Abstract
The t(10;11)(p12;q23) translocation and the t(10;11)(p12;q14) translocation, which encode the MLL-AF10 and CALM-AF10 fusion oncoproteins respectively, are two recurrent chromosomal rearrangements observed in patients with acute myeloid leukemia and acute lymphoblastic leukemia. Here we demonstrate that MLL-AF10 and CALM-AF10 mediated transformation is dependent on the H3K79 methyltransferase Dot1l using genetic and pharmacological approaches in mouse models. Targeted disruption of Dot1l using a conditional knockout mouse model abolished in vitro transformation of murine bone marrow cells and in vivo initiation and maintenance of MLL-AF10 or CALM-AF10 leukemia.. Treatment of MLL-AF10 and CALM-AF10 transformed cells with EPZ004777, a specific small-molecule inhibitor of Dot1l, suppressed expression of leukemogenic genes such as Hoxa cluster genes and Meis1, and selectively suppressed proliferation of MLL-AF10 and CALM-AF10 transformed cells. Pretreatment with EPZ004777 profoundly decreased the in vivo spleen-colony forming ability of MLL-AF10 or CALM-AF10 transformed bone marrow cells. These results show that patients with leukemias bearing chromosomal translocations that involve the AF10 gene may benefit from small molecule therapeutics that inhibit H3K79 methylation.
Keywords: MLL-AF10, CALM-AF10, MLL fusions, leukemia, Dot1l, EPZ004777
Introduction
Chromosomal translocations which encode fusion proteins are frequently associated with human leukemias. The t(10;11)(p12;q23) translocation, which encodes the MLL-AF10 fusion protein, is a recurrent chromosomal rearrangement observed mainly in patients with acute myelogenous leukemia (AML) (1, 2). Another translocation involving AF10, the t(10;11)(p12;q14) translocation, encodes a CALM-AF10 fusion protein and is observed in AML, acute lymphoblastic leukemia (ALL) and precursor T lymphoblastic lymphoma (3). Patients with AML harboring either MLL-AF10 or CALM-AF10 rearrangements have a particularly poor outcome compared to patients whose leukemia cells do not harbor these translocations (4). Thus new therapeutic approaches are clearly needed for patients with AF10-rearranged hematopoietic malignancies.
While similar motifs in the AF10 portion are retained in both MLL-AF10 as well as CALM-AF10 oncoproteins motifs, MLL and CALM are quite dissimilar proteins. The wild-type MLL (Mixed Lineage Leukemia) protein positively regulates expression of homeobox (Hox) genes, and is essential for hematopoietic development (5, 6). MLL possesses multiple N-terminal domains required for target gene recognition (7–9), which are retained in the oncogenic MLL-AF10 fusion protein. MLL also possesses a C-terminal H3 lysine 4 (H3K4) methyltransferase domain (10, 11), which activates Hox gene expression during normal development but is absent in the fusion protein. On the other hand, wild-type CALM (Clathrin Assembly Lymphoid Myeloid leukemia) protein, primarily localized in the cytoplasm, is involved in clathrin-mediated endocytosis and has been shown to be involved in erythropoiesis and iron metabolism (12, 13). The clathrin binding domain on the C-terminus of CALM is always retained in the CALM-AF10 fusion proteins and is sufficient for leukemogenesis when fused with AF10 (14). As a fusion partner of both MLL and CALM, wild-type AF10 (ALL-1 fused gene from chromosome 10) is a putative transcription factor containing N-terminal Plant Homeodomain (PHD) zinc finger motifs and a C-terminal octapeptide motif-leucine zipper (OM-LZ) domain (15). The AF10 OM-LZ domain is always retained in the MLL-AF10 and CALM-AF10 fusion proteins and has been identified as a domain that interacts with the histone H3 lysine 79 (H3K79)-specific methyltransferase DOT1L(16, 17). Although the mechanism by which the leukemogenic AF10 fusions transform hematopoietic cells has not been fully elucidated, it has been suggested that Dot1l interaction with the AF10 OM-LZ domain is critical for oncogenesis (16, 18).
H3K79 methylation, catalyzed solely by Dot1l, is a chromatin modification ubiquitously associated with actively transcribed genes (19, 20). In human and mouse MLL-AF10 and CALM-AF10 leukemia cells, dimethylated H3K79 is typically enriched in the promoter regions of leukemogenic genes, including the posterior Hoxa cluster genes (16, 18). Furthermore, the epigenetic deregulation of these specific leukemogenic genes correlates with aberrant overexpression in leukemia cells (14, 16, 18). Therefore, while DOT1L has not been found to be genetically altered in leukemia, its aberrant recruitment and/or activity may lead to epigenetic deregulation and overexpression of crucial fusion protein target genes.
Consistent with this model, it has been shown that hematopoietic progenitor cells cannot be transformed by MLL-AF10 after being treated with shRNA against Dot1l for one week (16). Moreover, bone marrow cells could not be transformed by CALM-AF10 when a dominant negative form of Dot1l was overexpressed (18). Although the knock-down and over-expression approaches have some limitations, these results raise the possibility that DOT1L may be a relevant therapeutic target for MLL-AF10 and CALM-AF10 leukemia. To determine whether inhibition of DOT1L represents a valid approach to treat MLL-AF10 and CALM-AF10 leukemias, we assessed the effects of genetic deletion and pharmacological inhibition of Dot1l in murine bone marrow cells immortalized by MLL-AF10 and CALM-AF10 oncoproteins. Using genetically defined models of human leukemia that bear specific epigenetic perturbations, our study demonstrates that such abnormal chromatin modifications can be specifically targeted using a small-molecule inhibitor. These observations are of particular interest in the light of mounting evidence for specific epigenetic alterations in several human tumors.
Materials and Methods
Mutant Mice
Mice engineered to harbour LoxP sites flanking exon 5 of Dot1l were generated in our laboratory and have been described previously (21). Bone marrow cells from 7–10 week old mice in Dot1l wild-type or homozygous floxed (Dot1lf/f) backgrounds were used for transformation assays and subsequent biochemical experiments.
Generation of Transformed Murine Cells and Leukemia
The MSCV based MLL-AF9-IRES-GFP, HoxA9-IRES-GFP, and Meis1a-PGK-Puromycin constructs were described previously (21). The FLAG-CALM-AF10 minimal fusion construct has been described in detail earlier (14) and the FLAG-MLL-AF10 construct was generated by fusing amino acids 1–1430 of MLL to amino acid 625–1027 of AF10 in an MSCV-IRES-GFP vector. The MSCV-IRES-Tomato (MiTomato, or MIT) plasmid was a kind gift from the lab of Hassan Jumaa (Max Planck Institute, Freiburg). The cDNA for Cre recombinase was sub-cloned into the MIT plasmid to generate the MSCV-Cre-IRES-Tomato (Cre-MiTomato, or Cre) construct. Retroviral supernatants were collected from 293-T cells separately transfected with the plasmids using standard protocols and used for retroviral spin infections. Sorted Lin−Sca−1+cKit+ (LSK) cells from mouse bone marrow were used for retroviral transduction experiments. The LSK cells were transduced with viruses carrying MLL-AF10, CALM-AF10, MLL-AF9, or HoxA9 and Meis1, and expanded for 2–5 days in methylcellulose M3234 (StemCell Technologies, Vancouver, Canada) supplemented with cytokines (6ng/ml IL3, 10ng/ml IL6 and 20ng/ml SCF). MLL-AF10 or CALM-AF10 transformed cells were then transduced with Cre or MIT and expanded in methylcellulose M3234 supplemented with cytokines. After two days, GFP+Tomato+ cells were sorted for in vitro colony forming assays, or sorted and transplanted into B6/129 syngeneic sublethally irradiated (550 rad) recipients at 5×105 cells/mouse. For secondary transplants, whole-bone marrow from leukemic mice was isolated and transduced with Cre or MIT on the same day; GFP+Tomato+ cells were sorted in two days, and transplanted into sublethally irradiated (550 rad) B6/129 syngeneic recipients at 5×105 cells/mouse for generation of secondary CALM-AF10 leukemia, or non-irradiated severe combined immunodeficiency (SCID) recipients at 2×105 cells/mouse for generation of secondary MLL-AF10 leukemia.
Colony Forming Assays
Colony forming cell (CFC) assays were performed by plating 1000 cells per ml of methylcellulose M3234 supplemented with cytokines (6ng/ml IL3, 10ng/ml IL6 and 20ng/ml SCF). On day 6–7 after plating, colonies were scored using a Nikon Eclipse TS100 microscope (Nikon, Tokyo, Japan) and classified into two categories - compact and hypercellular blast-like colonies or small and diffuse differentiated-type colonies. Colonies were then pooled and used for biochemical assays or replated for assessment of secondary replating potential at the same concentration. Cytospin preparations were performed from 50–100,000 cells. Pictures of colonies and Wright-Giemsa stained cytospin preparations were taken using a Nikon Eclipse E400 microscope (Nikon, Tokyo, Japan) and a SPOT RT color digital camera (Diagnostic Instruments, Sterling Heights, MI, USA).
EPZ004777
EPZ004777 was synthesized by Epizyme (Cambridge, MA). 50 mM stock solutions were prepared in DMSO and stored at −20 °C. Serial dilutions of stock solutions were carried out just prior to use in each experiment and final DMSO concentrations were kept at, or below 0.02%.
Cell Proliferation, Viability Assay and Colony-Forming Unit-Spleen (CFU-S) Assay
For assessment of cell proliferation and viability, cells from three independent transductions for each virus were plated, in duplicate, in 96-well plates at a density of 1.5×104 cells/well in a final volume of 150 µl. Cells were incubated in the presence of increasing concentrations of EPZ004777 up to 10 µM. Viable cell number was counted every 3–4 days for up to 17 days using Trypan blue staining. On days of cell counts, growth media and EPZ004777 were replaced and cells split to a density of 1.5×104 cells/well. Results were plotted as the percentage of split-adjusted viable cells in the presence of EPZ004777 compared to DMSO vehicle control. For CFU-S assays, cells from two independent transductions for each virus were plated in 24-well plates at a density of 1.5×104 cells/well with 10 µM EPZ004777 or DMSO control. Growth media and EPZ004777 were replaced on day 4. On day 8, viable cells were counted and injected into lethally irradiated (550rad twice) syngeneic recipients at 104 cells/mouse. After two weeks, mice were euthanized and spleens were collected and fixed using standard protocols.
Cell Cycle and Apoptosis Assays
MLL-AF10 and CALM-AF10 transformed bone marrow cells were plated in 12-well plates at a density of 2×105 cells/ml. Cells were incubated with 10 µM EPZ004777 or DMSO vehicle control in a final volume of 1 ml for up to 10 days during which media with inhibitor were changed every 3 to 4 days. Cell cycle analysis was performed after 30 min. of BrdU labeling using BrdU-APC/7AAD kit from BD-Pharmingen (San Jose, CA, USA) on day 0, 4, 8, and 10. Data was acquired on a 4-color Becton-Dickinson FACSCalibur flow cytometer and analyzed using BD FACS Diva (San Jose, CA, USA) and Modfit LT (Verity Software House, Topsham, ME, USA). Annexin V apoptosis assays were performed on day 10. Cells incubated with 10µM EPZ004777 or DMSO vehicle control were washed in PBS, resuspended in Ca/HEPES buffer (10 mM HEPES, pH7.4; 140 mM NaCl; 2.5 mM CaCl2) and incubated with Annexin V-PE (BioVision Inc., Mountain View, CA) for 20 mins. Data was acquired on a 4-color Becton-Dickinson FACSCalibur flow cytometer and analyzed using BD FACS Diva.
Western Blotting and Immunofluorescence
Histone purification was performed with triton extraction (1×PBS, 0.5% TritonX100 and 2 mM phenylmethylsulfonylfluoride) followed by acid extraction with 0.2 N HCl as previously described (21). Whole cell protein extracts were prepared for detection of MLL-AF10 and CALM-AF10 fusion proteins (see supplemental data for detailed protocol). Immunofluorescence was done following a previously established protocol (21). The following antibodies were used for detection: anti-H3K79me2 antibody ab3594 (Abcam, Cambridge, MA, USA), anti-total H3 antibody ab1791 (Abcam, Cambridge, MA, USA), anti-MLL antibody A300-086A (Bethyl Laboratories, Montgomery, Texas, USA), anti-FLAG m2 antibody F1804 (Sigma-Aldrich); secondary antibodies used: sheep anti-mouse ECL horseradish peroxidase linked NA931V, donkey anti-rabbit ECL horseradish peroxidase linked NA934V (GE healthcare UK limited, Little Chalfont Buckinghamshire, UK), and Alexa 594 conjugated goat anti-rabbit antibody A11072 (Invitrogen, Carlsbad, CA, USA).
Reverse transcription and Real Time PCR
Total RNA was isolated using Trizol (Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s instruction. The resultant cDNA was generated using the Tetro cDNA synthesis kit (Bioline, Taunton, MA, USA). Real time PCR was performed using Taqman probes (Applied Biosystems) on the ABI 7700 Sequence Detection System (Applied Biosystems). Expression levels (average values and standard deviations of triplicate determinations) were normalized to housekeeping gene GAPDH. All experiments were performed with technical duplicates from three individual experiments.
Results
H3K79me2 Is Abrogated by Genetic Inactivation of Dot1L in MLL-AF10 and CALM-AF10 Transformed Bone Marrow Cells
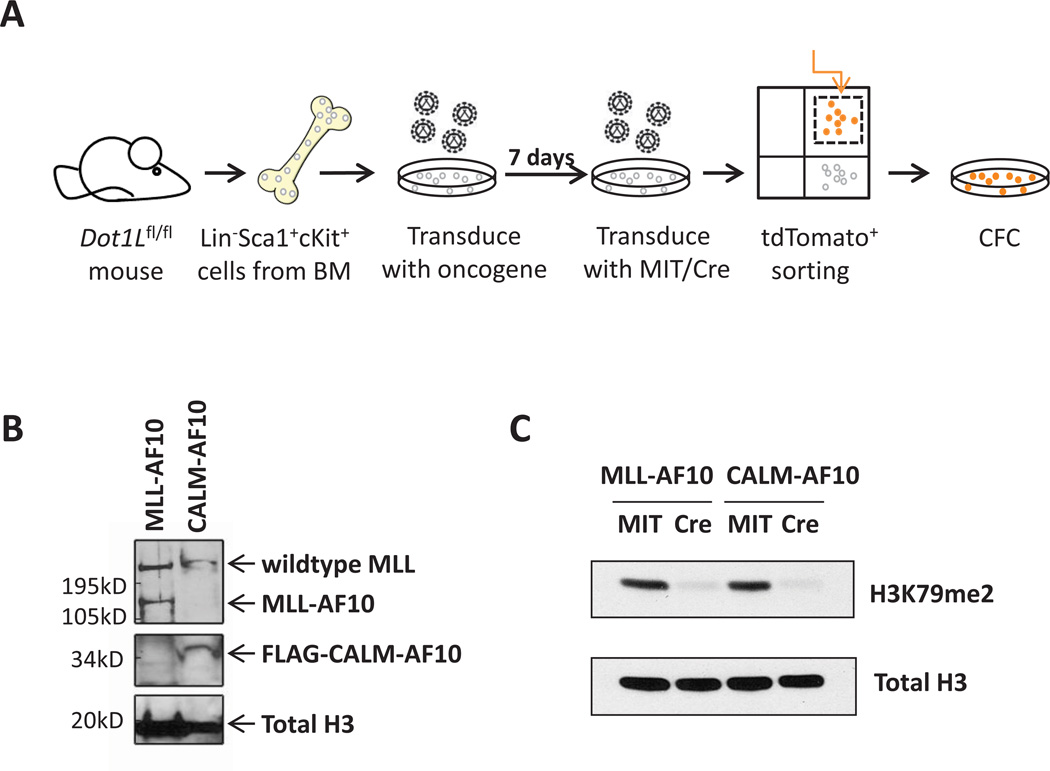
We used a Dot1l conditional knockout mouse model, in which the exon encoding the active site of Dot1l is flanked by LoxP sites (21), to determine whether Dot1l and H3K79 methylation are indeed required for transformation driven by MLL-AF10 and CALM-AF10. We sorted HSC-enriched Lin−Sca1+cKit+ (LSK) cells from Dot1l+/+ and Dot1lf/f mouse bone marrow cells, and transformed sorted cells with FLAG-MLL-AF10 or the FLAG-CALM-AF10 minimal fusion (14). The expression of MLL-AF10 and CALM-AF10 in transformed bone marrow cells was confirmed by western blotting using an antibody against the N-terminus of MLL and FLAG respectively (Figure 1B). Cells were then plated in cytokine-supplemented methylcellulose and expanded for 1–2 weeks. The MLL-AF10 or CALM-AF10 expressing cells were subsequently transduced with retroviruses encoding either the Cre-Mi-Tomato (Cre) or the control Mi-Tomato (MIT), sorted for tdTomato positive cells 2 days after transduction, and plated in methylcellulose (see Figure 1A). Genotyping of tdTomato+ cells right after sorting showed a complete deletion of Dot1l in cells transduced with Cre (Figure 2D). Consistent with deletion of Dot1l, H3K79 di-methylation was diminished in MLL-AF10 and CALM-AF10 transformed bone marrow cells after transduction with Cre (Figure 1C and 2B).
Fig.1. Cre-mediated deletion of Dot1l leads to loss of H3K79me2 in MLL-AF10 and CALM-AF10 immortalized murine bone marrow cells.
(A) Schematic representation of experimental design.
(B) Western blot showing the expression of MLL-AF10 using anti-MLL antibody and FLAG-CALM-AF10 using anti-FLAG antibody in transformed bone marrow cells. Total H3 was used as a control.
(C) Western blot showing the loss of H3K79 methylation 7 days after transduction with Cre or MIT.
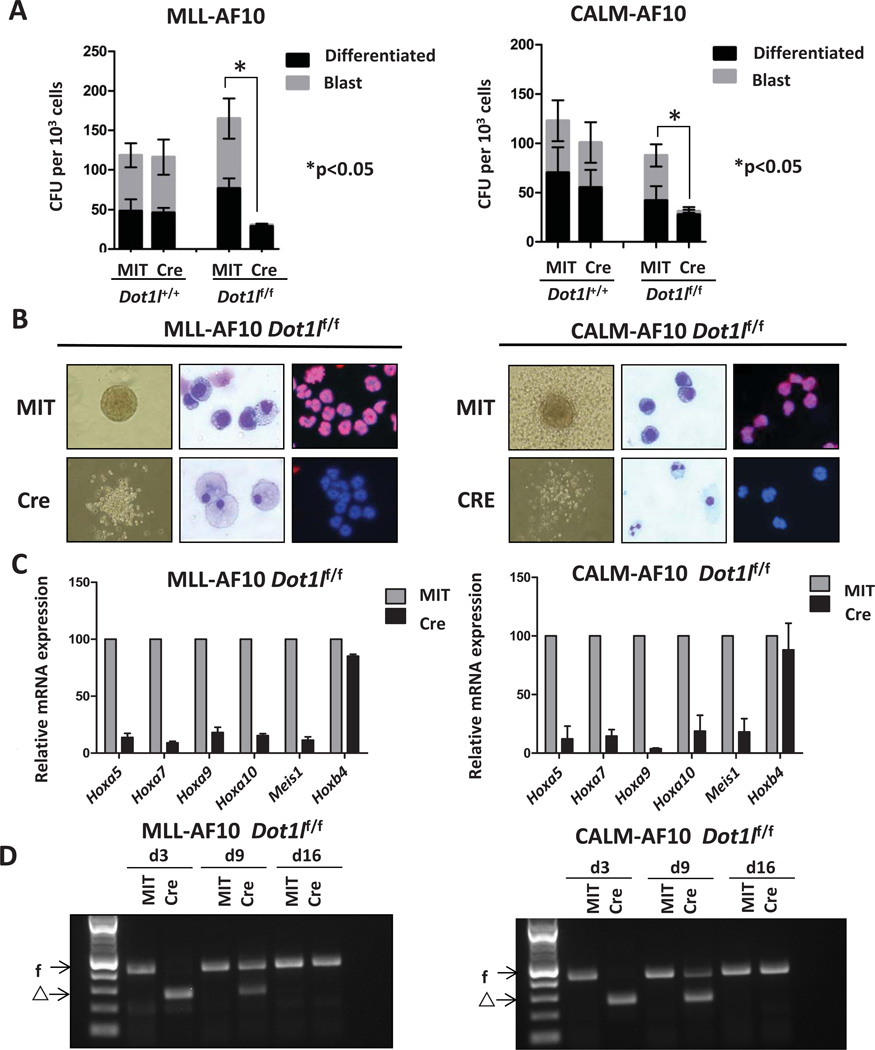
Fig.2. Loss of Dot1l leads to decreased colony forming potential and increased differentiation of MLL-AF10 or CALM-AF10 transformed cells.
(A) Blast and differentiated colony count of Dot1l-deleted MLL-AF10 (left) or CALM-AF10 (right) transformed cells in methylcellulose 9 days after transduction with Cre in comparison to controls (n=3 independent experiments).
(B) Morphologic changes (10X image of colony morphology in methylcellulose, 40X image of Wright-Giemsa stain) and H3K79me2 immunofluorescence (20X image, Alexa 674-H3K79me2 and DAPI nuclear stain) in MLL-AF10 (left) or CALM-AF10 (right) transformedpreleukemia cells 9 days after transduction with Cre.
(C) Relative expression levels of Hoxa5, Hoxa7, Hoxa9, Hoxa10, Meis1, and Hoxb4 on cells 5 days after transduction with Cre or MIT. Expression levels were normalized to Gapdh and expressed relative to MIT-transduced cells (set to 100%). Error bars indicate the SEM (n=3 independent experiments).
(D) Genotyping of transduced bone marrow cells on day 3, day 9, and day 16 after transduction of Cre. f: floxed allele. Δ: deleted allele.
Loss of Dot1l Inhibits Transformation by MLL-AF10 or CALM-AF10
We next evaluated the effects of genetic deletion of Dot1l on LSK cells transformed by MLL-AF10 or CALM-AF10. Deletion of Dot1l significantly reduced the colony forming potential in both cases (Figure 2A). The number of colonies was significantly decreased in the first week. In addition, Dot1l−/− colonies were morphologically distinct. While the majority of cells after transduction with MIT formed “blast-like” compact and hypercellular colonies, cells after transduction with Cre formed smaller and more diffuse colonies (Figure 2B). Moreover, Wright-Giemsa staining showed that the Cre-transduced cells have a larger cytoplasm and smaller nucleus, consistent with myeloid differentiation. We then verified the status of the Dot1l locus in the remaining cells harvested at different time points after Cre transduction. Figure 2D showed that 3 days after transduction only the excised allele could be detected. However, at the end of the first round of plating (day 9), the surviving cells had various degree of deletion, and at the end of the second round of plating (day 16), only the non-deleted allele could be detected. The fact that a small number of non-deleted cells outgrew in two weeks demonstrated a high selective pressure against Dot1l deleted cells.
We then assessed changes in expression of select MLL-fusion target genes in MLL-AF10 transformed bone marrow cells after the loss of Dot1l. The N-terminus of MLL binds to leukemogenic genes, including 5’ Hoxa cluster genes and Meis1, and the binding property is retained in MLL-AF10 fusion proteins (7, 8). The overexpression of MLL-fusion targets is a key feature of MLL-AF10 leukemia and the suppression of these leukemogenic genes, such as Hoxa9, is sufficient to abolish the leukemia (16, 18). As shown in Figure 2C, targets of MLL-AF10 were down-regulated after Cre-mediated deletion of Dot1l, while the expression of a gene that is not an MLL-fusion target, Hoxb4, was not changed. This finding demonstrates that Dot1l is required for continued expression of MLL-AF10 target genes which further supports the requirement of Dot1l in MLL-AF10 leukemia. We also assessed the expression changes of leukemia-related genes in CALM-AF10 transformed LSK cells after loss of Dot1l. It has been shown that leukemogenic genes, including 5’ Hoxa cluster genes and Meis1 are overexpressed in both CALM-AF10 mouse leukemias and CALM-AF10 positive patient leukemias (22, 23). The suppression of these leukemogenic genes, such as Hoxa5, is sufficient to abolish CALM-AF10 leukemia (18). As shown in Figure 2C, Hoxa5, Hoxa7, Hoxa9, Hoxa10, and Meis1 were all down-regulated after Cre-mediated deletion of Dot1l, while the expression of a control gene, Hoxb4, did not change (Figure 2C). This finding demonstrates that Dot1l is required for maintenance of the transformed phenotype in cells expressing the CALM-AF10.
Selective Anti-Proliferative Effect of MLL-AF10 and CALM-AF10 Transformed Cells by the Dot1l Inhibitor EPZ004777
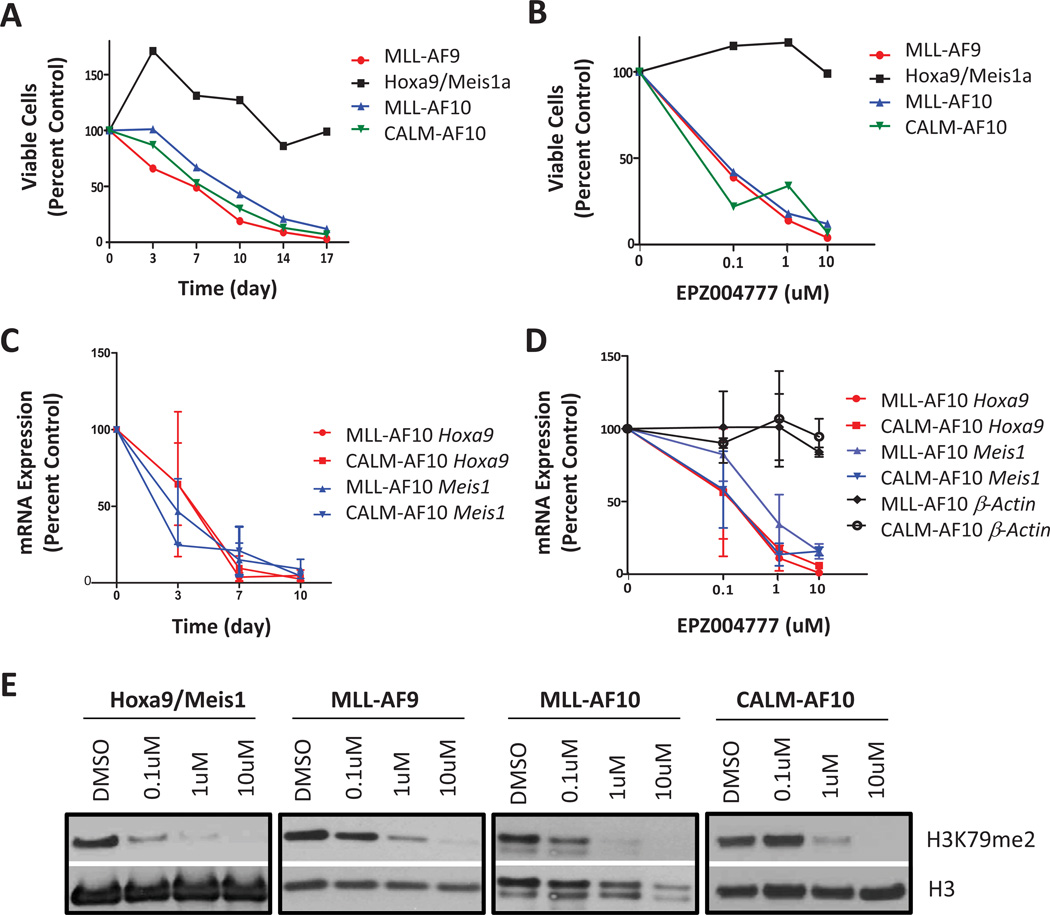
Having established that genetic inactivation of Dot1l inhibits H3K79 methylation and clonogenic potential of MLL-AF10 or CALM-AF10 transformed cells, we investigated the efficacy of Dot1l inhibitors against MLL-AF10 and CALM-AF10 transformed murine bone marrow cells. Recently it has been shown that the Dot1l inhibitor EPZ004777 selectively kills MLL-rearranged leukemia cells, including an MLL-AF4 leukemia cell line MV4-11 and an MLL-AF9 leukemia cell line MOLM13, but not MLL-germline leukemia cells, including Jurkat and HL-60 (24). However, the response of MLL-AF10 leukemia cells to a Dot1l inhibitor has not yet been evaluated, and the CALM-AF10 leukemia cell line U937 surprisingly showed minimal response to the inhibitor (24). Therefore, we performed proliferation assays over several days with three independently-transformed mouse LSK cell populations with MLL-AF10 or CALM-AF10 in the presence of increasing concentrations of EPZ004777 (up to 10 µM) or DMSO vehicle control. MLL-AF9 transformed cells were included as a positive control, and HoxA9 and Meis1 co-transformed (HoxA9/Meis1) cells were included as a non-MLL-rearranged cell line control which has been shown to be refractory to Dot1l inhibition (21). As shown in Figure 3A, the growth of MLL-AF10 and CALM-AF10 cells, as well as MLL-AF9 cells, was dramatically inhibited by EPZ004777, while the growth of HoxA9/Meis1 cells was unaffected. The anti-proliferative effect was dose-dependent, with an IC50 between 0.1 µM to 1 µM for MLL-AF10 and CALM-AF10 (Figure 3B). Similar to previous findings with human MLL-AF9 and MLL-AF4 leukemia cell lines, the anti-proliferative effect against MLL-AF10 and CALM-AF10-transformed primary murine hematopoietic progenitor cells only became apparent after 7 days. Nevertheless, when exposed to EPZ004777 longer, MLL-AF10 and CALM-AF10 transformed cells showed a dramatic decrease in cell number, while proliferation of the Hoxa9/Meis1 transformed cells was unaffected. Consistent with the proliferation and viability curve, Western blots showed a dose-dependent reduction of H3K79 dimethylation in all cell lines after incubation with EPZ004777 (Figure 3E).
Fig.3. EPZ004777 selectively inhibits proliferation of MLL-AF10 and CALM-AF10 transformed murine bone marrow cells.
(A) Growth of MLL-AF10, CALM-AF10, MLL-AF9, and Hoxa9/Meis1a transformed bone marrow cells during several days’ incubation with 10 µM EPZ004777. Viable cells were counted and replated at equal cell numbers in fresh media with fresh compound every 3–4 days. Results were plotted as percentage of split-adjusted viable cells in the presence of 10 µM EPZ004777 compared to DMSO vehicle control. Results are representative of three independent experiments.
(B) Dosage effect of EPZ004777 treatment on MLL-AF10, CALM-AF10, MLL-AF9, and HoxA9/Meis1a transformed bone marrow cells. Cells were counted and replated at equal cell numbers in fresh media with fresh compound every 3–4 days. Results were plotted as percentage of split-adjusted viable cells on day 17 in media with 0.1 µM, 1 µM, 10 µM of EPZ004777 compared to DMSO control (set as 100%). Results are representative of three independent experiments.
(C) Time course of Hoxa9 and Meis1 mRNA expression in MLL-AF10 and CALM-AF10 transformed cells over 10 days of incubation with 10 µM EPZ004777 as measured by quantitative real-time PCR. Expression levels were normalized to Gapdh and expressed relative to those at day 0 (set to 100%). Error bars indicate the SEM (n=3 independent experiments).
(D) Quantitative real-time PCR analysis of Hoxa9, Meis1, and β-Actin mRNA levels in MLL-AF10 and CALM-AF10 transformed cells following 7 days of incubation with EPZ004777. Relative mRNA expression levels are plotted as a percentage of those in vehicle-treated control cells. Error bars represent SEM (n=3 independent experiments).
(E) Inhibition of cellular H3K79me2 levels in MLL-AF10, CALM-AF10, MLL-AF9 or Hoxa9-Meis1a transformed bone marrow cells following 7 days of treatment with the indicated concentrations of EPZ004777 as measured by immunoblot analysis of extracted histones with an anti-H3K79me2 antibody.
We next tested whether the expression of MLL-AF10 and CALM-AF10 target genes was affected after EPZ004777 treatment. Hoxa9 and Meis1 overexpression is a hallmark of both MLL-AF10 and CALM-AF10 leukemia (16, 22, 23). We performed quantitative real-time PCR to examine the effect of EPZ004777 on Hoxa9 and Meis1 mRNA expression levels in MLL-AF10 and CALM-AF10 transformed cells. The mRNA expression levels of Hoxa9 and Meis1 started to decrease within 3 days and became significantly lower within 7 days’ treatment with 10 µM EPZ004777 (Figure 3C). Analysis of cells on day 7 of EPZ004777 treatment showed a concentration-dependent decrease of mRNA levels of Hoxa9 and Meis1, but not β-actin, which shows that the decrease in Hoxa9 and Meis1 expression is not caused by a general inhibition of gene expression (Figure 3D).
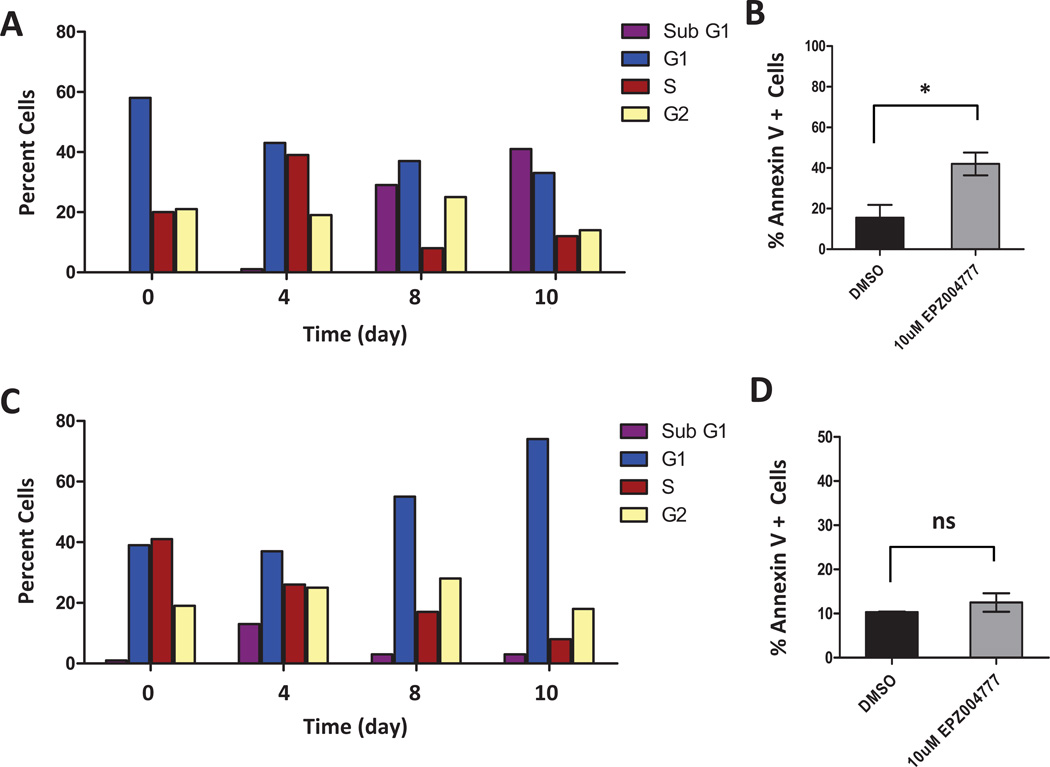
Next we determined if the decrease in cell number for EPZ004777 treated MLL-AF10 or CALM-AF10 transformed cells was due mostly to inhibition of cell proliferation or induction of apoptosis. Dot1l inhibition reduced the number of actively proliferating MLL-AF10 cells, with an increase in the percentage of cells in the G1 or sub G1fractions after 4 days of incubation with 10 µM EPZ004777 (Figure 4A). Moreover, a significant increase in the percentage of apoptotic cells after inhibition of Dot1l was observed in MLL-AF10 transformed cells compared to DMSO control on day 10 (Figure 4B). Similarly, Dot1l inhibition reduced the number of CALM-AF10 cells in S-phase, with the majority of cells found in G1 after 4 days of incubation with 10 µM EPZ004777 (Figure 4C). Interestingly, there was only a minimal increase in apoptotic cells throughout the length of this experiment (Figure 4D), suggesting the effect on CALM-AF10 may be more of a cell cycle arrest whereas MLL-AF10 cells respond with cell cycle arrest and more pronounced apoptosis.
Fig.4. EPZ004777 causes cell cycle arrest and apoptosis in MLL-AF10 and CALM-AF10 transformed bone marrow cells.
(A) Cell cycle changes (BrdU/7-AAD flow cytometry) in MLL-AF10 transformed bone marrow cells after being treated with 10 µM EPZ004777 for 0, 4, 8, or 10 days. Results are representative of two independent experiments.
(B) Annexin V staining in MLL-AF10 transformed bone marrow cells 10 days after treatment with 10 µM EPZ004777 or DMSO control (n = 2 independent experiments). Error bars represent standard error of the mean (SEM).
(C) Cell cycle changes (BrdU/7-AAD flow cytometry) in CALM-AF10 transformed bone marrow cells after being treated with 10 µM EPZ004777 for 0, 4, 8, or 10 days. Results are representative of two independent experiments.
(D) Annexin V staining in CALM-AF10 transformed bone marrow cells 10 days after treatment with 10µM EPZ004777 or DMSO control (n = 2 independent experiments). Error bars represent standard error of the mean (SEM).
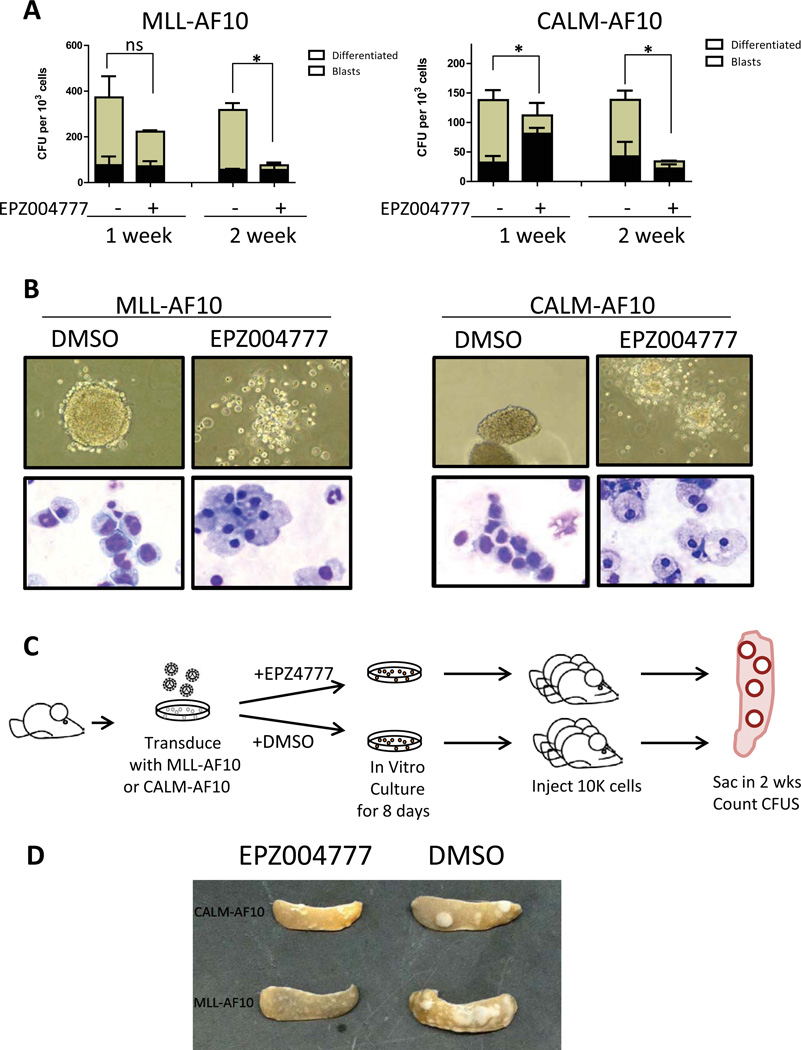
We went on to test whether EPZ004777 treatment affects the colony forming ability and serial replating capacity of MLL-AF10 and CALM-AF10 transformed LSK cells in in vitro CFC assays. We cultured MLL-AF10 or CALM-AF10 transformed mouse bone marrow cells in methylcellulose based medium containing DMSO or 10µM EZP004777. MLL-AF10 or CALM-AF10 transformed cells formed large, highly clonogenic compact blast-like colonies in methylcellulose based medium containing DMSO. However, MLL-AF10 or CALM-AF10 transformed cells formed significantly fewer compact colonies in medium containing 10µM EZP004777 starting from the first week (Figure 5A). The clonogenic potential of MLL-AF10 or CALM-AF10 transformed cells was abrogated after two weeks culture in the presence of inhibitor. Cytospin at the end of the first week showed that the majority of EPZ004777-treated cells have large cytoplasm and small or fragmented nucleus, consistent with myeloid differentiation.
Fig. 5. EPZ004777 decreased the colony forming potential and induced differentiation in MLL-AF10 and CALM-AF10 transformed bone marrow cells in vitro and EPZ004777 pretreatment diminished spleen-colony forming potential in vivo.
(A) Blast and differentiated colony count of MLL-AF10 or CALM-AF10 transformed cells cultured in methylcellulose based medium in the presence of 10uM EPZ004777 or DMSO vehicle control for 1 week or 2 weeks. *: p<0.05; ns: not significant (n=2 independent experiments).
(B) Morphologic changes (10X image of colony morphology in methylcellulose, 40X image of Wright-Giemsa stain) in MLL-AF10 (left) or CALM-AF10 (right) transformed cells cultured in methylcellulose based medium containing EPZ004777 for 7 days.
(C) Schematic representation of CFU-S experimental design.
(D) The morphology of spleen dissected from mice injected with pretreated MLL-AF10 or CALM-AF10 transformed cells.
To study the effect of pharmacological inhibition of Dot1l in vivo, we pretreated MLL-AF10 and CALM-AF10 transformed cells in liquid culture with either DMSO or 10µM EPZ004777 for 8 days before injecting into lethally irradiated recipients (Figure 5C). Two weeks after injection, we assessed the spleen-colony forming ability with CFU-S assay. MLL-AF10 or CALM-AF10 transformed cells without inhibitor treatment formed >10 large spleen colonies in two weeks. In contrast, cells pretreated with inhibitor failed to form any large spleen colonies, and instead formed several small and diffuse foci, similar to the colonies observed in the in vitro methylcellulose based cultures. These effects on the CFU-S forming activity suggest that EPZ004777 may show in vivo efficacy against AF10-fusion transformed cells (Figure 5D).
Dot1l is Indispensible for the Initiation and the Maintenance of MLL-AF10 or CALM-AF10 Leukemia Cells In Vivo
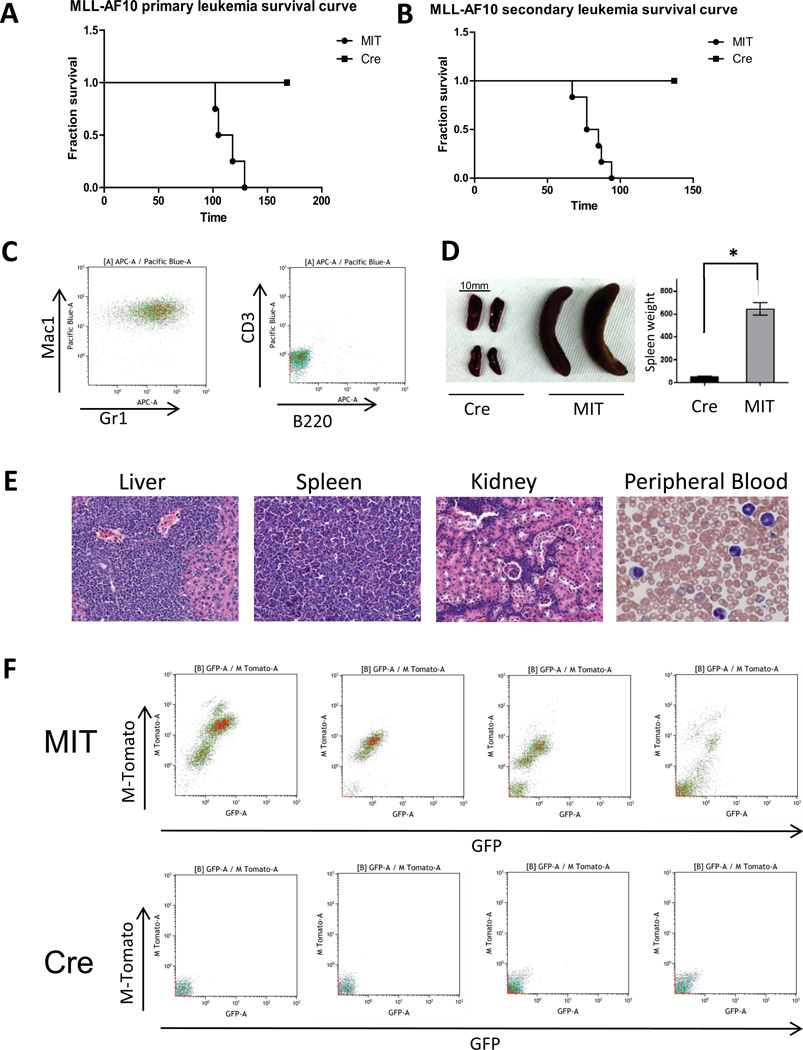
Since CFU-S activity does not directly test whether Dot1l inactivation impacts full-blown leukemogenesis, we sought to assess the impact of Dot1l deletion on in vivo leukemia initiation and maintenance by the leukemogenic AF10 fusions. Dot1lf/f bone marrow cells were transformed using retroviral MLL-AF10 or CALM-AF10. In two to three days, Dot1l was deleted in preleukemic-transformed cells through retroviral delivery of Cre-recombinase. Two days after MIT or Cre transduction, GFP+tdTomato+ cells were sorted and injected into sublethally irradiated recipients at 5×105 cells/mouse. Flow cytometric analysis of mouse peripheral blood 102 days after injection showed the propagation of Dot1lf/f MLL-AF10-transformed cells but not Dot1l−/− MLL-AF10-transformed cells in recipients (Figure 6F). Mice injected with preleukemic MLL-AF10-transformed Dot1lf/f cells all developed myeloid leukemia with a median of 112 days. However, no leukemia could be generated in mice injected with same doses of MLL-AF10-transformed Dot1l−/− cells (Figure 6A). Similar to MLL-AF10, preleukemic CALM-AF10-transformed Dot1lf/f cells represented an increasing population of GFP+tdTomato+ cells in recipients’ peripheral blood and ultimately caused myeloid leukemia in recipients, while CALM-AF10-transformed Dot1l−/− cells disappeared from the peripheral blood within 2 months (Figure 1s). This result shows that Dot1l is required for in vivo leukemogenesis of mouse bone marrow cells transformed by MLL-AF10 as well as CALM-AF10.
Figure 6. Dot1l is required for initiation and maintenance of MLL-AF10-driven leukemia in vivo.
(A) Survival curves for mice injected with 5X105 MLL-AF10 transformed bone marrow cells 2 days after transduction with Cre or MIT-control retrovirus and sorting for GFP+/tdTomato+ cells.
(B) Survival curves for secondary recipient mice that received 2X105 MLL-AF10 leukemia cells 2 days after transduction with Cre or MIT and sorting for GFP+/tdTomato+ cells.
(C) Immunophenotype of spleen cells of secondary MLL-AF10 leukemic mice (Mac1+Gr1+CD3−B220−).
(D) Spleen picture and spleen size in mice injected with Cre or MIT transduced Dot1l f/f MLL-AF10 cells (n=5.*: p<0.01).
(E) Morphology of peripheral blood smear (40X image of Wright-Giemsa stain) and pathology of organs from secondary MLL-AF10 leukemic mice (10X H&E stain).
(E) Peripheral blood chimerism in mice 102 days after injection of MIT or Cre transduced MLL-AF10 preleukemic cells. Donor cells are GFP+/tdTomato+ and recipient cells are GFP−/tdTomato−.
We then checked the effect of Dot1l deletion on established mouse leukemia cells. We collected bone marrow cells from primary MLL-AF10 or CALM-AF10 leukemia mice, performed MIT or Cre transduction, and sorted GFP+tdTomato+ cells for injection into mice. Dot1lf/f MLL-AF10 leukemic cells caused Mac1+Gr1+ secondary myeloid leukemia with a median of 81 days after transplantation, while MLL-AF10 leukemic cells lacking Dot1l failed to cause any leukemia in mice (Figure 6B and 6C). Histopathology study showed massive organ infiltration of leukemic cells in mice injected with Dot1lf/f MLL-AF10 (Figure 6E). The spleen weight of mice injected with Dot1lf/f MLL-AF10 was significantly higher than that of mice injected with Dot1l−/− MLL-AF10 (p<0.01, Figure 6D). Similar to MLL-AF10, Dot1lf/f CALM-AF10 leukemic cells resulted in an increasing population of GFP+tdTomato+ cells in recipients’ peripheral blood and ultimately caused myeloid leukemia in recipients, while Dot1l −/− CALM-AF10 leukemic cells appeared in peripheral blood initially, but disappeared from peripheral blood within 1 months (Figure 1s). This result shows that Dot1l is also required for the maintenance of MLL-AF10 or CALM-AF10 leukemia in vivo.
Discussion
A number of studies have recently demonstrated that DOT1L and H3K79 methylation plays an important role in MLL-AF9 (21, 25–27), MLL-GAS7 (25), and possibly MLL-AFX (25) transformation. In this study, we show that loss of Dot1l abrogates in vitro as well as in vivo transformation in MLL-AF10 and CALM-AF10 immortalized cells. This genetic approach circumvented the drawbacks of knock-down and overexpression approaches, such as off-target effects and problems of non-physiologic protein expression, thus strengthening the case for targeting Dot1l therapeutically in leukemias involving AF10 fusions.
Current advances in biology, biochemistry and pharmacology have raised the prospects of highly targeted therapeutics that maximize efficacy and minimize systemic toxicity. One outstanding example is targeting the fusion protein BCR-ABL kinase by imatinib in chronic myeloid leukemia (28). In our study, we tested the efficacy of targeting Dot1l in MLL-AF10 and CALM-AF10 leukemia. We show that the small-molecular inhibitor of Dot1l EPZ004777 selectively inhibits the proliferation of MLL-AF10 and CALM-AF10 transformed mouse bone marrow cells but has no effect on HoxA9/Meis1 transformed mouse bone marrow cells. The fact that Dot1l is dispensable for transformation driven by ectopically-expressed HoxA9 and Meis1 is in agreement with the model that the OM-LZ domain of AF10 present in leukemic AF10 fusions recruits Dot1l and activates Hox-Meis target genes through aberrant H3K79 methylation. Cells ectopically expressing retrovirally introduced HoxA9 and Meis1 genes are therefore immune to loss of H3K79 methylation. It not only demonstrates a strong rationale for inhibiting Dot1l as a strategy to target the AF10 rearranged leukemias, but also is in agreement with previous data that Dot1l is not absolutely required for cell proliferation (21). Recent studies showed that although conditional knockout of Dot1l leads to pancytopenia and failure of hematopoietic homeostasis in adult mice, the toxicity did not develop until 7–8 weeks after Dot1l inactivation (26). More importantly, in vivo administration of EPZ004777 for 2 weeks leads to extension of survival in a mouse MLL xenograft model with minimal hematopoietic side effects (24). These data provide further support for the continued development of DOT1L inhibitors as a potential therapeutic modality for MLL-rearranged and CALM-AF10 leukemias.
Interestingly, Daigle et al. recently demonstrated that the leukemia cell line U937, which harbors the CALM-AF10 fusion, was insensitive to EPZ004777 (24). The U937 cell line is a monocytic leukemia cell line in which the CALM-AF10 translocation was first identified in 1996 and it is the only readily available human leukemia cell line that carries the CALM-AF10 translocation (29). The long latency of CALM-AF10 leukemia in the murine bone marrow transplantation model and incomplete penetrance in the transgenic CALM-AF10 model strongly hints at additional collaborating mutations that need to be accumulated for CALM-AF10 leukemogenesis (22, 30). It is possible that the U937 cell line has accumulated other mutations, either during in-vivo leukemogenesis or during in-vitro passages, which may enable cells to circumvent the requirement of DOT1L for leukemia maintenance in vitro. The insensitivity of U937 to DOT1L inhibition is intriguing since both circumstantial evidence from human leukemias as well as experimental evidence presented here and from other studies points to a key role for the DOT1L methyltransferase in AF10 rearranged leukemias. The DOT1L interacting OM-LZ domain is consistently retained in both CALM-AF10 as well as MLL-AF10 patients, and the exclusion of the OM-LZ domain completely inhibits transforming activity of both the CALM –AF10 as well as MLL-AF10 fusions in murine models (16, 18). Moreover, the OM-LZ domain is also the minimal portion of AF10 required for the leukemogenesis for both the aforementioned AF10 fusions (14, 31). Importantly, our studies showing that Dot1l gene ablation as well as pharmacologic Dot1l inhibition demonstrates anti-leukemic activity demonstrate that DOT1L inhibition could be an attractive therapeutic target in human AF10 rearranged leukemias. Therefore, development of a panel of other AF10 rearranged cell lines or primary human xenograft models would appear to be warranted to assess the efficacy of DOT1L inhibition on human AF10 rearranged leukemia.
We and others have recently demonstrated that leukemogenesis mediated by a number of MLL fusions is dependent on abnormal H3K79 methylation (16, 21, 25–27). It was shown that the Dot1l inhibition in these MLL leukemias specifically interfered with the constitutive activation of MLL-target genes, resulting in abrogation of leukemogenesis mediated by these MLL-fusions. Interestingly, even though the CALM-AF10 fusion does not involve MLL as a fusion partner, gene expression studies have shown that the transcriptional profiles of CALM-AF10 patient samples bear strong similarities to those of MLL patient samples (23, 32). A simplistic explanation for this similarity could be the shared dependence of these fusions on aberrant H3K79 methylation for the activation of oncogenic programs. The relation between H3K79 methylation and AF10-rearranged leukemias is, however, more complicated. On one hand, aberrant H3K79 hypermethylation in MLL-AF10 and CALM-AF10 targets are required for activation of leukemogenic transcriptional programs and the maintenance of leukemia. On the other hand, MLL-AF10 and CALM-AF10 patient samples show a global hypomethylation of H3K79, possibly because the AF10 fusions disrupt normal AF10 function (33). Further studies will focus on the mechanisms of how abnormal H3K79 methylation patterns are established and how deregulation of a single epigenetic modification may act as a driver of leukemias with AF10 rearrangements.
Our observation that MLL-AF10 and CALM-AF10 fusions require Dot1l for initiation as well as maintenance of leukemia, strongly indicates that pharmacologic inhibition of aberrant H3K79 methylation could be of potential clinical benefit in the AF10-rearranged leukemias. Future studies will determine the effect of pharmacologic DOT1L inhibition on in vivo AF10-rearranged leukemias using syngenic or xenogenic leukemia models. At the moment, such studies are precluded by the poor pharmacokinetic properties of the DOT1L inhibitor used in our studies (24). These results could help inform future clinical trials with DOT1L inhibitors.
Supplementary Material
Acknowledgments
We would like to thank Ronald Mathieu and Mahnaz Pakhtinat for their excellent technical assistance with flow cytometry. This work was supported by grants from the National Cancer Institute, the American Cancer Society and the Leukemia and Lymphoma Society to S.A.A. A.J.D. was supported by a grant to the American Cancer Society and the NCI Howard Temin Pathway to Independence Award. K.M.B. was supported by NHLBI Career Development Award and funding from the William Lawrence and Blanche Hughes Foundation.
Footnotes
Conflict of interest
EJO, SRD, VMR and RMP are employees of Epizyme, Inc.. SAA is a consultant for Epizyme, Inc.
Supplementary information is available at the journal website.
References
- 1.Chaplin T, Bernard O, Beverloo HB, Saha V, Hagemeijer A, Berger R, et al. The t(10;11) translocation in acute myeloid leukemia (M5) consistently fuses the leucine zipper motif of AF10 onto the HRX gene. Blood. 1995 Sep 15;86(6):2073–2076. [PubMed] [Google Scholar]
- 2.Beverloo HB, Le Coniat M, Wijsman J, Lillington DM, Bernard O, de Klein A, et al. Breakpoint heterogeneity in t(10;11) translocation in AML-M4/M5 resulting in fusion of AF10 and MLL is resolved by fluorescent in situ hybridization analysis. Cancer Res. 1995 Oct 1;55(19):4220–4224. [PubMed] [Google Scholar]
- 3.Bohlander SK, Muschinsky V, Schrader K, Siebert R, Schlegelberger B, Harder L, et al. Molecular analysis of the CALM/AF10 fusion: identical rearrangements in acute myeloid leukemia, acute lymphoblastic leukemia and malignant lymphoma patients. Leukemia. 2000 Jan;14(1):93–99. doi: 10.1038/sj.leu.2401614. [DOI] [PubMed] [Google Scholar]
- 4.Dreyling MH, Schrader K, Fonatsch C, Schlegelberger B, Haase D, Schoch C, et al. MLL and CALM are fused to AF10 in morphologically distinct subsets of acute leukemia with translocation t(10;11): both rearrangements are associated with a poor prognosis. Blood. 1998 Jun 15;91(12):4662–4667. [PubMed] [Google Scholar]
- 5.Yu BD, Hess JL, Horning SE, Brown GA, Korsmeyer SJ. Altered Hox expression and segmental identity in Mll-mutant mice. Nature. 1995 Nov 30;378(6556):505–508. doi: 10.1038/378505a0. [DOI] [PubMed] [Google Scholar]
- 6.Yagi H, Deguchi K, Aono A, Tani Y, Kishimoto T, Komori T. Growth disturbance in fetal liver hematopoiesis of Mll-mutant mice. Blood. 1998 Jul 1;92(1):108–117. [PubMed] [Google Scholar]
- 7.Milne TA, Kim J, Wang GG, Stadler SC, Basrur V, Whitcomb SJ, et al. Multiple interactions recruit MLL1 and MLL1 fusion proteins to the HOXA9 locus in leukemogenesis. Mol Cell. 2010 Jun 25;38(6):853–863. doi: 10.1016/j.molcel.2010.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yokoyama A, Cleary ML. Menin critically links MLL proteins with LEDGF on cancer-associated target genes. Cancer Cell. 2008 Jul 8;14(1):36–46. doi: 10.1016/j.ccr.2008.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Thiel AT, Blessington P, Zou T, Feather D, Wu X, Yan J, et al. MLL-AF9-induced leukemogenesis requires coexpression of the wild-type Mll allele. Cancer Cell. 2010 Feb 17;17(2):148–159. doi: 10.1016/j.ccr.2009.12.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Milne TA, Briggs SD, Brock HW, Martin ME, Gibbs D, Allis CD, et al. MLL targets SET domain methyltransferase activity to Hox gene promoters. Mol Cell. 2002 Nov;10(5):1107–1117. doi: 10.1016/s1097-2765(02)00741-4. [DOI] [PubMed] [Google Scholar]
- 11.Nakamura T, Mori T, Tada S, Krajewski W, Rozovskaia T, Wassell R, et al. ALL-1 is a histone methyltransferase that assembles a supercomplex of proteins involved in transcriptional regulation. Mol Cell. 2002 Nov;10(5):1119–1128. doi: 10.1016/s1097-2765(02)00740-2. [DOI] [PubMed] [Google Scholar]
- 12.Tebar F, Bohlander SK, Sorkin A. Clathrin assembly lymphoid myeloid leukemia (CALM) protein: localization in endocytic-coated pits, interactions with clathrin, and the impact of overexpression on clathrin-mediated traffic. Mol Biol Cell. 1999 Aug;10(8):2687–2702. doi: 10.1091/mbc.10.8.2687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Klebig ML, Wall MD, Potter MD, Rowe EL, Carpenter DA, Rinchik EM. Mutations in the clathrin-assembly gene Picalm are responsible for the hematopoietic and iron metabolism abnormalities in fit1 mice. Proc Natl Acad Sci U S A. 2003 Jul 8;100(14):8360–8365. doi: 10.1073/pnas.1432634100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Deshpande AJ, Rouhi A, Lin Y, Stadler C, Greif PA, Arseni N, et al. The clathrin-binding domain of CALM and the OM-LZ domain of AF10 are sufficient to induce acute myeloid leukemia in mice. Leukemia. 2011 Nov;25(11):1718–1727. doi: 10.1038/leu.2011.153. [DOI] [PubMed] [Google Scholar]
- 15.Chaplin T, Ayton P, Bernard OA, Saha V, Della Valle V, Hillion J, et al. A novel class of zinc finger/leucine zipper genes identified from the molecular cloning of the t(10;11) translocation in acute leukemia. Blood. 1995 Mar 15;85(6):1435–1441. [PubMed] [Google Scholar]
- 16.Okada Y, Feng Q, Lin Y, Jiang Q, Li Y, Coffield VM, et al. hDOT1L links histone methylation to leukemogenesis. Cell. 2005 Apr 22;121(2):167–178. doi: 10.1016/j.cell.2005.02.020. [DOI] [PubMed] [Google Scholar]
- 17.Mohan M, Herz HM, Takahashi YH, Lin C, Lai KC, Zhang Y, et al. Linking H3K79 trimethylation to Wnt signaling through a novel Dot1-containing complex (DotCom) Genes Dev. 2010 Mar 15;24(6):574–589. doi: 10.1101/gad.1898410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Okada Y, Jiang Q, Lemieux M, Jeannotte L, Su L, Zhang Y. Leukaemic transformation by CALM-AF10 involves upregulation of Hoxa5 by hDOT1L. Nat Cell Biol. 2006 Sep;8(9):1017–1024. doi: 10.1038/ncb1464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Feng Q, Wang H, Ng HH, Erdjument-Bromage H, Tempst P, Struhl K, et al. Methylation of H3-lysine 79 is mediated by a new family of HMTases without a SET domain. Curr Biol. 2002 Jun 25;12(12):1052–1058. doi: 10.1016/s0960-9822(02)00901-6. [DOI] [PubMed] [Google Scholar]
- 20.Steger DJ, Lefterova MI, Ying L, Stonestrom AJ, Schupp M, Zhuo D, et al. DOT1L/KMT4 recruitment and H3K79 methylation are ubiquitously coupled with gene transcription in mammalian cells. Mol Cell Biol. 2008 Apr;28(8):2825–2839. doi: 10.1128/MCB.02076-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bernt KM, Zhu N, Sinha AU, Vempati S, Faber J, Krivtsov AV, et al. MLL-rearranged leukemia is dependent on aberrant H3K79 methylation by DOT1L. Cancer Cell. 2011 Jul 12;20(1):66–78. doi: 10.1016/j.ccr.2011.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Caudell D, Zhang Z, Chung YJ, Aplan PD. Expression of a CALM-AF10 fusion gene leads to Hoxa cluster overexpression and acute leukemia in transgenic mice. Cancer Res. 2007 Sep 1;67(17):8022–8031. doi: 10.1158/0008-5472.CAN-06-3749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dik WA, Brahim W, Braun C, Asnafi V, Dastugue N, Bernard OA, et al. CALM-AF10+ T-ALL expression profiles are characterized by overexpression of HOXA and BMI1 oncogenes. Leukemia. 2005 Nov;19(11):1948–1957. doi: 10.1038/sj.leu.2403891. [DOI] [PubMed] [Google Scholar]
- 24.Daigle SR, Olhava EJ, Therkelsen CA, Majer CR, Sneeringer CJ, Song J, et al. Selective killing of mixed lineage leukemia cells by a potent small-molecule DOT1L inhibitor. Cancer Cell. 2011 Jul 12;20(1):53–65. doi: 10.1016/j.ccr.2011.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chang MJ, Wu H, Achille NJ, Reisenauer MR, Chou CW, Zeleznik-Le NJ, et al. Histone H3 lysine 79 methyltransferase Dot1 is required for immortalization by MLL oncogenes. Cancer Res. 2010 Dec 15;70(24):10234–10242. doi: 10.1158/0008-5472.CAN-10-3294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jo SY, Granowicz EM, Maillard I, Thomas D, Hess JL. Requirement for Dot1l in murine postnatal hematopoiesis and leukemogenesis by MLL translocation. Blood. 2011 May 5;117(18):4759–4768. doi: 10.1182/blood-2010-12-327668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nguyen AT, Zhang Y. The diverse functions of Dot1 and H3K79 methylation. Genes Dev. 2011 Jul 1;25(13):1345–1358. doi: 10.1101/gad.2057811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Thompson CB. Attacking cancer at its root. Cell. 2009 Sep 18;138(6):1051–1054. doi: 10.1016/j.cell.2009.09.002. [DOI] [PubMed] [Google Scholar]
- 29.Dreyling MH, Martinez-Climent JA, Zheng M, Mao J, Rowley JD, Bohlander SK. The t(10;11)(p13;q14) in the U937 cell line results in the fusion of the AF10 gene and CALM, encoding a new member of the AP-3 clathrin assembly protein family. Proc Natl Acad Sci U S A. 1996 May 14;93(10):4804–4809. doi: 10.1073/pnas.93.10.4804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Deshpande AJ, Cusan M, Rawat VP, Reuter H, Krause A, Pott C, et al. Acute myeloid leukemia is propagated by a leukemic stem cell with lymphoid characteristics in a mouse model of CALM/AF10-positive leukemia. Cancer Cell. 2006 Nov;10(5):363–374. doi: 10.1016/j.ccr.2006.08.023. [DOI] [PubMed] [Google Scholar]
- 31.DiMartino JF, Ayton PM, Chen EH, Naftzger CC, Young BD, Cleary ML. The AF10 leucine zipper is required for leukemic transformation of myeloid progenitors by MLL-AF10. Blood. 2002 May 15;99(10):3780–3785. doi: 10.1182/blood.v99.10.3780. [DOI] [PubMed] [Google Scholar]
- 32.Mulaw MA, Krause AJ, Deshpande AJ, Krause LF, Rouhi A, La Starza R, et al. CALM/AF10-positive leukemias show upregulation of genes involved in chromatin assembly and DNA repair processes and of genes adjacent to the breakpoint at 10p12. Leukemia. 2011 Nov 8; doi: 10.1038/leu.2011.307. [DOI] [PubMed] [Google Scholar]
- 33.Lin YH, Kakadia PM, Chen Y, Li YQ, Deshpande AJ, Buske C, et al. Global reduction of the epigenetic H3K79 methylation mark and increased chromosomal instability in CALM-AF10-positive leukemias. Blood. 2009 Jul 16;114(3):651–658. doi: 10.1182/blood-2009-03-209395. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.