Abstract
As a platform for investigating the individual effects of substrate stiffness, permeability, and ligand density on cellular behavior, we developed a set of hydrogels with stiffness tuned by polymer backbone rigidity, independent of cross-link density and concentration. Previous studies report that poly(propargyl-L-glutamate) (PPLG), synthesized by ring-opening polymerization of the N-carboxy anhydride of γ-propargyl-L-glutamate (γpLglu), adopts a rigid a-helix conformation: we hypothesized that a random copolymer (PPDLG) with equal amounts of γpLglu and γ-propargyl-D-glutamate (γpDglu) monomers would exhibit a more flexible random coil conformation. The resulting macromers exhibited narrow molecular weight distributions (PDI = 1.15) and were grafted with ethylene glycol groups using a highly efficient “click” azide/alkyne cycloaddition reaction with average grafting efficiency of 97% for PPLG and 85% for PPDLG. The polypeptide secondary structure, characterized via circular dichroism spectroscopy, FTIR spectroscopy, and dynamic light scattering, is indeed dependent upon monomer chirality: PPLG exhibits an α-helix conformation while PPDLG adopts a random coil conformation. Hydrogel networks produced by cross-linking either helical or random coil polypeptides with poly(ethylene glycol) (PEG) were analyzed for amount of swelling, gelation efficiency, and permeability to a model protein. In addition, the elastic modulus of helical and coil polypeptide gels was determined by AFM indentation in fluid. Importantly, we found that helical and coil polypeptide gels exhibited similar swelling and permeability but different stiffnesses, which correspond to predictions from the theory of semi-flexible chains.
Introduction
Cellular phenotype, migration, and proliferation are dependent upon properties of the surrounding extracellular matrix (ECM) including stiffness and permeability to soluble signaling molecules such as cytokines, growth factors, and hormones. An important focus of biomedical materials research is parsing the roles of physiological and pathophysiological matrix stiffness in regulating cell behavior. Hydrogels are common substrates used for modeling the interplay between ECM properties and cellular phenomena with systems based on natural polymer gels such as collagen and matrigel, and synthetic polymer gels such as polyacrylamide and PEG.1–19 Hydrogel cross-link density is tuned to obtain gels with elastic moduli from 1 Pa to 100,000 Pa – notably, permeability to proteins the size of autocrine factors varies over this range (e.g. amphiregulin, interleukin-1)20–29 as does the extent of non-specific protein adsorption.30, 31 These studies provide valuable insight into mechanisms of cell-matrix interactions, but possess limitations in decoupling the contributions of individual properties because the permeability and ligand density of these models are dependent upon substrate cross-link density. Illuminating work with cells cultured on posts of differing flexibility32–34 partially bridges this gap but this strategy is not suited for three-dimensional cell culture, a crucial step forward.
Peptides have long been incorporated into hydrogels to facilitate cell adhesion,19, 35–37 provide substrates for enzymatic degradation,38–41 as structural components,42–46 and even to direct self-assembly processes.19, 47–49 Synthetic polypeptides, synthesized by ring-opening polymerization since the 1950s,50–54 introduce a powerful capability to generate macromolecular species using the amino acid backbone found in nature, thus providing a route to biocompatible polymeric systems with programmable function.55–58 Peptide sequences, like proteins, can fold into stable secondary structures such as beta sheets or helices. These structures facilitate the presentation of surface moieties that dictate cell signaling and molecular docking and provide unique opportunities to design supramolecular shape and function.59 Polypeptides produced from alkyne-modified glutamate monomers (i.e. γ-propargyl-L-glutamate, γpLglu)60–64 are impressively modular, as an almost unlimited range of molecules can be coupled to the peptide backbone through 1,3-dipolar cycloaddition,60 a type of highly efficient “click chemistry” used extensively in the development of biomedical materials since 2001.65–69
In order to design hydrogels with independently tunable permeability and stiffness, we produced a library of “click”-grafted polypeptides that exhibit either stiffer helical or more-flexible random coil conformations. Backbone rigidity (i.e. helix or coil) dictates the relative stiffness of gels made with an otherwise identical formulation: unlike cross-link density, backbone rigidity has a minimal effect on gel permeability. Poly(γ-propargyl-L-glutamate) (PPLG), synthesized by ring-opening polymerization of the N-carboxy anhydride of γpLglu,60 adopts a rigid a-helix conformation: we hypothesized that a random copolymer produced from a 50:50 mixture of γpLglu and γpDglu monomers (PPDLG) would exhibit a more flexible random coil conformation. Although insertion of opposite chirality residues into a peptide sequence is known to change or completely disrupt the resulting secondary structure,70, 71 early studies of poly(carboxybenzyl-D,L-glutamate) reported that the polymer consists of blocks of D and L monomers that form alternating left- and right-handed helices.72 We anticipate that this effect was partially due to a large discrepancy in monomer addition rates (i.e. the addition of L to L and of D to D occurred much more readily than the addition of L to D and of D to L) on account of the bulky nature of the carboxybenzyl protecting group.
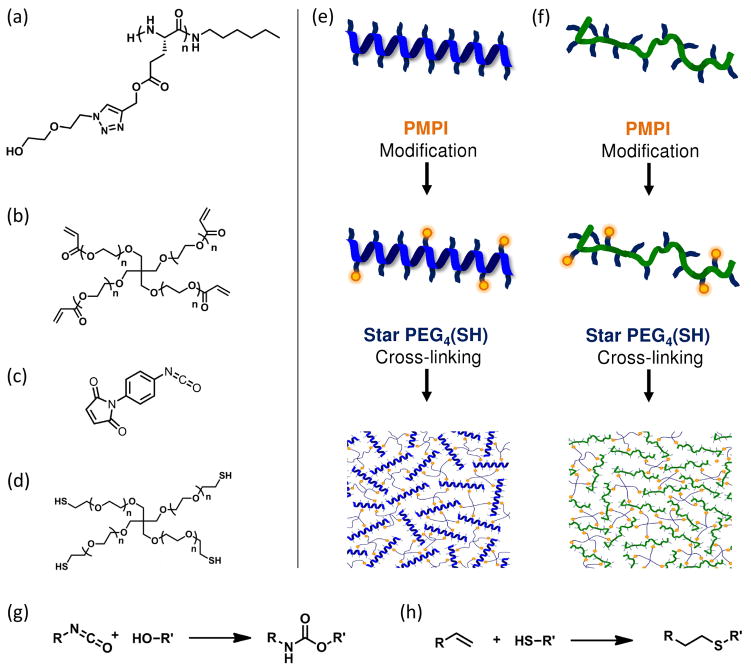
In this work, we investigate the structure/property relationships of a set of synthetic grafted polypeptides (Scheme 1) with tunable parameters including the degree of polymerization (n), monomer chirality (L, D), and grafting group size (m). The effect of monomer chirality on polypeptide conformation was studied by characterization of poly(γ-propargyl-L-glutamate)-graft-(ethylene glycol)2 (PPLGgEG2OH, 5a) and poly(γ-propargyl-D,L-glutamate)-graft-(ethylene glycol)2 (PPDLGgEG2OH, 5b) with circular dichroism (CD) spectroscopy, FTIR spectroscopy, and dynamic light scattering (DLS). Hydrogel networks were produced by cross-linking either helical or coil polypeptides with a coupling agent, 4-(maleinimido)phenyl isocyanate (PMPI), and star PEG thiol (PEG(SH)4) as shown in Figure 1: control gels were prepared by cross-linking star PEG acrylate (PEG(Acrl)4) and PEG(SH)4. In order to compare the helical polypeptide, coil polypeptide, and control hydrogels, we measured sample swelling, gelation efficiency, and permeability to a model protein. The hydrogel elastic moduli were measured via AFM indentation in fluid.
Scheme 1.
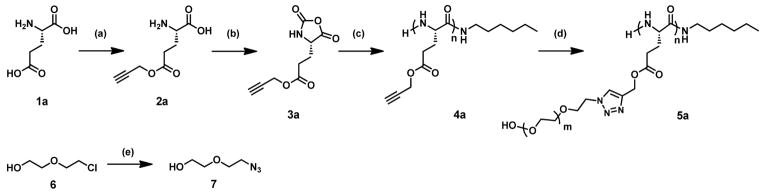
Polypeptide and grafting group synthesis. Reagents and conditions: (a) L-glutamic acid (1a), TMSCl, propargyl alcohol, 20 °C; (b) Î3pLglu (2a), triphosgene, EtOAc, 85 °C; (c) Î3pLglu NCA (3a), 1-aminohexane, DMF, 20 °C; (d) stereoregular PPLG (5a), azidoethoxyethanol (7), Cu(I)Br, PMDETA, DMF, 20 °C; (e) chloroethoxyethanol (6), NaN3, 50:50 water/ethanol, 85 °C. 5a = PPLGgEG2OH. An identical protocol (a–e) was used, starting from D-glutamic acid (1b, not shown), to produce the D-glutamate monomers Î3pDglu (2b), and Î3Dglu NCA (3b): a 50:50 mixture of 3a and 3b was polymerized to form PPDLG (4b), then grafted with 7 to form PPDLGgEG2OH (5b).
Figure 1.
Gels were produced by reacting polypeptide (a) with PMPI (c) and star PEG thiol (d); control gels were produced by reacting star PEG acrylate (b) with star PEG thiol (d). The PPLGgEG2OH homopolymer and PPDLGgEG2OH heteropolymer were cross-linked to form helical (e) and random coil (f) hydrogel networks, respectively. The alcohol groups along the polypeptide backbone reacted with isocyanate groups of PMPI to form urethane bonds (g) and the star PEG thiol reacted with the maleimide groups of PMPI (or acrylate groups of star PEG acrylate) to form thioether bonds (h).
Materials and methods
Materials
All chemicals were purchased from Sigma Aldrich, Acros Organics, or Invitrogen and used as received unless noted. The L-glutamic acid and D-glutamic acid starting materials (Sigma Aldrich) were re-crystallized in pure water before use. The coupling agent 4-(maleinimido)phenyl isocyanate (PMPI, Pierce/Thermo), PEG(SH)4, and PEG(Acrl)4 (Laysan Bio) were purchased in 50 mg, 1 g, and 1g sizes, respectively. These sensitive compounds were divided into small aliquots in a dry, nitrogen-filled glovebox using labeled, tared Eppendorf tubes: each aliquot was stored under nitrogen surrounded by desiccant at −20 °C until use. Further details of the materials and protocols used for polypeptide synthesis are located in the Supporting Information section.
Abbreviations
The following abbreviations are used throughout the text: atomic force microscopy (AFM), circular dichroism (CD), dichloromethane (DCM), dimethylformamide (DMF), dimethylsulfoxide (DMSO), Dulbecco’s phosphate-buffered saline (DPBS), dynamic light scattering (DLS), ethyl acetate (EtOAc), extracellular matrix (ECM), fluorescein-ovalbumin (fOA), fluorescence recovery after photo-bleaching (FRAP), Fourier-transform infrared (FTIR), γ-propargyl-L-glutamate (γpLglu), γ-propargyl-D-glutamate (γpDglu), γ-propargyl-L-glutamate NCA (γpLglu NCA), γ-propargyl-D-glutamate NCA (γpDglu NCA), gel permeation chromatography (GPC), molecular weight cut-off (MWCO), N-carboxy anhydride (NCA), nuclear magnetic resonance (NMR), poly(ethylene glycol) (PEG), star PEG acrylate (PEG(Acrl)4), star PEG thiol (PEG(SH)4), poly(γ-propargyl-L-glutamate) (PPLG), poly(γ-propargyl-D,L-glutamate) (PPDLG), poly(γ-propargyl-L-glutamate)-graft-(ethylene glycol)2 (PPLGgEG2OH), poly(γ-propargyl-D,L-glutamate)-graft-(ethylene glycol)2 (PPDLGgEG2OH), pentamethyldiethylenetriamine (PMDETA), 4-(maleinimido)phenyl-isocyanate (PMPI), phosphate-buffered saline (PBS), polydispersity index (PDI), trimethylsilyl chloride (TMSCl).
Monomer, polymer, and grafting group synthesis
A set of grafted polypeptides composed of either all L- or a mixture of D- and L- glutamate monomers was synthesized following methods adapted from a previous publication as shown in Scheme 1.60 Briefly, L-glutamate (1a) was suspended in propargyl alcohol and reacted with trimethylsilyl chloride for 36 hours at 25 °C. The crude mixture was purified by precipitation into diethyl ether followed by crystallization in a 10:1 mixture of acetonitrile and dimethylformamide (DMF) to yield γpLglu (2a) as fine, white crystals. Next, 2a was reacted with triphosgene in anhydrous ethyl acetate for 6 hours at reflux under argon. Warning: this reaction can generate toxic phosgene gas and should only be conducted by experienced personnel: please see the Supporting Information section for additional safety precautions and protocol details. The mixture was filtered, washed, dried, and then concentrated under vacuum to yield γpLglu NCA (3a) as a colorless oil.
Polypeptides were synthesized by reaction of NCA monomer (3a) with 1-aminohexane as initiator in anhydrous DMF for 72 hours at 25 °C under argon. The solvent was removed under vacuum and the crude product was precipitated from minimal DCM into diethyl ether to yield PPLG (4a) as a white powder. The stereoisomer forms of the L-glutamate monomer, namely γpDglu (2b) and γpDglu NCA (3b), were synthesized as described above using D-glutamic acid (1b) as the starting material. The heteropolymer PPDLG (4b) was synthesized by mixing equivalent amounts 3a and 3b in anhydrous DMF and initiating with 1-aminohexane, following the same reaction and purification protocol as described above for PPLG (4a).
Azide-functionalized ethylene glycol grafting groups were synthesized by reacting chloroethoxyethanol (6) with sodium azide in a water/ethanol mixture at reflux under nitrogen for 36 hours. The crude mixture was concentrated under vacuum, extracted into diethyl ether, and dried to yield azidoethoxyethanol (7) as a clear liquid. Finally, 4a was grafted by reaction with 7 catalyzed by PMDETA and Cu(I)Br in anhydrous DMF under argon for 8 hours at 20 °C. The solvent was removed under vacuum and the crude was dissolved in water and incubated with Dowex M4195 ion exchange resin for 20 minutes at 20 °C to remove the copper catalyst. The resin was removed by filtration and the filtrate was further purified by dialysis in pure water for 72 hours (1000 MWCO regenerated cellulose dialysis tubing, VWR): the polymer solution was then lyophilized to yield PPLGgEG2OH (5a) as a white powder. The heteropolymer PPDLG (4b) was grafted following an identical protocol to obtain PPDLGgEG2OH (5b).
Polymer composition and molecular weight
The structure and molecular weight of grafted polypeptides were evaluated via nuclear magnetic resonance (NMR) spectroscopy and gel permeation chromatography (GPC). Each compound produced in Scheme 1 was analyzed by 1H and 13C NMR spectroscopy (400 MHz, Bruker) in deuterated dimethyl sulfoxide (DMSO-d6, Cambridge Isotopes Lab). Polymer molecular weights were obtained from GPC analysis (Waters 1525 with refractive index detector) of polymer solutions (5 mg/mL in DMF) passed through a 0.45 μm filter and run at 75 °C with DMF as eluent. The molecular weight distributions were calculated by comparison to poly(methyl methacrylate) standards.
Polymer secondary structure and hydrodynamic radius
Polypeptide secondary structure was observed via CD spectroscopy (Aviv Model 202) of polymer samples (1 mg/mL in water) passed through a 0.45 μm filter and measured in a quartz cell (1 mm path length, New Era). Ellipticity was measured over the wavelength range of 195 nm to 250 nm at 25 °C. The effect of monomer chirality on polypeptide secondary structure was further analyzed using Fourier transform infrared (FTIR) spectroscopy. Polypeptide and FTIR-grade KBr (approximately 10 mg of each) were mixed and ground with a mortar and pestle into a fine, homogeneous powder. The powder was subsequently compacted in a sample press to form a thin, transparent, window and the vibrational spectrum of each sample was measured on an FTIR spectrometer (Bruker Alpha-E) over the range of 500 cm−1 to 4000 cm−1. Polymer hydrodynamic radii and diffusion coefficients were calculated from analysis of laser light scattering (830 nm, DynaPro NanoStar with Dynamics software, Wyatt Technology) from polymer samples (10 mg/mL in DMF, solutions centrifuged at 5000 g for 5 minutes) in a 12 μL quartz cuvette. The effect of molecular weight on polymer diffusivity was measured for PPLGgEG2OH, PPDLGgEG2OH, and linear PEG, a widely used random coil polymer.
Hydrogel formation
Gels were produced as shown in Figure 1 by cross-linking grafted polypeptides with the coupling agent PMPI and PEG(SH)4; control gels were prepared by cross-linking PEG(Acrl)4 with PEG(SH)4 (Laysan Bio). Briefly, stock solutions were produced by dissolving each compound separately in anhydrous dimethylsulfoxide (DMSO) as indicated: PPLGgEG2OH (200 mg/mL), PPDLGgEG2OH (200 mg/mL), PMPI (30 mg/mL), and PEG(SH)4 (110 mg/mL). PEG(Acrl)4 stock solution was prepared in DPBS (100 mg/mL, pH 7.4). Note: all solution preparation and gel mixing was conducted in a chemical fume hood to minimize exposure to PMPI, which has isocyanate groups that present an inhalation hazard. Gels with a variety of cross-link densities and polymer concentrations were easily prepared by mixing the requisite amount of each stock solution with additional DMSO as desired. For the gel comparison described herein, polypeptide gels containing 10 wt% polymer were produced by first mixing the requisite amount of polypeptide and DMSO, and then PMPI solution was added while vortexing to ensure even distribution. Next, the PEG solution was added while vortexing. Control PEG gels containing 10 wt% polymer were prepared in a similar fashion by mixing PEG(SH)4 and PEG(Acrl)4 in stoichiometric equivalents.
After mixing, the hydrogel sol was quickly pipetted into either a glass vial or an acrylate-modified glass bottom dish (Mattek). Glass bottom dishes were modified with acrylate groups by reaction with 3-acryloxypropyl trimethoxysilane (Gelest) in methanol (100 mM) for 3 hours then rinsed, dried, and stored at −20 °C with dessicant until use. After 18 hours, the resulting hydrogels were incubated in several changes of Dulbecco’s phosphate-buffered saline (DPBS, pH 7.4) for swelling and solvent exchange.
Hydrogel swelling, permeability, and elastic modulus
The hydrogels were formed in labeled, tared flasks. Each sample was weighed in the relaxed state (i.e. before swelling), then swelled to equilibrium with several changes of buffer (DPBS, pH 7.4). Samples were weighed at each buffer change and equilibrium swelling was assumed when the hydrogel mass no longer increased with time. Finally, gels were dried under vacuum and the residual solids were weighed. The hydrogel swelling ratio (Q), equilibrium water content (EWC), and gelation efficiency (ε) were calculated as follows:
| (1) |
| (2) |
| (3) |
where mr is the hydrogel mass in the relaxed state, ms is the hydrogel mass in the swollen state, mp,r is the ideal mass of polymer in the hydrogel, and mp,d is the actual mass of polymer that remains after swelling and drying the hydrogel.
The permeability of polypeptide hydrogels was determined by measuring the diffusivity of a fluorescent model protein with fluorescence recovery after photo-beaching (FRAP). Hydrogels were loaded with fluorescein-labeled ovalbumin (45 kDa, fOA) by incubation in fOA solution (1 mg/mL in DPBS pH 7.4) for 24 hours. The hydrogels were placed in a glass-bottom dish (Mattek, #1.5 coverslip bottom), surrounded by buffer, and covered with a coverslip to prevent evaporation during the experiment. A Deltavision microscope (Applied Precision) with a 488 nm argon ion laser was used to bleach sections of the hydrogel and then monitor the resulting recovery as non-bleached fluorescent protein diffused into the bleach area. The resulting images were analyzed with softWoRx software (Applied Precision) to solve the following equation:
| (4) |
where D is the diffusion coefficient, w is the bleach radius, and τ½ is the recovery half-life.73
Mechanical properties of hydrogels were measured on a commercial scanning probe microscope (Molecular Force Probe 3D, Asylum Research). Non-reflective silicon cantilevers modified with a 45-μm diameter polystyrene particle (Novascan Technologies, Ames, IA) were used to obtain the continuous force-displacement responses of the hydrogels in DPBS (pH 7.4) at 25 °C. Prior to indentation, the actual spring constant (kc) of each cantilever was determined experimentally as 12.96 N/m. Indentation force-displacement responses were analyzed in IGOR Pro software (WaveMetrics, Lake Oswego, OR). Application of the Hertz model for a spherical tip yielded the elastic modulus (E) of each sample as follows:
| (5) |
where F is applied force, R is the radius of the polystyrene particle, δ is the indentation depth, and v is the Poisson’s ratio, which was assumed to be 0.5 for the hydrogels.
Results and discussion
Grafted polypeptide synthesis
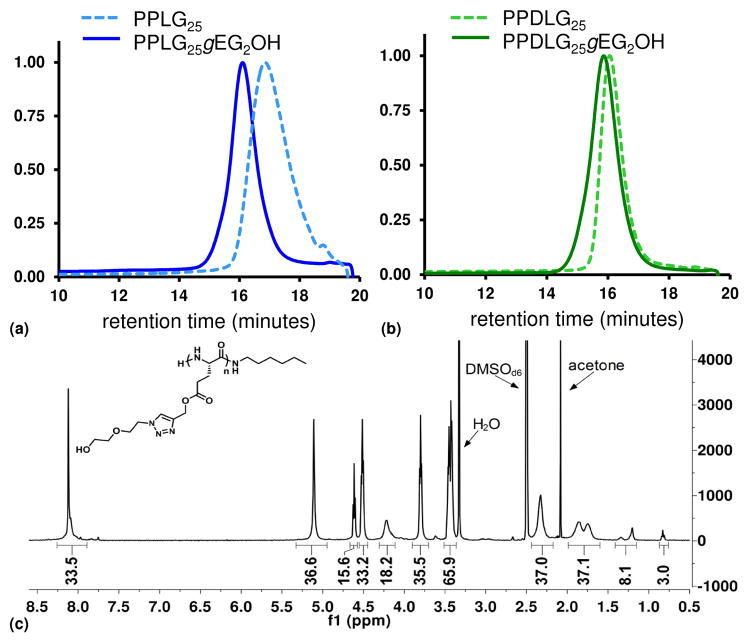
With the goal of obtaining a set of rigid and flexible polymers with otherwise identical composition, we synthesized glutamate-based polypeptides from either γpLglu or a 50:50 mixture of γpLglu and γpDglu to form the PPLG homopolymer and PPDLG heteropolymer, respectively (Scheme 1). The polymer composition and molecular weight were determined by NMR spectroscopy and GPC, as shown in Figure 2. Typical PDI values for the polypeptides are 1.15 before grafting and 1.25 after grafting. Importantly, the average grafting efficiency of PPLGgEG2OH was 97% while that of PPDLGgEG2OH was 85%, as determined via NMR spectroscopy. In an earlier publication, the high grafting efficiency of PPLG, in comparison to other polymer backbones grafted via azide-alkyne cycloaddition, was attributed to the helical nature of the polymer, which might reduce steric hindrance and result in increased accessibility of monomer side-chains for reaction.60 Our data not only confirm the very high grafting efficiency of PPLG but demonstrate that, under identical conditions, the heterochiral PPDLG polymer exhibits slightly lower, although still quite high, grafting efficiency. These results suggest that the PPDLG does indeed exhibit a different conformation than PPLG.
Figure 2.
Characterization of polypeptides: GPC traces of polymers before and after grafting with ethylene glycol groups (a, b), representative 1H NMR spectrum of polypeptide (PPLGgEG2OH) in DMSOd6 (c).
Secondary structure and hydrogen bonding of grafted polypeptides
The conformation of grafted polypeptides was investigated with CD and FTIR spectroscopy. The PPLGgEG2OH homopolymer adopts an α-helix conformation, as demonstrated by a CD spectrum with the characteristic minima at 222 nm and 208 nm (Figure 3).74 In contrast, the PPDLGgEG2OH heteropolymer exhibits a CD spectrum with a broad maximum at 215 nm and a minimum at 195 nm, suggesting that this species adopts a random coil conformation in solution.74 Importantly, the magnitude of the PPDLGgEG2OH spectrum is much lower than that of the homopolymer because the ellipticity induced by L and D monomers is opposite in sign. As a result, the CD spectrum of a sample containing 50% L and 50% D will sum to zero, regardless of what type of secondary structures are present. For this reason, we pursued additional confirmation of the grafted polypeptide secondary structure as described below.
Figure 3.
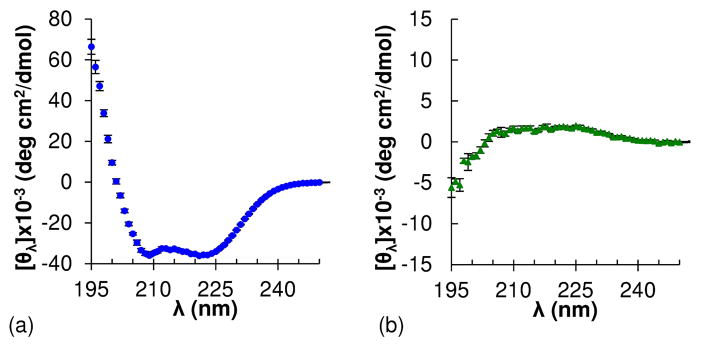
Circular dichroism spectra of (a) helical (PPLGgEG2OH) and (b) random coil (PPDLGgEG2OH) grafted polypeptides. Note: spectra are plotted on different y-axis scales for ease of view.
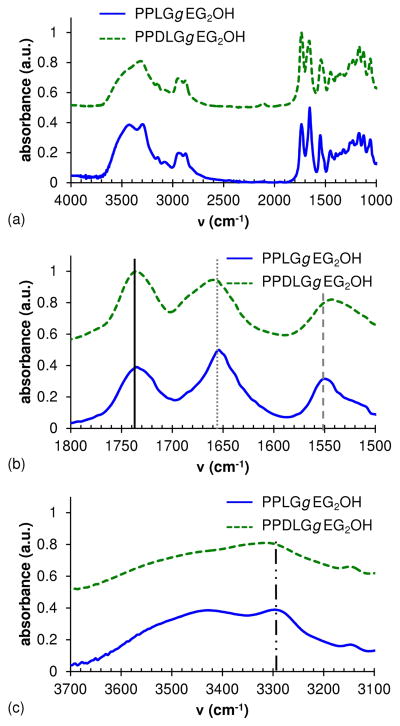
The solid phase FTIR spectra of polypeptides (Figure 4) provide further evidence of macromolecular secondary structure. Three relevant regions are those associated with amide bonds – specifically, the C=O stretch (amide I, 1665 ± 30 cm−1), N-H stretch (3170 – 3500 cm−1), and N-H bend (amide II, 1530 ± 30 cm−1): hydrogen bonding (or lack thereof) affects the frequency of these vibrational modes. The PPDLGgEG2OH heteropolymer exhibited peaks at 1660 cm−1 and 3315 cm−1, corresponding to amide I and N-H stretching vibrations, respectively. In contrast, the PPLGgEG2OH homopolymer exhibited amide I (1654 cm−1) and N-H stretching (3298 cm−1) frequencies that were lower than those of the PPDLGgEG2OH heteropolymer. Interestingly, the PPLGgEG2OH homopolymer exhibited a higher amide II frequency than the PPDLGgEG2OH heteropolymer (1550 cm−1 versus 1543 cm−1, respectively). Importantly, the frequency of the ester C=O stretch, typically found at 1740 ± 10 cm−1, occurs at 1736 cm−1 for both the PPLGgEG2OH and PPDLGgEG2OH polypeptides: this and other vibrational modes are highlighted in Figure 4. These data demonstrate that the PPLGgEG2OH homopolymer exhibits more hydrogen bonding than the PPDLGgEG2OH heteropolymer, as hydrogen bonding lowers stretching frequencies (reduced restoring force) but raises bending frequencies (increased restoring force).75 The results support our hypothesis that the PPLGgEG2OH homopolymers exist in predominantly α-helix conformation but that PPDLGgEG2OH heteropolymers containing both D- and L-glutamate monomers adopt random coil conformation. The shifts documented here for PPLGgEG2OH versus PPDLGgEG2OH polypeptides are similar to those observed by Doty et al.53 for poly(L-glutamic acid), the conformation of which can be shifted from α-helix to random coil by a change of solution pH. Specifically, the shift between helical and random coil polypeptides for the amide I peak was an identical 6 cm−1 for both the Doty et al. poly(L-glutamic acid) and our grafted poly(propargyl-glutamate). The amide II peak shift was slightly more pronounced at 10 cm−1 for Doty et al. in comparison to the 7.8 cm−1 measured for our grafted poly(propargyl-glutamate) (20% less). Part of this discrepancy may be due to the more complicated nature of the poly(L-glutamic acid) spectra in this region, which makes it difficult to assign the exact peak for amide II.
Figure 4.
FTIR spectra of PPLGgEG2OH (solid line) and PPDLGgEG2OH (dotted line) (a). The peak shifts of the amide I region (b) and the N-H stretch region (c) are due to the presence of hydrogen bonding in helical polypeptides and lack in random coil.
Effect of Chirality and Molecular Weight on Polypeptide Size and Diffusivity
The final confirmation of polypeptide secondary structure was obtained by measuring the size and diffusivity of PPLG, PPDLG, and linear PEG with dynamic light scattering. The relationship between diffusion coefficient and molecular weight of each type of polymer was fitted to the Mark-Houwink equation:
| (6) |
for which D is the diffusion coefficient, k″ is a constant, MN is the molecular weight, and ε is the scaling parameter. Values of ε are dependent upon the polymer shape in solution, ranging from 0.3 for compact spheres to 0.5 – 0.6 for random coils to 1 for rigid rods.76 We found that ε was 0.6 for linear PEG, 0.6 for PPDLGgEG2OH, and 0.9 for PPLGgEG2OH, as displayed in Table 1. The scaling parameters for PEG and PPDLGgEG2OH are similar, and indicate a random coil conformation, in contrast to the higher value found for PPLGgEG2OH, which denotes an extended rod conformation.
Table 1.
Comparison of scaling parameter (ε) and persistence length (LP) determined for polypeptides and linear PEG from dynamic light scattering data.
| Polymer | PEG | PPDLGgEG2OH | PPLGgEG2OH |
|---|---|---|---|
| ε | 0.6 | 0.6 | 0.9 |
| LP (nm) | 0.3±0.1 | 0.6±0.2 | 1.0±0.3 |
The polymer persistence length, which also varies according to conformation, was determined with the following equation:
| (7) |
where R is the hydrodynamic radius and L is the contour length, calculated by multiplying the length per monomer by the total number of monomers. The average persistence lengths for PEG, PPDLGgEG2OH, and PPLGgEG2OH were 0.3 nm, 0.6 nm, and 1.0 nm, respectively (Table 1). These values correspond to approximately 1.3 monomer units for PEG, 1.7 monomer units for PPDLGgEG2OH, and 6.8 monomer units for PPLGgEG2OH (assuming peptide random coil segments are about 3.8 Å long per residue and peptide helical segments are about 1.5 Å long per residue).77 These results are in good agreement with literature values for the persistence lengths of helical versus random coil peptides,78, 79 indicating that PPDLGgEG2OH adopts a random coil conformation similar to that of linear PEG, whereas PPLGgEG2OH adopts a rod-like conformation characteristic of helical polymers. This result supports our hypothesis that a random copolymer polymerized from a 50:50 mixture of γpLglu and γpDglu monomers would exhibit a random coil conformation. As such, we have developed a strategy that employs monomer chirality to create polypeptides with either rigid helix or flexible coil conformations that are otherwise chemically identical.
Hydrogel swelling and permeability
The effect of polypeptide backbone rigidity on gel network properties was evaluated by cross-linking either helical or random coil polypeptides with PMPI and PEG(SH)4 (PMPI:polypeptide ratio = 4:1, polypeptide:PEG ratio = 1:1) at a concentration of 10 wt% polymer overall (Figure 1). This cross-linking strategy was used due to its reproducibility and flexibility, as a variety of cross-linking concentrations can be produced by tuning the ratios of PMPI, peptide, and PEG. For comparison, control gels were produced by cross-linking PEG(Acrl)4 with a stoichiometric equivalent amount of PEG(SH)4 at 10 wt% polymer. Hydrogels formed within minutes of mixing and swelled to equilibrium within 24 hours. The equilibrium water content and gelation efficiency of all three types of hydrogel were not significantly different (Figure 5b), but the PEG and random coil polypeptide gels exhibited slightly higher swelling ratios than helical polypeptide gels (Figure 5a). Importantly, the diffusion coefficient of 45 kDa ovalbumin, measured with FRAP, was similar in all types of hydrogels (Figure 5c).
Figure 5.
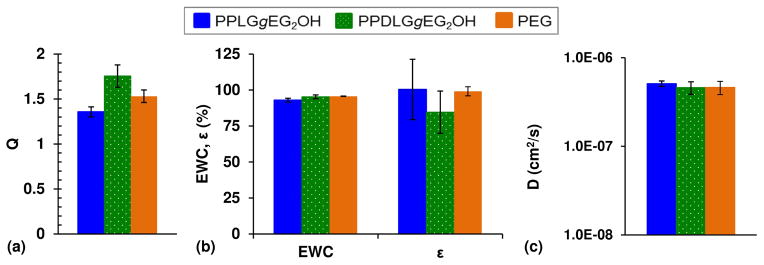
Swelling and permeability data for PPLGgEG2OH, PPDLGgEG2OH, and PEG hydrogels: (a) swelling ratio (Q), (b) equilibrium water content (EWC) and gelation efficiency (Îμ), and (c) diffusivity of 45 kDa ovalbumin.
Hydrogel mechanical properties
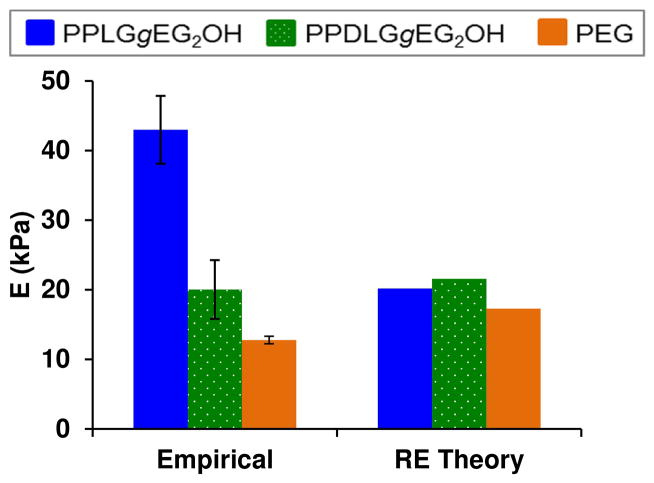
The elastic modulus of hydrogel samples, as measured by AFM indentation in fluid, is shown in Figure 6. The empirical elastic modulus for the heteropolymer PPDLGgEG2OH is similar to that of the control PEG gels: both exhibit moduli predicted by rubber elasticity theory,80 which describes properties for ideal gel networks with flexible chains as follows:
| (8) |
where νS is the concentration of elastic polymer chains in the swollen state, R is the gas constant, and T is the temperature. Although rubber elasticity predicts that the helical and random coil gels will have similar stiffness, the modulus of helical polypeptide gels is more than double that of the random coil polypeptide gels. The inability of rubber elasticity theory to predict properties of homopolymer PPLGgEG2OH hydrogels is due to the rigidity of the helical polypeptides: these macromers are more accurately described as semi-flexible chains. Semi-flexible polymer networks with different backbone rigidity that are otherwise identical should exhibit elastic moduli proportional to the square of the bending modulus, K (K = LPkT, where LP is persistence length, k is Boltzmann’s constant, and T is temperature).81 As such, the ratio of helical polypeptide to random coil polypeptide hydrogel elastic moduli calculated by the theory of semi-flexible chains is 2.17 (taking into account the slight differences in gelation efficiency, swelling, and ratio of polypeptide to PEG in the two types of gels). The empirical ratio from AFM data is 2.15, only 1.3% lower than the theoretical value. This finding supports our hypothesis that grafted polypeptides can be used to produce hydrogels with stiffness dependent upon backbone rigidity but similar permeability and composition.
Figure 6.
Hydrogel elastic moduli: empirical values determined by AFM indentation in fluid compared with values calculated from rubber elasticity theory.
Conclusions
We have shown that ring-opening polymerization of γpLglu produces helical polypeptides while polymerization of a 50:50 mixture of γpLglu and γpDglu produces random coil polypeptides that are otherwise chemically identical. As hypothesized, we can make hydrogels from either helical or coil polypeptides having the same formulation that exhibit similar swelling and permeability, but dramatically different stiffness due to rigidity of the polypeptide backbone. These results suggest that we can employ the grafted polypeptides as components of hydrogels with varying amounts of stiff rod-like segments or flexible coil segments, thus introducing the possibly of creating polymeric biomaterials with tunable and well-defined physicochemical properties. These macromers will be used to create a library of hydrogel substrates with systematically and independently varied stiffness, permeability, and ligand presentation with which to study fundamental parameters of cell-matrix interactions. Additional features will be introduced by application of “click chemistry” to graft functional groups, such as adhesion ligands, affinity probes, and macromolecular permeability modifiers, to the polypeptide backbone. We are also exploring alternative cross-linking strategies for all-aqueous hydrogel formation as well as self-assembled hydrogels.
Supplementary Material
Acknowledgments
This work was supported by NIH (R01 EB10246) and the NSF (EBICS STC). We gratefully acknowledge Deborah Pheasant and the Biophysical Instrumentation Facility for the Study of Complex Macromolecular Systems (NSF-0070319 and NIH GM68762) for assistance and instrumentation for the DLS and CD spectroscopy work. In addition, special thanks to Dr. Alan Schwartzman and the NanoMechanical Technology Laboratory in the Department of Materials Science and Engineering at MIT for AFM instrumentation and assistance. Thanks also to Zeynep Ilke Kalcioglu for advice regarding AFM indentation of hydrogels. Finally, thanks to Dr. Eliza Vasile and the Swanson Biotechnology Center at the MIT Koch Institute for microscopy assistance and instrumentation.
Footnotes
Electronic Supplementary Information (ESI) available: [details of any supplementary information available should be included here]. See DOI: 10.1039/b000000x/
Contributor Information
Abigail M. Oelker, Email: aoelker@mit.edu.
Shannon M. Morey, Email: smmorey@mit.edu.
Linda G. Griffith, Email: griff@mit.edu.
Paula T. Hammond, Email: hammond@mit.edu.
References
- 1.Pelham RJ, Wang Y-l. Proceedings of the National Academy of Sciences of the United States of America. 1997;94:13661–13665. doi: 10.1073/pnas.94.25.13661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pelham RJ, Wang YL. Molecular Biology of the Cell. 1999;10:935–945. doi: 10.1091/mbc.10.4.935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lo C-M, Wang H-B, Dembo M, Wang Y-l. Biophysical Journal. 2000;79:144–152. doi: 10.1016/S0006-3495(00)76279-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Deroanne CF, Lapiere CM, Nusgens BV. Cardiovascular Research. 2001;49:647–658. doi: 10.1016/s0008-6363(00)00233-9. [DOI] [PubMed] [Google Scholar]
- 5.Cukierman E, Pankov R, Stevens DR, Yamada KM. Science. 2001;294:1708–1712. doi: 10.1126/science.1064829. [DOI] [PubMed] [Google Scholar]
- 6.Marganski WA, Dembo M, Wang Y-L, Gerard M, Ian P. Methods in Enzymology. Vol. 361. Academic Press; 2003. pp. 197–211. [DOI] [PubMed] [Google Scholar]
- 7.Engler AJ, Griffin MA, Sen S, Bonnetnann CG, Sweeney HL, Discher DE. Journal of Cell Biology. 2004;166:877–887. doi: 10.1083/jcb.200405004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Engler A, Bacakova L, Newman C, Hategan A, Griffin M, Discher D. Biophysical Journal. 2004;86:617–628. doi: 10.1016/S0006-3495(04)74140-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yeung T, Georges PC, Flanagan LA, Marg B, Ortiz M, Funaki M, Zahir N, Ming WY, Weaver V, Janmey PA. Cell Motility and the Cytoskeleton. 2005;60:24–34. doi: 10.1002/cm.20041. [DOI] [PubMed] [Google Scholar]
- 10.Healy KE. Proceedings of the 26th Annual International Conference of the Ieee Engineering in Medicine and Biology Society, Vols 1–7; 2004. pp. 5035–5035. [Google Scholar]
- 11.Lampe KJ, Mooney RG, Bjugstad KB, Mahoney MJ. Journal of Biomedical Materials Research Part A. 2010;94A:1162–1171. doi: 10.1002/jbm.a.32787. [DOI] [PubMed] [Google Scholar]
- 12.Appelman TP, Mizrahi J, Elisseeff JH, Seliktar D. Biomaterials. In Press, Corrected Proof. [Google Scholar]
- 13.Zhang L, Furst EM, Kiick KL. Journal of Controlled Release. 2006;114:130–142. doi: 10.1016/j.jconrel.2006.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lindblad WJ, Schuetz EG, Redford KS, Guzelian PS. Hepatology. 1991;13:282–288. [PubMed] [Google Scholar]
- 15.Powers MJ, Rodriguez RE, Griffith LG. Biotechnology and Bioengineering. 1997;53:415–426. doi: 10.1002/(SICI)1097-0290(19970220)53:4<415::AID-BIT10>3.0.CO;2-F. [DOI] [PubMed] [Google Scholar]
- 16.Fassett J, Tobolt D, Hansen LK. Molecular Biology of the Cell. 2006;17:345–356. doi: 10.1091/mbc.E05-09-0871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cosgrove BD, Cheng C, Pritchard JR, Stolz DB, Lauffenburger DA, Griffith LG. Hepatology. 2008;48:276–288. doi: 10.1002/hep.22335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Genove E, Schmitmeier S, Sala A, Borros S, Bader A, Griffith LG, Semino CE. Journal of Cellular and Molecular Medicine. 2009;13:3387–3397. doi: 10.1111/j.1582-4934.2009.00970.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mehta G, Williams CM, Alvarez L, Lesniewski M, Kamm RD, Griffith LG. Biomaterials. 2010;31:4657–4671. doi: 10.1016/j.biomaterials.2010.01.138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Weber LM, Lopez CG, Anseth KS. Journal of Biomedical Materials Research Part A. 2009;90A:720–729. doi: 10.1002/jbm.a.32134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gilbert DL, Teruo O, Teruo M, Sung Wan K. International Journal of Pharmaceutics. 1988;47:79–88. [Google Scholar]
- 22.Peppas NA, Reinhart CT. Journal of Membrane Science. 1983;15:275–287. [Google Scholar]
- 23.Reinhart CT, Peppas NA. Journal of Membrane Science. 1984;18:227–239. [Google Scholar]
- 24.Levick JR. Experimental Physiology. 1987;72:409–437. doi: 10.1113/expphysiol.1987.sp003085. [DOI] [PubMed] [Google Scholar]
- 25.Karande TS, Ong JL, Agrawal CM. Annals of Biomedical Engineering. 2004;32:1728–1743. doi: 10.1007/s10439-004-7825-2. [DOI] [PubMed] [Google Scholar]
- 26.Wolf K, Alexander S, Schacht V, Coussens LM, von Andrian UH, van Rheenen J, Deryugina E, Friedl P. Seminars in Cell & Developmental Biology. 2009 doi: 10.1016/j.semcdb.2009.08.005. In Press, Corrected Proof. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Oelker AM, Berlin JA, Wathier M, Grinstaff MW. Biomacromolecules. 2011;12:1658–1665. doi: 10.1021/bm200039s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Oelker A, Grinstaff M. NanoBioscience, IEEE Transactions on. 2011:1–1. doi: 10.1109/TNB.2011.2166978. [DOI] [PubMed] [Google Scholar]
- 29.Oelker AM, Grinstaff MW. Journal of Materials Chemistry. 2008;18:2521–2536. [Google Scholar]
- 30.Bayramoglu G, Can Akcalı K, Gultekin S, Bengu E, Arica M. Macromolecular Research. 2011;19:385–395. [Google Scholar]
- 31.Jaiswal M, Koul V, Dinda AK, Mohanty S, Jain KG. Journal of Biomedical Materials Research Part B: Applied Biomaterials. 2011;98B:342–350. doi: 10.1002/jbm.b.31857. [DOI] [PubMed] [Google Scholar]
- 32.Kim DH, Wong PK, Park J, Levchenko A, Sun Y. Annual Review of Biomedical Engineering. 2009;11:203–233. doi: 10.1146/annurev-bioeng-061008-124915. [DOI] [PubMed] [Google Scholar]
- 33.Bershadsky AD, Balaban NQ, Geiger B. Annual Review of Cell and Developmental Biology. 2003;19:677–695. doi: 10.1146/annurev.cellbio.19.111301.153011. [DOI] [PubMed] [Google Scholar]
- 34.Bao G, Suresh S. Nat Mater. 2003;2:715–725. doi: 10.1038/nmat1001. [DOI] [PubMed] [Google Scholar]
- 35.Reinhart-King CA, Dembo M, Hammer DA. Langmuir. 2002;19:1573–1579. [Google Scholar]
- 36.Hersel U, Dahmen C, Kessler H. Biomaterials. 2003;24:4385–4415. doi: 10.1016/s0142-9612(03)00343-0. [DOI] [PubMed] [Google Scholar]
- 37.Shu XZ, Ghosh K, Liu Y, Palumbo FS, Luo Y, Clark RA, Prestwich GD. Journal of Biomedical Materials Research. 2004;68A:365–375. doi: 10.1002/jbm.a.20002. [DOI] [PubMed] [Google Scholar]
- 38.Gobin AS, West JL. FASEB Journal. 2002 doi: 10.1096/fj.01-0759fje. 01-0759fje. [DOI] [PubMed] [Google Scholar]
- 39.Tsurkan MV, Levental KR, Freudenberg U, Werner C. Chemical Communications. 2010;46:1141–1143. doi: 10.1039/b921616b. [DOI] [PubMed] [Google Scholar]
- 40.Park Y, Lutolf MP, Hubbell JA, Hunziker EB, Wong M. Tissue Engineering. 2004;10:515–522. doi: 10.1089/107632704323061870. [DOI] [PubMed] [Google Scholar]
- 41.Kraehenbuehl TP, Zammaretti P, Van der Vlies AJ, Schoenmakers RG, Lutolf MP, Jaconi ME, Hubbell JA. Biomaterials. 2008;29:2757–2766. doi: 10.1016/j.biomaterials.2008.03.016. [DOI] [PubMed] [Google Scholar]
- 42.Wathier M, Jung PJ, Carnahan MA, Kim T, Grinstaff MW. Journal of the American Chemical Society. 2004;126:12744–12745. doi: 10.1021/ja045870l. [DOI] [PubMed] [Google Scholar]
- 43.Wathier M, Johnson CS, Kim T, Grinstaff MW. Bioconjugate Chemistry. 2006;17:873–876. doi: 10.1021/bc060060f. [DOI] [PubMed] [Google Scholar]
- 44.Farmer R, Top A, Argust L, Liu S, Kiick K. Pharmaceutical Research. 2008;25:700–708. doi: 10.1007/s11095-007-9344-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Banwell EF, Abelardo ES, Adams DJ, Birchall MA, Corrigan A, Donald AM, Kirkland M, Serpell LC, Butler MF, Woolfson DN. Nature Materials. 2009;8:596–600. doi: 10.1038/nmat2479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Berdahl JP, Johnson CS, Proia AD, Grinstaff MW, Kim T. Archives of Ophthalmology. 2009;127:442–447. doi: 10.1001/archophthalmol.2008.582. [DOI] [PubMed] [Google Scholar]
- 47.Kisiday J, Jin M, Kurz B, Hung H, Semino C, Zhang S, Grodzinsky AJ. Proceedings of the National Academy of Sciences of the United States of America. 2002;99:9996–10001. doi: 10.1073/pnas.142309999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhang K, Diehl MR, Tirrell DA. Journal of the American Chemical Society. 2005;127:10136–10137. doi: 10.1021/ja051457h. [DOI] [PubMed] [Google Scholar]
- 49.Kopecek J, Yang J. Acta Biomaterialia. 2009;5:805–816. doi: 10.1016/j.actbio.2008.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Becker RR, Stahmann MA. Journal of the American Chemical Society. 1952;74:38–41. [Google Scholar]
- 51.Blout ER, Idelson M. Journal of the American Chemical Society. 1956;78:497–498. [Google Scholar]
- 52.Doty P, Holtzer AM, Bradbury JH, Blout ER. Journal of the American Chemical Society. 1954;76:4493–4494. [Google Scholar]
- 53.Doty P, Wada A, Yang JT, Blout ER. Journal of Polymer Science. 1957;23:851–861. [Google Scholar]
- 54.Goodman M, Schmitt EE, Yphantis D. Journal of the American Chemical Society. 1960;82:3483–3484. [Google Scholar]
- 55.Deming TJ. Adv Drug Deliv Rev. 2002;54:1145–1155. doi: 10.1016/s0169-409x(02)00062-5. [DOI] [PubMed] [Google Scholar]
- 56.Deming T. In: Klok H-A, Schlaad H, editors. Vol. 202. Springer; Berlin/Heidelberg: 2006. pp. 1–18. [Google Scholar]
- 57.Deming TJ. Progress in Polymer Science. 2007;32:858–875. [Google Scholar]
- 58.Patterson J, Martino MM, Hubbell JA. Materials Today. 2010;13:14–22. [Google Scholar]
- 59.Bromley EHC, Channon K, Moutevelis E, Woolfson DN. Acs Chemical Biology. 2008;3:38–50. doi: 10.1021/cb700249v. [DOI] [PubMed] [Google Scholar]
- 60.Engler AC, Lee H-i, Hammond PT. Angewandte Chemie. 2009;121:9498–9502. [Google Scholar]
- 61.Engler AC, Bonner DK, Buss HG, Cheung EY, Hammond PT. Soft Matter. 2011;7:5627–5637. [Google Scholar]
- 62.Xiao C, Zhao C, He P, Tang Z, Chen X, Jing X. Macromolecular Rapid Communications. 2010;31:991–997. doi: 10.1002/marc.200900821. [DOI] [PubMed] [Google Scholar]
- 63.Engler AC, Shukla A, Puranam S, Buss HG, Jreige N, Hammond PT. Biomacromolecules. 2011;12:1666–1674. doi: 10.1021/bm2000583. [DOI] [PubMed] [Google Scholar]
- 64.Zhao X, Poon Z, Engler AC, Bonner DK, Hammond PT. Biomacromolecules. 2012 doi: 10.1021/bm201873u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kolb HC, Finn MG, Sharpless KB. Angewandte Chemie International Edition. 2001;40:2004–2021. doi: 10.1002/1521-3773(20010601)40:11<2004::AID-ANIE2004>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- 66.Malkoch M, Vestberg R, Gupta N, Mespouille L, Dubois P, Mason AF, Hedrick JL, Liao Q, Frank CW, Kingsbury K, Hawker CJ. Chemical Communications. 2006:2774–2776. doi: 10.1039/b603438a. [DOI] [PubMed] [Google Scholar]
- 67.Moses JE, Moorhouse AD. Chemical Society Reviews. 2007;36:1249–1262. doi: 10.1039/b613014n. [DOI] [PubMed] [Google Scholar]
- 68.Van Dijk M, Nollet ML, Weijers P, Dechesne AC, van Nostrum CF, Hennink WE, Rijkers DTS, Liskamp RMJ. Biomacromolecules. 2008;9:2834–2843. doi: 10.1021/bm8005984. [DOI] [PubMed] [Google Scholar]
- 69.Hein C, Liu XM, Wang D. Pharmaceutical Research. 2008;25:2216–2230. doi: 10.1007/s11095-008-9616-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Hong SY, Oh JE, Lee KH. Biochem Pharmacol. 1999;58:1775–1780. doi: 10.1016/s0006-2952(99)00259-2. [DOI] [PubMed] [Google Scholar]
- 71.Brack A, Spach G. Journal of Molecular Evolution. 1979;13:35–46. doi: 10.1007/BF01732752. [DOI] [PubMed] [Google Scholar]
- 72.Lundberg RD, Doty P. Journal of the American Chemical Society. 1957;79:3961–3972. [Google Scholar]
- 73.Branco MC, Pochan DJ, Wagner NJ, Schneider JP. Biomaterials. 2009;30:1339–1347. doi: 10.1016/j.biomaterials.2008.11.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Greenfield N, Fasman GD. Biochemistry. 1969;8:4108. doi: 10.1021/bi00838a031. [DOI] [PubMed] [Google Scholar]
- 75.Colthup NB, Daly LH, Wiberley SE. Introduction to infrared and Raman spectroscopy. 2. Academic Press; New York: 1975. [Google Scholar]
- 76.Tsaih ML, Chen RH. Journal of Applied Polymer Science. 1999;73:2041–2050. [Google Scholar]
- 77.Miller WG, Flory PJ. Journal of Molecular Biology. 1966;15:298–314. doi: 10.1016/s0022-2836(66)80229-2. [DOI] [PubMed] [Google Scholar]
- 78.Danielsson J, Andersson A, Jarvet J, Gräslund A. Magnetic Resonance in Chemistry. 2006;44:S114–S121. doi: 10.1002/mrc.1814. [DOI] [PubMed] [Google Scholar]
- 79.Fitzkee NC, Rose GD. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:12497–12502. doi: 10.1073/pnas.0404236101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Flory PJ. Principles of Polymer Chemistry. Cornell University Press; Ithaca, NY: 1953. [Google Scholar]
- 81.MacKintosh FC, Kas J, Janmey PA. Physical Review Letters. 1995;75:4425. doi: 10.1103/PhysRevLett.75.4425. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.