Abstract
Cytoplasmic nucleophosmin (NPMc+) mutations and FMS-like tyrosine kinase 3 (FLT3) internal tandem duplication (ITD) mutations are two of the most common known molecular alterations in acute myeloid leukemia (AML), and they frequently occur together suggesting cooperative leukemogenesis. To explore the specific relationship between NPMc+ and FLT3/ITD in vivo, we crossed Flt3/ITD knock-in mice with transgenic NPMc+ mice. Mice with both mutations develop a transplantable leukemia of either myeloid or lymphoid lineage, definitively demonstrating cooperation between Flt3/ITD and NPMc+. In mice with myeloid leukemia, functionally significant loss of heterozygosity of the wild-type Flt3 allele is common, similar to what is observed in human FLT3/ITD+ AML, providing further in vivo evidence of the importance of loss of wild-type FLT3 in leukemic initiation and progression. Additionally, in vitro clonogenic assays reveal that the combination of Flt3/ITD and NPMc+ mutations causes a profound monocytic expansion, in excess of that seen with either mutation alone consistent with the predominance of myelomonocytic phenotype in human FLT3/ITD+/NPMc+ AML. This in vivo model of Flt3/ITD+/NPMc+ leukemia closely recapitulates human disease and will therefore serve as a tool for the investigation of the biology of this common disease entity.
Keywords: Acute myeloid leukemia, AML, mouse model, NPMc+, FLT3/ITD
Introduction
Patients with AML represent a large and heterogeneous group. Much work has been undertaken to identify unique molecular genetic alterations in AML that are clinically and functionally significant, work greatly facilitated recently by next generation sequencing technology. Internal tandem duplication (ITD) mutations of the FMS-like tyrosine kinase 3 (FLT3) gene and mutations of the nucleophosmin gene (NPM1) are two of the most frequently identified [1, 2].
NPM1 encodes nucleophosmin (NPM) which is a ubiquitously expressed nucleocytoplasmic shuttling phosphoprotein. Although the bulk of NPM resides in the nucleolus, it constantly exchanges between the nucleus and cytoplasm [3]. NPM plays a key role in several cellular functions including ribosome biogenesis and maintenance of genomic stability via regulation of centrosome duplication and control of DNA repair [4]. NPM also interacts with the oncosuppressors p53 and ARF and their partners thus controlling cell proliferation and apoptosis [4]. Acquired mutations in exon 12 of NPM1 were first reported by Falini et al in 2005 and are found frequently in AML patients, particularly in those with a normal karyotype (NK-AML) [5]. These mutations are characteristically heterozygous with the mutated allele encoding a protein that aberrantly localizes to the cytoplasm, thus the designation NPM-cytoplasmic positive (NPMc+) AML. NPM1 exon 12 mutations target 30–35% of all adult AML and up to 50–60% of adult NK-AML [ 5]. In childhood AML, the prevalence is significantly less with approximately 8% of all AML and approximately 20% of NK-AML [6, 7]. In most studies, NPMc+ mutation is associated with improved prognosis, with a significantly higher CR rate [5, 8–10] and, in many studies, longer OS and EFS [7–10]. Given its distinctive biologic and clinical features and its clear clinical relevance, NPMc+ AML is included as a provisional entity in the 2008 World Health Organization classification of myeloid malignancies [11].
FLT3 is a receptor tyrosine kinase that together with its ligand, FL plays important roles in the proliferation, survival, and differentiation of hematopoietic stem/progenitor cells [12, 13]. Upon binding FL the receptor dimerizes, activating its tyrosine kinase domain resulting in autophosphorylation [14]. Several important signaling proteins such as Ras-GAP, PLC-b, PI3-kinase, STAT5, PIM1, and MAP kinase have been linked to FLT3 activation [14, 15]. Mutations in FLT3 have been reported in approximately 20–35% of AML patients [14, 16–19]. These mutations are either internal tandem duplication (ITD) mutations, most commonly occurring in the juxtamembrane domain or point mutations in the kinase domain, which result in the constitutive dimerization and activation of FLT3, independent of FL. FLT3/ITD mutations confer a poor prognosis in studies of pediatric and adult AML [14, 19, 20].
Importantly, NPMc+ mutations and FLT3/ITD mutations coexist frequently in AML. FLT3/ITD mutations are approximately two-fold more frequent in NPMc+ leukemia compared to leukemia lacking NPM1 mutation [5, 6, 8]. Given the frequency with which these two mutations coexist in AML, we hypothesized that they cooperate to cause leukemia. To specifically investigate the relationship between NPMc+ and FLT3/ITD mutations, we crossed mice with Flt3/ITD constitutively knocked-in with NPMc+ transgenic mice. Flt3/ITD knock-in mice develop a fatal myeloproliferative neoplasm with a relatively long latency, but do not develop leukemia [21, 22]. NPMc+ transgenic mice develop a non-fatal myeloproliferation and also do not develop overt leukemia [23]. Indeed, combination of Flt3/ITD and NPMc+ resulted in the development of leukemia in mice, providing an in vivo model of Flt3/ITD+/NPMc+ leukemia which closely recapitulates human disease, thus making an in depth investigation of disease biology possible.
Methods
Mice
Mice with an 18bp-ITD mutation knocked into the juxtamembrane domain of the murine Flt3 gene (FLT3wt/ITD) and transgenic mice expressing Flag-tagged human NPMc+ mutant A driven by human MRP8 promoter (hMRP8-NPMc+) were generated as previously reported [22, 23]. Mice were categorized as wild-type (wt), positive for the NPMc+ mutation alone (NPMc+), positive for the Flt3/ITD mutation alone (ITD), or positive for both mutations (ITD/NPMc+) based on PCR of germline DNA using the primers mITD-5F + mITD-3R, NPM874F + MRP8R (Sequences in Supplementary table 1s).
For transplantation experiments, CD45.1+ mice received 700cGy of gamma irradiation. Then 1×106 whole bone marrow cells isolated from leukemic ITD/NPMc+ CD45.2 mice were injected via retro-orbital injection. Engraftment was evaluated by flow cytometry determination of the percentage of CD45.2+ cells in the peripheral blood. All animal experiments were reviewed and approved by the Johns Hopkins IACUC.
Flow cytometry analysis
Flow cytometric analysis of murine BM cells, splenocytes, and cells of other infiltrated organs was performed with the following monoclonal antibodies: lineage mixture (Caltag laboratories/Invitrogen, Carlsbad, CA), Ly-6A/E (Sca-1) (Invitrogen, Carlsbad, CA), CD135, CD117 (c-Kit), CD41, Ter119, CD11b, Gr1, CD24, CD43, CD19, CD45R/B220, CD4, CD3, and CD8a (BD Pharmingen, San Jose, CA). For intracellular flow cytometry BM cells were obtained, washed, fixed and permeabilized. Alexa Fluor 488 conjugated anti-phosphoSTAT5 (pY694) (BD biosciences) was used to detect phophorylated STAT5. Analysis was performed using FlowJo software (TreeStar, Ashland, OR).
Reverse-transcriptase (RT)PCR
RNA was isolated from murine tissues and was reverse transcribed and amplified using the following primers (primer sequences found in supplementary table 1s): For Flag-tagged NPMc+: exon 1F + NPM500R; for HPRT: HPRT1 + HPRT2. For Flt3: mFLT3-RT-5F + mFLT3-RT-3R.
In vitro clonogenic assay
Lineage negative murine BM cells from 2-month-old mice were placed in liquid culture with RPMI-1640 plus 20% FBS, murine SCF (50ng/mL), IL-3 (10ng/mL) and IL-6 (10ng/mL) or plated in Methocult M3434 medium supplemented with recombinant murine SCF (50ng/mL), IL-3 (10ng/mL), IL-6 (10ng/mL), GM-CSF (10ng/mL), and EPO (3U/mL). Cells in liquid culture were counted daily and on day 7 were analyzed by flow cytometry for lineage commitment. On methylcellulose, all colonies were scored 9–11 days after plating. Cells were then isolated from the plates and analyzed by flow cytometry for cKit, Mac1, Gr1, and Ter119. Experiments were performed in triplicate.
In vitro cytotoxicity assay
Murine BM cells were cultured in RPMI 1640 (Gibco/Invitrogen, Carlsbad, CA) plus 10% heat inactivated fetal bovine serum (FBS), 1% L-glutamine, 50ng/mL SCF, 10ng/mL IL3, and 10ng/mL IL6 (Pepro Tech, Rocky Hill, NJ). Cells were treated with increasing concentrations of lestaurtinib (Cephalon, Inc., Malvern, PA) for 48 hours. WST-1 reagent (Roche Diagnostics, Manheim, Germany) was added to the culture medium (1:10 dilution) and absorbance was measured at 450nm using a Bio-Rad model 680 microplate reader (BioRad). Assays were performed in triplicate.
Statistics
Kaplan-Meier survival curve was generated and analyzed using Graph Pad Prism version 4.0 software (GraphPad, San Diego, CA). Log-rank test was performed to compare the generated survival curves. Anova followed by pairwise t-tests were used where appropriate (GraphPad).
Results
Flt3/ITD and NPMc+ mutations cooperate to cause acute leukemia
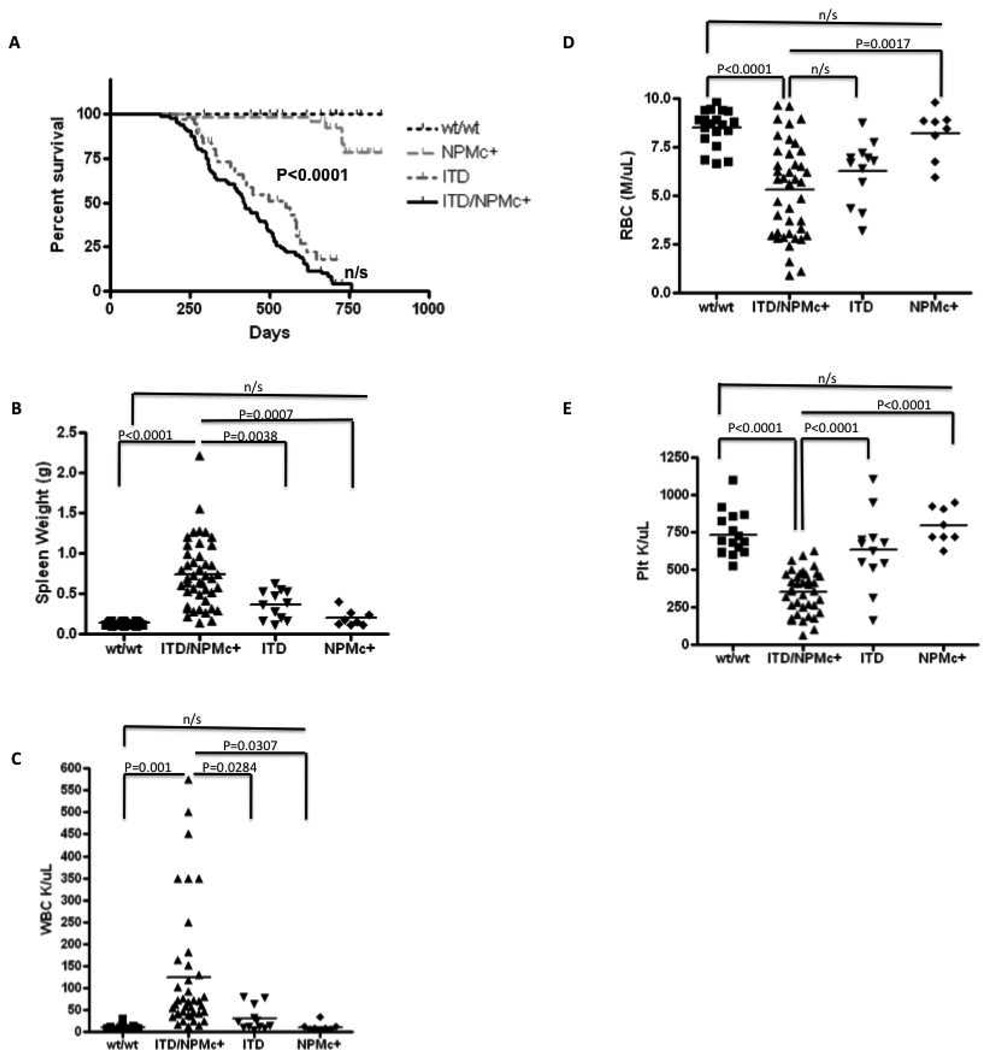
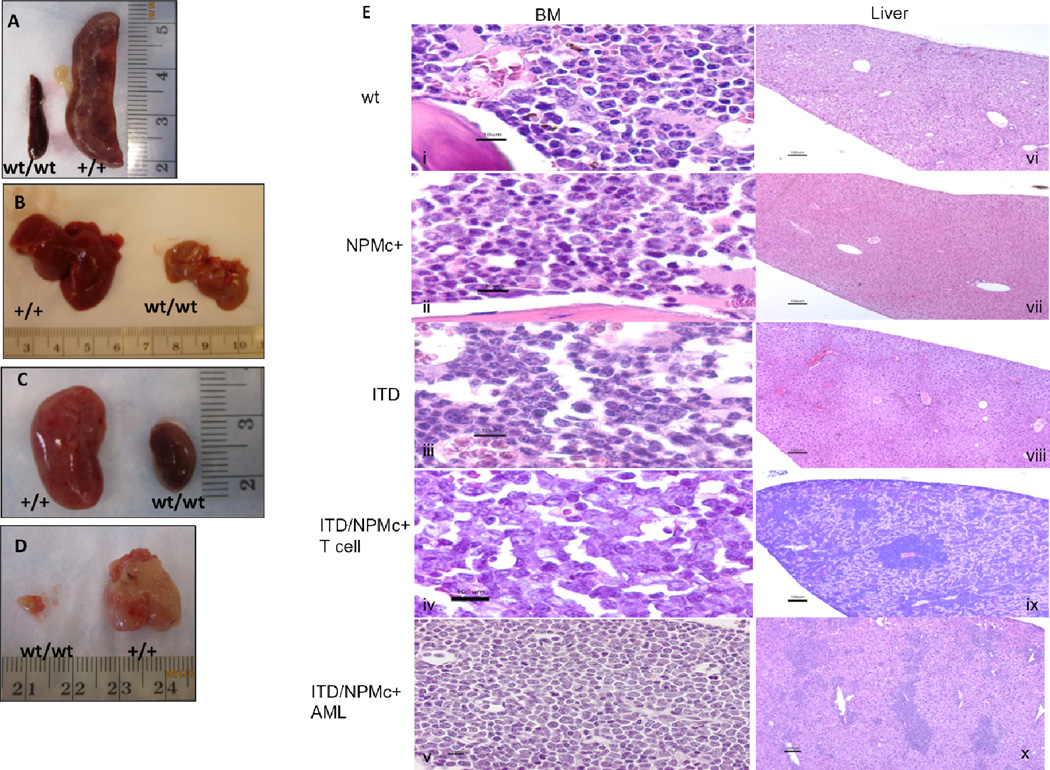
Mice with both Flt3/ITD and NPMc+ mutations (ITD/NPMc+) die between 6 and 18 months with a median survival of 420 days (Figure 1A). Their survival is significantly shorter than the survival of wild-type (wt) mice and NPMc+ mice (P<0.0001). Mice with Flt3/ITD mutation alone (ITD) succumb to a fatal myeloproliferative neoplasm (MPN) between 6 and 20 months [22] and in our cohort had a median survival of 550 days, which was longer than ITD/NPMc+ mice, though this difference did not reach statistical significance. ITD/NPMc+ mice become moribund prior to death. Moribund mice were sacrificed (N=40) and found to have splenomegaly (mean spleen weight 0.741±0.06g vs 0.13±0.005g for wt mice 0.2±0.04g in NPMc+ mice, and 0.37±0.05g in ITD mice), leukocytosis (mean WBC 124.5±22.5K/µL vs 10.7±1.3K/µL in wt mice, 11.1±3.4K/µL in NPMc+ mice, and 30.4±8K/µL in ITD mice), anemia (mean RBC 5.3±0.4M/µL vs 8.5±0.2M/µL in wt mice, 8.2±0.4M/µL in NPMc+ mice, and 6.3±0.5M/µL in ITD mice), and thrombocytopenia (mean platelet count 352.3±23K/µL vs 735±37.1K/µL in wt mice, 795.5±42K/µL in NPMc+ mice, and 629.3±72.2K/µL in ITD mice) (Figure 1B–D). All of these parameters were statistically significant compared to wt littermate controls (N=19), ill ITD mice (N=12) and littermate control NPMc+ mice (N=8) except RBC which trended towards more severe in the ITD/NPMc+ mice but did not reach statistical significance compared to ill ITD mice. On gross examination there was pathologic enlargement of multiple organs including spleens and livers of all ill ITD/NPMc+ mice and kidneys and thymuses of a subset of ill ITD/NPMc+ mice (Figure 2A–D). On histologic examination, large monotonous cells were seen infiltrating the bone marrow (BM) and infiltrating and disrupting the normal architecture of the livers of leukemic mice with both mutations (Figure 2E). Additionally, the malignant cells infiltrating the BM are found in high numbers circulating in the peripheral blood of ill ITD/NPMc+ mice (Supplemental Figure 1s). Further, the spleens of all ill ITD/NPMc+ mice and the kidneys, and meninges of a subset of ill ITD/NPMc+ mice were found to be infiltrated with large monotonous cells on H&E staining indicating a very aggressive, infiltrative disease (Supplemental Figure 1s).
Figure 1. ITD/NPMc+ mice have a shortened survival and a clinical phenotype consistent with acute leukemia.
A) Kaplan-Meier survival curve demonstrating a shortened overall survival for mice with both a Flt3/ITD mutation and a NPMc+ mutation (ITD/NPMc+, N=87) compared to mice of other genotypes (wild type controls (wt/wt, N=18), Flt3/ITD alone (ITD, N=50), NPMc+ alone (NPMc+, N=30)). These mice become moribund prior to death and have clinical characteristic of acute leukemia including: B) splenomegaly, C) leukocytosis, D) anemia, and E) thrombocytopenia. For B–E, number of mice included: wt, N=20; ITD/NPMc+, N=40; ITD, N=15; NPMc+, N=8.
n/s, not statistically significant.
Figure 2. The leukemia that develops in ITD/NPMc+ mice is aggressive, infiltrating multiple organs.
Gross evaluation of representative leukemic ITD/NPMc+ mice, revealing involvement of the A) spleen, B) liver, C) kidneys, and in some cases the D) thymus. E) H&E stains of BM (i–v) and liver (vi–x) of representative mice of each genotype. Scale bars are as follows: i–v 10µm, vi–x 100µm.
Mice harboring both NPMc+ and FLT3/ITD mutations develop transplantable myeloid and lymphoid leukemias
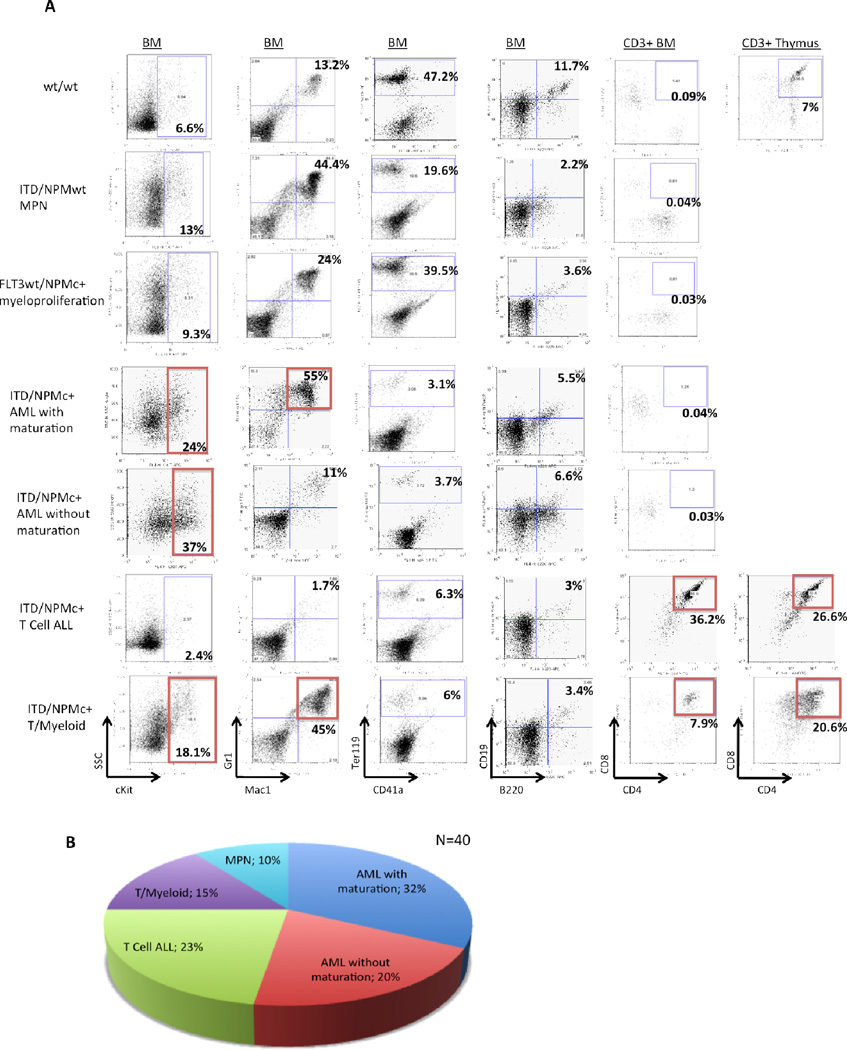
To characterize the disease of ill ITD/NPMc+ mice, flow cytometry was performed on BM cells, splenocytes, and cells of other infiltrated tissue. Wild-type littermate controls were sacrificed and analyzed at the same time as ill ITD/NPMc+ mice (Figure 3A, row 1). A number of ill mice with Flt3/ITD alone or littermate control mice with NPMc+ alone were also analyzed in the same fashion (Figure 3A, rows 2 and 3 respectively). As anticipated, there was a high frequency of myeloid leukemia in ITD/NPMc+ mice. Most commonly, ITD/NPMc+ mice developed myeloid leukemia with maturation, characterized by high cKit positivity, and Mac1+/Gr1+ myeloblasts (Figure 3A, row 4) [24]. We also observed a number of myeloid leukemias without maturation, characterized by high cKit positivity, but myeloblasts lacking cell surface markers indicative of maturation (Figure 3A, row 5) [24]. In addition, we observed a number of T cell acute lymphoblastic leukemias (ALLs) characterized by cKit−, CD3+/CD4+/CD8+ lymphoblasts (Figure 3A, row 6) or less commonly CD3+/CD4−/CD8+ lymphoblasts (Supplementary Figure 2s). A number of ITD/NPMc+ mice also developed a mixed lineage T/myeloid disease with high cKit positivity and both Mac1+/Gr1+ myeloblasts and CD3+/CD4+/CD8+ lymphoblasts (Figure 3A, rows 7). All the leukemic mice also had a paucity of normal maturing erythrocytes as demonstrated by decreased Ter119+ cells in the BM and maturing B lymphocytes indicated by decreased B220+/CD19+ cell in the BM (Figure 3, columns 3 and 4 respectively). The relative frequency with which each of the leukemic subtypes occurred in the ITD/NPMc+ mice is summarized in Figure 3B with over 50% of ill mice having a myeloid leukemia.
Figure 3. ITD/NPMc+ mice develop myeloid and lymphoid leukemias.
A) Flow cytometry plots demonstrating the characteristic phenotype of wt mice (row 1), ITD mice with myeloproliferative neoplasm (row 2), NPMc+ mice with myeloproliferation (row 3) and each of the 4 most common types of acute leukemia diagnosed in ITD/NPMc+ mice (rows 4–7). All the leukemic mice also have a paucity of normal maturing erythrocytes as demonstrated by decreased Ter119+ cells in the BM and maturing B lymphocytes indicated by decreased B220+/CD19+ cell in the BM. B) Disease distribution of the 40 fully characterized ITD/NPMc+ mice.
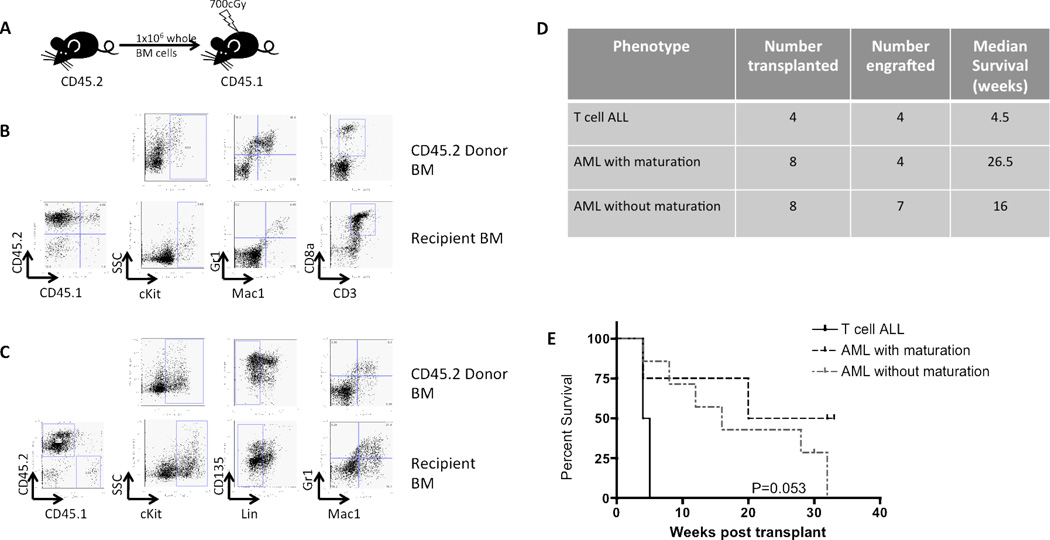
In transplantation experiments, sub-lethally irradiated syngeneic mice injected with 1×106 whole BM cells from leukemic mice with either myeloid or T lineage disease engrafted by 4 weeks post-transplant and develop disease with the same phenotypic characteristics as the leukemic donor mouse (Figure 4A–D). The kinetics of the transplanted leukemias were variable, with transplanted T cell ALL causing more rapid decline than transplanted AML (Figure 4D and E).
Figure 4. Both T cell and myeloid leukemias are transplantable.
A) Experimental design of transplantation experiment. B) Phenotype of the donor mouse with T cell ALL (row 1). Engraftment (as determined by %CD45.2 positivity) and phenotype of a representative recipient (row 2). C) Phenotype of the leukemic donor mouse with AML without maturation (row 1). Engraftment and phenotype of a representative recipient (row 2). D) Table summarizing transplantation experiments. E) Kaplan-meier survival curves demonstrating disease kinetics engrafted mice of the three disease phenotypes (2/4 mice that engrafted with AML with maturation had not succumbed to illness when the experiment was terminated at 32 weeks, and were therefore censored).
Expression of both Flt3/ITD and the flag-tagged NPMc+ transgene mRNA in the BM and infiltrated non-hematopoietic tissue of leukemic mice was demonstrated by reverse-transcriptase (RT)-PCR (Supplementary Figure 3s). Given the likely importance of NPMc+ localization to the cytoplasm in its role in leukemogenesis, we confirmed mutant transgenic NPMc+ protein localization to the cytoplasm by immunofluorescence and immunohistochemistry (Supplementary Figure 3s). To demonstrate functional effects of NPMc+ mutation, we examined expression of Hox cluster genes known to be overexpressed in human NPMc+ AML and other NPMc+ animal models [4, 25, 26]. By RT-qPCR we found increased expression of both HoxA9 and HoxA10 in whole BM cells from 2-month-old NPMc+, and ITD/NPMc+ mice (Figure 3S). We also found increased expression of HoxA9 in mice with Flt3/ITD mutation alone, as has been reported previously [27]. However the ITD/NPMc+ mice had higher levels of HoxA9 expression than mice with either mutation alone suggesting an additive effect. HoxA10 expression was increased only in mice with NPMc+ mutations, and to the same degree both in the presence and absence of concomitant Flt3/ITD mutation, suggesting this increased expression was due solely to the effects of the NPMc+ mutation.
The occurrence of T lineage leukemia was an unexpected finding because NPMc+ mutations are not associated with T cell ALL, and activating FLT3 mutations rarely occur in T cell ALL [5, 28]. Further, the hMRP8 promoter which drives expression of NPMc+ is expected to only be expressed in myeloid lineage Mac1+/Gr1+ cells. We hypothesized that there may be a degree of infidelity of the hMRP8 promoter allowing for NPMc+ expression in the T cells of mice with the hMRP8-NPMc+ transgene. We therefore isolated T cells from the spleens of 2-month-old mice of each genotype. By RT-PCR, NPMc+ expression was demonstrated in T cells of mice with the NPMc+ transgene (Supplementary Figure 4s). Further, similar T cell disease has been documented in other mouse models utilizing the hMPR8 promoter [29].
Loss of heterozygosity (LOH) of the wild-type Flt3 allele occurs with a high frequency in myeloid leukemia and is functionally significant
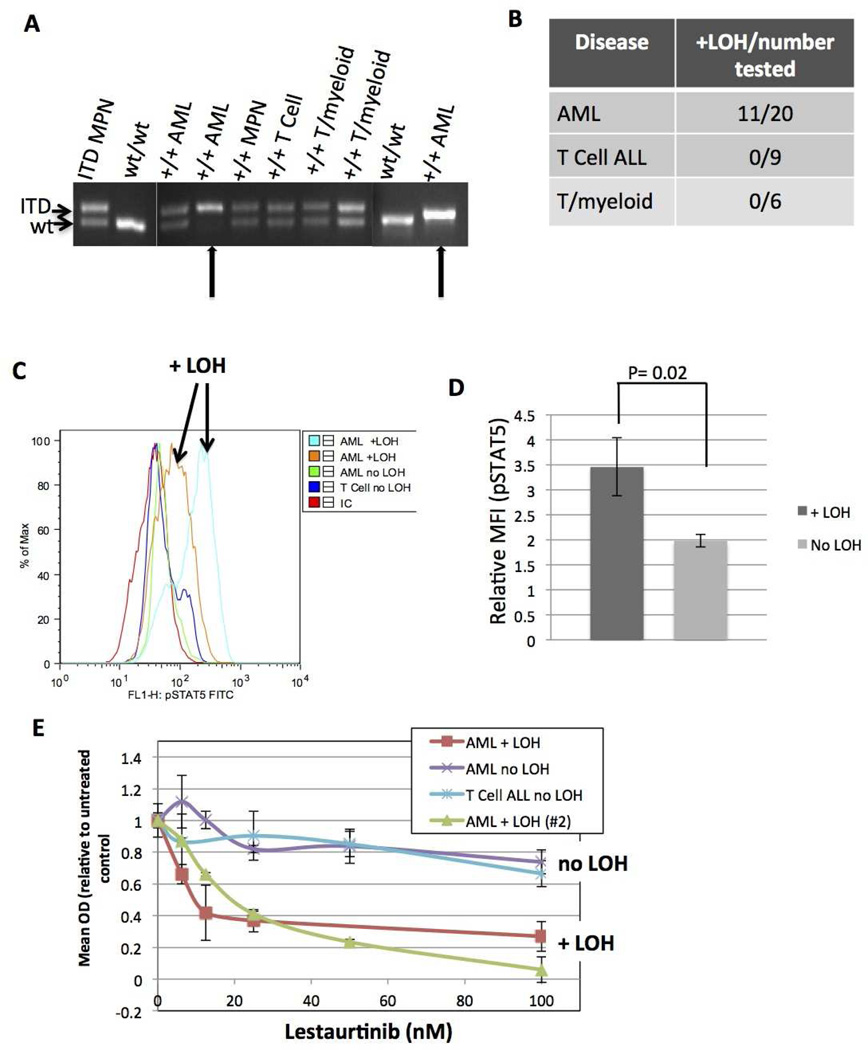
In human AML, loss of the wild-type allele of FLT3 is observed in patients with FLT3/ITD+ AML and is associated with worsened survival [20, 30, 31]. To determine if this phenomenon occurs in leukemic ITD/NPMc+ mice DNA was extracted from the BM of leukemic ITD/NPMc+ mice and PCR amplification of the portion of the juxtamembrane domain of Flt3 harboring the ITD mutation was performed (Figure 5A). In 11 of 20 evaluated mice with AML, only the ITD allele was detected, indicating a loss of the wild-type allele. By contrast, none of the 15 mice with either T cell ALL or mixed lineage T/myeloid disease had LOH of the wild-type Flt3 allele (Figures 5A and 5B). This indicates a strong selective pressure for LOH of wild-type Flt3 in myeloid disease, but no such pressure in T cell disease.
Figure 5. Loss of heterozygosity (LOH) of the wild-type Flt3 allele is common in myeloid leukemia and is functionally significant.
A) PCR of DNA extracted from BM showing examples of mice that remain heterozygous for wild-type Flt3 and 2 mice with AML that have LOH of the wild-type allele (arrows). Image shows 3 separate gels, separated by white lines. B) Table of leukemic mice showing proportion of each disease type with LOH. C) Representative histogram demonstrating measurement of intracellular pSTAT5 detected by intracellular flow cytometry in mice with and without LOH. D) Quantification of all pSTAT5 experiments comparing mice with (N=8) and without (N=9) LOH. E) WST1 cytotoxicity assay dose-response curves comparing sensitivity to Flt3 inhibition by the Flt3 inhibitor lestaurtinib in mice with and without LOH.
LOH, loss of heterozygosity of wild-type Flt3; pSTAT5, phosphorylated STAT5.
To explore the functional significance of loss of wild-type Flt3, intracellular flow cytometry for phosphorylated (activated) STAT5 (pSTAT5) was performed on BM cells of leukemic mice. STAT5 is a critical downstream target of activated FLT3 in leukemia and its activation status serves as a useful surrogate for FLT3 activity [14, 32]. Bone marrow cells grown in liquid culture from leukemic mice with LOH were found to have statistically higher levels of pSTAT5 compared to BM from leukemic mice without LOH (Figure 5C and 5D) indicating increased Flt3/ITD-induced signaling. Further, murine BM cells were treated in vitro with the FLT3 inhibitor lestaurtinib. BM cells with LOH of wild-type Flt3 were more sensitive to FLT3 inhibition, indicating a higher level of dependence on Flt3/ITD-induced signaling for survival and proliferation (Figure 5E).
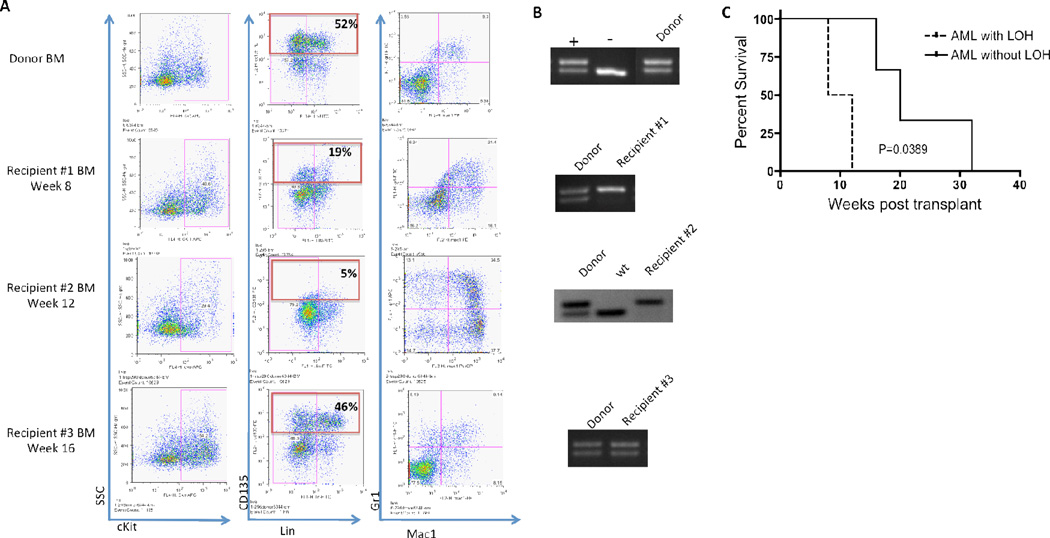
In addition, after transplantation of ITD/NPMc+ AML blasts without LOH of wild-type Flt3, in 2/5 engrafted animals, the AML that developed in recipient mice was found to have LOH of the wild-type allele (Figure 6B). The AML that developed in these transplanted mice was similar phenotypically to that of the donor, with the exception of decreased expression of surface Flt3 (Figure 6A). This would be expected with loss of the wild-type copy of Flt3, because unlike wild-type Flt3 the mutated version does not localize to the cell surface, but accumulates in the perinuclear region [33]. The transplanted mice that developed LOH developed overt disease more rapidly than those mice that retained wild-type Flt3 (Figure 6C). These data indicate that LOH of the wild-type Flt3 is important in the progression of myeloid leukemia. Such LOH did not occur after transplantation of ITD/NPMc+ T cell ALL.
Figure 6. Loss of heterozygosity of wild-type Flt3 occurs after transplantation of Flt3/ITD+/NPMc+ AML.
A) Flow cytometry analysis comparing surface expression of cKit (column 1), lineage and surface Flt3 (CD135) (column 2) and Mac1 and Gr1 (column 3) of donor mouse with AML heterozygous for wt-Flt3 and Flt3/ITD with 3 representative recipient mice. B) PCR of donor leukemic mouse BM without LOH of wt-Flt3 compared to 3 recipients. (Top gel: controls on separate gel than donor, gel for recipient #2 is an inverted image). C) Kaplan-meier survival curve comparing mice transplanted with AML cells from a donor mouse without LOH who secondarily lost heterozygousity of wild-type Flt3 (AML with LOH) to recipients who did not lose heterozygousity (AML without LOH).
The combination of FLT3/ITD and NPMc+ mutations lead to the abnormal expansion of monocytic cells in in vitro clonogenic assays
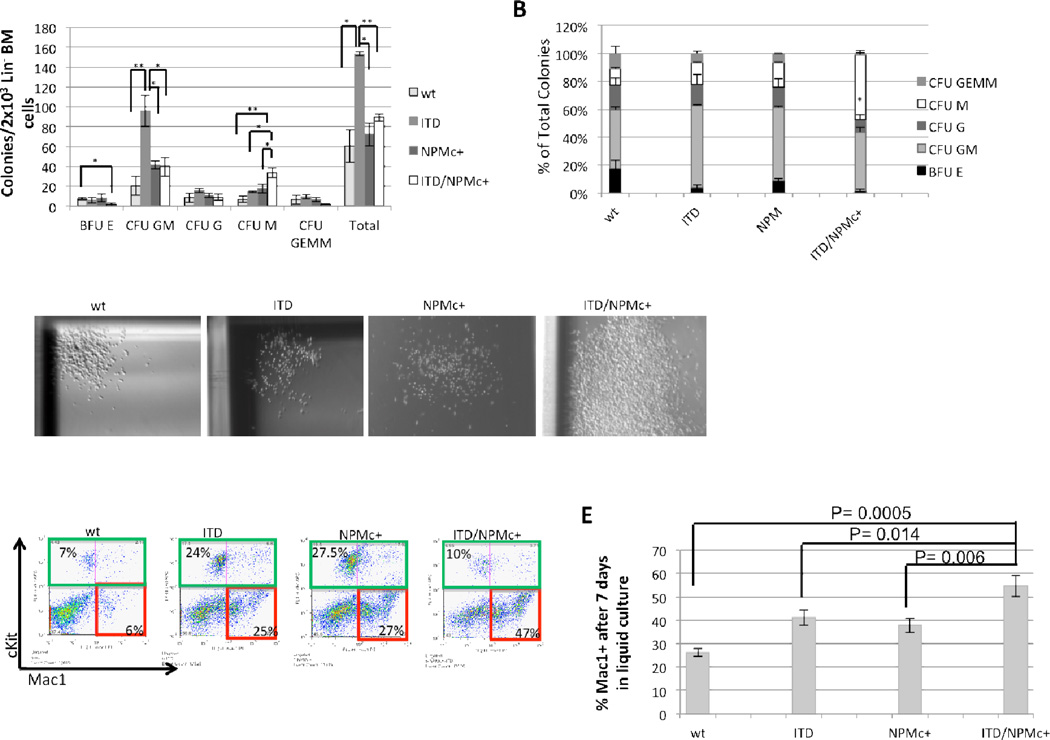
In methylcellulose-based assays, lineage-depleted BM cells from 2-month-old ITD/NPMc+ mice demonstrated an enhanced ability to generate CFU-M colonies compared to other genotypes (Figure 7A). This difference was statistically significant compared to all other genotypes both in absolute count of CFU-M and in percentage of total colonies that were CFU-M (Figure 7A and 7B). The CFU-M colonies generated by the ITD/NPMc+ lineage-depleted BM cells were aberrantly large compared to normal wt CFU-M colonies, and compared to ITD and NPMc+ CFU-M colonies which were increased in number but not uniformly increased in size (Figure 7C). ITD/NPMc+ lineage-depleted BM also generated fewer BFU-E colonies compared to other genotypes, and this difference was statistically significant comparing ITD/NPMc+ to wt whereas ITD and NPMc+ alone were not significantly reduced compared to wt controls (Figure 7A). After scoring the methylcellulose plates, cells were isolated, washed, stained for lineage-specific cell surface markers then analyzed by flow cytometry. On flow cytometric analysis ITD/NPMc+ cells were large based on forward scatter (data not shown) and highly positive for Mac1, confirming the monocytic phenotype (Figure 7D). This indicates that lineage-depleted ITD/NPMc+ BM cells have a greater ability to expand the monocytic lineage compared to FLT3/ITD or NPMc+ alone. This was also seen when the same lineage-depleted BM cells were cultured in liquid medium with a significantly greater proportion of ITD/NPMc+ cells being Mac1 positive by the 7th day in culture compared to other genotypes (Figure 7E). In addition to the expansion of the monocytic lineage we also observed a depletion of more primitive lin−, cKit+ cells likely secondary to the aberrant partial differentiation toward the monocytic lineage (Figure 7D). Additionally, ITD/NPMc+ cells were not able to replate serially (data not shown), likely secondary to the depletion of this more primitive population. This data indicates that the presence of both mutations causes enhanced and significant monocytic expansion, which is consistent with the prevalence of a monocytic morphology in human ITD/NPMc+ AML. In pre-illness mice between the ages of 2 to 6 months, we observed only a trend towards more monocytes and neutrophils in the peripheral blood of ITD, NPMc+, and ITD/NPMc+ mice that was not statistically significant (Figure 5S), suggesting that in vivo, this expansion may be a more slowly evolving process given the presence of only physiologic concentrations of growth promoting cytokines.
Figure 7. BM from ITD/NPMc+ mice demonstrates enhanced monocytic colony-forming activity in vitro.
Methylcellulose in vitro colony-forming assay of lineage-depleted BM cells from 2-month-old mice. A) Total colony counts and B) percentage distribution of colony type by genotype. Data are the results from 3 independent experiments. Data are expressed as means (bars) plus or minus SEM (error bars). ** P<0.01, * P<0.02. C) Representative CFU-M colonies generated from wt, ITD, NPMc+, and ITD/NPMc+ lineage-depleted BM. Original magnification 20×. D) Representative flow cytometric analysis of cells collected from plates 10 days after the first plating with percentage of cKit+ (green outline) and Mac1+/cKit− (red outline). E) Flow cytometry of lineage-depleted BM cells from 2-month-old mice after 7 days of culture in RPMI 1640 plus SCF, IL3, and IL6. Data are the results from 3 independent experiments. Data are expressed as means (bars) plus or minus SEM (error bars).
Discussion
There is accumulating evidence that leukemogenesis is a process that requires multiple genetic and/or epigenetic “hits” [34, 35]. Numerous mouse models suggest that no singular genetic mutation is sufficient to cause acute leukemia, and that cooperating events are required [22, 23, 36–39]. We hypothesized that NPMc+ and FLT3/ITD mutations are two such cooperating events, given the fact that they occur together with a very high frequency in AML. When we crossed Flt3/ITD knock-in mice with NPMc+ transgenic mice, acute leukemia developed, demonstrating cooperation. As expected, most of the resultant leukemias were AML recapitulating human disease. In in vitro clonogenic assays, lineage-depleted BM cells from pre-leukemic ITD/NPMc+ mice demonstrate an aberrant expansion of the monocytic lineage. This is consistent with reports that human NPMc+-mutant progenitor cells are capable of differentiating into more mature cells of the monocytic lineage [40, 41]. Additionally, when Flt3/ITD is expressed in progenitor cells with lymphoid and myeloid potential, myeloid differentiation is favored [42]. Thus, both mutations individually cause expansion of the myelomonocytic lineage. But while neither mutation alone is sufficient to cause an overt leukemia, perhaps the combined effects result in an accumulation of cells with aberrant partial monocytic differentiation, which are prone to the acquisition of additional leukemogenic mutations. This finding demonstrates that our model closely mimics human disease, as this monocytic expansion is consistent with the phenotype of human FLT3/ITD+/NPMc+ AML, which is most commonly of the M4/M5 myelomonocytic morphology [43]. A subset of mice also developed T cell malignancies. While this was unexpected, aberrant NPMc+ expression was demonstrated in the T cell compartment and both mutations are associated with T cell malignancies. FLT3/ITD mutations are known to occur in T cell ALL [28, 44], and NPM may contribute to lymphomagenesis in T cell anaplastic large cell lymphoma with NPM-ALK fusion [45].
Frequently in human FLT3/ITD+ AML loss of heterozygosity (LOH) of the wild-type FLT3 allele occurs [20, 31]. LOH of the wild-type FLT3 allele has also been noted in mouse models crossing Flt3/ITD with other collaborating genetic mutations [27, 46] We therefore examined leukemic murine BM for LOH of wild-type Flt3 and found a preponderance of LOH in mice with myeloid leukemia suggesting a strong selective pressure to lose the wild-type copy of Flt3. The presence of LOH at diagnosis suggests an important role in leukemic initiation and the loss of wild-type Flt3 in our transplantation model of myeloid disease indicates a role of LOH in leukemic progression. Additionally we found that LOH is associated with increased Flt3/ITD-induced downstream signaling and increased sensitivity to FLT3 inhibition compared to samples without LOH. Therefore, when LOH of wild-type Flt3 occurs, Flt3/ITD is crucial for leukemic maintenance. Given the long latency to disease onset in our model it is likely that while Flt3/ITD and NPMc+ cooperate, they require additional cooperating oncogenic events. For most myeloid disease, perhaps LOH of the wild-type Flt3 allele serves as such an event. Both an increase in Flt3/ITD gene dosage and loss of the wild-type Flt3 allele results in an enhanced myeloproliferative phenotype compared to mice with heterozygous Flt3/ITD mutations, suggesting a tumor suppressor effect of wild-type Flt3 [47, 48] The frequent LOH in myeloid leukemia and resultant functional changes observed in our model further implicate a tumor suppressor effect of wild-type Flt3, and indicated that loss of wild-type Flt3 could represent an additional collaborating event in myeloid disease. In contrast, as we observed no LOH in T lineage disease it is likely that other unrelated contributing events must accumulate. It is possible that in T cell disease, aberrant NPMc+ expression in the T cell compartment leads to functional changes that ultimately result in the accumulation of other collaborating mutations. Alternatively, perhaps the effects of wild-type FLT3 are context specific; exerting tumor suppressive effects only in the myeloid compartment. Therefore loss of the wild-type allele in other hematopoietic compartments would offer no proliferative advantage upon transformation. Supporting this possibility is the fact that other mouse models of FLT3/ITD induced leukemia found LOH only in myeloid leukemia [27, 46]
Another factor likely contributing to the long disease latency observed in our model is the relatively low level of expression of the NPMc+ transgene and the presence of two wild-type copies of native Npm [23]. NPM is known to be a haploinsufficient tumor suppressor, with myeloid and lymphoid leukemias developing in hemizygous (Npm+/−) mice [49]. In NPMc+ leukemia, not only is there loss of one wild-type allele, but mutant NPMc+ interacts with and delocalizes much of the produced wild-type NPM to the cytoplasm further reducing the function of NPM [50, 51]. In our model, wild-type NPM protein is present in higher amounts compared to patients with NPMc+ AML because two wild-type alleles are present. In a recently reported murine model, conditionally knocked-in NPMc+ was found to be capable of significantly perturbing hematopoiesis [52]. Further, when a similar knock-in model was combined with constitutively knocked-in Flt3/ITD, a severe MPN or MPN-like myeloid leukemia with a short latency developed. In these models, only one copy of wild-type NPM was present, thus the more severe disruption of hematopoiesis in the NPMc+ alone mice and the shorter latency to disease onset in the NPMc+ knock-in plus Flt3/ITD model compared to our model, suggests a dosage effect of wild-type NPM in terms of its tumor suppressive effects. The development of leukemia in our model does demonstrate that leukemogenesis in NPMc+ leukemia is due not only to a loss of wild-type NPM function but also other effects of the mutated NPMc+ protein.
In conclusion, utilizing a mouse model we have documented that FLT3/ITD mutations cooperate with NPMc+ mutations to cause leukemia. This is also further definitive in vivo evidence of the leukemogenic activity of NPMc+, corroborating other recent murine data [25, 26, 53]. This model can be utilized to advance the understanding of the biology of this common disease entity and identify and test the efficacy of potential therapeutic targets in vivo.
Supplementary Material
Acknowledgments
The authors would like to thank the members of the Brown lab: Eric Schafer, Edward Allan Sison, Sandeep Negi, Allison Kaeding, as well as all the members of the Small lab for providing informative discussion and helpful suggestions. This work was funded in part through: The St. Baldrick’s Foundation (Post-Doctoral Fellowship for Childhood Cancer Research Award, R.R.); Leukemia and Lymphoma Society (Translational Research Program Grant, P.B.; Scholar in Clinical Research Award, P.B.); National Cancer Institute (K23 CA111728, P.B. .; R01CA90668, P01CA70970, D.S.), Damon Runyon Cancer Research Foundation (Clinical Investigator Award, P.B.) and Giant Food Children’s Cancer Research Fund.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Authorship
R.R., P.B. and D.S. designed the study; R.R., D.M., S.G., and C.A. performed the mouse experiments; R.R., and C.A. performed the in vitro experiments; S.G. and L.L., assisted in training R.R. to perform mouse experiments and helped with experimental design; E. M., and S.G. assisted in training R.R. to perform in vitro experiments; D.H. prepared and photographed histologic sections of mouse tissues; A.S.D performed immunohistochemisty and photographed histologic sections of mouse tissues; J.G.C., M.R., and P.P.P, provided the NPMc+ transgenic mice as well as expert guidance and informative discussion; D.S. provided the FLT3/ITD knock-in mice as well as invaluable expertise and guidance; R.R. and P.B. prepared the manuscript.
Disclosures
All authors approved the final manuscript. None of the authors have anything to disclose.
References
- 1.Patel JP, Gonen M, Figueroa ME, et al. Prognostic relevance of integrated genetic profiling in acute myeloid leukemia. N Engl J Med. 2012;366:1079–1089. doi: 10.1056/NEJMoa1112304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.The Cancer Genome Atlas Research Network. Genomic and Epigenomic Landscapes of Adult De Novo Acute Myeloid Leukemia. N Engl J Med. 2013 doi: 10.1056/NEJMoa1301689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cordell JL, Pulford KA, Bigerna B, et al. Detection of normal and chimeric nucleophosmin in human cells. Blood. 1999;93:632–642. [PubMed] [Google Scholar]
- 4.Falini B, Nicoletti I, Martelli MF, Mecucci C. Acute myeloid leukemia carrying cytoplasmic/mutated nucleophosmin (NPMc+ AML): biologic and clinical features. Blood. 2007;109:874–885. doi: 10.1182/blood-2006-07-012252. [DOI] [PubMed] [Google Scholar]
- 5.Falini B, Mecucci C, Tiacci E, et al. Cytoplasmic nucleophosmin in acute myelogenous leukemia with a normal karyotype. N Engl J Med. 2005;352:254–266. doi: 10.1056/NEJMoa041974. [DOI] [PubMed] [Google Scholar]
- 6.Brown P, McIntyre E, Rau R, et al. The incidence and clinical significance of nucleophosmin mutations in childhood AML. Blood. 2007;110:979–985. doi: 10.1182/blood-2007-02-076604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hollink IH, Zwaan CM, Zimmermann M, et al. Favorable prognostic impact of NPM1 gene mutations in childhood acute myeloid leukemia, with emphasis on cytogenetically normal AML. Leukemia. 2009;23:262–270. doi: 10.1038/leu.2008.313. [DOI] [PubMed] [Google Scholar]
- 8.Gale RE, Green C, Allen C, et al. The impact of FLT3 internal tandem duplication mutant level, number, size, and interaction with NPM1 mutations in a large cohort of young adult patients with acute myeloid leukemia. Blood. 2008;111:2776–2784. doi: 10.1182/blood-2007-08-109090. [DOI] [PubMed] [Google Scholar]
- 9.Schnittger S, Schoch C, Kern W, et al. Nucleophosmin gene mutations are predictors of favorable prognosis in acute myelogenous leukemia with a normal karyotype. Blood. 2005;106:3733–3739. doi: 10.1182/blood-2005-06-2248. [DOI] [PubMed] [Google Scholar]
- 10.Thiede C, Koch S, Creutzig E, et al. Prevalence and prognostic impact of NPM1 mutations in 1485 adult patients with acute myeloid leukemia (AML) Blood. 2006;107:4011–4020. doi: 10.1182/blood-2005-08-3167. [DOI] [PubMed] [Google Scholar]
- 11.Vardiman JW, Thiele J, Arber DA, et al. The 2008 revision of the World Health Organization (WHO) classification of myeloid neoplasms and acute leukemia: rationale and important changes. Blood. 2009;114:937–951. doi: 10.1182/blood-2009-03-209262. [DOI] [PubMed] [Google Scholar]
- 12.Small D, Levenstein M, Kim E, et al. STK-1, the human homolog of Flk-2/Flt-3, is selectively expressed in CD34+ human bone marrow cells and is involved in the proliferation of early progenitor/stem cells. Proc Natl Acad Sci U S A. 1994;91:459–463. doi: 10.1073/pnas.91.2.459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rosnet O, Schiff C, Pebusque MJ, et al. Human FLT3/FLK2 gene: cDNA cloning and expression in hematopoietic cells. Blood. 1993;82:1110–1119. [PubMed] [Google Scholar]
- 14.Levis M, Small D. FLT3: ITDoes matter in leukemia. Leukemia. 2003;17:1738–1752. doi: 10.1038/sj.leu.2403099. [DOI] [PubMed] [Google Scholar]
- 15.Kim KT, Baird K, Ahn JY, et al. Pim-1 is up-regulated by constitutively activated FLT3 and plays a role in FLT3-mediated cell survival. Blood. 2005;105:1759–1767. doi: 10.1182/blood-2004-05-2006. [DOI] [PubMed] [Google Scholar]
- 16.Nakao M, Yokota S, Iwai T, et al. Internal tandem duplication of the flt3 gene found in acute myeloid leukemia. Leukemia. 1996;10:1911–1918. [PubMed] [Google Scholar]
- 17.Meshinchi S, Woods WG, Stirewalt DL, et al. Prevalence and prognostic significance of Flt3 internal tandem duplication in pediatric acute myeloid leukemia. Blood. 2001;97:89–94. doi: 10.1182/blood.v97.1.89. [DOI] [PubMed] [Google Scholar]
- 18.Stirewalt DL, Radich JP. The role of FLT3 in haematopoietic malignancies. Nat Rev Cancer. 2003;3:650–665. doi: 10.1038/nrc1169. [DOI] [PubMed] [Google Scholar]
- 19.Gilliland DG, Griffin JD. The roles of FLT3 in hematopoiesis and leukemia. Blood. 2002;100:1532–1542. doi: 10.1182/blood-2002-02-0492. [DOI] [PubMed] [Google Scholar]
- 20.Meshinchi S, Alonzo TA, Stirewalt DL, et al. Clinical implications of FLT3 mutations in pediatric AML. Blood. 2006;108:3654–3661. doi: 10.1182/blood-2006-03-009233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lee BH, Williams IR, Anastasiadou E, et al. FLT3 internal tandem duplication mutations induce myeloproliferative or lymphoid disease in a transgenic mouse model. Oncogene. 2005;24:7882–7892. doi: 10.1038/sj.onc.1208933. [DOI] [PubMed] [Google Scholar]
- 22.Li L, Piloto O, Nguyen HB, et al. Knock-in of an internal tandem duplication mutation into murine FLT3 confers myeloproliferative disease in a mouse model. Blood. 2008;111:3849–3858. doi: 10.1182/blood-2007-08-109942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cheng K, Sportoletti P, Ito K, et al. The cytoplasmic NPM mutant induces myeloproliferation in a transgenic mouse model. Blood. 2010;115:3341–3345. doi: 10.1182/blood-2009-03-208587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kogan SC, Ward JM, Anver MR, et al. Bethesda proposals for classification of nonlymphoid hematopoietic neoplasms in mice. Blood. 2002;100:238–245. doi: 10.1182/blood.v100.1.238. [DOI] [PubMed] [Google Scholar]
- 25.Mallardo M, Caronno A, Pruneri G, et al. NPMc+ and FLT3_ITD mutations cooperate in inducing acute leukaemia in a novel mouse model. Leukemia. 2013 doi: 10.1038/leu.2013.114. [DOI] [PubMed] [Google Scholar]
- 26.Vassiliou GS, Cooper JL, Rad R, et al. Mutant nucleophosmin and cooperating pathways drive leukemia initiation and progression in mice. Nat Genet. 2011;43:470–475. doi: 10.1038/ng.796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Greenblatt S, Li L, Slape C, et al. Knock-in of a FLT3/ITD mutation cooperates with a NUP98-HOXD13 fusion to generate acute myeloid leukemia in a mouse model. Blood. 2012;119:2883–2894. doi: 10.1182/blood-2011-10-382283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Paietta E, Ferrando AA, Neuberg D, et al. Activating FLT3 mutations in CD117/KIT(+) T-cell acute lymphoblastic leukemias. Blood. 2004;104:558–560. doi: 10.1182/blood-2004-01-0168. [DOI] [PubMed] [Google Scholar]
- 29.Jaiswal S, Traver D, Miyamoto T, Akashi K, Lagasse E, Weissman IL. Expression of BCR/ABL and BCL-2 in myeloid progenitors leads to myeloid leukemias. Proc Natl Acad Sci U S A. 2003;100:10002–10007. doi: 10.1073/pnas.1633833100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Whitman SP, Archer KJ, Feng L, et al. Absence of the wild-type allele predicts poor prognosis in adult de novo acute myeloid leukemia with normal cytogenetics and the internal tandem duplication of FLT3: a cancer and leukemia group B study. Cancer Res. 2001;61:7233–7239. [PubMed] [Google Scholar]
- 31.Thiede C, Steudel C, Mohr B, et al. Analysis of FLT3-activating mutations in 979 patients with acute myelogenous leukemia: association with FAB subtypes and identification of subgroups with poor prognosis. Blood. 2002;99:4326–4335. doi: 10.1182/blood.v99.12.4326. [DOI] [PubMed] [Google Scholar]
- 32.Choudhary C, Brandts C, Schwable J, et al. Activation mechanisms of STAT5 by oncogenic Flt3-ITD. Blood. 2007;110:370–374. doi: 10.1182/blood-2006-05-024018. [DOI] [PubMed] [Google Scholar]
- 33.Koch S, Jacobi A, Ryser M, Ehninger G, Thiede C. Abnormal localization and accumulation of FLT3-ITD, a mutant receptor tyrosine kinase involved in leukemogenesis. Cells Tissues Organs. 2008;188:225–235. doi: 10.1159/000118788. [DOI] [PubMed] [Google Scholar]
- 34.Speck NA, Gilliland DG. Core-binding factors in haematopoiesis and leukaemia. Nat Rev Cancer. 2002;2:502–513. doi: 10.1038/nrc840. [DOI] [PubMed] [Google Scholar]
- 35.Frohling S, Scholl C, Gilliland DG, Levine RL. Genetics of myeloid malignancies: pathogenetic and clinical implications. J Clin Oncol. 2005;23:6285–6295. doi: 10.1200/JCO.2005.05.010. [DOI] [PubMed] [Google Scholar]
- 36.Castilla LH, Garrett L, Adya N, et al. The fusion gene Cbfb-MYH11 blocks myeloid differentiation and predisposes mice to acute myelomonocytic leukaemia. Nat Genet. 1999;23:144–146. doi: 10.1038/13776. [DOI] [PubMed] [Google Scholar]
- 37.Chan IT, Kutok JL, Williams IR, et al. Conditional expression of oncogenic K-ras from its endogenous promoter induces a myeloproliferative disease. J Clin Invest. 2004;113:528–538. doi: 10.1172/JCI20476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.MacKenzie KL, Dolnikov A, Millington M, Shounan Y, Symonds G. Mutant N-ras induces myeloproliferative disorders and apoptosis in bone marrow repopulated mice. Blood. 1999;93:2043–2056. [PubMed] [Google Scholar]
- 39.Grisendi S, Bernardi R, Rossi M, et al. Role of nucleophosmin in embryonic development and tumorigenesis. Nature. 2005;437:147–153. doi: 10.1038/nature03915. [DOI] [PubMed] [Google Scholar]
- 40.Pasqualucci L, Liso A, Martelli MP, et al. Mutated nucleophosmin detects clonal multilineage involvement in acute myeloid leukemia: Impact on WHO classification. Blood. 2006;108:4146–4155. doi: 10.1182/blood-2006-06-026716. [DOI] [PubMed] [Google Scholar]
- 41.Martelli MP, Pettirossi V, Thiede C, et al. CD34+ cells from AML with mutated NPM1 harbor cytoplasmic mutated nucleophosmin and generate leukemia in immunocompromised mice. Blood. 2010;116:3907–3922. doi: 10.1182/blood-2009-08-238899. [DOI] [PubMed] [Google Scholar]
- 42.Mead AJ, Kharazi S, Atkinson D, et al. FLT3-ITDs Instruct a Myeloid Differentiation and Transformation Bias in Lymphomyeloid Multipotent Progenitors. Cell Rep. 2013;3:1766–1776. doi: 10.1016/j.celrep.2013.04.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dohner K, Schlenk RF, Habdank M, et al. Mutant nucleophosmin (NPM1) predicts favorable prognosis in younger adults with acute myeloid leukemia and normal cytogenetics: interaction with other gene mutations. Blood. 2005;106:3740–3746. doi: 10.1182/blood-2005-05-2164. [DOI] [PubMed] [Google Scholar]
- 44.Zhang J, Ding L, Holmfeldt L, et al. The genetic basis of early T-cell precursor acute lymphoblastic leukaemia. Nature. 2012;481:157–163. doi: 10.1038/nature10725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Naoe T, Suzuki T, Kiyoi H, Urano T. Nucleophosmin: a versatile molecule associated with hematological malignancies. Cancer Sci. 2006;97:963–969. doi: 10.1111/j.1349-7006.2006.00270.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zorko NA, Bernot KM, Whitman SP, et al. Mll partial tandem duplication and Flt3 internal tandem duplication in a double knock-in mouse recapitulates features of counterpart human acute myeloid leukemias. Blood. 2012;120:1130–1136. doi: 10.1182/blood-2012-03-415067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Li L, Bailey E, Greenblatt S, Huso D, Small D. Loss of the wild-type allele contributes to myeloid expansion and disease aggressiveness in FLT3/ITD knockin mice. Blood. 2011;118:4935–4945. doi: 10.1182/blood-2011-01-328096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kharazi S, Mead AJ, Mansour A, et al. Impact of gene dosage, loss of wild-type allele, and FLT3 ligand on Flt3-ITD-induced myeloproliferation. Blood. 2011;118:3613–3621. doi: 10.1182/blood-2010-06-289207. [DOI] [PubMed] [Google Scholar]
- 49.Sportoletti P, Grisendi S, Majid SM, et al. Npm1 is a haploinsufficient suppressor of myeloid and lymphoid malignancies in the mouse. Blood. 2008;111:3859–3862. doi: 10.1182/blood-2007-06-098251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Falini B, Bolli N, Shan J, et al. Both carboxy-terminus NES motif and mutated tryptophan(s) are crucial for aberrant nuclear export of nucleophosmin leukemic mutants in NPMc+ AML. Blood. 2006;107:4514–4523. doi: 10.1182/blood-2005-11-4745. [DOI] [PubMed] [Google Scholar]
- 51.Bolli N, Nicoletti I, De Marco MF, et al. Born to be exported: COOH-terminal nuclear export signals of different strength ensure cytoplasmic accumulation of nucleophosmin leukemic mutants. Cancer Res. 2007;67:6230–6237. doi: 10.1158/0008-5472.CAN-07-0273. [DOI] [PubMed] [Google Scholar]
- 52.Sportoletti P, Varasano E, Rossi R, et al. The human NPM1 mutation A perturbs megakaryopoiesis in a conditional mouse model. Blood. 2013;121:3447–3458. doi: 10.1182/blood-2012-08-449553. [DOI] [PubMed] [Google Scholar]
- 53.Mupo A, Celani L, Dovey O, et al. A powerful molecular synergy between mutant Nucleophosmin and Flt3-ITD drives acute myeloid leukemia in mice. Leukemia. 2013 doi: 10.1038/leu.2013.77. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.