenic Arabidopsis plants. Plant Cell. (2009);21:622–641. doi: 10.1105/tpc.108.061994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lee J.H., Yoon H.J., Terzaghi W., Martinez C., Dai M., Li J., Byun M.O., Deng X.W. DWA1 and DWA2, two Arabidopsis DWD protein components of CUL4-based E3 ligases, act together as negative regulators in ABA signal transduction. Plant Cell. (2010);22:1716–1732. doi: 10.1105/tpc.109.073783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lee J.H., Terzaghi W., Deng X.W. DWA3, an Ara-bidopsis DWD protein, acts as a negative regulator in ABA signal transduction. Plant Sci. (2011);180:352–357. doi: 10.1016/j.plantsci.2010.10.008. [DOI] [PubMed] [Google Scholar]
- 45.Liu Q., Kasuga M., Sakuma Y., Abe H., Miura S., Yamaguchi-Shinozaki K., Shinozaki K. Two transcription factors, DREB1 and DREB2, with an EREBP/AP2 DNA binding domain separate two cellular signal transduction pathways in drought- and low-temperature-responsive gene expression, re-spectively, in Arabidopsis. Plant Cell. (1998);10:1391–1406. doi: 10.1105/tpc.10.8.1391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lopez-Molina L., Mongrand S., Chua N.H. A postgermination developmental arrest checkpoint is mediated by abscisic acid and requires the ABI5 transcription factor in Ara-bidopsis. Proc. Natl. Acad. Sci. USA. (2001);98:4782–4787. doi: 10.1073/pnas.081594298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Luo J., Shen G., Yan J., He C., Zhang H. AtCHIP functions as an E3 ubiquitin ligase of protein phosphatase 2A subunits and alters plant response to abscisic acid treatment. Plant J. (2006);46:649–657. doi: 10.1111/j.1365-313X.2006.02730.x. [DOI] [PubMed] [Google Scholar]
- 48.Lyapina S., Cope G., Shevchenko A., Serino G., Tsuge T., Zhou C., Wolf D.A., Wei N., Shevchenko A., Deshaies R.J. Promotion of NEDD-CUL1 conjugate cleavage by COP9 signalosome. Science. (2001);292:1382–1385. doi: 10.1126/science.1059780. [DOI] [PubMed] [Google Scholar]
- 49.Miao Y., Zentgraf U. A HECT E3 ubiquitin ligase nega-tively regulates Arabidopsis leaf senescence through degradation of the transcription factor WRKY53. Plant J. (2010);63:179–188. doi: 10.1111/j.1365-313X.2010.04233.x. [DOI] [PubMed] [Google Scholar]
- 50.Mukhopadhyay D., Riezman H. Proteasome-independent functions of ubiquitin in endocytosis and signaling. Science. (2007);315:201–205. doi: 10.1126/science.1127085. [DOI] [PubMed] [Google Scholar]
- 51.Nakashima K., Ito Y., Yamaguchi-Shinozaki K. Transcriptional regulatory networks in response to abiotic stresses in Arabidopsis and grasses. Plant Physiol. (2009);149:88–95. doi: 10.1104/pp.108.129791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Novillo F., Alonso J.M., Ecker J.R., Salinas J. CBF2/ DREB1C is a negative regulator of CBF1/DREB1B and CBF3/ DREB1A expression and plays a central role in stress tolerance in Arabidopsis. Proc. Natl. Acad. Sci. USA. (2004);101:3985–3990. doi: 10.1073/pnas.0303029101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ohi M.D., Vander Kooi C.W., Rosenberg J.A., Chazin W.J., Gould K.L. Structural insights into the U-box, a domain associated with multi-ubiquitination. Nat. Struct. Biol. (2003);10:250–255. doi: 10.1038/nsb906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Oono Y., Seki M., Nanjo T., Narusaka M., Fujita M., Satoh R., Satou M., Sakurai T., Ishida J., Akiyama K., et al. Monitoring expression profiles of Arabidopsis gene expression during rehydration process after dehydration using ca. Plant J. (2003);34:868–887. doi: 10.1046/j.1365-313x.2003.01774.x. [DOI] [PubMed] [Google Scholar]
- 55.Oravecz A., Baumann A., Máté Z., Brzezinska A., Molinier J., Oakeley E.J., Adám E., Schäfer E., Nagy F., Ulm R. CONSTITUTIVELY PHOTOMORPHOGENIC1 is required for the UV-B response in Arabidopsis. Plant Cell. (2006);18:1975–1990. doi: 10.1105/tpc.105.040097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Petroski M.D., Deshaies R.J. Function and regulation of cullin-RING ubiquitin ligases. Nat. Rev. Mol. Cell Biol. (2005);6:9–20. doi: 10.1038/nrm1547. [DOI] [PubMed] [Google Scholar]
- 57.Pickart C.M. Ubiquitin enters the new millennium. Mol. Cell. (2001);8:499–504. doi: 10.1016/s1097-2765(01)00347-1. [DOI] [PubMed] [Google Scholar]
- 58.Pickart C.M., Eddins M.J. Ubiquitin: structures, functions, mechanisms. Biochim. Biophys. Acta. (2004);1695:55–72. doi: 10.1016/j.bbamcr.2004.09.019. [DOI] [PubMed] [Google Scholar]
- 59.Pintard L., Willems A., Peter M. Cullin-based ubiquitin ligases: Cul3-BTB complexes join the family. EMBO J. (2004);23:1681–1687. doi: 10.1038/sj.emboj.7600186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Read M.A., Brownell J.E., Gladysheva T.B., Hottelet M., Parent L.A., Coggins M.B., Pierce J.W., Podust V.N., Luo R.S., Chau V., et al. Nedd8 modification of cul-1 activates SCFβ-TrCP-dependent ubiquitination of IκBα. Mol. Cell. Biol. (2000);20:2326–2333. doi: 10.1128/mcb.20.7.2326-2333.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Risseeuw E.P., Daskalchuk T.E., Banks T.W., Liu E., Cotelesage J., Hellmann H., Estelle M., Somers D.E., Crosby W.L. Protein interaction analysis of SCF ubiquitin E3 ligase subunits from Arabidopsis. Plant J. (2003);34:753–767. doi: 10.1046/j.1365-313x.2003.01768.x. [DOI] [PubMed] [Google Scholar]
- 62.Ryu M.Y., Cho S.K., Kim W.T. The Arabidopsis C3H2C3-Type RING E3 ubiquitin ligase AtAIRP1 is a positive regulator of an ABA-dependent response to drought stress. Plant Physiol. (2010);154:1983–1997. doi: 10.1104/pp.110.164749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Santner A., Estelle M. The ubiquitin-proteasome system regulates plant hormone signaling. Plant J. (2010);61:1029–1040. doi: 10.1111/j.1365-313X.2010.04112.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Schroeder D.F., Gahrtz M., Maxwell B.B., Cook R.K., Kan J.M., Alonso J.M., Ecker J.R., Chory J. De-etiolated 1 and damaged DNA binding protein 1 interact to regulate Ara-bidopsis photomorphogenesis. Curr. Biol. (2002);12:1462–1472. doi: 10.1016/s0960-9822(02)01106-5. [DOI] [PubMed] [Google Scholar]
- 65.Schulman B.A., Carrano A.C., Jeffrey P.D., Bowen Z., Kinnucan E.R., Finnin M.S., Elledge S.J., Harper J.W., Pagano M., Pavletich N.P. Insights into SCF ubiquitin ligases from the structure of the Skp1-Skp2 complex. Nature. (2000);408:381–386. doi: 10.1038/35042620. [DOI] [PubMed] [Google Scholar]
- 66.Schwechheimer C., Calderon Villalobos L.I. Cullin-containing E3 ubiquitin ligases in plant development. Curr. Opin. Plant Biol. (2004);7:677–686. doi: 10.1016/j.pbi.2004.09.009. [DOI] [PubMed] [Google Scholar]
- 67.Schwechheimer C., Serino G., Callis J., Crosby W.L., Lyapina S., Deshaies R.J., Gray W.M., Estelle M., Deng X.W. Interactions of the COP9 signalosome with the E3 ubiquitin ligase SCFTIR1 in mediating auxin response. Science. (2001);292:1379–1382. doi: 10.1126/science.1059776. [DOI] [PubMed] [Google Scholar]
- 68.Shinozaki K., Yamaguchi-Shinozaki K. Molecular responses to dehydration and low temperature: Differences and crosstalk between two stress signaling pathways. Curr. Opin. Plant Biol. (2000);3:217–223. [PubMed] [Google Scholar]
- 69.Smalle J., Viestra R.D. The ubiquitin 26S proteasome proteolytic pathway. Annu. Rev. Plant Physiol. Plant Mol. Biol. (2004);55:555–590. doi: 10.1146/annurev.arplant.55.031903.141801. [DOI] [PubMed] [Google Scholar]
- 70.Son O., Cho S.K., Kim E.Y., Kim W.T. Characterization of three Arabidopsis homologs of human RING membrane anchor E3 ubiquitin ligase. Plant Cell Rep. (2009);28:561–569. doi: 10.1007/s00299-009-0680-8. [DOI] [PubMed] [Google Scholar]
- 71.Son O., Cho S.K., Kim S.J., Kim W.T. In vitro and in vivo interaction of AtRma2 E3 ubiquitin ligase and auxin binding protein 1. Biochem. Biophys. Res. Commun. (2010);393:492–497. doi: 10.1016/j.bbrc.2010.02.032. [DOI] [PubMed] [Google Scholar]
- 72.Stockinger E.J., Gilmour S.J., Thomashow M.F. Ara-bidopsis thaliana CBF1 encodes an AP2 domain-containing transcriptional activator that binds to the C-repeat/DRE, a cis-acting DNA regulatory element that stimulates transcription in response to low temperature and water deficit. Proc. Natl. Acad. Sci. USA. (1997);94:1035–1040. doi: 10.1073/pnas.94.3.1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Stone S.L., Hauksdóttir H., Troy A., Herschleb J., Kraft E., Callis J. Functional analysis of the RING-type ubiquitin ligase family of Arabidopsis. Plant Physiol. (2005);137:13–30. doi: 10.1104/pp.104.052423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Takahashi S., Seki M., Ishida J., Satou M., Sakurai T., Narusaka M., Kamiya A., Nakajima M., Enju A., Akiyama K., et al. Monitoring the expression profiles of genes induced by hyper-osmotic, high salinity, and oxidative stress and abscisic acid treatment in Arabidopsis cell culture using a full-length cDNA microarray. Plant Mol. Biol. (2004);56:29–55. doi: 10.1007/s11103-004-2200-0. [DOI] [PubMed] [Google Scholar]
- 75.Thomann A., Dieterle M., Genschik P. Plant CULLIN-based E3s: Phytohormones come first. FEBS Lett. (2005);579:3239–3245. doi: 10.1016/j.febslet.2005.02.068. [DOI] [PubMed] [Google Scholar]
- 76.Uno Y., Furihata T., Abe H., Yoshida R., Shinozaki K., Yama-guchi-Shinozaki K. Arabidopsis basic leucine zipper transcription factors involved in an abscisic acid-dependent signal transduction pathway under drought and high-salinity conditions. Proc. Natl. Acad. Sci. USA. (2000);97:11632–11637. doi: 10.1073/pnas.190309197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Vierstra R.D. The ubiquitin/26S proteasome pathway, the complex last chapter in the life of many plant proteins. Trends Plant Sci. (2003);8:135–142. doi: 10.1016/S1360-1385(03)00014-1. [DOI] [PubMed] [Google Scholar]
- 78.Vierstra R.D. The ubiquitin-26S proteasome system at the nexus of plant biology. Nat. Rev. Mol. Cell Biol. (2009);110:385–397. doi: 10.1038/nrm2688. [DOI] [PubMed] [Google Scholar]
- 79.Vij S., Tyagi A.K. Emerging trends in the functional genomics of the abiotic stress response in crop plants. Plant Biotechnol. J. (2007);5:361–380. doi: 10.1111/j.1467-7652.2007.00239.x. [DOI] [PubMed] [Google Scholar]
- 80.Wiborg J., O’Shea C., Skriver K. Biochemical function of typical and variant Arabidopsis thaliana U-box E3 ubiquitin-protein ligases. Biochem. J. (2008);413:447–457. doi: 10.1042/BJ20071568. [DOI] [PubMed] [Google Scholar]
- 81.Wojcik C. Ubiquitin - more than just a signal for protein degradation. Trends Cell Biol. (2001);11:397–399. doi: 10.1016/s0962-8924(01)02084-0. [DOI] [PubMed] [Google Scholar]
- 82.Wu K., Chen A., Pan Z.Q. Conjugation of Nedd8 to CUL1 enhances the ability of the ROC1-CUL1 complex to promote ubiquitin polymerization. J. Biol. Chem. (2000);275:32317–32324. doi: 10.1074/jbc.M004847200. [DOI] [PubMed] [Google Scholar]
- 83.Yamaguchi-Shinozaki K., Shinozaki K. A novel cis-acting element in an Arabidopsis gene is involved in responsiveness to drought, low-temperature, or high-salt stress. Plant Cell. (1994);6:251–264. doi: 10.1105/tpc.6.2.251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Yan J., Wang J., Li Q., Hwang J.R., Patterson C., Zhang H. AtCHIP, a U-box-containing E3 ubiquitin ligase, plays a critical role in temperature stress tolerance in Arabidopsis. Plant Physiol. (2003);132:861–869. doi: 10.1104/pp.103.020800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Yee D., Goring D.R. The diversity of plant U-box E3 ubiquitin ligases: from upstream activators to downstream target substrates. J. Exp. Bot. (2009);60:1109–1121. doi: 10.1093/jxb/ern369. [DOI] [PubMed] [Google Scholar]
- 86.Zhang Y., Yang C., Li Y., Zheng N., Chen H., Zhao Q., Gao T., Guo H., Xie Q. SDIR1 is a RING finger E3 ligase that positively regulates stress-responsive abscisic acid signaling in Arabidopsis. Plant Cell. (2007);19:1912–1929. doi: 10.1105/tpc.106.048488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Zhang Y., Feng S., Chen F., Chen H., Wang J., McCall C., Xiong Y., Deng X.W. Arabidopsis DDB1-CUL4 ASSOCIATED FACTOR1 forms a nuclear E3 ubiquitin ligase with DDB1 and CUL4 that is involved in multiple plant develop-mental processes. Plant Cell. (2008a);20:1437–1455. doi: 10.1105/tpc.108.058891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Zhang Y.Y., Li Y., Gao T., Zhu H., Wang D.J., Zhang H.W., Ning Y.S., Liu L.J., Wu Y.R., Chu C.C., et al. Arabidopsis SDIR1 enhances drought tolerance in crop plants. Biosci. Biotechnol. Biochem. (2008b);72:2251–2254. doi: 10.1271/bbb.80286. [DOI] [PubMed] [Google Scholar]
- 89.Zhang Y., Xu W., Li Z., Deng X.W., Wu W., Xue Y. F-box protein DOR functions as a novel inhibitory factor for abscisic acid-induced stomatal closure under drought stress in Arabidopsis. Plant Physiol. (2008c);148:2121–2133. doi: 10.1104/pp.108.126912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Zhang C., Guo H., Zhang J., Guo G., Schumaker K.S., Guo Y. Arabidopsis cockayne syndrome A-like proteins 1A and 1B form a complex with CULLIN4 and damage DNA binding protein 1A and regulate the response to UV irradiation. Plant Cell. (2010);22:2353–2369. doi: 10.1105/tpc.110.073973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Zheng N., Schulman B.A., Song L., Miller J.J., Jeffrey P.D., Wang P., Chu C., Koepp D.M., Elledge S.J., Pagano M., et al. Structure of the Cul1-Rbx1-Skp1-F boxSkp2 SCF ubiquitin lig-ase complex. Nature. (2002);416:703–709. doi: 10.1038/416703a. [DOI] [PubMed] [Google Scholar]
- 92.Zhu J.K. Salt and drought stress signal transduction in plants. Annu. Rev. Plant Biol. (2002);53:247–273. doi: 10.1146/annurev.arplant.53.091401.143329. [DOI] [PMC free article] [PubMed] [Google Scholar]


