Abstract
The circadian clock in plants regulates many important physiological and biological processes, including leaf movement. We have used an imaging system to genetically screen Arabidopsis seedlings for altered leaf movement with the aim of identifying a circadian clock gene. A total of 285 genes were selected from publicly available microarrays that showed an expression pattern similar to those of the Arabidopsis core oscillator genes. We subsequently isolated 42 homozygous recessive mutants and analyzed their leaf movements. We also analyzed leaf movements of activation tagging mutants that showed altered flowering time. We found that agl6-1D plants, in which AGAMOUS-LIKE 6 (AGL6) was activated by the 35S enhancer, showed a shortened period of leaf movement as well as a high level of ZEITLUPE (ZTL) expression, reduced amplitude of LATE ELONGATED HYPOCOTYL (LHY) expression, and arrhythmic TIMING OF CAB EXPRESSION1 (TOC1)/CIRCADIAN CLOCK ASSOCIATED1 (CCA1) expression. A shortened period of leaf movement was also seen in 35S-AGL6-myc plants, although 35S-amiRAGL6 plants, transgenic plants overexpressing an artificial miRNA (amiR) targeting AGL6, showed unaltered leaf movement. The amplitude of CHLOROPHYLL A/B BINDING PROTEIN 2 (CAB2) expression, a circadian output gene, was also reduced in agl6-1D plants. Taken together, these results suggest that AGL6 plays a potential role in the regulation of the circadian clock by regulating ZTL mRNA level in Arabidopsis.
Keywords: AGL6, CCA1, circadian clock, leaf movement, ZTL
INTRODUCTION
Plants occur in a wide range of climates and environments that have necessitated the evolution of different survival strategies and ingenious adaptations. Many biological processes in higher plants, such as leaf movement, diurnal changes in photosynthetic activities, and photoperiod-dependent flowering, are regulated by the circadian clock (Barak et al., 2000; McClung, 2001). Plants demonstrating circadian resonance will accumulate more chlorophyll and fix more carbon. Consequently, plants that correctly match their circadian clock period with the external light-dark cycle increase photosynthesis, growth, and survival rates, suggesting that the clock confers a higher level of fitness to plants (Green et al., 2002).
The circadian system has been categorized into three main components: oscillator, input pathway, and output pathway. The oscillator is the pacemaker that is responsible for generating self-sustained rhythmicity (Barak et al., 2000; McClung, 2008). In Arabidopsis, the central oscillator is proposed to consist of elements in multiple feedback loops (Edwards et al., 2006). The two proteins CIRCADIAN CLOCK ASSOCIATED1 (CCA1) and LATE ELONGATED HYPOCOTYL (LHY), which belong to the superfamily of MYB transcription factors, play a central role in one loop by repressing TIMING OF CAB EXPRESSION1 (TOC1), which subsequently induces CCA1 and LHY transcription. In a second loop, Pseudo-Response Regulator PRR7 and PRR9 are induced by CCA1 and LHY, which are subsequently repressed by CCA1 and LHY (Farre et al., 2005; Nakamichi et al., 2005; Salome and McClung, 2005). In a third loop, GIGANTEA (GI) positively regulates TOC1 (Locke et al., 2006). TOC1, CCA1, and LHY together negatively regulate GI. The input pathways connect the clock with the environmental signals, enabling the latter to synchronize with the central oscillator.
Light is a predominant entrainment cue in plants (Devlin, 2002). Phytochromes (PHYs) and Cryptochromes (CRYs) are major core components of the circadian clock through their contribution to light input (Somers et al., 1998). In addition, phototropins (PHOTs), ZEITLUPE (ZTL), and LOV (LIGHT OXYGEN OR VOLTAGE)/KELCH PROTEIN 2 (LKP2) may also actively mediate additional light input. ZTL is a circadian photoreceptor stabilized by GI in blue light; it has the ability to degrade TOC1 protein (Alonso et al., 2003; Kim et al., 2007). The circadian clock can be also entrained by temperature cycles according to the temperature difference between daytime and nighttime (Alonso et al., 2003; Devlin, 2002; Millar, 2004). Genes in the output pathway are the clock-regulated genes whose function is not necessary for endogenous circadian oscillation. CHLOROPHYLL A/B BINDING PROTEIN 2 (CAB2) is a hallmark gene of the downstream signaling activities of the circadian clock (Millar et al., 1995).
Although the components of the circadian clock system have been extensively studied, many components have not yet been identified and/or defined, leaving many questions still to be solved. In the study reported here, we used an imaging system to genetically screen mutants with an altered leaf movement phenotype. We identified that activation of AGL6 either by the 35S enhancer (agl6-1D) or by the 35S promoter (35S-AGL6- myc) caused a shortened period in leaf movement. In Arabidopsis agl6-1D mutants, ZTL expression was elevated, and the expression of oscillator genes and an output pathway gene was altered. Based on our results, we suggest that AGL6 is potentially functional in the regulation of the circadian clock in Arabidopsis.
MATERIALS AND METHODS
Plant materials
Experiments were performed using wild-type Arabidopsis (eco-type Columbia) seedlings, unless otherwise noted. The plants were grown under long-day conditions (16:8 h, light : dark) at 23℃ with an intensity of 120 μmol/m2/s. For RNA sampling in the circadian rhythm experiment, plants grown for 9 days under long-day conditions were transferred to continuous light conditions. After entrainment for 24 h, the plants were harvested at 4-h intervals for RNA extraction. For the leaf movement measurements, Arabidopsis seeds sown on solidified MS medium supplemented with 1.5% sucrose were incubated for 2 days in the dark at 4℃, at which time the germinated seedlings were transferred to a light-dark cycle of 12:12 h and grown for 3 days. These seedlings were then transferred to a regime of continuous white light for measurement.
Measurement of leaf movement and calculation of the period
The movement of the cotyledon tip was recorded at 30-min intervals using video cameras (AXIS 206M; http://www.axis.com) for 3 or 4 days and the time-lapse images were saved to produce a movie. The pixel positions of each cotyledon tip were detected by the NKTRACE program (Onai et al., 2004) based on software IGOR PRO (release 4.0.7.0; WaveMetrics, USA). Circadian periods, phase, and the relative amplitude error (RAE) were calculated using BRASS and fast Fourier transform- nonlinear least squares (FFT-NLLS) analysis (Plautz et al., 1997). The statistical significance of the observed circadian rhythm was assessed using the RAE value, which is defined as the ratio of the amplitude error to the most probable derived amplitude magnitude (Millar et al., 1995). A RAE cut-off value of 0.5 was used to identify robust rhythms. The time-lapse frames taken over 3 or 4 days were compiled to produce a movie, which was studied in detail in an attempt to determine the difference in rhythmic leaf movement between mutant and wild-type plants.
Gene expression analysis
Gene expression levels were determined by a semi-quantitative reverse transcriptase-mediated PCR (RT-PCR) (Yoo et al., 2005) or northern hybridization analysis. Total RNA was isolated from whole seedlings with Trizol reagent (Invitrogen Life Technologies, USA) in accordance with the manufacturer’s instructions. In brief, 1 μg of total RNA was first treated with DNaseI to remove any genomic DNA contaminants and then first-stranded cDNA was synthesized from the total RNA. The oligonucleotide sequences utilized in the measurements of clock-associated gene expression levels are shown in Supplementary Table 2. Northern hybridization was performed, as described elsewhere (Yamaguchi-Shinozaki and Shinozaki, 1994). UBIQUITIN10 (UBQ10) was used as an internal positive control for both analyses.
RESULTS
An imaging system for analyzing leaf movement
To screen for circadian clock mutants with an altered leaf movement phenotype, we set up an imaging system (Supplementary Figs. 1A and 1C) consisting of nine cameras, a switching hub, and a computer in a configuration similar to that described by Dowson-Day (Dowson-Day and Millar, 1999). As many as 432 seedlings could be analyzed at any one time using this system. We first analyzed the leaf movement of two known circadian clock mutants, toc1-1 and lhy-22 (SALK_ 149287), to evaluate the imaging system. Tracing leaf movements with this system revealed that both toc1-1 and lhy-22 plants had a shortened period compared to wild-type plants (21.70 ± 1.00 vs. 24.54 ± 0.66 h, respectively, Supplementary Figs. 1D and 1E; 22.64 ± 0.28 vs. 25.14 ± 1.65 h, respectively, Supplementary Figs. 1F and 1G). These results were consistent with previously reported data (Alonso et al., 2003; Mizoguchi et al., 2002; Somers et al., 1998), suggesting that our imaging system is in fact an accurate method for screening leaf movement mutants.
Measurement of the leaf movement of T-DNA insertion mutants
In this study, two different strategies were adopted for isolating the circadian clock mutants. The first consisted of analyzing loss-of-function alleles of genes whose expression levels were oscillating. Since genes that play an important role in circadian clock control commonly show robust diurnal expression patterns, we selected candidate genes that showed a diurnal expression pattern similar to those of core oscillator genes (TOC1, LHY, CCA1, and PCL1) from publicly available microarrays (Edwards et al., 2006). The loss-of-function alleles of these genes were isolated from the Salk T-DNA library (Alonso et al., 2003) and their leaf movement phenotypes were analyzed. The second strategy consisted of analyzing leaf movement of activation tagging mutants that we previously generated (Ahn et al., 2007).
From microarrays, we selected 285 candidate genes (Supplementary Table 1) that showed an expression profile similar (more than 85% in their expression pattern) to those of core oscillator genes (Supplementary Fig. 1H). Classification of these candidate genes according to the guidelines of the Functional Catalogue of the Munich Information Center for Protein Sequences (MIPS) (Table 1) (Ruepp et al., 2004) revealed that many of them play a role in metabolism (22.46%) and subcellular localization (13.68%). Among them, we selected 84 genes that are involved in transcriptional regulation, RNA modification, and photosynthesis. In order to find their T-DNA insertional mutants, we searched the Salk T-DNA library (Alonso et al., 2003) and found that T-DNA mutants of 55 genes were available. We obtained these T-DNA mutants and genotyped them to isolate homozygous lines (data not shown). We were unable to identify any homozygous mutants of 13 genes. Possible explanations for this failure to find a homozygous mutant line are that the homozygous lines were lethal or prediction of T-DNA insertion site was incorrect. Finally, T-DNA mutants of 42 genes were isolated and used for further analysis.
Table 1.
Functional catalog of candidate genes for screening leaf movement based on MIPS
| Catalogue | Percentage |
|---|---|
| METABOLISM | 22.46 |
| SUBCELLULAR LOCALIZATION | 13.68 |
| PROTEIN WITH BINDING FUNCTION AND COFACTOR REQUIREMENT | 12.63 |
| CELLULAR TRANSPORT, TRANSPORT FACILITIES AND TRANSPORT ROUTES | 7.37 |
| TRANSCRIPTION | 7.37 |
| PROTEIN RATE | 6.32 |
| CELL CYCLE AND DNA PROCESSING | 3.86 |
| ENERGY | 2.46 |
| INTERACTION WITH THE ENVIRONMENT | 2.11 |
| DEVELOPMENT | 1.05 |
| REGULATION OF METABOLISM AND PROTEIN FUNCTION | 1.05 |
| BIOGENESIS OF CELLULAR COMPONENTS | 0.70 |
| CELL FATE | 0.35 |
| CELLULAR COMMUNICATION/SIGNAL TRANSDUCTION MECHANISM | 0.35 |
| UNCLASSIFIED PROTEINS | 18.25 |
We analyzed the leaf movement phenotypes of 42 mutants. Our initial analysis of leaf movement using these 42 mutants suggested that the period of a few mutants (e.g., AT1G10200 and AT1G13270) was longer than that of the wild-type plants (Fig. 1). However, further studies revealed that there was no significant difference in terms of the phase of leaf movement between these mutants and the wild-type plants (AT1G10200: 13.75 ± 0.95 h vs. wild-type: 15.75 ± 1.85 h; AT1G13270: 10.95 ± 2.16 h vs. wild-type: 9.48 ± 1.60 h). No significant difference in their leaf movement was observed from the analysis of timelapse movies (data not shown). These results suggested that we failed to detect significant changes in the leaf movement from the T-DNA insertion mutants of genes having expression profile similar to those of core oscillator genes.
Fig. 1. Leaf movements and distribution of periods in T-DNA insertion mutants of the candidate genes. Black line and solid squares indicate leaf movements and scatter plots of period versus RAE for wild-type plants, respectively. White line and open circles indicate leaf movements and scatter plots of period versus RAE for each T-DNA mutant. Solid boxes indicate subjective nighttime. ZT, zeitbeger time.

agl6-1D plants showed a shortened period of leaf movement
We identified many mutants with developmental abnormalities from an activation tagging library (Ahn et al., 2007). In order to isolate a circadian clock mutant, we selected mutants with an altered flowering time and analyzed their leaf movements. We found that agl6-1D, in which AGL6 expression was enhanced by the 35S enhancer (Yoo et al., 2010), showed a shortened period (approximately 1 h difference) in leaf movement compared with that of wild-type plants (agl6-1D: 22.74 ± 0.37 h vs. wild-type: 23.75 ± 0.47 h) (Figs. 2A and 2B). The altered leaf movement of agl6-1D plants was also confirmed in a time-lapse movie (Supplementary Fig. 2). The expression analysis of genes close to a T-DNA insertion in agl6-1D plants suggested that AGL6 was likely the only gene that was activated by the 35S enhancer (Supplementary Fig. 3). To confirm again that the altered leaf movement seen in agl6-1D plants was resulted from the activation of AGL6, we generated 35S-AGL6-myc plants, in which myc-tagged AGL6 was overexpressed (Yoo et al., 2010), and measured the period in leaf movement. Consistent with the results in agl6-1D plants, 35S-AGL6-myc plants showed a shortened period length (approximately 1 h difference) in leaf movement compared with that of wild-type plants (35S-AGL6-myc: 23.08 ± 0.33 h vs. wild-type: 23.77 ± 0.68 h) (Figs. 2C and 2D). Furthermore, a time-lapse movie clearly showed the changed leaf movement in 35S-AGL6-myc plants compared with wild-type plants (Supplementary Fig. 4). These results indicated that AGL6 activation was responsible for the altered leaf movement.
Fig. 2. Changes in leaf movement in various AGL6 alleles. (A) Leaf movements in agl6-1D (gray line) and wild-type plants (black line). Solid boxes indicate subjective nighttime. (B) Scatter plot of period versus RAE for agl6-1D (open circles) and wild-type (WT) plants (solid squares). (C) Leaf movements in 35S-AGL6-myc (gray line) and wild-type plants (black line). Solid boxes indicate subjective nighttime. (D) Scatter plot of period versus RAE for 35S-AGL6-myc (open circles) and wild-type plants (solid squares). (E) Leaf movements in 35S-amiRAGL6 (gray line) and wild-type plants (black line). Solid boxes indicate subjective nighttime. (F) Scatter plot of period versus RAE for 35S-amiRAGL6 (open circles) and wild-type plants (solid squares). ZT, zeitbeger time. Open and solid boxes indicate subjective daytime and nighttime, respectively.
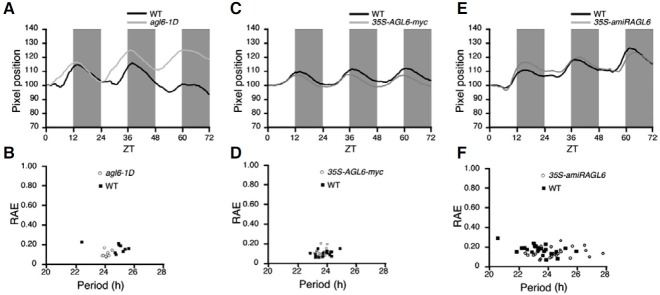
To investigate a loss-of-function phenotype of AGL6, we analyzed transgenic plants overexpressing an artificial miRNA targeting AGL6 (Yoo et al., 2010), since a reliable AGL6 null allele was not available. 35S-amiRAGL6 plants showed a similar period in leaf movement compared with wild-type plants (35S-amiRAGL6: 23.92 ± 0.86 h vs. wild-type: 23.55 ± 0.91 h) (Figs. 2E and 2F) (Supplementary Fig. 5). This result indicated that knock-down of AGL6 did not resulted in a visible phenotype, suggesting that AGL6 has a redundant partner(s) in circadian clock regulation. Collectively, our data raised a possibility that activation of AGL6 alters the genes functioning in circadian clock regulation.
Expression analysis of clock-associated genes in agl6-1D plants
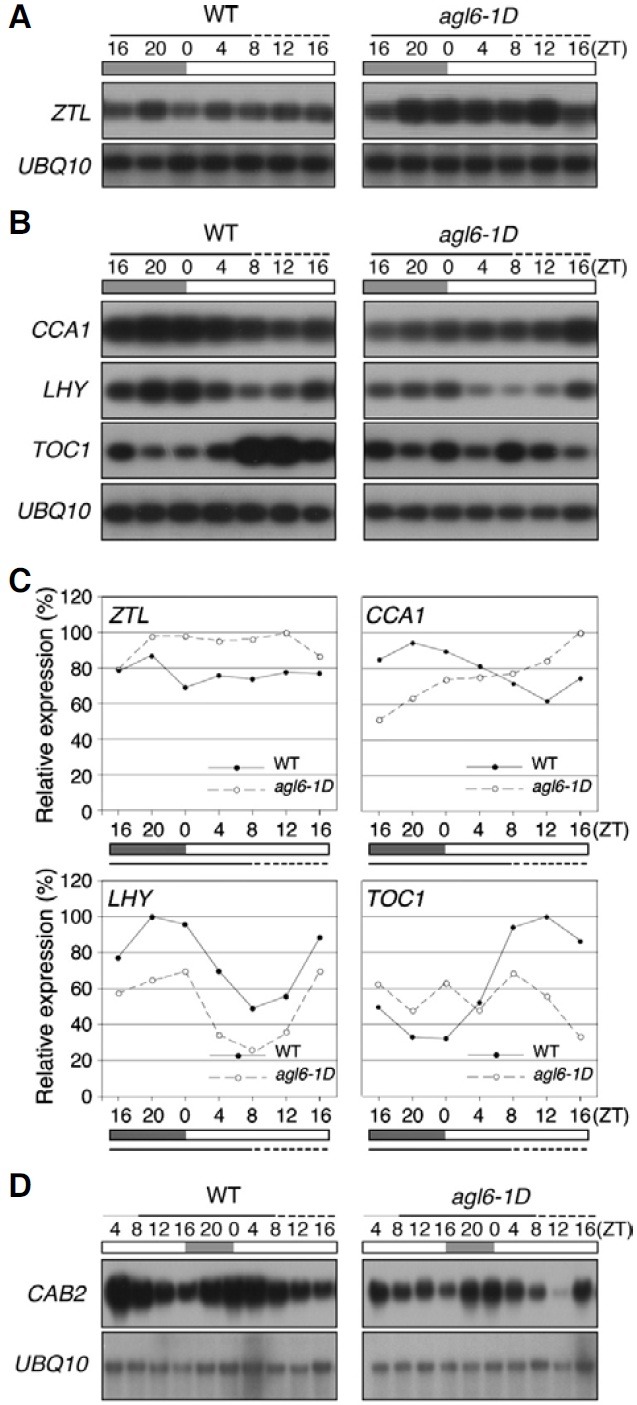
To identify the molecular basis of the shortened period of the leaf movement in agl6-1D plants, we analyzed the expression pattern of the core oscillator genes and found that the overall expression levels of ZTL mRNA were high in agl6-1D plants (Figs. 3A and 3C). We then determined whether the altered expression of ZTL in agl6-1D plants affected the expression of the downstream components in the circadian clock. TOC1 and CCA1, TOC1 and LHY showed an opposite expression pattern in wild-type plants (Figs. 3B and 3C). CCA1 and LHY showed a typical circadian rhythmic expression pattern (24 h period) in wild-type plants (Makino et al., 2002; Schaffer et al., 1998; Wang et al., 1997). However, in agl6-1D plants, the CCA1 transcripts level did not seem to be regulated in a circadian rhythm, with the CCA1 mRNA levels being strongly reduced at the early stages of the rhythm without a typical rhythmic pattern and increasing gradually thereafter. Although the overall amplitude and expression level of LHY were reduced in agl6-1D plants, the period length and phase were similar to those of wild-type plants. In addition, the clear circadian expression pattern of TOC1, an evening gene (Alabadi et al., 2001; Strayer et al., 2000), in wild-type plants had disappeared in agl6-1D plants. Overall TOC1 expression seemed to be arrhythmic in agl6-1D plants. LHY and CCA1 play an important role in maintaining the period of CAB2 expression, an output gene (Gould et al., 2006). The alteration in LHY and CCA1 expression led us to determine CAB2 expression in agl6-1D plants with the aim of testing the effect of regulating the circadian clock of AGL6 (Fig. 3D). Northern hybridization analysis revealed that the amplitude of CAB2 expression level had notably decreased in agl6-1D plants, although the phase was not significantly changed. The altered level of an output gene by AGL6 activation also supported the hypothesis that AGL6 is a regulator of the circadian clock. When all of these results are taken together, it is apparent that the circadian rhythms of the core oscillator genes are defective in agl6-1D plants, suggesting that the circadian clock is impaired by AGL6 activation.
Fig. 3. Expression of ZTL, CCA1, LHY, TOC1, and CAB2 in 10- (solid line) and 11-(dotted line) day-old agl6-1D and wild-type plants. (A) RT-PCR analysis of expression of ZTL in 10- (solid line) and 11- (dotted line) day-old agl6-1D and wild-type plants. (B) RT-PCR analysis of expression of CCA1, LHY, and TOC1 in 10- (solid line) and 11- (dotted line) day-old agl6-1D and wild-type plants. (C) Quantification of ZTL, CCA1, LHY, and TOC1 expression in (A, B). Expression levels were normalized against UBQ10, which was used as an internal positive control. (D) Expression of CAB2 in 10- (solid line)- and 11- (dotted line) day-old agl6-1D and wild-type plants by northern hybridization. Open and solid boxes indicate subjective daytime and nighttime, respectively. ZT, zeitbeger time.
DISCUSSION
In this study, we used an imaging system to screen for mutants that show aberrant cotyledon movement. We also analyzed activation tagging mutants and ultimately identified agl6-1D plants that showed a short period of leaf movement. We were able to demonstrate that AGL6 activation altered ZTL mRNA levels, which subsequently affected the phase of circadian oscillators and an output gene.
In wild-type plants, ZTL mRNA abundance is neither circadian- regulated (Mizoguchi and Coupland, 2000) nor lightregulated (Somers et al., 2000). ZTL does, however, affect the expression of the circadian-clock associated genes, especially that of the main circadian oscillators. In agl6-1D mutants, we found that both the overall level of ZTL mRNA abundance (Figs. 3A and 3C) and the expression of the core oscillators were altered (Figs. 3B and 3C). These results suggest that the alteration of ZTL expression, caused by AGL6 activation, affects the circadian clock through the core oscillators. This proposed mechanism is consistent with the finding that the CCA1 mRNA level cycle is out of phase in strong ZTL overexpressor plants and that ztl mutations cause a delay in peak phase of CCA1 mRNA under continuous light conditions (Somers et al., 2004). The TOC1 expression pattern in ZTL overexpressor plants showed an advanced phase on the first day, whereas TOC1 expression became arrhythmic in agl6-1D plants. Taken together, these data suggest that the enhanced ZTL mRNA level controls the expression of CCA1, LHY, and TOC1 at the transcriptional level in agl6-1D plants.
agl6-1D plants with high ZTL mRNA levels showed a short period length in leaf movement, which leads to the important question of whether leaf movement and ZTL abundance are closely related. ZTL overexpressor plants were found to have a very short period (13.1 h) in leaf movement (Somers et al., 2004). In addition, seven different lines that had different ZTL mRNA levels displayed different waveforms and amplitudes of the circadian rhythm. Consistent with the phenotype of ZTL overexpressor plants, ztl-1 plants exhibited a lengthened free-running rhythm of leaf movement (Somers et al., 2000). These data suggest that altered ZTL mRNA levels are closely associated with the leaf movement phenotype in agl6-1D plants. They also suggest that AGL6 may play a role in the regulation of the circadian clock by regulating ZTL mRNA level.
In this study, we have shown that AGL6 activation alters leaf movement and expression of core oscillator genes. This suggests that AGL6 plays a role in the circadian clock control, although investigation using a knock-down allele suggested that there may be a redundant partner of AGL6. Thus further genetic studies using a complete knock-out allele of AGL6 should provide data demonstrating a clear link between the role of AGL6 and the regulation of the circadian clock.
Note: Supplementary information is available on the Molecules and Cells website (www.molcells.org).
Acknowledgments
We are very grateful to Steve Kay (University of California, USA) for providing toc1-1 seeds and M. Ishiura (Nagoya University, Japan) for sharing the NKTRACE program. S.K. Yoo and S.M. Hong were supported by the Brain Korea 21 program. S.M. Hong was supported by Seoul Science Fellowship. This work was supported by Creative Research Initiatives (R16- 2008-106-01001-0) of the Ministry of Education, Science and Technology (MEST)/Korea Science and Engineering Foundation (KOSEF) to J.H.A.We are very grateful to Steve Kay (University of California, USA) for providing toc1-1 seeds and M. Ishiura (Nagoya University, Japan) for sharing the NKTRACE program. S.K. Yoo and S.M. Hong were supported by the Brain Korea 21 program. S.M. Hong was supported by Seoul Science Fellowship. This work was supported by Creative Research Initiatives (R16- 2008-106-01001-0) of the Ministry of Education, Science and Technology (MEST)/Korea Science and Engineering Foundation (KOSEF) to J.H.A.
References
- 1.Ahn J.H., Kim J., Yoo S.J., Yoo S.Y., Roh H., Choi J.H., Choi M.S., Chung K.S., Han E.J., Hong S.M., et al. Isolation of 151 mutants that have developmental defects from T-DNA tagging. Plant Cell Physiol. (2007);48:169–178. doi: 10.1093/pcp/pcl052. [DOI] [PubMed] [Google Scholar]
- 2.Alabadi D., Oyama T., Yanovsky M.J., Harmon F.G., Mas P., Kay S.A. Reciprocal regulation between TOC1 and LHY/CCA1 within the Arabidopsis circadian clock. Science. (2001);293:880–883. doi: 10.1126/science.1061320. [DOI] [PubMed] [Google Scholar]
- 3.Alonso J.M., Stepanova A.N., Leisse T.J., Kim C.J., Chen H., Shinn P., Stevenson D.K., Zimmerman J., Barajas P., Cheuk R., et al. Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science. (2003);301:653–657. doi: 10.1126/science.1086391. [DOI] [PubMed] [Google Scholar]
- 4.Barak S., Tobin E.M., Andronis C., Sugano S., Green R.M. All in good time: the Arabidopsis circadian clock. Trends Plant Sci. (2000);5:517–522. doi: 10.1016/s1360-1385(00)01785-4. [DOI] [PubMed] [Google Scholar]
- 5.Devlin P.F. Signs of the time: environmental input to the circadian clock. J. Exp. Bot. (2002);53:1535–1550. doi: 10.1093/jxb/erf024. [DOI] [PubMed] [Google Scholar]
- 6.Dowson-Day M.J., Millar A.J. Circadian dysfunction causes aberrant hypocotyl elongation patterns in Arabidopsis. Plant J. (1999);17:63–71. doi: 10.1046/j.1365-313x.1999.00353.x. [DOI] [PubMed] [Google Scholar]
- 7.Edwards K.D., Anderson P.E., Hall A., Salathia N.S., Locke J.C., Lynn J.R., Straume M., Smith J.Q., Millar A.J. FLOWERING LOCUS C mediates natural variation in the hightemperature response of the Arabidopsis circadian clock. Plant Cell. (2006);18:639–650. doi: 10.1105/tpc.105.038315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Farre E.M., Harmer S.L., Harmon F.G., Yanovsky M.J., Kay S.A. Overlapping and distinct roles of PRR7 and PRR9 in the Arabidopsis circadian clock. Curr. Biol. (2005);15:47–54. doi: 10.1016/j.cub.2004.12.067. [DOI] [PubMed] [Google Scholar]
- 9.Gould P.D., Locke J.C., Larue C., Southern M.M., Davis S.J., Hanano S., Moyle R., Milich R., Putterill J., Millar A.J., et al. The molecular basis of temperature compensation in the Arabidopsis circadian clock. Plant Cell. (2006);18:1177–1187. doi: 10.1105/tpc.105.039990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Green R.M., Tingay S., Wang Z.Y., Tobin E.M. Circadian rhythms confer a higher level of fitness to Arabidopsis plants. Plant Physiol. (2002);129:576–584. doi: 10.1104/pp.004374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kim W.Y., Fujiwara S., Suh S.S., Kim J., Kim Y., Han L., David K., Putterill J., Nam H.G., Somers D.E. ZEITLUPE is a circadian photoreceptor stabilized by GIGANTEA in blue light. Nature. (2007);449:356–360. doi: 10.1038/nature06132. [DOI] [PubMed] [Google Scholar]
- 12.Locke J.C., Kozma-Bognar L., Gould P.D., Feher B., Kevei E., Nagy F., Turner M.S., Hall A., Millar A.J. Experimental validation of a predicted feedback loop in the multioscillator clock of Arabidopsis thaliana. Mol. Syst. Biol. (2006);2:59. doi: 10.1038/msb4100102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Makino S., Matsushika A., Kojima M., Yamashino T., Mizuno T. The APRR1/TOC1 quintet implicated in circadian rhythms of Arabidopsis thaliana: I. Characterization with APRR1-overexpressing plants. Plant Cell Physiol. (2002);43:58–69. doi: 10.1093/pcp/pcf005. [DOI] [PubMed] [Google Scholar]
- 14.McClung C.R. Circadian rhythms in plants. Annu. Rev. Plant Physiol. Plant Mol. Biol. (2001);52:139–162. doi: 10.1146/annurev.arplant.52.1.139. [DOI] [PubMed] [Google Scholar]
- 15.McClung C.R. Comes a time. Curr. Opin. Plant Biol. (2008);11:514–520. doi: 10.1016/j.pbi.2008.06.010. [DOI] [PubMed] [Google Scholar]
- 16.Millar A.J. Input signals to the plant circadian clock. J. Exp. Bot. (2004);55:277–283. doi: 10.1093/jxb/erh034. [DOI] [PubMed] [Google Scholar]
- 17.Millar A.J., Carre I.A., Strayer C.A., Chua N.H., Kay S.A. Circadian clock mutants in Arabidopsis identified by luciferase imaging. Science. (1995);267:1161–1163. doi: 10.1126/science.7855595. [DOI] [PubMed] [Google Scholar]
- 18.Mizoguchi T., Coupland G. ZEITLUPE and FKF1: novel connections between flowering time and circadian clock control. Trends Plant Sci. (2000);5:409–411. doi: 10.1016/s1360-1385(00)01747-7. [DOI] [PubMed] [Google Scholar]
- 19.Mizoguchi T., Wheatley K., Hanzawa Y., Wright L., Mizoguchi M., Song H.R., Carre I.A., Coupland G. LHY and CCA1 are partially redundant genes required to maintain circadian rhythms in Arabidopsis. Dev. Cell. (2002);2:629–641. doi: 10.1016/s1534-5807(02)00170-3. [DOI] [PubMed] [Google Scholar]
- 20.Nakamichi N., Kita M., Ito S., Yamashino T., Mizuno T. PSEUDO-RESPONSE REGULATORS, PRR9, PRR7 and PRR5, together play essential roles close to the circadian clock of Arabidopsis thaliana. Plant Cell Physiol. (2005);46:686–698. doi: 10.1093/pcp/pci086. [DOI] [PubMed] [Google Scholar]
- 21.Onai K., Okamoto K., Nishimoto H., Morioka C., Hirano M., Kami-Ike N., Ishiura M. Large-scale screening of Arabidopsis circadian clock mutants by a high-throughput realtime bioluminescence monitoring system. Plant J. (2004);40:1–11. doi: 10.1111/j.1365-313X.2004.02191.x. [DOI] [PubMed] [Google Scholar]
- 22.Plautz J.D., Straume M., Stanewsky R., Jamison C.F., Brandes C., Dowse H.B., Hall J.C., Kay S.A. Quantitative analysis of Drosophila period gene transcription in living animals. J. Biol. Rhythms. (1997);12:204–217. doi: 10.1177/074873049701200302. [DOI] [PubMed] [Google Scholar]
- 23.Ruepp A., Zollner A., Maier D., Albermann K., Hani J., Mokrejs M., Tetko I., Guldener U., Mannhaupt G., Munsterkotter M., et al. The FunCat, a functional annotation scheme for systematic classification of proteins from whole genomes. Nucleic Acids Res. (2004);32:5539–5545. doi: 10.1093/nar/gkh894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Salome P.A., McClung C.R. PSEUDO-RESPONSE REGULATOR 7 and 9 are partially redundant genes essential for the temperature responsiveness of the Arabidopsis circadian clock. Plant Cell. (2005);17:791–803. doi: 10.1105/tpc.104.029504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schaffer R., Ramsay N., Samach A., Corden S., Putterill J., Carre I.A., Coupland G. The late elongated hypocotyl mutation of Arabidopsis disrupts circadian rhythms and the photoperiodic control of flowering. Cell. (1998);93:1219–1229. doi: 10.1016/s0092-8674(00)81465-8. [DOI] [PubMed] [Google Scholar]
- 26.Somers D.E., Devlin P.F., Kay S.A. Phytochromes and cryptochromes in the entrainment of the Arabidopsis circadian clock. Science. (1998);282:1488–1490. doi: 10.1126/science.282.5393.1488. [DOI] [PubMed] [Google Scholar]
- 27.Somers D.E., Schultz T.F., Milnamow M., Kay S.A. ZEITLUPE encodes a novel clock-associated PAS protein from Arabidopsis. Cell. (2000);101:319–329. doi: 10.1016/s0092-8674(00)80841-7. [DOI] [PubMed] [Google Scholar]
- 28.Somers D.E., Kim W.Y., Geng R. The F-box protein ZEITLUPE confers dosage-dependent control on the circadian clock, photomorphogenesis, and flowering time. Plant Cell. (2004);16:769–782. doi: 10.1105/tpc.016808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Strayer C., Oyama T., Schultz T.F., Raman R., Somers D.E., Mas P., Panda S., Kreps J.A., Kay S.A. Cloning of the Arabidopsis clock gene TOC1, an autoregulatory response regulator homolog. Science. (2000);289:768–771. doi: 10.1126/science.289.5480.768. [DOI] [PubMed] [Google Scholar]
- 30.Wang Z.Y., Kenigsbuch D., Sun L., Harel E., Ong M.S., Tobin E.M. A Myb-related transcription factor is involved in the phytochrome regulation of an Arabidopsis Lhcb gene. Plant Cell. (1997);9:491–507. doi: 10.1105/tpc.9.4.491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yamaguchi-Shinozaki K., Shinozaki K. A novel cisacting element in an Arabidopsis gene is involved in responsiveness to drought, low-temperature, or high-salt stress. Plant Cell. (1994);6:251–264. doi: 10.1105/tpc.6.2.251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yoo S.K., Chung K.S., Kim J., Lee J.H., Hong S.M., Yoo S.J., Yoo S.Y., Lee J.S., Ahn J.H. CONSTANS activates SUPPRESSOR OF OVEREXPRESSION OF CONSTANS 1 through FLOWERING LOCUS T to promote flowering in Arabidopsis. Plant Physiol. (2005);139:770–778. doi: 10.1104/pp.105.066928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yoo S.K., Wu X., Lee J.S., Ahn J.H. AGAMOUSLIKE 6 is a floral promoter that negatively regulates the FLC/ MAF clade genes and positively regulates FT in Arabidopsis. Plant J. (2010);65:62–76. doi: 10.1111/j.1365-313X.2010.04402.x. [DOI] [PubMed] [Google Scholar]


