Abstract
Electron microscopic images of mitochondrial nucleoids isolated from mung bean seedlings revealed a relatively homogeneous population of particles, each consisting of a chromatin-like structure associated with a membrane com-ponent. Association of F-actin with mitochondrial nucleoids was also observed. The mitochondrial nucleoid structure identified in situ showed heterogeneous genomic organization. After pulsed-field gel electrophoresis (PFGE), a large proportion of the mitochondrial nucleoid DNA remained in the well, whereas the rest migrated as a 50-200 kb smear zone. This PFGE migration pattern was not af-fected by high salt, topoisomerase I or latrunculin B treatments; however, the mobility of a fraction of the fast-moving DNA decreased conspicuously following an in-gel ethidium-enhanced UV-irradiation treatment, suggesting that molecules with intricately compact structures were present in the 50-200 kb region. Approximately 70% of the mitochondrial nucleoid DNA molecules examined via electron microscopy were open circles, supercoils, complex forms, and linear molecules with interspersed sigma-shaped structures and/or loops. Increased sensitivity of mtDNA to DNase I was found after mitochondrial nucleoids were pretreated with high salt. This result indicates that some loosely bound or peripheral DNA binding proteins protected the mtDNA from DNase I degradation.
Keywords: mitochondrial DNA, mitochondrial nucleoid, mtDNA binding protein, mtDNA complexity, plant mitochondria
INTRODUCTION
In living cells, organellar DNA molecules are compactly folded with specific proteins into a highly organized structure most commonly referred to as organelle nucleoids. Initially, nucleoids were defined morphologically as DNA-containing foci visualized inside mitochondria (Kuroiwa, 1982; Nass et al., 1965; Stevens, 1982) or chloroplasts (Eva and Chiang, 1984; Kuroiwa et al., 1982). Later, mitochondrial nucleoids were isolated at a defined density in gradient centrifugation and appeared to retain their morphological structure in vitro (Dai et al., 2005; Miyakawa et al., 1987; Newman et al., 1996; Suzuki et al., 1982). Spelbrink et al. (2001) showed that nucleoids in mammalian cells are stable assemblies of multiple mitochondrial proteins with mtDNA. In so far as DNA compaction, replication, and RNA transcription are concerned, mitochondrial nucleoids are roughly analogous to cell nuclei (Dai et al., 2005; Kuroiwa, 1982; Miyakawa et al., 1996; Sakai et al., 2004).
Compared with other groups of organisms, higher plants are known to have much larger mitochondrial genomes, ranging from about 200 to 2,400 Kbp (Levings and Brown, 1989; Ward et al., 1981). Several studies have documented drastic differences in mtDNA amount per mitochondrial nucleoid in cells of different function and/or different divisional or developmental states (Kuroiwa, 1982; Kuroiwa et al., 1992; Sakai et al., 2004). In addition to the DNA content variation of mitochondrial nucleoids, the structural organization of the plant mitochondrial (mt) genome is complex and requires further investigation re-viewed by Mackenzie, 2007). Regardless of its genomic size or its restriction-endonuclease-generated genomic map (circular or linear), when fractionated by pulsed-field gel electrophoresis (PFGE) most plant mtDNA migrates as a smear zone corresponding to 50 to 200 kb linear DNA molecules. However, much of the mtDNA from all plants remains in the well at the origin of electrophoresis (André and Walbot, 1995; Backert et al., 1997; Bendich, 1996; Dai et al., 2005; Maleszka et al., 1991; Narayanan et al., 1993; Oldenburg and Bendich, 1996; Sakai et al., 1999).
In the course of this study, we elucidated the structure and DNA complexity of mung bean mitochondrial nucleoids by using physical/chemical reagent treatments capable of altering the organization of mitochondrial nucleoid DNA and by electron microscopic ultrastructural analysis, which provides direction evidence of mtDNA complexity.
MATERIALS AND METHODS
Isolation of mitochondrial nucleoids from mitochondria
Mitochondria were isolated from 3-day old etiolated mung bean seedlings (Vigna radiata L. [Wilzed] cv. Tainan No. 5) as described (Dai et al., 1991). Mitochondrial nucleoids were then purified exactly as described in our previous report (Dai et al., 2005).
Electron microscopic studies
For freeze fracture, mitochondrial nucleoid samples were rapidly frozen on the specimen carrier with liquid nitrogen slush and fractured in a Balzers BAF 400D freeze etch unit (Balzers Union, Liechtenstein) at -105℃. Fractured specimens werethen replicated by evaporation of platinum-carbon by an electron-beam gun positioned at a 45° angle, followed by carbon coating at a 90° angle. The replicas were floated on 70% sulfuric acid for 24 h followed by three water washes of 15 min each. The replicas were transferred to 15% Na-hypochlorite and incubated for 24 h. After three water washes, the replicas were transferred to a 300 mesh grid and examined in a Philips CM 100 electron microscope.
Isolated mitochondrial nucleoid samples were prepared for negatively-stained whole-mount electron microscopy following the same procedures previously described (Miyakawa et al., 1987). Fine structure analysis of mung bean tissue was done as described (Dai et al., 1998).
Pulsed-field gel electrophoresis
PFGE analyses of mitochondria and mitochondrial nucleoids were conducted exactly as described in our previous reports (Dai et al., 2005). The amounts of mitochondria and mitochondrial nucleoids used for PFGE were 200 μg and 20 μg, respectively. Protein content was measured by the Micro BCA protein assay kit from Pierce #23235. In mitochondria/mitochondrial nucleoid DNA synthesis analysis, an autoradiogram exposure was performed after the gel was dry. To count α-[32p]-dCTP incorporation into newly-synthesized wb- and fm-DNA, respectively, after PFGE the dried gel was sliced into wb and fm fractions according to the autoradiogram and then counted in a scintillation counter (Hewlett Packard).
In vitro DNA synthesis of mitochondrial nucleoids and in organello DNA synthesis of mitochondria were performed exactly as in our previous report (Dai et al., 2005).
For Southern blot analysis, the probe used was either pure mtDNA or a PCR-generated cox 3 gene fragment, and the same results were obtained with both probes.
Topoisomerase I treatment of mitochondrial nucleoid DNA
Mitochondrial nucleoid (isolated from 2 mg of mitochondrial protein as determined by the Micro BCA protein assay kit from Pierce #23235) DNA synthesis was carried out for 1 h. Mitochondrial nucleoids were then precipitated and dissolved in buffer [20 mM Tris-HCl (pH 7.5), 50 mM NaCl, 0.1 mM EDTA, 1 mM DTT]. Different concentrations of topoisomerase I (Promega; 0, 0.4, 2 and 20 units) were added to the mitochondrial nucleoid samples with newly-synthesized mtDNA. Topoisomerase I treatment was carried out at 37℃ for one more hour. After the reaction was stopped, agarose plug embedding and PFGE analysis were carried out. An identical experiment was done exactly as above except that topoisomerase I treatment (0, 1, 5 and 20 units/reaction) was added to the mitochondrial nucleoids without processing for DNA synthesis. A second PFGE analysis was used to assess the change in mobility of fm DNA treated with topoisomerase I (0 and 20 units/reaction), as described in the following section.
High salt/latrunculin B treatment followed by DNase I digestion of mitochondrial nucleoids
Purified mitochondrial nucleoids were treated with 1 M KCl in buffer (20 mM HEPES, pH 7.4, 2 mM MgCl2, 1 mM PMSF) on ice for 15 min. Lat B treatment (60 μM) was carried out in buffer (2 mM Tris-HCl, pH 8.0, 0.2 mM CaCl2, 0.2 mM ATP, 0.25 mM sucrose, 1 mM PMSF) for 30 min on ice. Mitochondrial nucleoids were then centrifuged at 20,000 × g twice for 20 min each.
Sequential DNase I (0, 0.05, 0.25 and 1.25 μg/ml) treatment was then performed in buffer (10 mM TES, pH 7.2, 10 mM MgCl2) at 25℃ for 30 min.
Changes in the mobility of fast-moving (fm) DNA caused by ethidium bromide plus UV treatment as detected by PFGE
Mitochondrial nucleoid DNA was fractionated by PFGE as described above. The UV/EtBr treatment was similar to the method applied by Brugère et al. (2000) with some modifications. After the conventional one-dimensional PFGE, gel strips containing the wb and fm fractions were sliced out and treated in-gello with EtBr and UV-irradiation either alone or in combination. Gel slices were immersed in EtBr (13 mM) for 60 min followed by UV irradiation for 7.5 min (UV-366 nm, 115 volts, 60 Hz, 0.16 amps, UVP, Inc). The UV source was 25 mm above the gel. After EtBr/UV treatment, the treated gel strips were then subjected to a second PFGE run with an electrophoretic direction 90° to the first run. The second dimension of pulsed field gel electrophoresis was carried out with the same program as described above. Southern blot analysis was then performed to detect mitochondrial DNA as described above.
Mitochondrial nucleoid DNA preparation and EM examination
Mitochondrial nucleoids were prepared as described. The mitochondria nucleoids were lysed with 0.1 mg of proteinase K and 1% sarkosyl followed by incubation at 50℃ for 1 h. MtDNA was purified by phenol-chloroform extraction and ethanol precipitation. The whole process was carried out using extremely gentle motions. The EM analysis of mtDNA employed the droplet method (Lang and Mitani, 1970).
Fluorescent visualization of mitochondrial nucleoids
A mitochondrial nucleoid sample was double-stained with the DNA-specific dye YOYO-3 (5 μg/ml) and the cardiolipin-specific dye NAO (10 N-nonyl acridine orange, 5 μM) or double stained with the DNA-specific dye YOYO-1 (0.5 μM) and the F-actin-specific dye Alexa TM 594 Phalloidin (5 units/ml). The resulting double-stained sample was excited using a 488 nm argon laser for YOYO-1/NAO, and fluorescence was collected with a 510-525 nm band pass filter; the fluorescence of YOYO-3/Alexa TM 594 Phalloidin was excited using a 543 nm line of a He-Ne laser, and fluorescence was collected with a 565-615 nm band pass filter.
RESULTS
Fine structure of mitochondrial nucleoids
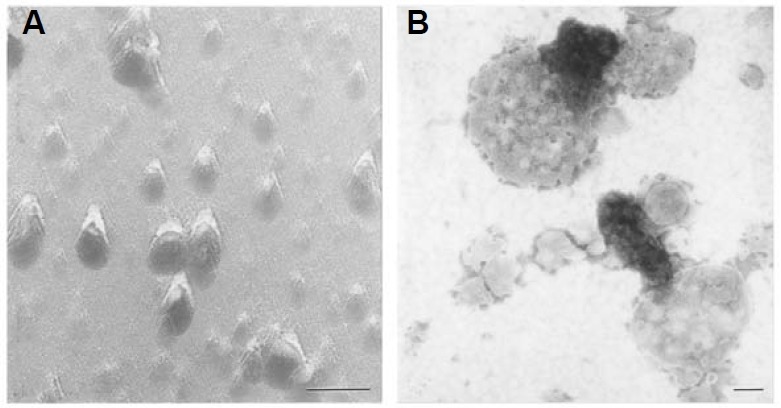
Mung bean mitochondrial nucleoid preparations appeared as a relatively homogeneous population of ~70 nm particles by freeze fracture electron microscopy (Fig. 1A). In negatively-stained whole-mount analysis, an electron-dense chromatin-like structure and a vesicle component were visualized (Fig. 1B); most of the chromatin-like elements appeared to be either linked or located immediately next to the membrane vesicles.
Fig. 1. Visualization of mitochondrial nucleoids by electron microscopy. Freshly isolated mitochondrial nucleoid samples from a highly purified orthodox mitochondrial population (Dai et al., 1998) were freeze fractured (A), or negatively stained with 1% sodium phospho-tungstate at pH 7.0 (B) for electron microscopy. Bars: 100 nm.
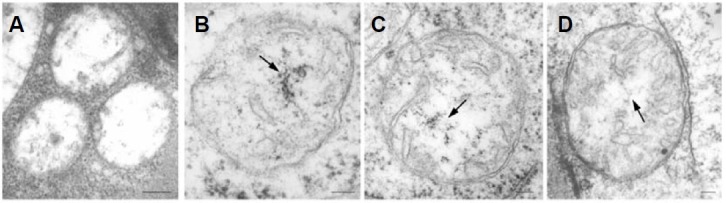
This chromatin-like structure of mitochondrial nucleoids was also identified in plant tissues in situ as shown in Fig. 2. During early seed germination, cotyledon mitochondrial nucleoids underwent a distinct change in fine structure characteristic of their dynamic nature in vivo. The conspicuous structural changes observed in mitochondrial nucleoids as dormant seeds sprout into rapidly-growing seedlings appear to provide in vivo evidence of the progressive development of the nucleoids’ functional competence (Figs. 2A-2C). This change of cotyledon mtDNA organization during early seed germination was also reported previously (Dai 2005, Table 1). No visible chromatin- or fibril-like structures were observed in 12 h-4℃ immersed cotyledon mitochondria (Fig. 2A). However, chromatin-like structures were observed in 12 h-27℃ cotyledon mitochondria (Fig. 2B, see arrow). Fibril-like mitochondrial nucleoids instead of chromatin-like structures were visualized when cotyledons reached day 1 of seed germination (Fig. 2C, see arrow). Figure 2D shows in situ mitochondrial nucleoids from a hook region of a day 3 mung bean seedling, and they exhibit a fibril-like mtDNA organization phase similar to that of the day-1 cotyledon mitochondrial nucleoids shown in Fig. 2C.
Fig. 2. Chromatin-like DNA structures in mitochondria from different plant tissues. Cotyledons of mung bean seeds immersed in 4℃ water for 12 h (A), 27℃ water for 12 h (B), or 27℃ water for 12 h followed by 12 h of growth on vermiculite (C), and the hook region of 3-day-old etiolated mung bean seedlings were fixed, embedded, sectioned and stained for electron microscopy as described. No visible chromatin- or fibril-like structures is observed in 12 h-4℃ immersed cotyledon mitochondria (A). Chromatin-like structures are observed in 12 h-27℃ cotyledon mitochondria (B, see arrow). Fibril-like mitochondrial nucleoids instead of chromatin-like structure are visualized when cotyledons reached day 1 of seed germination (C, see arrow). In situ mitochondrial nucleoids from a hook region of a day 3 mung bean seedling exhibiting a fibril-like mtDNA organization phase similar to that of the day-1 cotyledon mitochondrial nucleoids is shown in Fig. 2D. Bars:100 nm.
Confocal microscopic analysis of mitochondrial nucleoids
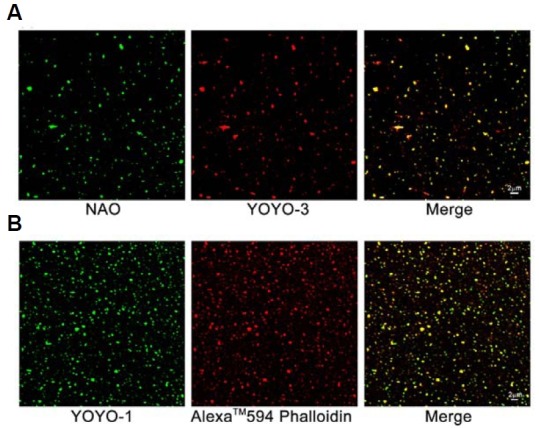
Mitochondrial nucleoids visualized by confocal microscopy after double staining with NAO and YOYO-3 (Fig. 3A) showed the co-occurrence of mtDNA (stained by YOYO-3) and cardiolipin (stained by NAO, a mitochondrial inner membrane-specific dye that binds specifically to cardiolipin) in isolated mitochondrial nucleoids. Associations between mtDNA and F-actin were also revealed by double staining of mitochondrial nucleoids with YOYO-1 and Alexa TM 594 Phalloidin (F-actin-specific dye) and subsequent confocal microscopic analysis (Fig. 3B).
Fig. 3. Mitochondrial nucleoids associated with the inner mitochondrial membrane and F-actin were detected by fluorescent image analysis. Mitochondrial nucleoid samples were double stained with the DNA-specific dye YOYO-3 and the cardiolipin-specific dye NAO (A) or were double stained with the DNA-specific dye YOYO-1 and the F-actin-specific dye Alexa TM 594 Phalloidin (B) followed by confocal microscopic analysis. The merged yellow fluorescence shown in (A, B) indicates that mitochondrial nucleoids are composed of both mtDNA and mitochondrial membrane (A), and an association of F-actin with mtDNA is also evidenced (B).
Analysis of mitochondrial nucleoid DNA by pulsed field gel electrophoresis
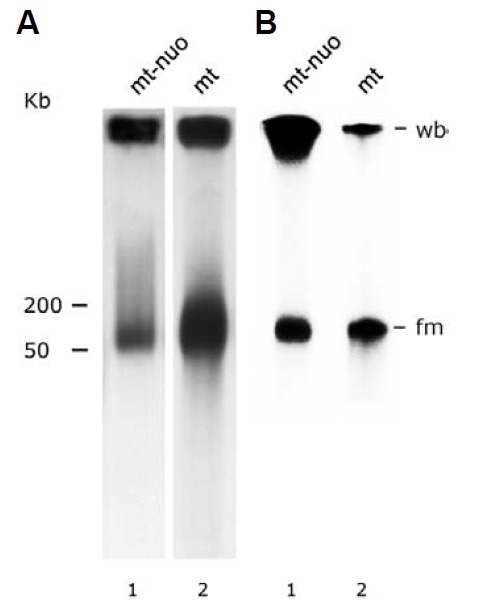
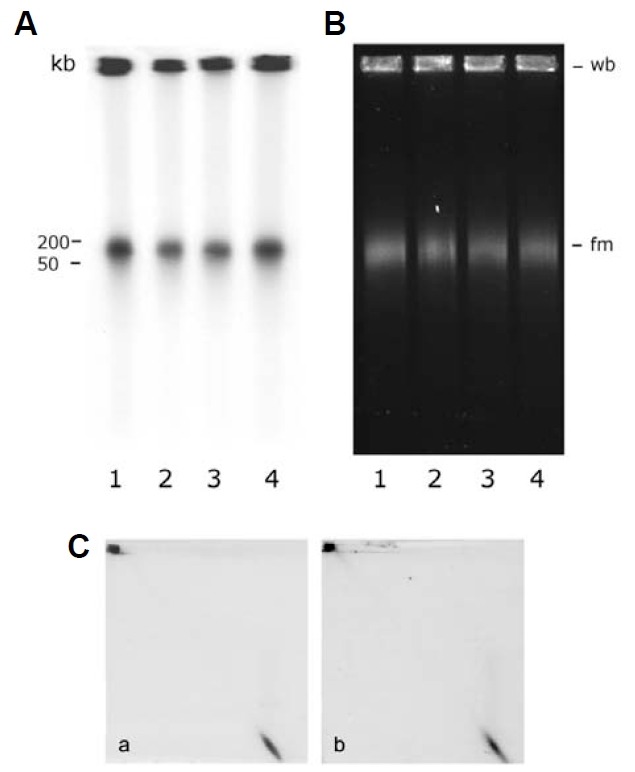
Mung bean mitochondrial nucleoids isolated by the classical method (Dai et al., 2005; Miyakawa et al., 1987) at the 15-30% sucrose-gradient interface possess heterogeneous constituent DNA molecules. A bimodal migration pattern in pulsed field gel electrophoresis (PFGE) characterizes the gross heterogeneity of these molecules. The majority of mitochondrial nucleoid DNA remained in the sample-loading well (about 68% of the total mitochondrial nucleoid DNA by counting, as described in “Mate-rials and Methods”). In addition to this well-bound (wb) popula-tion, a fast-moving (fm) population of DNA molecules migrated as a smear zone corresponding to 50-200 kb double-stranded DNA (Fig. 4A, lane 1). This result indicates that conformationally complex DNA molecules with a wide range of molecular masses are present in isolated mung bean mito-chondrial nucleoids. Total DNA from intact mitochondria exhibited a PFGE fractionation pattern similar to that of mitochondrial nucleoids except that the proportion of the fm fraction was con-siderably greater (about 67% of the total by counting, Fig. 4A, lane 2). Mitochondrial nucleoids’ DNA synthesis was assessed via pulse labeling with [32P]-dCTP. Similar to what was found regarding pre-existing mtDNA in mitochondrial nucleoids and mitochondria (shown in Fig. 4A), in vitro newly synthesized mitochondrial nucleoid DNA contained 59% newly synthesized wb DNA (Fig. 4B, lane 1) compared to only 33% newly synthe-sized wb DNA in mitochondria (Fig. 4B, lane 2).
Fig. 4. Fractionation of mitochondrial nucleoid DNA by PFGE. (A) Southern hybridization analysis of DNA from mitochondrial nucleoids (mt-nuo, lane 1) and from mitochondria (mt, lane 2) after size fractionation by pulsed field gel electrophoresis. The amounts of mitochondria and mitochondrial nucleoids used for PFGE were 200 μg and 20 μg, respectively. The probe used was pure mtDNA. The well-bound fraction (wb) remained at the top of the gel. The molecular weight (kb) of the DNA is indicated at the left side of the figure. (B) Autoradiogram of newly-synthesized mitochondrial nucleoid DNA (mt-nuo, lane 1) and mitochondrial DNA (mt, lane 2) fraction-ated by PFGE. The X-ray film was exposed for a very short period (~5 h).
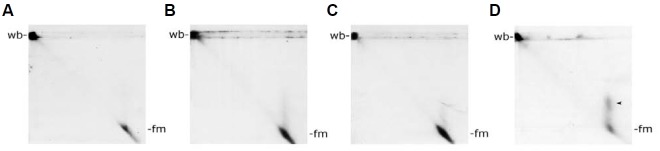
To characterize the heterogeneity of mitochondrial nucleoid DNA further, we carried out a two-dimensional PFGE analysis. After the conventional one-dimensional PFGE, gel strips con-taining the immobile wb-fraction and fractionated fm molecules were sliced out and treated in-gello with EtBr and UV-irradiation either alone or in combination. These treated gel strips were then subjected to a second PFGE run with an electrophoretic direction 90° to the first run. Upon EtBr treatment, a minor frac-tion of the fm population of the first PFGE run now appeared asa faint streak with varying degrees of migration retardation which were absent in the control (compare Fig. 5B to Fig. 5A). Under our experimental conditions, UV irradiation alone was less effective at retarding the migration of the fm population (Fig. 5C). Combined EtBr-UV treatment synergistically retarded a sizable fraction of the fm population (Fig. 5D). No significant effect on the wb population was observed irrespective of the treatment employed (Figs. 5A-5D). Ethidium bromide is an intercalating agent. By inserting between the stacked bases in double-stranded DNA, EtBr may cause DNA to become less compact and lead to migration changes of some fm DNA molecules during second-dimension gel electrophoresis. UV treatment may nick DNA in the agarose gel which alter DNA migration in the second-dimension PFGE analysis. The combined effects of EtBr and UV can cause serious mtDNA nicking medi-ated by ethidium-enhanced UV irradiation. Changes in mtDNA complexity and configuration cause the obvious migration alterations shown in D (see arrowheads).
Fig. 5. Two-dimensional pulsed field gel electrophoretic analysis of the mobility of mitochondrial nucleoid fm-DNA with and without EtBr/UV treatment. Mitochondrial nucleoid DNA was fractionated by first-dimension PFGE followed by in-gel EtBr/UV treatment as described in “Materials and Methods”. Second-dimension PFGE was then carried out with the same program as the first-dimension PFGE. Mitochondrial nucleoid DNA in the gel was then detected by Southern blot analysis as described above. No EtBr/UV treatment, only EtBr treatment, only UV treatment, and EtBr/UV treatment are shown in (A, B, C and D), respectively. After second- dimension PFGE analysis, a minor fraction of the fm population appears as a faint streak with varying degrees of migration retardation upon EtBr and UV treatment, respectively (Figs. 5B and 5C). Combined EtBr-UV treatment synergistically retarded a sizable fraction of the fm population (Fig. 5D).
EM analysis of mitochondrial nucleoid DNA
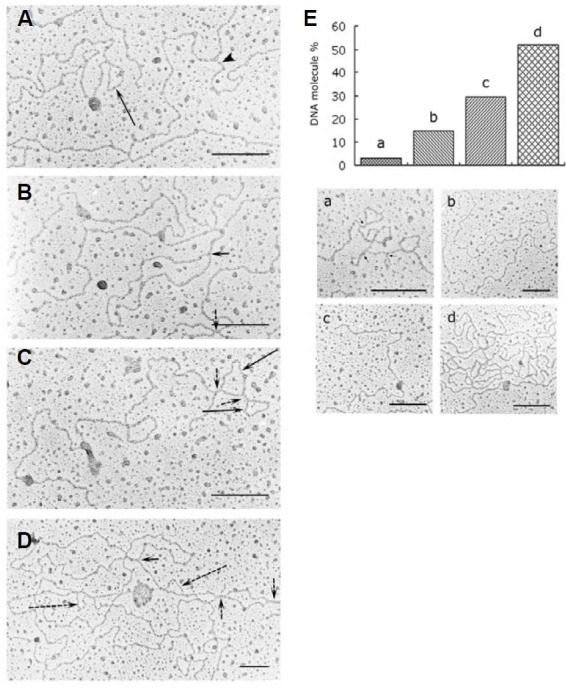
By summing restriction fragments of total DNA isolated from intact mitochondria, the mung bean mitochondrial genome was estimated to be 312 kb, or about 106 μm in length (unpublished data). Despite attempting multiple methods under a variety of experimental conditions, only short DNA fragments (~2-9 kb) were recovered from agarose gel slices containing wb-DNA in our preliminary experiments (unpublished data). These molecules are not very informative. We report here an EM study of DNA purified from mitochondrial nucleoids. As shown in Fig. 6, approximately 70% of visualized mitochondrial nucleoid DNA molecules have a sigma-like structure (Figs. 6B and 6D, short arrows), a circular structure of varying contour lengths (Figs. 6A and 6C, long arrows), a D-loop bubble (Fig. 6D, long dashed arrows) or a branched structure (Fig. 6A, arrowhead). Multiple such structures are often seen interspersed along a molecule. Even more complex molecules with combinations of intertwined circular and branching structures have also been visualized. In addition to these complex molecules, which comprise more than half of the total molecules analyzed on a random basis, simple linear, open circular, and supercoiled molecules have also been observed (Fig. 6E). The relative frequencies of these different types of mitochondrial nucleoid DNA molecules are shown in Fig. 6E. Molecules with a four-way branching segment spanning roughly 70-260 nm (~25 to 90 bp) were fre quently encountered (Figs. 6B-6D, short dashed arrows).
Fig. 6. Electron micrographs of mitochondrial nucleoid DNA. (A-D) present four examples of different conformational structures of mitochondrial nucleoid DNA. The long arrow in (A) indicates a small circular mtDNA structure. The arrowhead in (A) shows a branch from a linear mtDNA molecule. Sigma-like structures are exhibited in (B, D) as indicated by short arrows. The dotted short arrows in (B, C, and D) indicate possible “four-stranded joint regions.” The long arrows in (C) indicate two continuous small circles of mtDNA. The dotted long arrows in (D) indicate D-loop bubble structures. More than 50% of the mitochondrial nucleoid DNA molecules we examined showed highly complex structures, as exhibited in the histogram of Fig. 6E (d). The distributions of supercoiled DNA (a), open circular DNA (b), and linear form DNA (c) among the mitochondrial nucleoid population we examined are presented in histogram of (E). EM pictures of supercoiled DNA, open circular DNA, linear DNA and complex DNA forms are presented as a, b, c and d, respectively, at the bottom section of (E). The total number of DNA molecules we examined was 121. Bars: 200 nm in (A) through (E).
Effect of topoisomerase I on mitochondrial nucleoid DNA
From the results presented above, the presence of molecular heterogeneity in both the wb and the fm populations of mitochondrial nucleoid DNA molecules appears evident (Figs. 4-6). We carried out an experiment with exogenously supplemented topoisomerase I to ascertain whether its activity would affect the conformational heterogeneity of either newly-synthesized or pre-existing mitochondrial nucleoid DNA. Mitochondrial nucleoids were first allowed to label for an hour before administering topoisomerase I at different concentrations. As shown in Fig. 7, even at the highest concentrations employed, topoisomerase I had no detectable effect on the [32P]-dCTP-labeled newly-synthesized wb- or fm-form DNA (Fig. 7A, lanes 2-4). The same was true for EtBr-stained total mitochondrial nucleoid DNA of both forms (Fig. 7B, lanes 2-4). That there was no effect of topoisomerase I on the conformational heterogeneity of mitochondrial nucleoid DNA was also revealed by second-dimen-sion PFGE analysis (Fig. 7C). These results indicate that under our experimental conditions, fm and wb DNA are insensitive to topoisomerase I in so far as gross conformation alterations are concerned, whereas EtBr-UV treatment did so alter fm-DNA (compare Fig. 7C to Fig. 5).
Fig. 7. The effect of topoisomerase I treatment on mitochondrial nucleoid DNA. (A) In vitro mitochondrial nucleoid DNA synthesis was carried out for 1 h as described in “Materials and Methods”. The newly-synthesized mitochondrial nucleoid DNA was then treated with different concentration of topoisomerase I. Lanes 1, 2, 3 and 4 show the newly synthesized mitochondrial nucleoid DNA PFGE migration pattern after treatment with 0, 0.4, 2 and 20 units of topoisomerase I, respectively. (B) presents the PFGE fractionation pattern of mitochondrial nucleoid DNA after topoisomerase I treatment. Mitochondrial nucleoids not processed for DNA synthesis were treated with 0, 1, 5 and 20 units of topoisomerase I and are presented in lanes 1, 2, 3 and 4, respectively. No migration alteration of well-bound (wb) or fast-moving (fm) DNA caused by topoisomerase I treatment was detected in a second dimensional PFGE analysis [(C), treatment with 0 and 20 units topoisomerase I/reaction is shown in a and b, respectively].
Effects of high salt and latrunculin B (Lat B) on mitochondrial nucleoid DNA
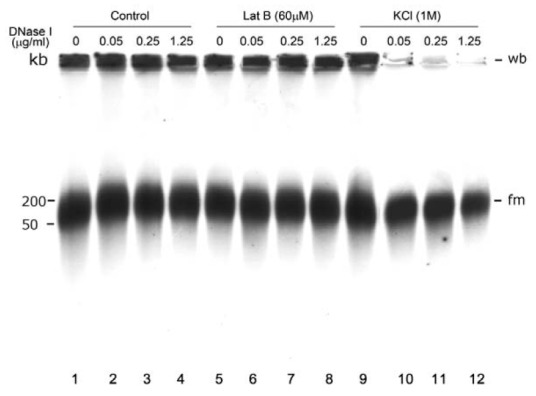
To assess the potential roles of mitochondrial nucleoid-associated actin and mitochondrial nucleoid DNA-binding proteins in protecting mtDNA, mitochondrial nucleoids were treated with high salt or the actin depolymerization reagent latrunculin B, followed by DNase I treatment. As shown in Fig. 8, neither KCl (1 M) nor latrunculin B (60 μM) treatment had any effect on the heterogeneous migration of the wb and fm forms (compare lanes 1, 5 and 9). However, high-salt treatment may change the sensitivity of mitochondrial nucleoid DNA to DNase I degrada-tion (compare lanes 10, 11, 12 to lanes 2, 3, 4 and lanes 6, 7, 8, respectively).
Fig. 8. Effects of high salt and latrunculin B treatments on the sensitivity of mitochondrial nucleoid DNA to DNase I degradation. Purified mitochondrial nucleoids were treated with 1 M KCl or 60 μM latrunculin B (Lat B) followed by different concentration of DNase I. No change in the mtDNA PFGE migration pattern was detected in response to high salt or latrunculin B treatment of mitochondrial nucleoid DNA (compare lanes 1, 5, and 9). However, the sensitivity of mtDNA to DNase I significantly increased after mitochondrial nucleoids were stripped by KCl. The same effect was not found with latrunculin B-treated mitochondrial nucleoids. DNase I was applied at 0 μg/ml (lanes 1, 5, and 9), 0.05 μg/ml (lanes 2, 6, and 10), 0.25 μg/ml (lanes 3, 7, and 11) and 1.25 μg/ml (lanes 4, 8, and 12).
DISCUSSION
Our freeze fracture-electron microscopic analysis of mitochondrial nucleoids isolated by conventional methods from mung bean seedlings revealed a relatively homogeneous population of particles (~70 nm). The ultrastructure of the mitochondrial nucleoids was studied in vitro and compared with those visualized in situ. We found by electron microscope that negatively-stained mitochondrial nucleoids manifested as a chromatin-like structure linked to a membrane component (Fig. 1). This phenomenon was further characterized by fluorescent analysis. YOYO-3-stained mtDNA (in red) merged with NAO-stained cardiolipin (in green) and appeared as yellow specks in Fig. 3A. This mtDNA-membrane relationship has not been extensively investigated heretofore, but it is consistent with findings showing that a fast-sedimenting plant mtDNA-containing fraction exhibiting nucleic acid synthesis activity colocalizes with membranous vesicles derived from the inner mitochondrial membrane (Echeverria et al., 1991; Fey et al., 1999, our unpublished data). Most studies of mitochondrial nucleoids have focused on mtDNA binding proteins and their functions, including mtDNA replication, transcription and translation (Bogenhagen et al., 2008; Chen and Butow, 2005; Dai et al., 2005; Iborra et al., 2004; Sakai et al., 2004; Spelbrink, 2010; Wang and Begenhagen, 2006). How membranes associate with mtDNA/ mitochondrial nucleoids and the functional role of the attached membranes in plant mitochondrial nucleoids remain unclear. These are essential and fundamental questions that merit further investigation.
Similar chromatin-like DNA structures were also identified within mitochondrial profiles of thin tissue sections (Fig. 2). The fact that the ultrastructure of mitochondrial nucleoids appears to vary in different mitochondria during cotyledon development suggests that the organization of DNA in mitochondrial nucleoids is not static but is capable of dynamic changes. This phe-nomenon probably reflects their changing functional status in vivo, and it was also reported in our previous study (Dai et al., 2005). Under the etiolated conditions employed in this study, respiration is the only energy conversion pathway available for mung bean seedling development. Thus, the activation of mitochondrial genes in preparation for rapid growth is mandatory. Mitochondrial DNA in cotyledons transformed from invisibility to chromatin-like and then to fibril-like structures as the cotyledon aged (Fig. 2 and Table 1; Dai et al., 2005). The synchronized dynamic changes in the mtDNA organization during cotyledon mitochondrial development reflects the progressive changes in mitochondrial gene expression (e.g., from transcriptionally inactive chromatin form to transcriptionally active fibril form) that takes place during this period. Our unpublished data also showed that the respiration activity of cotyledon mitochondria of seeds immersed at 4℃ for 12 h was undetectable. The respiration activity of cotyledon mitochondria increased gradually during seed germination, became obvious at day 1 and peaked at day 3 of seed germination. Thus, the observed structural changes of mitochondrial nucleoids appear to be coordinated with mitochondrial biogenesis.
It was proposed that plant mtDNA may use a T4 phage-like recombination-dependent mechanism of replication and/or an alternative rolling-circle mode of replication (Backert and Börner, 2000; Oldenburg and Bendich, 2001). The predicted replication-intermediate structures from these two replication mechanisms have been proposed as the probable cause of the immobile wb form in PFGE fractionation analysis. In addition, the ratio of wbform to fm-form is significantly higher in mitochondrial nucleoid DNA than in mitochondrial DNA detected by blot hybridization (Fig. 4A). The same phenomenon was also found between newly synthesized mitochondrial nucleoid DNA (in vitro) and mitochondrial DNA (in organelle), as shown in Fig. 4B. These results suggest that the conversion of wb form DNA to fm form is more prominent in mitochondria than in mitochondrial nucleoids, as we reported previously (Dai, 2005). Obviously, the surrounding environment essential for wb to fm conversion is lacking in isolated mitochondrial nucleoids. These results suggest that not all mtDNA molecules, including some capable of in organelle replication, are organized into a structure with a defined conformation and density identical to that of mitochondrial nucleoids isolatable by conventional procedures.
Although the size range of the mitochondrial nucleoid DNA molecules that we observed was less than optimal, nonetheless our finding of various irregular and complex DNA structures (Fig. 6) is consistent with other plant mtDNA molecules that have been visualized (Backert and Börner, 2000; Backert et al., 1996, 1997; Bendich, 1996; Lilly and Havey, 2001; Oldenburg and Bendich, 1998). It is likely that these irregular and complex DNA structures are temporary intra-/inter-molecular pairings of short repetitive sequences and/or single-stranded regions abundant in plant mtDNA genomes. Alternatively, they could be unresolved Holliday-type cross-over intermediate structures. Among 121 mitochondrial nucleoid DNA molecules we examined, the length ranged from 1 to 56 μm, but the most abundant ones were 6-8 μm long. At present, no conclusions can be drawn from these data alone due to the sample size and the lengths of the molecules observed.
Linear molecules contained in the 50-200 kb smear zone have also been observed (Backert and Börner, 2000; Backert et al., 1997; Oldenburg and Bendich, 1996; 1998; 2001). Our finding that ethidium-UV treatment retarded a fraction of mung bean mtDNA molecules present in the 50-200 kb zone (Fig. 5) indicates that this zone contains not only linear subgenomic fragments but also supercoils or other intricately compact structures that can be relaxed by ethidium-enhanced UV irradiation-mediated nicking. The fact that topoisomerase I did not retard the mobility of the 50-200 kb molecules (Fig. 7C) suggests that the compact structures relaxed by EtBr-UV treatment are not likely to consist of regular negative supercoils from covalently closed circular molecules.
The binding of actin to mitochondrial nucleoids has been reported by us and by the other investigators (Boldogh et al., 2003; Dai et al., 2005; Wang and Bogenhagen, 2006). We suggest here that mtDNA-bound actin may mediate the movement and segregation of mtDNA directly or indirectly, and it may play an important role in mtDNA inheritance. Boldogh et al. (2003) concluded that mtDNA is linked to cytosolic actin through the mitochondrial protein complex known as the mitochore, and that this interaction between mtDNA and actin is critical for mtDNA inheritance.
To determine whether actin or other mitochondrial nucleoproteins protect mtDNA from endonucleases, mitochondrial nucleoids were treated with high salt or latrunculin B followed by DNase I digestion (Fig. 8). Neither high salt nor latrunculin B treatment altered the PFGE migration pattern of mitochondrial nucleoid DNA. However, stripping away some loosely bound or peripheral proteins associated with mitochondrial nucleoids may significantly increase the sensitivity of mtDNA to DNase I digestion. This finding provides evidence that some KCl-removable proteins associated with mtDNA in mitochondrial nucleoids are involved in protecting mtDNA from endonucleases.
Acknowledgments
This research was supported by research grants from the Na-tional Science Council (NSC 91-2311-B-001-100) and from Academia Sinica, ROC. We deeply thank Emeritus Professor Kwen-Sheng Chiang (The University of Chicago) for his most cogent advice and insightful discussions. AL was supported by an NSC post-doctoral fellowship. We thank Dr. Wann-Neng Jane for his help with the EM study.
References
- 1.Andre C.P., Walbot V. Pulsed-field gel mapping of maize mitochondrial chromosomes. Mol. Gen. Genet. (1995);247:255–263. doi: 10.1007/BF00705657. [DOI] [PubMed] [Google Scholar]
- 2.Backert S., Börner T. Phage T-4-like intermediates of DNA replication and recombination in the mitochondria of the higher plant Chenopodium album (L.). Curr. Genet. (2000);37:304–314. doi: 10.1007/s002940050532. [DOI] [PubMed] [Google Scholar]
- 3.Backert S., Lurz R., Börner T. Electron micro-scopic investigation of mitochondrial DNA from Chenopodium album (L.). Curr. Genet. (1996);29:427–436. doi: 10.1007/BF02221510. [DOI] [PubMed] [Google Scholar]
- 4.Backert S., Lurz R., Oyarzabal O.A., Borner T. High content, size and distribution of single-stranded DNA in the mitochondria of Chenopodium album (L.). Plant Mol. Biol. (1997);33:1037–1050. doi: 10.1023/a:1005791310886. [DOI] [PubMed] [Google Scholar]
- 5.Bendich A.J. Structural analysis of mitochondrial DNA molecules from fungi and plants using moving pictures and pulsed-field gel electrophoresis. J. Mol. Biol. (1996);255:564–588. doi: 10.1006/jmbi.1996.0048. [DOI] [PubMed] [Google Scholar]
- 6.Bogenhagen D.F., Rousseau D., Burke S. The layered structure of human mitochondrial DNA nucleoids. J. Biol. Chem. (2008);283:3665–3675. doi: 10.1074/jbc.M708444200. [DOI] [PubMed] [Google Scholar]
- 7.Boldogh I.R., Nowakowski D.W., Yang H.C., Chung H.S., Karmon S., Royes P., Pon L.A. A protein complex containing Mdm10p, Mdm12p, and Mmm1p links mitochondrial membranes and DNA to cytoskeleton-based segregation machinery. Mol. Biol. Cell. (2003);14:4618–4627. doi: 10.1091/mbc.E03-04-0225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brugère J.-F., Cornillot E., Méténier G., Bivares C.P. In-gel DNA radiolabelling and two-dimensional pulsed field gel electrophoresis procedures suitable for fingerprinting and mapping small eukaryotic genomes. Nucleic Acids Res. (2000);28:e48. doi: 10.1093/nar/28.10.e48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen X.J., Butow R.A. The organization and inheritance of the mitochondrial genome. Nat. Rev. Genet. (2005);6:815–825. doi: 10.1038/nrg1708. [DOI] [PubMed] [Google Scholar]
- 10.Dai H., Lo Y.S., Jane W.N., Lee L.W., Chiang K.S. Population heterogeneity of higher-plant mitochondria in structure and function. Eur. J. Cell Biol. (1998);75:198–209. doi: 10.1016/S0171-9335(98)80062-9. [DOI] [PubMed] [Google Scholar]
- 11.Dai H., Lo Y.S., Wu C.Y., Tsou C.L., Hsu G.S., Chern C.G., Ruddat M., Chiang K.S. Protein synthesis in isolated mitochondria of rice (Oryza sativa L.) seedlings. Plant Physiol. (1991);96:319–323. doi: 10.1104/pp.96.1.319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dai H., Lo Y.S., Litvinchuk A., Wang Y.T., Jane W.N., Hsiao L.J., Chiang K.S. Structural and functional characterizations of mung bean mitochondrial nucleoids. Nucleic Acids Res. (2005);33:4725–4739. doi: 10.1093/nar/gki783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Echeverria M., Robert D., Carde J.P., Litvak S. Isolation from wheat mitochondria of a membrane-associated high molecular weight complex involved in DNA synthesis. Plant Mol. Biol. (1991);16:301–315. doi: 10.1007/BF00020561. [DOI] [PubMed] [Google Scholar]
- 14.Eves E.M., Chiang K.S. Genetics of Chlamydomonas reinhardtii diploids. Genetics. (1984);107:563–576. doi: 10.1093/genetics/107.4.563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fey J., Vermel M., Grienenberger J.-M., Marechal-Drouard L., Gualberto J.M. Characterization of a plant mitochondrial active chromosome. FEBS Lett. (1999);458:124–128. doi: 10.1016/s0014-5793(99)01140-0. [DOI] [PubMed] [Google Scholar]
- 16.Iborra F.J., Kimura H., Cook P.R. The functional organization of mitochondrial genomes in human cells. BMC Biol. (2004);2:9. doi: 10.1186/1741-7007-2-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kuroiwa T. Mitochondrial nuclei. Internat. Rev. Cyto. (1982);75:1–59. doi: 10.1016/s0074-7696(08)61001-3. [DOI] [PubMed] [Google Scholar]
- 18.Kuroiwa T., Kawano S., Nishibayashi S., Sato C. Epifluorescent microscopic evidence for maternal inheritance of chloroplast DNA. Nature. (1982);289:481–483. doi: 10.1038/298481a0. [DOI] [PubMed] [Google Scholar]
- 19.Kuroiwa T., Fujie M., Kuroiwa J. Synthesis of mitochondrial DNA occurs actively in a specific region just above the quiescent center in the root meristem of Pelargonium sonale. J. Cell Sci. (1992);101:483–493. [Google Scholar]
- 20.Lang D., Mitani M. Simplified quantitative electron microscopy of biopolymers. Miopolymers. (1970);9:373–379. doi: 10.1002/bip.1970.360090310. [DOI] [PubMed] [Google Scholar]
- 21.Levings III C.S., Brown G.G. Molecular biology of plant mitochondria. Cell. (1989);56:171–179. doi: 10.1016/0092-8674(89)90890-8. [DOI] [PubMed] [Google Scholar]
- 22.Lilly J.W., Havey M.J. Small, repetitive DNAs contribute significantly to the expanded mitochondrial genome of cucumber. Genetics. (2001);159:317–328. doi: 10.1093/genetics/159.1.317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mackenzie S.A., Logan D.C. The unique biology of mitochondrial genome instability in plants. In Plant Mitochondria. Blackwell Pub.; Oxford, UK: (2007). pp. 36–49. [Google Scholar]
- 24.Maleszka T., Skelly P.J., Clark-Walker G.D. Rolling circle replication of DNA in yeast mitochondria. EMBO J. (1991);10:3923–3929. doi: 10.1002/j.1460-2075.1991.tb04962.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Miyakawa I., Sando N., Kawano S., Nakamura S., Kuroiwa T. Isolation of morphologically intact mitochondrial nicleoids from the yeast, Saccharomyces cerevisiae. J. Cell Sci. (1987);88:431–439. doi: 10.1242/jcs.88.4.431. [DOI] [PubMed] [Google Scholar]
- 26.Miyakawa I., Okazaki-Higashi C., Higashi T., Furutani Y., Sando N. Isolation and characterization of mitochondrial nucleoids from the yeast Pichia jadinii. Plant Cell Physiol. (1996);37:816–824. doi: 10.1093/oxfordjournals.pcp.a029017. [DOI] [PubMed] [Google Scholar]
- 27.Narayanan K.K., André C.P., Yang J., Walbot V. Organization of a 117-kb circular mitochondrial chromosome in IR36 rice. Curr. Genet. (1993);23:248–254. doi: 10.1007/BF00351503. [DOI] [PubMed] [Google Scholar]
- 28.Nass M.M.K., Nass S., Afzelius B.A. The general occurrence of mitochondrial DNA. Exp. Cell Res. (1965);37:516–539. doi: 10.1016/0014-4827(65)90204-1. [DOI] [PubMed] [Google Scholar]
- 29.Newman S.M., Olga Z.-T., Periman P.S., Butow R.A. Analysis of mitochondrial DNA nucleoids in wild-type and a mutant strain of Saccharomyces cerevisiae that lacks the mitochondrial HMG box protein Abf2p. Nucleic Acids Res. (1996);24:386–393. doi: 10.1093/nar/24.2.386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Oldenburg D.J., Bendich A.J. Size and structure of replicating mitochondrial DNA in cultured tobacco cells. Plant Cell. (1996);8:447–461. doi: 10.1105/tpc.8.3.447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Oldenburg D.J., Bendich A.J. The structure of mitochondrial DNA from the liverwort, Marchantia polymorpha. J. Mol. Biol. (1998);276:745–758. doi: 10.1006/jmbi.1997.1581. [DOI] [PubMed] [Google Scholar]
- 32.Oldenburg D.J., Bendich A.J. Mitochondrial DNA from the Liverwort Marchantia polymorpha: circularly permuted linear molecules, head-to-tail concatemers, and a 5′ protein. J. Mol. Biol. (2001);310:549–562. doi: 10.1006/jmbi.2001.4783. [DOI] [PubMed] [Google Scholar]
- 33.Sakai A., Suzuki T., Nagata N., Sasaki N., Miyazawa Y., Saito C., Inade N., Nishimura Y., Kuroiwa T. Comparative analysis of DNA synthesis activity in plastid-nuclei and mitochondrial-nuclei simultaneously isolated from cultured tobacco cells. Plant Sci. (1999);140:9–19. [Google Scholar]
- 34.Sakai A., Takano H., Kuroiwa T. Organelle nuclei in higher plants: structure, composition, function, and evolution. In International Review of Cytology - a Survey of Cell Biology, Vol. 238. Elsevier Academic Press Inc.; San Diego, USA: (2004). pp. 59–118. [DOI] [PubMed] [Google Scholar]
- 35.Spelbrink J.N. Functional organization of mammalian mitochondrial DNA in nucleoids: history, recent developments, and future challenges. IUBMB Life. (2010);62:19–32. doi: 10.1002/iub.282. [DOI] [PubMed] [Google Scholar]
- 36.Spelbrink J.N., Li F.Y., Tiranti V., Nikali K., Yuan Q.P., Tariq M., Wanrooij S., Garrido N., Comi G., Morandi L., et al. Human mitochondrial DNA deletions associated with mutations in the gene encoding Twinkle, a phage T7 gene LF-like protein localized in mitochondria. Nat. Genet. (2001);28:223–231. doi: 10.1038/90058. [DOI] [PubMed] [Google Scholar]
- 37.Stevens B., Strathern J.N., Jones E.W., Broach J.R. Mitochondrial structure. In The Molecular Biology of the Yeast Saccharomyces. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: (1982). pp. 471–504. [Google Scholar]
- 38.Suzuki T., Kawano S., Kuroiwa T. Structure of three-dimensionnally rod-shaped mitochondrial nucleoids isolated from the slime mould, Physarum polycephalum. J. Cell Sci. (1982);58:241–261. doi: 10.1242/jcs.58.1.241. [DOI] [PubMed] [Google Scholar]
- 39.Wang Y., Bogenhagen D.F. Human mitochondrial DNA nucleoids are linked to protein folding machinery and metabolic enzymes at the mitochondrial inner membrane. J. Biol. Chem. (2006);281:25791–25802. doi: 10.1074/jbc.M604501200. [DOI] [PubMed] [Google Scholar]
- 40.Ward B.L., Anderson R.S., Bendich A.J. The mitochondrial genome is large and variable in a family of plants (Cucubitaceae). Cell. (1981);25:792–803. doi: 10.1016/0092-8674(81)90187-2. [DOI] [PubMed] [Google Scholar]


