Abstract
Tumor suppressor p53 plays a pivotal role in tumor suppression. p53 is the most frequently mutated gene in cancer. As a transcription factor, p53 mainly exerts its role in tumor suppression through transcriptional regulation of its downstream target genes. Thus, p53 and its target genes form a complex p53 signaling pathway to regulate a wide variety of biological processes to prevent tumorigenesis. Recent studies have revealed that in addition to apoptosis, cell cycle arrest and senescence, p53's functions in the regulation of energy metabolism and anti-oxidant defense contribute significantly to its role in tumor suppression. Studies further show that many tumor-associated mutant p53 proteins not only lose tumor suppressive functions of wild-type p53, but also gain new oncogenic activities that are independent of wild-type p53, including promoting tumor cell proliferation, survival, metabolic changes, angiogenesis, and metastasis, which are defined as mutant p53 gain-of-function. The frequent loss of wild-type p53 function and the gain-of-function of mutant p53 in human tumors make p53 an extremely attractive target for cancer therapy. Different strategies and many small-molecule drugs are being developed for the p53-based tumor therapy. Here, we review the mechanisms of p53 in tumor suppression and gain-of-function mutant p53 in tumor development, as well as the recent advances in the development of the p53-based tumor therapy.
Keywords: tumor suppressor, p53, mutant p53, gain-of-function, tumor therapy
Introduction
p53 is a key tumor suppressor [1–3]. p53 is the most frequently mutated gene in human tumors. p53 mutations occur in almost every type of tumor and in over 50% of all tumors. p53 mutations are found in ∼30%–50% of lung, esophageal, colorectal, head and neck, and ovarian cancers, and in ∼5% of leukemia, sarcoma, melanoma, testicular cancer, and cervical cancer [4,5]. In those cancers with low p53 mutation rates, p53 is often inactivated by alternative mechanisms. For instance, p53 is often inactivated and degraded by human papillomavirus E6 protein (HPV-E6) in cervical cancer [6,7]. MDM2 (mouse double minute 2 homolog), the most critical p53 negative regulator, is frequently amplified and/or overexpressed in sarcoma, which leads to the degradation of p53 protein [8]. It was estimated that around 80% of human tumors have dysfunctional p53. Germline p53 mutations are the cause of Li–Fraumeni syndrome, a hereditary cancer predisposition syndrome [9]. p53 knockout in mice leads to the development of tumors, including lymphomas and sarcomas, at young ages [10]. As a transcription factor, p53 transcribes its target genes to regulate various cellular biological processes, including cell cycle arrest, apoptosis, senescence, energy metabolism, and anti-oxidant defense, to prevent tumorigenesis [1–3]. Interestingly, majority of p53 mutations found in human tumors are missense mutations, which usually result in the expression of full-length mutant p53 proteins. Recent studies have demonstrated that many tumor-associated mutant p53 proteins not only lose tumor suppressive functions of wild-type p53, but also gain new oncogenic functions that are independent of wild-type p53, including promoting tumor cell proliferation, anti-apoptosis, angiogenesis, metastasis, and metabolic changes, which are defined as mutant p53 gain-of-function [11,12]. In this review, we present an overview of the mechanisms of p53 in tumor suppression as well as the gain-of-function oncogenic activities of mutant p53 in cancers.
The p53 Signaling Pathway
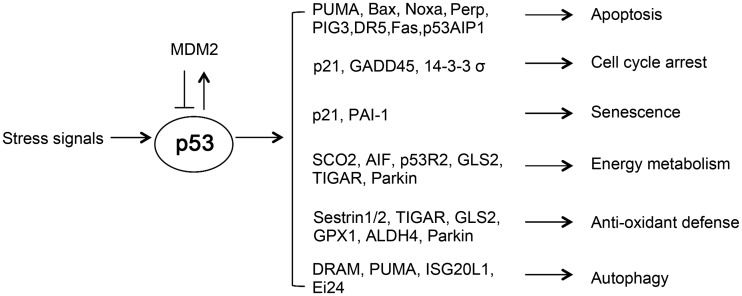
As a transcription factor, p53 mainly exerts its function through transcriptional regulation of its target genes. Under the non-stressed condition, the p53 protein is maintained at a low level in cells by the proteasome degradation pathway. MDM2, an E3 ubiquitin ligase, is the most critical negative regulator for p53 [13,14]. In response to a wide variety of stress signals, including DNA damage, ribonucleotide depletion, nutritional starvation, hypoxia, and oncogene activation, p53 is stabilized through post-translational modifications by a wide variety of enzymes. These enzymes include kinases, phosphatases, acetyltransferases, deacetylases, ubiquitin ligases, deubiquitinases, methylases, and sumoylases [2,3]. Once activated, p53 binds to a specific DNA sequence, termed the p53-responsive element, in its target genes to regulate their expression. p53-responsive element is composed of RRRCWWGYYY (spacer of 0–21 nucleotides) RRRCWWGYYY, where R is a purine, W is A or T, and Y is a pyrimidine [15]. Through the transcriptional regulation of its target genes, p53 regulates a wide range of cellular biological processes to maintain genomic integrity and prevent tumor formation, including cell cycle arrest, apoptosis, senescence, energy metabolism, anti-oxidant defense, autophagy, etc. (Fig. 1).
Figure 1.
p53 transactivates its target genes to regulate various cellular biological processes for tumor suppression In normal unstressed cells, the p53 protein is maintained at a low level in cells by its negative regulators, such as MDM2. In response to a wide variety of stress signals, activated p53 transcriptionally regulates the expression of its target genes to regulate various cellular biological processes, including apoptosis, cell cycle arrest, senescence, energy metabolism, anti-oxidant defense, and autophagy, to exert its role as a tumor suppressor.
Tumor Suppressive Functions of p53
Among these cellular biological processes regulated by p53, apoptosis, cell cycle arrest, and senescence have been widely accepted as the main mechanisms for p53's tumor suppressive function. Apoptosis is the most intensively studied function of p53. It was first reported in mouse thymocytes in response to irradiation [16,17]. Since then the p53-dependant apoptosis has been reported in a wide range of cells in response to many different stress signals. Once activated by these stress signals, p53 transcriptionally induces a group of target genes involved in apoptosis, including PUMA (p53 up-regulated modulator of apoptosis), Bax (BCL2-associated X protein), Noxa (PMAIP1), PIG3 (tumor protein p53 inducible protein 3), Killer/DR5 (tumor necrosis factor receptor superfamily, member 10b), Fas (Fas cell surface death receptor), Perp (p53 apoptosis effector related to PMP-22), and p53AIP1 (tumor protein p53 regulated apoptosis inducing protein 1), leading to apoptosis [18]. Among these p53 targets involved in apoptosis, the PUMA seems to play a more crucial role since only loss of PUMA displays similar apoptotic changes as loss of p53 in irradiated T-lymphocytes in mouse models [19]. Recent studies have shown that p53 can also regulate apoptosis through a transcription-independent pathway. In response to stress, a fraction of the p53 protein translocates to mitochondria, where p53 interacts with anti-apoptotic Bcl-xL and Bcl-2 to inhibit their functions, resulting in the release of cytochrome c from the mitochondria, and thereby induces apoptosis [20,21].
Another extensively studied function of p53 is to induce cell cycle arrest. In response to various stress signals, p53 transactivates some special target genes, resulting in cellular growth arrest at different cell cycle checkpoints to prevent the propagation and accumulation of DNA damage and mutations. It is well-established that p53 can induce G1 arrest through transcriptional induction of p21, a cyclin-dependent kinase inhibitor [22,23]. p53 was also reported to transcriptionally activate GADD45 (growth arrest and DNA-Damage-inducible 45) and 14-3-3σ (tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, sigma polypeptide), which in turn leads to G2 arrest [24,25]. The temporary G1 or G2 arrest induced by stress, especially mild stress signals, allows cells to survive until damage has been repaired or stress signals have been removed.
Inducing senescence is another important function of p53. Many DNA-damaging agents used in chemotherapy can activate p53 and induce senescence. Interestingly, it was recently reported that reactivation of p53 in p53-deficient tumors completely represses tumor growth through senescence in a mouse liver tumor model [26]. However, the mechanism by which p53 induces senescence is not as clear as the mechanisms for apoptosis and cell cycle arrest. Many senescence signals activate p53, which in turn transactivates p21 and induces p53-dependent senescence. Recently, PAI-1 (plasminogen activator inhibitor-1) was reported to be another p53 target gene involved in the p53-dependent senescence [27].
In addition to apoptosis, cell cycle arrest, and senescence, recent studies have revealed some additional mechanisms for p53 in tumor suppression, including regulation of cellular metabolism, anti-oxidant defense, autophagy, and microRNAs (miRNAs) [3,28]. Recently, metabolic changes have been regarded as a hallmark of tumor cells, which could be a key contributor to tumorigenesis [29,30]. p53 up-regulates mitochondrial oxidative phosphorylation and down-regulates glycolysis in cells to maintain the homeostasis of energy metabolism. p53 transcriptionally induces its target SCO2 (synthesis of cytochrome c oxidase 2), AIF (apoptosis-inducing factor), and p53R2 (ribonucleotide reductase M2 B) to maintain the mitochondrial integrity and promote mitochondrial oxidative phosphorylation [31–33]. p53 also induces the expression of mitochondrial glutaminase GLS2 to promote oxidative phosphorylation [34]. At the same time, p53 reduces glucose uptake through repressing the expression of GLUT1, 3, and 4 (glucose transporter 1, 3, and 4) [35,36]. Furthermore, p53 transcriptionally induces TIGAR (TP53-inducible glycolysis and apoptosis regulator) and Parkin (parkin RBR E3 ubiquitin protein ligase) to inhibit glycolysis [37,38]. p53 was also reported to bind to and reduce the activity of glucose-6-phosphate dehydrogenase, a rate-limiting enzyme in the pentose phosphate pathway, to down-regulate glucose metabolism [39].
Recent studies also showed that p53 protects cells from oxidation by reducing intracellular reactive oxygen species (ROS), a major cause of DNA damage and genetic instability, which contributes greatly to p53's role as a tumor suppressor [40,41]. p53 deficiency in cells and mouse tissues results in the elevation of intracellular ROS levels, which in turn leads to the increased DNA oxidation and mutation rates in cells. These effects can be substantially reversed by ectopic expression of Sestrins, p53 targets involved in anti-oxidant defense, in p53 deficiency cells. Furthermore, dietary supplementation with anti-oxidant N-acetylcysteine prevents the early onset tumors in p53 knockout mice [40,41]. To exert its anti-oxidant function, p53 transcriptionally induces a group of anti-oxidant genes, including sestrins 1/2, TIGAR, GPX1, ALDH4, GLS2, and Parkin, especially under conditions of non-stress or low stress, to reduce the intracellular levels of ROS and prevent DNA damage induced by ROS [42,43].
Autophagy is an important cellular catabolic process characterized by the formation of double-membrane autophagosomes around cytoplasmic components targeted for degradation, such as long-lived proteins and old/damaged organelles. Recently, it has been suggested that autophagy may play a dual role in tumorigenesis; autophagy plays an important role in maintaining genomic stability and tumor prevention in normal cells and tissues, whereas autophagy can promote tumor cell survival and tumor progression in tumors [44]. p53 has been reported to promote autophagy through different mechanisms, which may contribute to the role of p53 in tumor prevention. p53 promotes autophagy through inhibition of the mTOR (mammalian target of rapamycin) pathway, which is a critical negative regulator of autophagy [45]. p53 also induces the expression of several genes, including DRAM (DNA-damage regulated autophagy modulator 1), PUMA, ISG20L1 (interferon-stimulated exonuclease gene 20 kDa-like 1), and Ei24 (etoposide-induced 2.4), to promote autophagy [46–48]. Interestingly, p53 was also reported to inhibit autophagy under certain circumstances. For example, cytoplasmic p53 inhibits autophagy without activation of p53 target genes in some types of cells [49]. Similarly, tumor-associated mutant forms of p53, especially those located in the cytoplasm, were also reported to inhibit autophagy [50] (Fig. 1).
In addition to transcriptional regulation of protein-coding genes, recent studies have shown that p53 can transcriptionally regulate the expression of miRNAs as a new mechanism for p53 to exert its tumor suppressive functions [51,52]. miRNAs are a class of small (20–25 nucleotide) non-coding RNAs, which play a key role in the post-transcriptional regulation of gene expression. miRNAs bind to the 3′-untranslated regions (3′-UTRs) of target mRNAs, leading to the inhibition of translation and degradation of mRNAs. The miR-34 family members, miR-34a/b/c, were the first group of miRNAs that were identified as direct p53 target genes [53–55]. p53 regulates the expression of miR34-a/b/c through direct binding to the p53-responsive elements in their promoters. miR-34 family members repress the expression of several targets involved in the regulation of cell cycle, cell proliferation, and survival, including cyclin E2, CDK4/6, and BCL2. Ectopic expression of miR-34 family members promotes p53-mediated apoptosis, cell cycle arrest, and senescence [53–55]. Since then, a group of miRNAs has been reported to be directly induced by p53, including miR-145, miR-107, miR-192/194/215, miR-15a/16-1, miR-215, and let-7, to mediate the function of p53 in regulating different biological processes, including cell cycle arrest, senescence, apoptosis, metabolism, mesenchymal–epithelial transition, and differentiation [51,52]. In addition to the transcriptional regulation of specific miRNAs, p53 also promotes the post-transcriptional maturation of specific miRNAs. It has been reported that p53 promotes the Drosha-mediated processing of certain miRNAs, including miR-16-1, miR-143, and miR-145, which display the growth-suppressive function in cells. This function of p53 is mediated by the interaction of p53 with Drosha, and furthermore, this interaction requires p68 and p72 [56]. Furthermore, p53 affects the miRNA target selection by regulating RNA-binding proteins, such as RBM38 (RNA-binding-motif protein 38), which competes with miRNAs for binding to 3′-UTRs of mRNAs of target genes [57].
With the identification of more and more functions of p53, an important question has been raised: which function(s) is crucial for p53's role in tumor suppression. Many studies have been carried out to address this question, and many interesting observations have been made. However, so far, there is no clear answer to this question, and some observations even appear to be contradictory. For instance, while it is well-established that p21 plays a critical role in mediating p53's role in inducing cell cycle arrest in response to stress, unlike p53-null mice, p21-null animals are not prone to early onset tumorigenesis [23], suggesting that the function of p53 in inducing cell cycle arrest does not contribute significantly to its role in tumor suppression. Disruption of apoptosis by Bcl-2 overexpression or loss of PUMA promoted Eμ-myc-induced lymphomagenesis in mice [58,59]. However, the Bcl-2 transgenic or PUMA knockout mice were not as tumor-prone as p53 knockout mice [60], suggesting that inducing apoptosis alone cannot mediate the tumor suppressive function of p53. Mice expressing a mutant p53 (p53R172P) deficient for p53-mediated apoptosis but not cell cycle arrest and senescence were resistant to early onset tumorigenesis [61–63]. Mice expressing p53(25,26), a mutant p53 which contains two mutations at codons 25 and 26 and is deficient for cell cycle arrest and apoptosis but not senescence, retained the ability to inhibit KrasG12D-induced lung carcinogenesis [64]. Interestingly, p53 mutations in three acetylation sites (K117R+K161R+K162R) in mice impaired the p53-mediated apoptosis, cell cycle arrest, and senescence; however, these mutations did not affect the activities of p53 to regulate energy metabolism and ROS production in mice [65]. Notably, these mice did not develop early onset lymphomas as p53 knockout mice, suggesting that the regulation of energy metabolism and ROS production by p53 contributes significantly to the role of p53 in tumor suppression [65]. While it is still unclear which function(s) of p53 is critical for p53 in tumor suppression, these findings suggested one possibility that p53 might exert its role as a tumor suppressor with distinct mechanisms in different contexts, including different types of tissues and cells, different genetic background and microenvironment of cells, and in response to different types of stress signals.
It still remains largely unclear how p53 selectively regulates different groups of target genes and initiates different cellular responses to exert its tumor suppressive function in different types of cells and tissues in response to different stress signals. Interestingly, recent studies have reported that a group of proteins are involved in modulating the selection of p53 target genes. For instance, hCAS/CSE1L (human cellular apoptosis susceptibility protein) was reported to associate with the promoters of a subset of p53 target genes, such as pro-apoptotic PIG3, but not p21. This effect is achieved through the regulation of histone methylation and chromatin modification of p53 target genes by hCAS/CSE1L [66]. ASSP1 and ASSP2 (apoptosis-stimulating of p53 proteins 1 and 2) bind to p53 protein and selectively stimulate the binding of p53 to the promoters of p53 target genes involved in apoptosis, such as PIG3 and Bax. This effect was not observed for p53 target genes involved in cell cycle arrest, such as p21, although the mechanism is unclear [67]. Hzf (hematopoietic zinc finger), a zinc-finger protein, directly interacts with the DNA-binding domain of p53, and preferentially induces p53 target genes involved in cell cycle arrest, such as p21 and 14-3-3σ. In response to prolonged stress signals, Hzf is degraded by the proteasome degradation pathway, which in turn leads to the transcriptional activation of p53 targets involved in apoptosis, such as Bax, Noxa, and Perp [68]. SLUG (snail family zinc finger 2) is induced by p53 and antagonizes p53-mediated apoptosis triggered by DNA damage. SLUG exerts this protective role by repressing p53 target PUMA, a pro-apoptotic protein [69]. In addition, lincRNA-p21, a large intergenic non-coding RNA (lincRNA) was recently reported to serve as a repressor in p53-dependent transcriptional responses, which is mediated through the physical association with hnRNP-K [70]. This interaction is required for proper genomic localization of hnRNP-K at repressed genes and regulation of p53-mediated apoptosis. Future studies will further elucidate the precise mechanism by which p53 selectively regulates different cellular responses and coordinates these responses in different contexts to exert its role as a tumor suppressor.
Gain-of-Function of Mutant p53 in Cancers
Majority of tumor suppressor genes, such as RB (retinoblastoma-associated protein), APC (adenomatous polyposis coli), and VHL (Von Hippel-Lindau tumor suppressor), are frequently inactivated by deletion or truncation mutations in tumors, resulting in the decreased or loss of expression of their proteins. Interestingly, the majority of p53 mutations in human cancer are missense mutations, which usually result in the expression of full-length mutant p53 proteins. Although p53 mutations have been found in all coding exons of the p53 gene, the majority of the missense mutations are clustered in exons 4–9, which is a p53 DNA-binding domain, resulting in the loss of DNA-binding activity of mutant p53. Furthermore, ∼25% of p53 mutations occur at six ‘mutational hotspots’ in the DNA-binding domain of p53, including residues R175, G245, R248, R249, R273, and R282 [5,71]. When wild-type and mutant p53 alleles exist in a heterozygous status in tumor cells, mutant p53 can block the function of wild-type p53 through the dominant negative effect. However, p53 mutations are usually followed by loss of heterozygosity in human cancer, leading to the deletion or mutation of the rest wild-type p53 allele. While wild-type p53 protein is kept at a low level in cells by the proteasome degradation pathway under non-stressed conditions, mutant p53 protein usually accumulates to a high level in tumors, and the underlying mechanisms are not fully understood [11,12].
It has been well-documented that many tumor-associated mutant p53 proteins not only lose their tumor suppression functions, but also gain new oncogenic functions, which is termed the gain-of-function of mutant p53. The first evidence came from the findings that transfection of mutant p53 in p53-null tumor cells greatly increased the tumorigenicity of those cells in nude mice [72,73]. Since then, by ectopic expression of mutant p53 in p53-null tumor cells or by knockdown of endogenous mutant p53 in tumor cells that have lost the wild-type p53 allele, many studies have demonstrated different gain-of-function activities of mutant p53, including promoting cell proliferation, anti-apoptosis, metabolic changes, migration, invasion, angiogenesis, and metastasis [11,12,74,75]. Recently, the gain-of-function oncogenic activities of mutant p53 were also clearly demonstrated in two mutant p53 knock-in mouse models. Mice expressing R172H or R270H mutp53 (equivalent to human R175H and R273H, respectively) develop an altered spectrum of tumors and more metastatic tumors compared with p53−/− mice [76,77].
Mechanisms of Mutant p53 Gain-of-Function
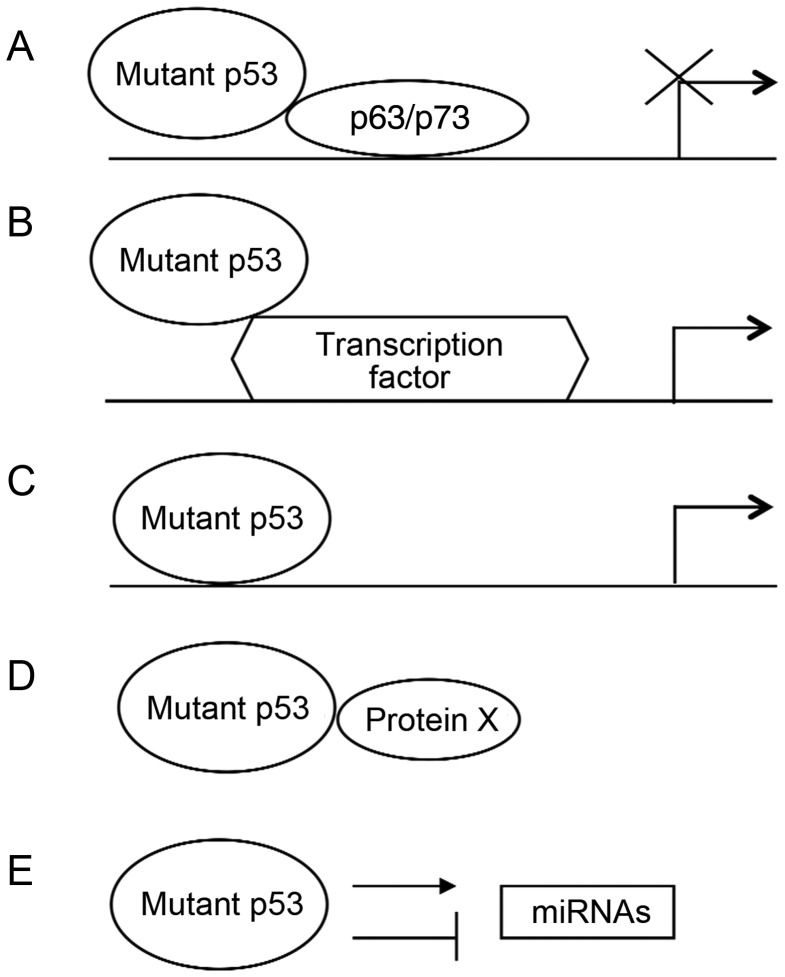
Recent studies have proposed the following several mechanisms by which mutant p53 gains new oncogenic activities in tumor cells (Fig. 2).
Figure 2.
Models of proposed mechanisms for mutant p53 gain-of-function Recent studies have suggested the following several mechanisms for mutant p53 gain-of-function. (A) Mutant p53 decreases transcriptional activities of p63 and p73 by binding to p63 and p73. (B) Mutant p53 regulates transcription of genes by interacting with other transcription factors. (C) Mutant p53 binds to DNA to regulate gene expression. (D) Mutant p53 interacts with other proteins to regulate their functions. (E) Mutant p53 influences the expression and processing of miRNAs.
Mutant p53 interacts with p63 and p73
p63 and p73 are two structural and functional homologs of p53 [78]. p63 and p73 bind to and activate many p53 target genes, and mediate cell cycle arrest, apoptosis, and senescence in response to stress. p63 and p73 were shown to form homotetramers and heterotetramers with each other, but they do not form heterotetramers with wild-type p53. Interestingly, several forms of mutant p53 were reported to interact with p63 and p73 through their DNA-binding domains to inhibit the transcriptional activities of p63 and p73 [79,80]. The interaction between mutant p53 and p63/p73 are related with many aspects of the gain-of-function of mutant p53, such as chemoresistance, migration, invasion, and metastasis.
Mutant p53 binds to transcription factors to regulate their functions
It has been reported that mutant p53 can interact with other transcription factors and be recruited to their binding sites to modulate the expression of their target genes. For example, mutant p53 has been shown to interact with transcription factor NF-Y, and up-regulate the expression of NF-Y target genes [81]. Mutant p53 was also reported to bind to vitamin D receptor (VDR) and be recruited to VDR-regulated genes to modulate their expression [82]. In addition, mutant p53 enhances sp1 transcriptional activity when it interacts with sp1 at the consensus sp1 responsive elements in the HIV-LTR [83]. Recently, mutant p53 was reported to interact with SREBP (sterol regulatory element-binding protein) family of transcription factors to regulate the expression of genes in the mevalonate pathway to disrupt tissue architecture in breast cancer cells [84].
Mutant p53 interacts with proteins to change their function
Mutant p53 can form complex with some other proteins and affect their functions, which contributes to the gain-of-function of mutant p53. For example, mutant p53 interacts with MRE11, a DNA nuclease required for homologous recombination DNA repair. The interaction between mutant p53 and MRE11 promotes genomic instability and tumor progression [85]. In addition, mutant p53 interacts with and co-localizes with PML (promyelocytic leukemia) protein, activating mutant p53 transcriptional activity in cells [86]. Mutant p53 was also reported to interact with topoisomerase 1, which maintains DNA topology, resulting in hyper-recombination and genomic instability [87]. Prolyl isomerase Pin1, which regulates conformational changes of proteins to affect protein stability and activity, was reported to be an additional mutant p53-binding protein. Pin1 cooperates with mutant p53 in Ras-dependent transformation. In breast cancer cells, Pin1 enhances the oncogenic activity of mutant p53 to promote aggressiveness through mutant p53-dependent inhibition of p63 and induction of a mutant p53 transcriptional program [88].
Mutant p53 binds to DNA to alter gene expression
Like wild-type p53 which functions as a transcription factor, mutant p53 has been reported to up-regulate or down-regulate a number of genes involved in different aspects of tumorigenesis, including Myc, Fos, PCNA, IGF1R, EGR1, NF-κB2, BCL-xL, IGF2, VEGFA, etc. [74]. Through the transcriptional regulation of these genes, mutant p53 promotes proliferation, anti-apopotosis, inflammation, and angiogenesis. The ability of mutant p53 to bind directly to DNA appears to be important for mutant p53 in regulating transcription of these genes. However, unlike wild-type p53, which is a DNA sequence-specific transcription factor, no defined mutant p53-responsive element has been characterized so far. Interestingly, it has been reported that mutant p53 binds directly to DNA in a DNA structure-selective mode. For instance, mutant p53 has a high affinity for nuclear matrix attachment regions, which are highly AT-rich regions that mediate structural organization of the chromatin and often adopt non-B DNA conformations [89,90]. Furthermore, mutant p53 was shown to bind selectively and with high affinity to non-B DNA [91].
Mutant p53 regulates miRNAs
Recent studies also demonstrated that mutant p53 regulates miRNAs, contributing to its gain-of-function. Mutant p53 induces or represses the expression of certain miRNAs to gain new oncogenic activities. For instance, mutant p53 directly binds to the promoter of miR-130b and inhibits its transcription [92]. As a negative regulator of ZEB1, miR-130b promotes epithelial–mesenchymal transition and cancer cell invasion in endometrial cancer [92]. Mutant p53 induces miR-155 to drive invasion in breast cancer. MiR-155 represses the expression of zinc-finger transcriptional repressor ZNF652, which represses the expression of proteins that promote invasion and metastasis, such as TGFB1/2, EGFR, and SMAD2 [93]. Mutant p53 binds to the miR-27a promoter region and represses its expression. Since EGFR is a direct target of miR-27a, through repressing miR-27a, mutant p53 promotes a sustained EGF-induced ERK1/2 activation, thereby promoting cell proliferation and tumorigenesis [94]. Furthermore, mutant p53 induces miR-128-2, which targets E2F5, to enhance chemoresistance in lung cancer cells [95]. In addition to regulating the expression of miRNAs, mutant p53 also affects the processing of miRNAs. For example, mutant p53 inhibits the processing of pri-miRNAs by Drosha, and thereby decreases the levels of certain mature miRNAs in cells, including miR-16-1, miR-143, and miR-145 [56]. These miRNAs have been shown to negatively regulate cell cycle and cell proliferation. In addition, mutant p53 was also reported to suppress DICER1 expression through binding and inactivation of p63 [96] (Fig. 2).
p53 and Cancer Therapy
The p53 signaling pathway is estimated to be dysfunctional in ∼80% of human tumors through mutations and other mechanisms, which makes p53 an extremely attractive target for cancer therapy. Numerous studies have shown that reactivation of p53 is detrimental for cancer cells. For instance, re-expression of wild-type p53 in p53-deficient cancer cells leads to apoptosis or senescence in cultured cells. In mouse models, re-introduction of wild-type p53 into p53-deficient tumors leads to tumor regression [26,97], whereas re-introduction of wild-type p53 into mutant p53-harboring tumors suppresses tumor growth [98]. Tremendous efforts have been made to develop the p53-based cancer therapy during the past decade. Several strategies have been developed, including ectopic expression of wild-type p53, activation of dysfunctional wild-type p53, destabilization or inactivation of mutant p53, and reactivation of mutant p53 in tumors.
p53-based gene therapy
Adenovirus-mediated p53 gene transfer to treat non-small cell lung carcinoma was first reported in 1996 [99]. Due to the consideration of biosafety, the replication-defective recombinant adenovirus expressing p53 (rAd-p53) was developed later, which has a better transduction efficiency and lower toxicity. The rAd-p53, under the brand name of gendicine, has been approved for clinical use for the treatment of head and neck cancer in China [100].
Small molecules that activate wild-type p53 function
As a key negative regulator for p53, MDM2 is frequently amplified and/or overexpressed in various tumors, which leads to the dysfunction of p53. Through binding to the pocket of MDM2 to block the interaction between MDM2 and p53, a non-genotoxic small-molecule Nutlin-3 has been developed, which can inhibit MDM2 and activate wild-type p53 in tumor cells. Nutlin-3 induces p53-mediated cell cycle arrest, apoptosis, and other antitumor activities in various cultured tumor cells and xenograft tumors in mice in a wild-type p53-dependent manner [101]. Nutlin-3 is currently being tested in phase 1 clinical trail. MI-219 is another small-molecule MDM2 inhibitor. It can disrupt p53-MDM2 binding, leading to the activated p53 signaling and suppression of tumor growth in animal models [102]. Small-molecule RITA was reported to bind to p53, which disrupts the binding of p53 with its negative regulators, including MDM2. It has been shown that RITA can activate p53 signaling and suppress tumor growth in vivo [103]. CP-31398 was reported to be another small molecule that stabilizes the wild-type p53 and enhances its transcriptional activity in cells [104]. However, a recent study reported that CP-31398 causes toxicity in liver and other tissues in animal models, suggesting the necessity to modify the structure of CP-31398 to reduce its toxicity [105].
Small molecules that inactivate or destabilize mutant p53
Mutant p53 is frequently accumulated to high levels in tumors and displays gain-of-function oncogenic activities. Therefore, inactivation or destabilization of mutant p53 is being developed as an important strategy for cancer therapy. Recent studies showed that HDAC6/Hsp90 signaling plays an important role in stabilizing mutant p53 in tumor cells. Inhibition of HDAC6 or Hsp90 has been shown to destabilize mutant p53 in cancer cells and decrease tumorigenicity of cancer cells. In addition, SAHA, a HDAC inhibitor, was shown to destabilize mutant p53 in tumor cells [106,107]. A small-molecule RETRA has been reported to inhibit the mutant p53–p73 interaction, and thereby releases p73 and restores p73's function in transcriptional activation [108]. RETRA was shown to transactivate p53 target genes in mutant p53-bearing tumors and prevent the growth of xenograft tumors in mice [108].
The reactivation of wild-type function in mutant p53
Recent studies have also led to the identification of a class of small molecules that converts mutant p53 proteins into forms that exhibit wild-type p53 functions, thereby allowing p53 to induce cell cycle arrest or apoptosis in cancer cells. PRIMA-1 is such a molecule that can restore sequence-specific DNA binding and convert mutant p53 conformation to wild-type, thereby leading to the transactivation of p53 target genes. PRIMA-1 was reported to sensitize cancer cells to chemotherapy and inhibit tumor growth in vivo [109]. The PRIMA-1 analog APR-246 is being tested in phase I clinical trials in liver or prostate cancer patients [110]. In addition to its function to stabilize and activate wild-type p53, CP-31398 can also restore DNA-binding activity and therefore the wild-type p53 function to mutant p53 to inhibit tumor cell growth in culture and tumor growth in animal models [111]. Recently, an allele-specific p53-reactivating compound NSC-319725 was identified, which can restore the wild-type structure and function to the R175H mutant p53. This compound induces extensive apoptosis in R172H mutant p53 knock-in mice and inhibits the growth of xenograft tumors containing R175H mutant p53 in mice [112].
Conclusion
p53 has been one of the most extensively studied proteins since its discovery. In this review, we focused on the functions of wild-type p53 and its gain-of-function mutants in cancer. In addition to the well-known functions of p53 in inducing cell cycle arrest, apoptosis, and senescence, recent studies have revealed additional novel functions of p53 in tumor suppression, including the regulation of metabolism and anti-oxidant defense. However, it still remains largely unclear how p53 selectively and/or coordinately regulates these functions to exert its role in tumor prevention and tumor therapy in different types of tissues and cells. Although the hypothesis of gain-of-function of mutant p53 has existed almost since the beginning of p53 research, only in recent years, tremendous efforts have been made to demonstrate that mutant p53 promotes tumorigenesis through regulating many different aspects of oncogenetic processes. However, the underlying mechanisms for mutant p53 gain-of-function are not fully understood. Furthermore, it is unclear how different forms of mutant p53 impact upon tumorigenesis. Further understanding the mechanisms of p53 in tumor suppression and mechanisms of gain-of-function of mutant p53 in tumor development will provide novel targets and approaches for cancer therapy. As our understanding of p53 continues to grow, future studies on p53 will lead to the development of cancer-specific p53-based therapy and novel chemicals to significantly improve cancer therapy.
Funding
This work was supported by the grants from NIH (R01CA143204) the New Jersey Commission on Cancer Research, and Rutgers Cancer Institute of New Jersey Foundation.
References
- 1.Vogelstein B, Lane D, Levine AJ. Surfing the p53 network. Nature. 2000;408:307–310. doi: 10.1038/35042675. [DOI] [PubMed] [Google Scholar]
- 2.Feng Z, Levine AJ. The regulation of energy metabolism and the IGF-1/mTOR pathways by the p53 protein. Trends Cell Biol. 2010;20:427–434. doi: 10.1016/j.tcb.2010.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Vousden KH, Prives C. Blinded by the light: the growing complexity of p53. Cell. 2009;137:413–431. doi: 10.1016/j.cell.2009.04.037. [DOI] [PubMed] [Google Scholar]
- 4.Hollstein M, Sidransky D, Vogelstein B, Harris CC. p53 mutations in human cancers. Science. 1991;253:49–53. doi: 10.1126/science.1905840. [DOI] [PubMed] [Google Scholar]
- 5.Olivier M, Hussain SP, Caron de Fromentel C, Hainaut P, Harris CC. TP53 mutation spectra and load: a tool for generating hypotheses on the etiology of cancer. IARC Sci Publ. 2004;157:247–270. [PubMed] [Google Scholar]
- 6.Tommasino M, Accardi R, Caldeira S, Dong W, Malanchi I, Smet A, Zehbe I. The role of TP53 in Cervical carcinogenesis. Hum Mutat. 2003;21:307–312. doi: 10.1002/humu.10178. [DOI] [PubMed] [Google Scholar]
- 7.Scheffner M, Werness BA, Huibregtse JM, Levine AJ, Howley PM. The E6 oncoprotein encoded by human papillomavirus types 16 and 18 promotes the degradation of p53. Cell. 1990;63:1129–1136. doi: 10.1016/0092-8674(90)90409-8. [DOI] [PubMed] [Google Scholar]
- 8.Wade M, Li YC, Wahl GM. MDM2, MDMX and p53 in oncogenesis and cancer therapy. Nat Rev Cancer. 2013;13:83–96. doi: 10.1038/nrc3430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Strong LC. General keynote: hereditary cancer: lessons from Li-Fraumeni syndrome. Gynecol Oncol. 2003;88:S4–S7. doi: 10.1006/gyno.2002.6673. discussion S11–13. [DOI] [PubMed] [Google Scholar]
- 10.Donehower LA, Harvey M, Slagle BL, McArthur MJ, Montgomery CA, Jr, Butel JS, Bradley A. Mice deficient for p53 are developmentally normal but susceptible to spontaneous tumours. Nature. 1992;356:215–221. doi: 10.1038/356215a0. [DOI] [PubMed] [Google Scholar]
- 11.Muller PA, Vousden KH. p53 mutations in cancer. Nat Cell Biol. 2013;15:2–8. doi: 10.1038/ncb2641. [DOI] [PubMed] [Google Scholar]
- 12.Freed-Pastor WA, Prives C. Mutant p53: one name, many proteins. Genes Dev. 2012;26:1268–1286. doi: 10.1101/gad.190678.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Harris SL, Levine AJ. The p53 pathway: positive and negative feedback loops. Oncogene. 2005;24:2899–2908. doi: 10.1038/sj.onc.1208615. [DOI] [PubMed] [Google Scholar]
- 14.Brooks CL, Gu W. p53 ubiquitination: Mdm2 and beyond. Mol Cell. 2006;21:307–315. doi: 10.1016/j.molcel.2006.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.el-Deiry WS, Kern SE, Pietenpol JA, Kinzler KW, Vogelstein B. Definition of a consensus binding site for p53. Nat Genet. 1992;1:45–49. doi: 10.1038/ng0492-45. [DOI] [PubMed] [Google Scholar]
- 16.Clarke AR, Purdie CA, Harrison DJ, Morris RG, Bird CC, Hooper ML, Wyllie AH. Thymocyte apoptosis induced by p53-dependent and independent pathways. Nature. 1993;362:849–852. doi: 10.1038/362849a0. [DOI] [PubMed] [Google Scholar]
- 17.Lowe SW, Schmitt EM, Smith SW, Osborne BA, Jacks T. p53 is required for radiation-induced apoptosis in mouse thymocytes. Nature. 1993;362:847–849. doi: 10.1038/362847a0. [DOI] [PubMed] [Google Scholar]
- 18.Riley T, Sontag E, Chen P, Levine A. Transcriptional control of human p53-regulated genes. Nat Rev Mol Cell Biol. 2008;9:402–412. doi: 10.1038/nrm2395. [DOI] [PubMed] [Google Scholar]
- 19.Jeffers JR, Parganas E, Lee Y, Yang C, Wang J, Brennan J, MacLean KH, et al. Puma is an essential mediator of p53-dependent and -independent apoptotic pathways. Cancer Cell. 2003;4:321–328. doi: 10.1016/s1535-6108(03)00244-7. [DOI] [PubMed] [Google Scholar]
- 20.Green DR, Kroemer G. Cytoplasmic functions of the tumour suppressor p53. Nature. 2009;458:1127–1130. doi: 10.1038/nature07986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vaseva AV, Moll UM. The mitochondrial p53 pathway. Biochim Biophys Acta. 2009;1787:414–420. doi: 10.1016/j.bbabio.2008.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Brugarolas J, Chandrasekaran C, Gordon JI, Beach D, Jacks T, Hannon GJ. Radiation-induced cell cycle arrest compromised by p21 deficiency. Nature. 1995;377:552–557. doi: 10.1038/377552a0. [DOI] [PubMed] [Google Scholar]
- 23.Deng C, Zhang P, Harper JW, Elledge SJ, Leder P. Mice lacking p21CIP1/WAF1 undergo normal development, but are defective in G1 checkpoint control. Cell. 1995;82:675–684. doi: 10.1016/0092-8674(95)90039-x. [DOI] [PubMed] [Google Scholar]
- 24.Hermeking H, Lengauer C, Polyak K, He TC, Zhang L, Thiagalingam S, Kinzler KW, et al. 14–3–3 sigma is a p53-regulated inhibitor of G2/M progression. Mol Cell. 1997;1:3–11. doi: 10.1016/s1097-2765(00)80002-7. [DOI] [PubMed] [Google Scholar]
- 25.Kastan MB, Zhan Q, el-Deiry WS, Carrier F, Jacks T, Walsh WV, Plunkett BS, et al. A mammalian cell cycle checkpoint pathway utilizing p53 and GADD45 is defective in ataxia-telangiectasia. Cell. 1992;71:587–597. doi: 10.1016/0092-8674(92)90593-2. [DOI] [PubMed] [Google Scholar]
- 26.Xue W, Zender L, Miething C, Dickins RA, Hernando E, Krizhanovsky V, Cordon-Cardo C, et al. Senescence and tumour clearance is triggered by p53 restoration in murine liver carcinomas. Nature. 2007;445:656–660. doi: 10.1038/nature05529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kortlever RM, Higgins PJ, Bernards R. Plasminogen activator inhibitor-1 is a critical downstream target of p53 in the induction of replicative senescence. Nat Cell Biol. 2006;8:877–884. doi: 10.1038/ncb1448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Levine AJ, Oren M. The first 30 years of p53: growing ever more complex. Nat Rev Cancer. 2009;9:749–758. doi: 10.1038/nrc2723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vousden KH, Ryan KM. p53 and metabolism. Nat Rev Cancer. 2009;9:691–700. doi: 10.1038/nrc2715. [DOI] [PubMed] [Google Scholar]
- 30.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 31.Matoba S, Kang JG, Patino WD, Wragg A, Boehm M, Gavrilova O, Hurley PJ, et al. p53 regulates mitochondrial respiration. Science. 2006;312:1650–1653. doi: 10.1126/science.1126863. [DOI] [PubMed] [Google Scholar]
- 32.Stambolsky P, Weisz L, Shats I, Klein Y, Goldfinger N, Oren M, Rotter V. Regulation of AIF expression by p53. Cell Death Differ. 2006;13:2140–2149. doi: 10.1038/sj.cdd.4401965. [DOI] [PubMed] [Google Scholar]
- 33.Bourdon A, Minai L, Serre V, Jais JP, Sarzi E, Aubert S, Chretien D, et al. Mutation of RRM2B, encoding p53-controlled ribonucleotide reductase (p53R2), causes severe mitochondrial DNA depletion. Nat Genet. 2007;39:776–780. doi: 10.1038/ng2040. [DOI] [PubMed] [Google Scholar]
- 34.Hu W, Zhang C, Wu R, Sun Y, Levine A, Feng Z. Glutaminase 2, a novel p53 target gene regulating energy metabolism and antioxidant function. Proc Natl Acad Sci USA. 2010;107:7455–7460. doi: 10.1073/pnas.1001006107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schwartzenberg-Bar-Yoseph F, Armoni M, Karnieli E. The tumor suppressor p53 down-regulates glucose transporters GLUT1 and GLUT4 gene expression. Cancer Res. 2004;64:2627–2633. doi: 10.1158/0008-5472.can-03-0846. [DOI] [PubMed] [Google Scholar]
- 36.Kawauchi K, Araki K, Tobiume K, Tanaka N. p53 regulates glucose metabolism through an IKK-NF-kappaB pathway and inhibits cell transformation. Nat Cell Biol. 2008;10:611–618. doi: 10.1038/ncb1724. [DOI] [PubMed] [Google Scholar]
- 37.Bensaad K, Tsuruta A, Selak MA, Vidal MN, Nakano K, Bartrons R, Gottlieb E, et al. TIGAR, a p53-inducible regulator of glycolysis and apoptosis. Cell. 2006;126:107–120. doi: 10.1016/j.cell.2006.05.036. [DOI] [PubMed] [Google Scholar]
- 38.Zhang C, Lin M, Wu R, Wang X, Yang B, Levine AJ, Hu W, et al. Parkin, a p53 target gene, mediates the role of p53 in glucose metabolism and the Warburg effect. Proc Natl Acad Sci USA. 2011;108:16259–16264. doi: 10.1073/pnas.1113884108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jiang P, Du W, Wang X, Mancuso A, Gao X, Wu M, Yang X. p53 regulates biosynthesis through direct inactivation of glucose-6-phosphate dehydrogenase. Nat Cell Biol. 2011;13:310–316. doi: 10.1038/ncb2172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Budanov AV, Sablina AA, Feinstein E, Koonin EV, Chumakov PM. Regeneration of peroxiredoxins by p53-regulated sestrins, homologs of bacterial AhpD. Science. 2004;304:596–600. doi: 10.1126/science.1095569. [DOI] [PubMed] [Google Scholar]
- 41.Sablina AA, Budanov AV, Ilyinskaya GV, Agapova LS, Kravchenko JE, Chumakov PM. The antioxidant function of the p53 tumor suppressor. Nat Med. 2005;11:1306–1313. doi: 10.1038/nm1320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Liang Y, Liu J, Feng Z. The regulation of cellular metabolism by tumor suppressor p53. Cell Biosci. 2013;3:9. doi: 10.1186/2045-3701-3-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bensaad K, Vousden KH. p53: new roles in metabolism. Trends Cell Biol. 2007;17:286–291. doi: 10.1016/j.tcb.2007.04.004. [DOI] [PubMed] [Google Scholar]
- 44.Kimmelman AC. The dynamic nature of autophagy in cancer. Genes Dev. 2011;25:1999–2010. doi: 10.1101/gad.17558811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Feng Z, Zhang H, Levine AJ, Jin S. The coordinate regulation of the p53 and mTOR pathways in cells. Proc Natl Acad Sci USA. 2005;102:8204–8209. doi: 10.1073/pnas.0502857102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Crighton D, Wilkinson S, O'Prey J, Syed N, Smith P, Harrison PR, Gasco M, et al. DRAM, a p53-induced modulator of autophagy, is critical for apoptosis. Cell. 2006;126:121–134. doi: 10.1016/j.cell.2006.05.034. [DOI] [PubMed] [Google Scholar]
- 47.Maiuri MC, Galluzzi L, Morselli E, Kepp O, Malik SA, Kroemer G. Autophagy regulation by p53. Curr Opin Cell Biol. 2010;22:181–185. doi: 10.1016/j.ceb.2009.12.001. [DOI] [PubMed] [Google Scholar]
- 48.Zhao YG, Zhao H, Miao L, Wang L, Sun F, Zhang H. The p53-induced gene Ei24 is an essential component of the basal autophagy pathway. J Biol Chem. 2012;287:42053–42063. doi: 10.1074/jbc.M112.415968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tasdemir E, Maiuri MC, Galluzzi L, Vitale I, Djavaheri-Mergny M, D'Amelio M, Criollo A, et al. Regulation of autophagy by cytoplasmic p53. Nat Cell Biol. 2008;10:676–687. doi: 10.1038/ncb1730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Morselli E, Tasdemir E, Maiuri MC, Galluzzi L, Kepp O, Criollo A, Vicencio JM, et al. Mutant p53 protein localized in the cytoplasm inhibits autophagy. Cell Cycle. 2008;7:3056–3061. doi: 10.4161/cc.7.19.6751. [DOI] [PubMed] [Google Scholar]
- 51.Feng Z, Zhang C, Wu R, Hu W. Tumor suppressor p53 meets microRNAs. J Mol Cell Biol. 2011;3:44–50. doi: 10.1093/jmcb/mjq040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hermeking H. MicroRNAs in the p53 network: micromanagement of tumour suppression. Nat Rev Cancer. 2012;12:613–626. doi: 10.1038/nrc3318. [DOI] [PubMed] [Google Scholar]
- 53.Chang TC, Wentzel EA, Kent OA, Ramachandran K, Mullendore M, Lee KH, Feldmann G, et al. Transactivation of miR-34a by p53 broadly influences gene expression and promotes apoptosis. Mol Cell. 2007;26:745–752. doi: 10.1016/j.molcel.2007.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.He L, He X, Lim LP, de Stanchina E, Xuan Z, Liang Y, Xue W, et al. A microRNA component of the p53 tumour suppressor network. Nature. 2007;447:1130–1134. doi: 10.1038/nature05939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Raver-Shapira N, Marciano E, Meiri E, Spector Y, Rosenfeld N, Moskovits N, Bentwich Z, et al. Transcriptional activation of miR-34a contributes to p53-mediated apoptosis. Mol Cell. 2007;26:731–743. doi: 10.1016/j.molcel.2007.05.017. [DOI] [PubMed] [Google Scholar]
- 56.Suzuki HI, Yamagata K, Sugimoto K, Iwamoto T, Kato S, Miyazono K. Modulation of microRNA processing by p53. Nature. 2009;460:529–533. doi: 10.1038/nature08199. [DOI] [PubMed] [Google Scholar]
- 57.Leveille N, Elkon R, Davalos V, Manoharan V, Hollingworth D, Oude Vrielink J, le Sage C, et al. Selective inhibition of microRNA accessibility by RBM38 is required for p53 activity. Nat Commun. 2011;2:513. doi: 10.1038/ncomms1519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Schmitt CA, Fridman JS, Yang M, Baranov E, Hoffman RM, Lowe SW. Dissecting p53 tumor suppressor functions in vivo. Cancer Cell. 2002;1:289–298. doi: 10.1016/s1535-6108(02)00047-8. [DOI] [PubMed] [Google Scholar]
- 59.Hemann MT, Zilfou JT, Zhao Z, Burgess DJ, Hannon GJ, Lowe SW. Suppression of tumorigenesis by the p53 target PUMA. Proc Natl Acad Sci USA. 2004;101:9333–9338. doi: 10.1073/pnas.0403286101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Michalak EM, Villunger A, Adams JM, Strasser A. In several cell types tumour suppressor p53 induces apoptosis largely via Puma but Noxa can contribute. Cell Death Differ. 2008;15:1019–1029. doi: 10.1038/cdd.2008.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Cosme-Blanco W, Shen MF, Lazar AJ, Pathak S, Lozano G, Multani AS, Chang S. Telomere dysfunction suppresses spontaneous tumorigenesis in vivo by initiating p53-dependent cellular senescence. EMBO Rep. 2007;8:497–503. doi: 10.1038/sj.embor.7400937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Liu G, Parant JM, Lang G, Chau P, Chavez-Reyes A, El-Naggar AK, Multani A, et al. Chromosome stability, in the absence of apoptosis, is critical for suppression of tumorigenesis in Trp53 mutant mice. Nat Genet. 2004;36:63–68. doi: 10.1038/ng1282. [DOI] [PubMed] [Google Scholar]
- 63.Van Nguyen T, Puebla-Osorio N, Pang H, Dujka ME, Zhu C. DNA damage-induced cellular senescence is sufficient to suppress tumorigenesis: a mouse model. J Exp Med. 2007;204:1453–1461. doi: 10.1084/jem.20062453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Brady CA, Jiang D, Mello SS, Johnson TM, Jarvis LA, Kozak MM, Kenzelmann Broz D, et al. Distinct p53 transcriptional programs dictate acute DNA-damage responses and tumor suppression. Cell. 2011;145:571–583. doi: 10.1016/j.cell.2011.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Li T, Kon N, Jiang L, Tan M, Ludwig T, Zhao Y, Baer R, et al. Tumor suppression in the absence of p53-mediated cell-cycle arrest, apoptosis, and senescence. Cell. 2012;149:1269–1283. doi: 10.1016/j.cell.2012.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Tanaka T, Ohkubo S, Tatsuno I, Prives C. hCAS/CSE1L associates with chromatin and regulates expression of select p53 target genes. Cell. 2007;130:638–650. doi: 10.1016/j.cell.2007.08.001. [DOI] [PubMed] [Google Scholar]
- 67.Samuels-Lev Y, O'Connor DJ, Bergamaschi D, Trigiante G, Hsieh JK, Zhong S, Campargue I, et al. ASPP proteins specifically stimulate the apoptotic function of p53. Mol Cell. 2001;8:781–794. doi: 10.1016/s1097-2765(01)00367-7. [DOI] [PubMed] [Google Scholar]
- 68.Das S, Raj L, Zhao B, Kimura Y, Bernstein A, Aaronson SA, Lee SW. Hzf determines cell survival upon genotoxic stress by modulating p53 transactivation. Cell. 2007;130:624–637. doi: 10.1016/j.cell.2007.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wu WS, Heinrichs S, Xu D, Garrison SP, Zambetti GP, Adams JM, Look AT. Slug antagonizes p53-mediated apoptosis of hematopoietic progenitors by repressing puma. Cell. 2005;123:641–653. doi: 10.1016/j.cell.2005.09.029. [DOI] [PubMed] [Google Scholar]
- 70.Huarte M, Guttman M, Feldser D, Garber M, Koziol MJ, Kenzelmann-Broz D, Khalil AM, et al. A large intergenic noncoding RNA induced by p53 mediates global gene repression in the p53 response. Cell. 2010;142:409–419. doi: 10.1016/j.cell.2010.06.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Harris CC, Hollstein M. Clinical implications of the p53 tumor-suppressor gene. N Eng J Med. 1993;329:1318–1327. doi: 10.1056/NEJM199310283291807. [DOI] [PubMed] [Google Scholar]
- 72.Wolf D, Harris N, Rotter V. Reconstitution of p53 expression in a nonproducer Ab-MuLV-transformed cell line by transfection of a functional p53 gene. Cell. 1984;38:119–126. doi: 10.1016/0092-8674(84)90532-4. [DOI] [PubMed] [Google Scholar]
- 73.Dittmer D, Pati S, Zambetti G, Chu S, Teresky AK, Moore M, Finlay C, et al. Gain of function mutations in p53. Nat Genet. 1993;4:42–46. doi: 10.1038/ng0593-42. [DOI] [PubMed] [Google Scholar]
- 74.Brosh R, Rotter V. When mutants gain new powers: news from the mutant p53 field. Nat Rev Cancer. 2009;9:701–713. doi: 10.1038/nrc2693. [DOI] [PubMed] [Google Scholar]
- 75.Zhang C, Liu J, Liang Y, Wu R, Zhao Y, Hong X, Lin M, et al. Tumor-associated mutant p53 drives the Warburg effect. Nat Commun. 2013;4:2935. doi: 10.1038/ncomms3935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Lang GA, Iwakuma T, Suh YA, Liu G, Rao VA, Parant JM, Valentin-Vega YA, et al. Gain of function of a p53 hot spot mutation in a mouse model of Li-Fraumeni syndrome. Cell. 2004;119:861–872. doi: 10.1016/j.cell.2004.11.006. [DOI] [PubMed] [Google Scholar]
- 77.Olive KP, Tuveson DA, Ruhe ZC, Yin B, Willis NA, Bronson RT, Crowley D, et al. Mutant p53 gain of function in two mouse models of Li-Fraumeni syndrome. Cell. 2004;119:847–860. doi: 10.1016/j.cell.2004.11.004. [DOI] [PubMed] [Google Scholar]
- 78.Levrero M, De Laurenzi V, Costanzo A, Gong J, Wang JY, Melino G. The p53/p63/p73 family of transcription factors: overlapping and distinct functions. J Cell Sci. 2000;113:1661–1670. doi: 10.1242/jcs.113.10.1661. (Pt 10) [DOI] [PubMed] [Google Scholar]
- 79.Li Y, Prives C. Are interactions with p63 and p73 involved in mutant p53 gain of oncogenic function? Oncogene. 2007;26:2220–2225. doi: 10.1038/sj.onc.1210311. [DOI] [PubMed] [Google Scholar]
- 80.Gaiddon C, Lokshin M, Ahn J, Zhang T, Prives C. A subset of tumor-derived mutant forms of p53 down-regulate p63 and p73 through a direct interaction with the p53 core domain. Mol Cell Biol. 2001;21:1874–1887. doi: 10.1128/MCB.21.5.1874-1887.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Di Agostino S, Strano S, Emiliozzi V, Zerbini V, Mottolese M, Sacchi A, Blandino G, et al. Gain of function of mutant p53: the mutant p53/NF-Y protein complex reveals an aberrant transcriptional mechanism of cell cycle regulation. Cancer Cell. 2006;10:191–202. doi: 10.1016/j.ccr.2006.08.013. [DOI] [PubMed] [Google Scholar]
- 82.Stambolsky P, Tabach Y, Fontemaggi G, Weisz L, Maor-Aloni R, Siegfried Z, Shiff I, et al. Modulation of the vitamin D3 response by cancer-associated mutant p53. Cancer Cell. 2010;17:273–285. doi: 10.1016/j.ccr.2009.11.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Chicas A, Molina P, Bargonetti J. Mutant p53 forms a complex with Sp1 on HIV-LTR DNA. Biochem Biophys Res Commun. 2000;279:383–390. doi: 10.1006/bbrc.2000.3965. [DOI] [PubMed] [Google Scholar]
- 84.Freed-Pastor WA, Mizuno H, Zhao X, Langerod A, Moon SH, Rodriguez-Barrueco R, Barsotti A, et al. Mutant p53 disrupts mammary tissue architecture via the mevalonate pathway. Cell. 2012;148:244–258. doi: 10.1016/j.cell.2011.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Song H, Hollstein M, Xu Y. p53 gain-of-function cancer mutants induce genetic instability by inactivating ATM. Nat Cell Biol. 2007;9:573–580. doi: 10.1038/ncb1571. [DOI] [PubMed] [Google Scholar]
- 86.Haupt S, di Agostino S, Mizrahi I, Alsheich-Bartok O, Voorhoeve M, Damalas A, Blandino G, et al. Promyelocytic leukemia protein is required for gain of function by mutant p53. Cancer Res. 2009;69:4818–4826. doi: 10.1158/0008-5472.CAN-08-4010. [DOI] [PubMed] [Google Scholar]
- 87.Restle A, Farber M, Baumann C, Bohringer M, Scheidtmann KH, Muller-Tidow C, Wiesmuller L. Dissecting the role of p53 phosphorylation in homologous recombination provides new clues for gain-of-function mutants. Nucleic Acids Res. 2008;36:5362–5375. doi: 10.1093/nar/gkn503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Girardini JE, Napoli M, Piazza S, Rustighi A, Marotta C, Radaelli E, Capaci V, et al. A Pin1/mutant p53 axis promotes aggressiveness in breast cancer. Cancer Cell. 2011;20:79–91. doi: 10.1016/j.ccr.2011.06.004. [DOI] [PubMed] [Google Scholar]
- 89.Muller BF, Paulsen D, Deppert W. Specific binding of MAR/SAR DNA-elements by mutant p53. Oncogene. 1996;12:1941–1952. [PubMed] [Google Scholar]
- 90.Will K, Warnecke G, Wiesmuller L, Deppert W. Specific interaction of mutant p53 with regions of matrix attachment region DNA elements (MARs) with a high potential for base-unpairing. Proc Natl Acad Sci USA. 1998;95:13681–13686. doi: 10.1073/pnas.95.23.13681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Gohler T, Jager S, Warnecke G, Yasuda H, Kim E, Deppert W. Mutant p53 proteins bind DNA in a DNA structure-selective mode. Nucleic Acids Res. 2005;33:1087–1100. doi: 10.1093/nar/gki252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Dong P, Karaayvaz M, Jia N, Kaneuchi M, Hamada J, Watari H, Sudo S, et al. Mutant p53 gain-of-function induces epithelial-mesenchymal transition through modulation of the miR-130b-ZEB1 axis. Oncogene. 2013;32:3286–3295. doi: 10.1038/onc.2012.334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Neilsen PM, Noll JE, Mattiske S, Bracken CP, Gregory PA, Schulz RB, Lim SP, et al. Mutant p53 drives invasion in breast tumors through up-regulation of miR-155. Oncogene. 2013;32:2992–3000. doi: 10.1038/onc.2012.305. [DOI] [PubMed] [Google Scholar]
- 94.Wang W, Cheng B, Miao L, Mei Y, Wu M. Mutant p53-R273H gains new function in sustained activation of EGFR signaling via suppressing miR-27a expression. Cell Death Dis. 2013;4:e574. doi: 10.1038/cddis.2013.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Donzelli S, Fontemaggi G, Fazi F, Di Agostino S, Padula F, Biagioni F, Muti P, et al. MicroRNA-128–2 targets the transcriptional repressor E2F5 enhancing mutant p53 gain of function. Cell Death Differ. 2012;19:1038–1048. doi: 10.1038/cdd.2011.190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Su X, Chakravarti D, Cho MS, Liu L, Gi YJ, Lin YL, Leung ML, et al. TAp63 suppresses metastasis through coordinate regulation of Dicer and miRNAs. Nature. 2010;467:986–990. doi: 10.1038/nature09459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Ventura A, Kirsch DG, McLaughlin ME, Tuveson DA, Grimm J, Lintault L, Newman J, et al. Restoration of p53 function leads to tumour regression in vivo. Nature. 2007;445:661–665. doi: 10.1038/nature05541. [DOI] [PubMed] [Google Scholar]
- 98.Wang Y, Suh YA, Fuller MY, Jackson JG, Xiong S, Terzian T, Quintas-Cardama A, et al. Restoring expression of wild-type p53 suppresses tumor growth but does not cause tumor regression in mice with a p53 missense mutation. J Clin Invest. 2011;121:893–904. doi: 10.1172/JCI44504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Roth JA, Nguyen D, Lawrence DD, Kemp BL, Carrasco CH, Ferson DZ, Hong WK, et al. Retrovirus-mediated wild-type p53 gene transfer to tumors of patients with lung cancer. Nat Med. 1996;2:985–991. doi: 10.1038/nm0996-985. [DOI] [PubMed] [Google Scholar]
- 100.Tazawa H, Kagawa S, Fujiwara T. Advances in adenovirus-mediated p53 cancer gene therapy. Expert Opin Biol Ther. 2013;13:1569–1583. doi: 10.1517/14712598.2013.845662. [DOI] [PubMed] [Google Scholar]
- 101.Vassilev LT, Vu BT, Graves B, Carvajal D, Podlaski F, Filipovic Z, Kong N, et al. In vivo activation of the p53 pathway by small-molecule antagonists of MDM2. Science. 2004;303:844–848. doi: 10.1126/science.1092472. [DOI] [PubMed] [Google Scholar]
- 102.Azmi AS, Philip PA, Beck FW, Wang Z, Banerjee S, Wang S, Yang D, et al. MI-219-zinc combination: a new paradigm in MDM2 inhibitor-based therapy. Oncogene. 2011;30:117–126. doi: 10.1038/onc.2010.403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Issaeva N, Bozko P, Enge M, Protopopova M, Verhoef LG, Masucci M, Pramanik A, et al. Small molecule RITA binds to p53, blocks p53-HDM-2 interaction and activates p53 function in tumors. Nat Med. 2004;10:1321–1328. doi: 10.1038/nm1146. [DOI] [PubMed] [Google Scholar]
- 104.Wang W, Takimoto R, Rastinejad F, El-Deiry WS. Stabilization of p53 by CP-31398 inhibits ubiquitination without altering phosphorylation at serine 15 or 20 or MDM2 binding. Mol Cell Biol. 2003;23:2171–2181. doi: 10.1128/MCB.23.6.2171-2181.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Johnson WD, Muzzio M, Detrisac CJ, Kapetanovic IM, Kopelovich L, McCormick DL. Subchronic oral toxicity and metabolite profiling of the p53 stabilizing agent, CP-31398, in rats and dogs. Toxicology. 2011;289:141–150. doi: 10.1016/j.tox.2011.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Li D, Marchenko ND, Schulz R, Fischer V, Velasco-Hernandez T, Talos F, Moll UM. Functional inactivation of endogenous MDM2 and CHIP by HSP90 causes aberrant stabilization of mutant p53 in human cancer cells. Mol Cancer Res. 2011;9:577–588. doi: 10.1158/1541-7786.MCR-10-0534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Li D, Marchenko ND, Moll UM. SAHA shows preferential cytotoxicity in mutant p53 cancer cells by destabilizing mutant p53 through inhibition of the HDAC6-Hsp90 chaperone axis. Cell Death Differ. 2011;18:1904–1913. doi: 10.1038/cdd.2011.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Kravchenko JE, Ilyinskaya GV, Komarov PG, Agapova LS, Kochetkov DV, Strom E, Frolova EI, et al. Small-molecule RETRA suppresses mutant p53-bearing cancer cells through a p73-dependent salvage pathway. Proc Natl Acad Sci USA. 2008;105:6302–6307. doi: 10.1073/pnas.0802091105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Bykov VJ, Issaeva N, Shilov A, Hultcrantz M, Pugacheva E, Chumakov P, Bergman J, et al. Restoration of the tumor suppressor function to mutant p53 by a low-molecular-weight compound. Nat Med. 2002;8:282–288. doi: 10.1038/nm0302-282. [DOI] [PubMed] [Google Scholar]
- 110.Farnebo M, Bykov VJ, Wiman KG. The p53 tumor suppressor: a master regulator of diverse cellular processes and therapeutic target in cancer. Biochem Biophys Res Commun. 2010;396:85–89. doi: 10.1016/j.bbrc.2010.02.152. [DOI] [PubMed] [Google Scholar]
- 111.Tang X, Zhu Y, Han L, Kim AL, Kopelovich L, Bickers DR, Athar M. CP-31398 restores mutant p53 tumor suppressor function and inhibits UVB-induced skin carcinogenesis in mice. J Clin Invest. 2007;117:3753–3764. doi: 10.1172/JCI32481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Yu X, Vazquez A, Levine AJ, Carpizo DR. Allele-specific p53 mutant reactivation. Cancer Cell. 2012;21:614–625. doi: 10.1016/j.ccr.2012.03.042. [DOI] [PMC free article] [PubMed] [Google Scholar]