Abstract
DNA methylation is an important epigenetic mechanism that ensures correct gene expression and maintains genetic stability. DNA methyltransferase 1 (DNMT1) is the primary enzyme that maintains DNA methylation during replication. Dysregulation of DNMT1 is implicated in a variety of diseases. DNMT1 protein stability is regulated via various post-translational modifications, such as acetylation and ubiquitination, but also through protein–protein interactions. These mechanisms ensure DNMT1 is properly activated during the correct time of the cell cycle and at correct genomic loci, as well as in response to appropriate extracellular cues. Further understanding of these regulatory mechanisms may help to design novel therapeutic approaches for human diseases.
Keywords: DNA (cytosine-5-)-methyltransferase, epi-genetics, protein stability, post-translational modification, neoplasms
Introduction
Epigenetic changes—inheritance of genetic traits that cannot be explained by changes in the DNA sequence—rely heavily on DNA methylation to propagate responses to environmental and/or developmental cues [1]. Enzymes that catalyze this modification are part of the DNA methyltransferase family. Among them, DNA (cytosine-5-)-methyltransferase 3 alpha and DNA (cytosine-5-)-methyltransferase 3 beta are thought to be de novo DNA methyltransferases, which methylate genes to regulate them in a developmental context. Conversely, DNA (cytosine-5-)-methyltransferase 1 (DNMT1) specifically methylates DNA during replication to maintain correct methylation patterns on the new DNA strand [2].
DNA methylation via DNMT1 ensures correct formation of heterochromatin [3,4] and promoter repression via histones [5]. Interactions between histone deacetylases and DNMT1 further underscore the relationship between chromatin and DNMT1 [6], but also belie a mechanism for DNMT1 regulation [7].
Correct regulation of DNMT1 and its relationship with histone modifications becomes especially important in the case of ‘bivalent chromatin’—those promoters with both activating and repressive histone marks. The latter modifications could inadvertently recruit DNMT1 to aberrantly methylate promoters, permanently turning off a gene that should otherwise be temporarily silenced [8].
Dysregulation of DNMT1 activity causes human diseases, such as cancer [9] and various genetic disorders [10,11]. DNMT1 mutations are found in patients with hereditary sensory neuropathy [12,13] and in human cancers [14]. Cancer epigenetic landscapes are generally defined by global DNA hypomethylation with localized promoter hypermethylation at tumor suppressors [8]. These tumor suppressors include Cadherin 1, Type 1, E-Cadherin [15], adenomatous polyposis coli [16], Ras association domain family member 1 [17], p16 and TIMP metallopeptidase inhibitor 3 [18] among many others. As a corollary, both overexpression and developmental disruption of DNMT1 will lead to tumorigenesis in both experimental models and human cancer studies [19–24].
As such, given the complications of bivalent chromatin and the ramifications of incorrect DNMT1 protein levels, regulation of DNMT1 becomes especially vital and occurs through a variety of pathways [9,25] (summarized in Table 1). Although DNMT1 is also regulated at the transcriptional level [34–37], this review will focus on emerging mechanisms of post-translational regulation of DNMT1 protein stability.
Table 1.
List of modifications to DNMT1, the enzymes that catalyze them, and the resultant effects on DNMT1 stability
Regulation of DNMT1 Protein Stability by Acetylation and Ubiquitination
DNMT1 protein abundance is tightly regulated during the cell cycle: it peaks at the early S phase, decreases after the S phase, and reaches lowest level at the G1 phase [26]. The primary function of DNMT1 is to methylate newly synthesized DNA during the S phase [1]. A crucial component of the replication machinery, proliferating cell nuclear antigen (PCNA), interacts with DNMT1 to ensure its correct positioning at the replication fork and at the appropriate time during replication [38,39]. The chromatin-associated ubiquitinase ubiquitin-like with PHD and ring finger domains 1 (UHRF1) [40,41] binds to hemimethylated loci that are formed during DNA replication, helping DNMT1 co-localize with the replication fork [42] as well as chromatin [43]. A recent study using an Xenopus interphase egg extract system suggests an alternative mechanism that DNMT1 is recruited to the replication forks through binding to ubiquitinated histone H3, whereas the ubiquitination of histone is catalyzed by UHRF1 [44].
In addition to localizing DNMT1 correctly, UHRF1 also causes its degradation, as it ubiquitinates acetylated DNMT1. The acetyltransferase Tip60 protein levels are elevated in the late S phase, which, in turn, leads to DNMT1 acetylation. Acetylation of DNMT1 enhances its binding affinity with UHRF1 and thus triggers DNMT1 ubiquitination and degradation (Fig. 1) [26]. Conversely, DNMT1 is deacetylated by histone deacetylase 1 (HDAC1) and deubiquitnated by ubiquitin specific peptidase 7 (HAUSP), thereby stabilizing DNMT1 (Fig. 1) [26,29,30]. This model bears out in vivo, as DNMT1 levels correlate with HAUSP levels in human tumors [26]. Furthermore, the role of Tip60 and UHRF1 in DNMT1 stability connects with the DNA damage response via ataxia telangiectasia mutated (ATM). When phosphorylated ATM interacts with DNMT1, it coordinates its acetylation and subsequent ubiquitination via Tip60 and UHRF1 [45], consistent with previous reports [26]; accordingly, Rb antagonizes ATM and helps stabilize DNMT1 [45]. Tip60 acetylation can be regulated via extracellular cues, as the autocrine/paracrine signaling molecule gastrokine 1 decreases DNMT1 levels via upregulation of Tip60 and concomitant DNMT1 acetylation [27]. Other enzymes that modulate DNMT1 acetylation include K (lysine) acetyltransferase 2B for acetylation or sirtuin 1 for deacetylation [28]. Sirt1 deacetylation is especially notable for modulating DNMT1's methyltransferase activity as well [28].
Figure 1.
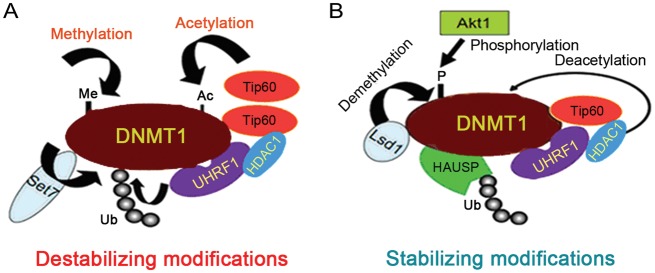
Post-translational modifications regulate DNMT1 protein stability (A) Destabilizing modifications. Methylation catalyzed by Set7 and Tip60 acetylation activity result in UHRF1 ubiquitination of DNMT1. (B) Stabilizing modifications. Demethylation by Lsd1 and/or Akt1 phosphorylation counteracts Set7 methylation. Deacetylation via HDAC1 stabilizes DNMT1, and HAUSP/USP7 deubiquitinates DNMT1.
Regulation of DNMT1 Protein Stability by Phosphorylation and Methylation
DNMT1 stability is also affected by lysine methylation. Specifically, DNMT1 binds with the histone methyltransferase Set7 and becomes methylated, especially during the S/G2 phase of the cell cycle. This mark catalyzes DNMT1 ubiquitination and subsequent degradation [31]. Conversely, the histone demethylase lysine (K)-specific demethylase 1A (LSD1) interacts with and demethylates DNMT1; when LSD1 is knocked out, cells experience global DNA hypomethylation and decreased DNMT1 levels (Fig. 1) [32].
Moreover, DNMT1 methylation at K142 occurs in opposition to its phosphorylation. While phosphorylation of DNMT1 had been shown to affect its methyltransferase activity [46] and its interaction with PCNA and UHRF1 [47], it also plays a role in DNMT1 stability [33]. When v-akt murine thymoma viral oncogene homolog 1 phosphorylates DNMT1 at S143, Set7 becomes unable to methylate DNMT1 at K142 and DNMT1 is stabilized. Much like ubiquitination, DNMT1 phosphorylation changes during the cell cycle: another example of the relationship between DNMT1 stability and appropriate timing of DNA methylation.
Regulation of DNMT1 Protein Stability by Associated Proteins
DNMT1 interacts with a multitude of proteins that have varying functions within the cell [48]. These proteins variously affect DNMT1 stability, activity, and localization. As an example, we recently identified an interaction between DNMT1 protein and the wingless-type mouse mammary tumor virus integration site family (Wnt) pathway effector, β-catenin. DNMT1 levels increase upon stimulation with Wnt3a in a β-catenin-dependent manner; moreover, the interaction between β-catenin and DNMT1 is mutually stabilizing (J. Song, Z. Wang and R. Ewing, unpublished data). Given the crucial role of Wnt signaling in development, these findings suggest a mechanism by which developmental cues can have an impact on the epigenetic landscape of a developing organism.
Therapeutic Implications
Several USA Food and Drug Administration approved epigenetic drugs have been shown to target DNMT1 protein stability. DNMT1 inhibitors (DNMTi), azacitidine and decitabine, are used to treat myelodysplastic syndromes. Canonically, they act via targeting DNMT1 [49], covalently binding with DNA methyltransferases to inactivate them [50–52]. However, recent research demonstrates that DNMTi also induce the proteosomal degradation of DNMT1 [53–55]. Interestingly, HDAC inhibitors (HDACi), another kind of epigenetic drug, also cause proteasome-mediated DNMT1 degradation [7,26]. HDACi, such as vorinostat, are currently on the market for treating cutaneous T-cell lymphoma [56]. However, neither HDACi nor DNMTi as single agent has an effect on solid tumors. Several ongoing clinical trials indicate that at least a subset of lung cancer patients is responsive to combination treatment of HDACi and DNMTi [57].
The HDAC1-HAUSP axis of DNMT1 regulation provides rationale for novel combinatorial therapy options. Knockout of HAUSP in colon cancer cells potentiates HDACi-induced DNMT1 degradation and thus enhances the tumor inhibitory effect of HDACi in a xenograft tumor model. [26]. These observations suggest that combination of HDACi with a HAUSP inhibitor (HAUSPi) may be an effective approach to treat solid tumors. In fact, several distinct HAUSPi have been developed. Some, such as HBX 19,818 [58,59] and P5091 [60,61] were derived from high-throughput compound screens. Another group rationally derived a peptide based on the interaction between HAUSP and a viral protein that inhibits its deubiquitinase activity [62]. Moreover, a compound isolated from sponges also has anti-HAUSP activity [63]. Many of these demonstrate cytotoxicity in vitro and in animal models of disease, particularly through potentiating p53-mediated apoptosis. As predicted by the interaction between HDAC1 and HAUSP, two different HAUSPis demonstrate synergistic effects with HDACi in cell culture [61, and P. Zhang and Z. Wang, unpublished data].
Perspectives and Future Directions
As increasing emphasis is placed on epigenetic mechanisms of gene expression, understanding DNMT1 stability and regulation becomes paramount. Like many other proteins, post-translational modifications of DNMT1 play a crucial role in how and when it is activated. When this protein becomes dysregulated, aberrant methylation patterns and subsequent tumor initiation can occur. Anti-cancer therapies increasingly emphasize the release of tumor suppressor genes from epigenetic mechanisms of repression [64,65]. One important way to effect this goal is to choose compounds that can affect DNMT1 stability.
An important outstanding question in the field of DNMT1 and cancer development is the mechanism by which specific loci become silenced, especially in the context of global demethylation. Compounds that target specific loci for demethylation rather than causing global hypomethylation have an advantage from a therapeutic perspective, preventing chromosomal instability while restoring tumor suppressor expression; one such compound is the DNMT1 inhibitor RG108 [66]. Long non-coding RNAs (lincRNAs) are proposed as a way for gene silencing machinery to target-specific gene loci [9,67,68]. It has been shown that the lincRNA, KCNQ1 opposite strand/antisense transcript 1, interacts with DNMT1 and mediates imprinting at imprinted loci. A recent study demonstrated that a nuclear RNA transcribed from the CCAAT/enhancer binding protein alpha (CEBPA) locus interacts with DNMT1, blocks it from that locus and thus inhibits methylation around CEBPA's promoter [69]. It is possible that other DNMT1-associated RNAs target DNMT1 around other gene loci or specific chromatin regions. In addition, the DNMT1 binding partner UHRF1 contains domains that bind specific H3 tail modifications and therefore can target DNMT1 to specific loci via the histone code [70]. Protein–protein interactions are also implicated, as the DNMT1-binding partner nibrin helps DNMT1 localize and repress survivin during DNA damage [71]. Other evidence suggests that the N-terminal domains of DNMT1, while not necessary for catalytic methyltransferase activity, are still important to target the protein to correct genomic locations [72]. Extending these models to tumor suppressor loci will be useful in expanding the role of DNMT1 in tumorigenesis and may belie alternate mechanisms for treatment.
Funding
This work was supported by the NIH grants (R01CA127590, R21CA160060, R21 CA181859, P50CA150964, and P30 CA043703).
References
- 1.Bird A. Perceptions of epigenetics. Nature. 2007;447:396–398. doi: 10.1038/nature05913. [DOI] [PubMed] [Google Scholar]
- 2.Jones PA, Baylin SB. The epigenomics of cancer. Cell. 2007;128:683–692. doi: 10.1016/j.cell.2007.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Damelin M, Bestor TH. Biological functions of DNA methyltransferase 1 require its methyltransferase activity. Mol Cell Biol. 2007;27:3891–3899. doi: 10.1128/MCB.00036-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cedar H, Bergman Y. Linking DNA methylation and histone modification: patterns and paradigms. Nat Rev Genet. 2009;10:295–304. doi: 10.1038/nrg2540. [DOI] [PubMed] [Google Scholar]
- 5.Robertson KD, Ait-Si-Ali S, Yokochi T, Wade PA, Jones PL, Wolffe AP. DNMT1 forms a complex with Rb, E2F1 and HDAC1 and represses transcription from E2F-responsive promoters. Nat Genet. 2000;25:338–342. doi: 10.1038/77124. [DOI] [PubMed] [Google Scholar]
- 6.Rountree MR, Bachman KE, Baylin SB. DNMT1 binds HDAC2 and a new co-repressor, DMAP1, to form a complex at replication foci. Nat Genet. 2000;25:269–277. doi: 10.1038/77023. [DOI] [PubMed] [Google Scholar]
- 7.Zhou Q, Agoston AT, Atadja P, Nelson WG, Davidson NE. Inhibition of histone deacetylases promotes ubiquitin-dependent proteasomal degradation of DNA methyltransferase 1 in human breast cancer cells. Mol Cancer Res. 2008;6:873–883. doi: 10.1158/1541-7786.MCR-07-0330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ting AH, McGarvey KM, Baylin SB. The cancer epigenome—components and functional correlates. Genes Dev. 2006;20:3215–3231. doi: 10.1101/gad.1464906. [DOI] [PubMed] [Google Scholar]
- 9.Denis H, Ndlovu MN, Fuks F. Regulation of mammalian DNA methyltransferases: a route to new mechanisms. EMBO Rep. 2011;12:647–656. doi: 10.1038/embor.2011.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Robertson KD. DNA methylation and human disease. Nat Rev Genet. 2005;6:597–610. doi: 10.1038/nrg1655. [DOI] [PubMed] [Google Scholar]
- 11.Feinberg AP. Phenotypic plasticity and the epigenetics of human disease. Nature. 2007;447:433–440. doi: 10.1038/nature05919. [DOI] [PubMed] [Google Scholar]
- 12.Klein CJ, Botuyan MV, Wu Y, Ward CJ, Nicholson GA, Hammans S, Hojo K, et al. Mutations in DNMT1 cause hereditary sensory neuropathy with dementia and hearing loss. Nat Genet. 2011;43:595–600. doi: 10.1038/ng.830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Winkelmann J, Lin L, Schormair B, Kornum BR, Faraco J, Plazzi G, Melberg A, et al. Mutations in DNMT1 cause autosomal dominant cerebellar ataxia, deafness and narcolepsy. Hum Mol Genet. 2012;21:2205–2210. doi: 10.1093/hmg/dds035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vogelstein B, Papadopoulos N, Velculescu VE, Zhou S, Diaz LA, Jr, Kinzler KW. Cancer genome landscapes. Science. 2013;339:1546–1558. doi: 10.1126/science.1235122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Graff JR, Herman JG, Lapidus RG, Chopra H, Xu R, Jarrard DF, Isaacs WB, et al. E-cadherin expression is silenced by DNA hypermethylation in human breast and prostate carcinomas. Cancer Res. 1995;55:5195–5199. [PubMed] [Google Scholar]
- 16.Jain S, Chang TT, Hamilton JP, Lin SY, Lin YJ, Evans AA, Selaru FM, et al. Methylation of the CpG sites only on the sense strand of the APC gene is specific for hepatocellular carcinoma. PLoS ONE. 2011;6:e26799. doi: 10.1371/journal.pone.0026799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Donninger H, Vos MD, Clark GJ. The RASSF1A tumor suppressor. J Cell Sci. 2007;120:3163–3172. doi: 10.1242/jcs.010389. [DOI] [PubMed] [Google Scholar]
- 18.Peng DF, Kanai Y, Sawada M, Ushijima S, Hiraoka N, Kitazawa S, Hirohashi S. DNA methylation of multiple tumor-related genes in association with overexpression of DNA methyltransferase 1 (DNMT1) during multistage carcinogenesis of the pancreas. Carcinogenesis. 2006;27:1160–1168. doi: 10.1093/carcin/bgi361. [DOI] [PubMed] [Google Scholar]
- 19.Holm TM, Jackson-Grusby L, Brambrink T, Yamada Y, Rideout WM, III, Jaenisch R. Global loss of imprinting leads to widespread tumorigenesis in adult mice. Cancer Cell. 2005;8:275–285. doi: 10.1016/j.ccr.2005.09.007. [DOI] [PubMed] [Google Scholar]
- 20.De Marzo AM, Marchi VL, Yang ES, Veeraswamy R, Lin X, Nelson WG. Abnormal regulation of DNA methyltransferase expression during colorectal carcinogenesis. Cancer Res. 1999;59:3855–3860. [PubMed] [Google Scholar]
- 21.Robertson KD, Uzvolgyi E, Liang G, Talmadge C, Sumegi J, Gonzales FA, Jones PA. The human DNA methyltransferases (DNMTs) 1, 3a and 3b: coordinate mRNA expression in normal tissues and overexpression in tumors. Nucleic Acids Res. 1999;27:2291–2298. doi: 10.1093/nar/27.11.2291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Etoh T, Kanai Y, Ushijima S, Nakagawa T, Nakanishi Y, Sasako M, Kitano S, et al. Increased DNA methyltransferase 1 (DNMT1) protein expression correlates significantly with poorer tumor differentiation and frequent DNA hypermethylation of multiple CpG islands in gastric cancers. Am J Pathol. 2004;164:689–699. doi: 10.1016/S0002-9440(10)63156-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wu J, Issa JP, Herman J, Bassett DE, Jr, Nelkin BD, Baylin SB. Expression of an exogenous eukaryotic DNA methyltransferase gene induces transformation of NIH 3T3 cells. Proc Natl Acad Sci USA. 1993;90:8891–8895. doi: 10.1073/pnas.90.19.8891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Agoston AT, Argani P, Yegnasubramanian S, De Marzo AM, Ansari-Lari MA, Hicks JL, Davidson NE, et al. Increased protein stability causes DNA methyltransferase 1 dysregulation in breast cancer. J Biol Chem. 2005;280:18302–18310. doi: 10.1074/jbc.M501675200. [DOI] [PubMed] [Google Scholar]
- 25.Bronner C. Control of DNMT1 abundance in epigenetic inheritance by acetylation, ubiquitylation, and the histone code. Sci Signal. 2011;4:pe3. doi: 10.1126/scisignal.2001764. [DOI] [PubMed] [Google Scholar]
- 26.Du Z, Song J, Wang Y, Zhao Y, Guda K, Yang S, Kao HY, et al. DNMT1 stability is regulated by proteins coordinating deubiquitination and acetylation-driven ubiquitination. Sci Signal. 2010;3:ra80. doi: 10.1126/scisignal.2001462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yoon JH, Choi YJ, Choi WS, Ashktorab H, Smoot DT, Nam SW, Lee JY, et al. GKN1-miR-185-DNMT1 axis suppresses gastric carcinogenesis through regulation of epigenetic alteration and cell cycle. Clin Cancer Res. 2013;19:4599–4610. doi: 10.1158/1078-0432.CCR-12-3675. [DOI] [PubMed] [Google Scholar]
- 28.Peng L, Yuan Z, Ling H, Fukasawa K, Robertson K, Olashaw N, Koomen J, et al. SIRT1 deacetylates the DNA methyltransferase 1 (DNMT1) protein and alters its activities. Mol Cell Biol. 2011;31:4720–4734. doi: 10.1128/MCB.06147-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Felle M, Joppien S, Nemeth A, Diermeier S, Thalhammer V, Dobner T, Kremmer E, et al. The USP7/Dnmt1 complex stimulates the DNA methylation activity of DNMT1 and regulates the stability of UHRF1. Nucleic Acids Res. 2011;39:8355–8365. doi: 10.1093/nar/gkr528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Qin W, Leonhardt H, Spada F. Usp7 and Uhrf1 control ubiquitination and stability of the maintenance DNA methyltransferase Dnmt1. J Cell Biochem. 2011;112:439–444. doi: 10.1002/jcb.22998. [DOI] [PubMed] [Google Scholar]
- 31.Esteve PO, Chin HG, Benner J, Feehery GR, Samaranayake M, Horwitz GA, Jacobsen SE, et al. Regulation of DNMT1 stability through SET7-mediated lysine methylation in mammalian cells. Proc Natl Acad Sci USA. 2009;106:5076–5081. doi: 10.1073/pnas.0810362106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang J, Hevi S, Kurash JK, Lei H, Gay F, Bajko J, Su H, et al. The lysine demethylase LSD1 (KDM1) is required for maintenance of global DNA methylation. Nat Genet. 2009;41:125–129. doi: 10.1038/ng.268. [DOI] [PubMed] [Google Scholar]
- 33.Esteve PO, Chang Y, Samaranayake M, Upadhyay AK, Horton JR, Feehery GR, Cheng X, et al. A methylation and phosphorylation switch between an adjacent lysine and serine determines human DNMT1 stability. Nat Struct Mol Biol. 2011;18:42–48. doi: 10.1038/nsmb.1939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.He S, Wang F, Yang L, Guo C, Wan R, Ke A, Xu L, et al. Expression of DNMT1 and DNMT3a are regulated by GLI1 in human pancreatic cancer. PLoS ONE. 2011;6:e27684. doi: 10.1371/journal.pone.0027684. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 35.Bakin AV, Curran T. Role of DNA 5-methylcytosine transferase in cell transformation by fos. Science. 1999;283:387–390. doi: 10.1126/science.283.5400.387. [DOI] [PubMed] [Google Scholar]
- 36.Jinawath A, Miyake S, Yanagisawa Y, Akiyama Y, Yuasa Y. Transcriptional regulation of the human DNA methyltransferase 3A and 3B genes by Sp3 and Sp1 zinc finger proteins. Biochem J. 2005;385:557–564. doi: 10.1042/BJ20040684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kimura H, Nakamura T, Ogawa T, Tanaka S, Shiota K. Transcription of mouse DNA methyltransferase 1 (Dnmt1) is regulated by both E2F-Rb-HDAC-dependent and -independent pathways. Nucleic Acids Res. 2003;31:3101–3113. doi: 10.1093/nar/gkg406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chuang LS, Ian HI, Koh TW, Ng HH, Xu G, Li BF. Human DNA-(cytosine-5) methyltransferase-PCNA complex as a target for p21WAF1. Science. 1997;277:1996–2000. doi: 10.1126/science.277.5334.1996. [DOI] [PubMed] [Google Scholar]
- 39.Schneider K, Fuchs C, Dobay A, Rottach A, Qin W, Wolf P, Álvarez-Castro JM, et al. Dissection of cell cycle-dependent dynamics of Dnmt1 by FRAP and diffusion-coupled modeling. Nucleic Acids Res. 2013;41:4860–4876. doi: 10.1093/nar/gkt191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Citterio E, Papait R, Nicassio F, Vecchi M, Gomiero P, Mantovani R, Di Fiore PP, et al. Np95 is a histone-binding protein endowed with ubiquitin ligase activity. Mol Cell Biol. 2004;24:2526–2535. doi: 10.1128/MCB.24.6.2526-2535.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Karagianni P, Amazit L, Qin J, Wong J. ICBP90, a novel methyl K9 H3 binding protein linking protein ubiquitination with heterochromatin formation. Mol Cell Biol. 2008;28:705–717. doi: 10.1128/MCB.01598-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sharif J, Muto M, Takebayashi S, Suetake I, Iwamatsu A, Endo TA, Shinga J, et al. The SRA protein Np95 mediates epigenetic inheritance by recruiting Dnmt1 to methylated DNA. Nature. 2007;450:908–912. doi: 10.1038/nature06397. [DOI] [PubMed] [Google Scholar]
- 43.Bostick M, Kim JK, Esteve PO, Clark A, Pradhan S, Jacobsen SE. UHRF1 plays a role in maintaining DNA methylation in mammalian cells. Science. 2007;317:1760–1764. doi: 10.1126/science.1147939. [DOI] [PubMed] [Google Scholar]
- 44.Nishiyama A, Yamaguchi L, Sharif J, Johmura Y, Kawamura T, Nakanishi K, Shimamura S, et al. Uhrf1-dependent H3K23 ubiquitylation couples maintenance DNA methylation and replication. Nature. 2013;502:249–253. doi: 10.1038/nature12488. [DOI] [PubMed] [Google Scholar]
- 45.Shamma A, Suzuki M, Hayashi N, Kobayashi M, Sasaki N, Nishiuchi T, Doki Y, et al. ATM mediates pRB function to control DNMT1 protein stability and DNA methylation. Mol Cell Biol. 2013;33:3113–3124. doi: 10.1128/MCB.01597-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lavoie G, Esteve PO, Laulan NB, Pradhan S, St-Pierre Y. PKC isoforms interact with and phosphorylate DNMT1. BMC Biol. 2011;9:31. doi: 10.1186/1741-7007-9-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hervouet E, Lalier L, Debien E, Cheray M, Geairon A, Rogniaux H, Loussouarn D, et al. Disruption of Dnmt1/PCNA/UHRF1 interactions promotes tumorigenesis from human and mice glial cells. PLoS ONE. 2010;5:e11333. doi: 10.1371/journal.pone.0011333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Qin W, Leonhardt H, Pichler G. Regulation of DNA methyltransferase 1 by interactions and modifications. Nucleus. 2011;2:392–402. doi: 10.4161/nucl.2.5.17928. [DOI] [PubMed] [Google Scholar]
- 49.Juttermann R, Li E, Jaenisch R. Toxicity of 5-aza-2′-deoxycytidine to mammalian cells is mediated primarily by covalent trapping of DNA methyltransferase rather than DNA demethylation. Proc Natl Acad Sci USA. 1994;91:11797–11801. doi: 10.1073/pnas.91.25.11797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Santi DV, Garrett CE, Barr PJ. On the mechanism of inhibition of DNA-cytosine methyltransferases by cytosine analogs. Cell. 1983;33:9–10. doi: 10.1016/0092-8674(83)90327-6. [DOI] [PubMed] [Google Scholar]
- 51.Santi DV, Norment A, Garrett CE. Covalent bond formation between a DNA-cytosine methyltransferase and DNA containing 5-azacytosine. Proc Natl Acad Sci USA. 1984;81:6993–6997. doi: 10.1073/pnas.81.22.6993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Goffin J, Eisenhauer E. DNA methyltransferase inhibitors-state of the art. Ann Oncol. 2002;13:1699–1716. doi: 10.1093/annonc/mdf314. [DOI] [PubMed] [Google Scholar]
- 53.Ghoshal K, Datta J, Majumder S, Bai S, Kutay H, Motiwala T, Jacob ST, et al. 5-Aza-deoxycytidine induces selective degradation of DNA methyltransferase 1 by a proteasomal pathway that requires the KEN box, bromo-adjacent homology domain, and nuclear localization signal. Mol Cell Biol. 2005;25:4727–4741. doi: 10.1128/MCB.25.11.4727-4741.2005. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 54.Patel K, Dickson J, Din S, Macleod K, Jodrell D, Ramsahoye B. Targeting of 5-aza-2′-deoxycytidine residues by chromatin-associated DNMT1 induces proteasomal degradation of the free enzyme. Nucleic Acids Res. 2010;38:4313–4324. doi: 10.1093/nar/gkq187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bhalla KN. Epigenetic and chromatin modifiers as targeted therapy of hematologic malignancies. J Clin Oncol. 2005;23:3971–3993. doi: 10.1200/JCO.2005.16.600. [DOI] [PubMed] [Google Scholar]
- 56.Karberg S. Switching on epigenetic therapy. Cell. 2009;139:1029–1031. doi: 10.1016/j.cell.2009.11.038. [DOI] [PubMed] [Google Scholar]
- 57.Kaiser J. Epigenetic drugs take on cancer. Science. 2010;330:576–578. doi: 10.1126/science.330.6004.576. [DOI] [PubMed] [Google Scholar]
- 58.Reverdy C, Conrath S, Lopez R, Planquette C, Atmanene C, Collura V, Harpon J, et al. Discovery of specific inhibitors of human USP7/HAUSP deubiquitinating enzyme. Chem Biol. 2012;19:467–477. doi: 10.1016/j.chembiol.2012.02.007. [DOI] [PubMed] [Google Scholar]
- 59.Colland F, Formstecher E, Jacq X, Reverdy C, Planquette C, Conrath S, Trouplin V, et al. Small-molecule inhibitor of USP7/HAUSP ubiquitin protease stabilizes and activates p53 in cells. Mol Cancer Ther. 2009;8:2286–2295. doi: 10.1158/1535-7163.MCT-09-0097. [DOI] [PubMed] [Google Scholar]
- 60.Nicholson B, Suresh Kumar KG. The multifaceted roles of USP7: new therapeutic opportunities. Cell Biochem Biophys. 2011;60:61–68. doi: 10.1007/s12013-011-9185-5. [DOI] [PubMed] [Google Scholar]
- 61.Chauhan D, Tian Z, Nicholson B, Kumar KG, Zhou B, Carrasco R, McDermott JL, et al. A small molecule inhibitor of ubiquitin-specific protease-7 induces apoptosis in multiple myeloma cells and overcomes bortezomib resistance. Cancer Cell. 2012;22:345–358. doi: 10.1016/j.ccr.2012.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lee HR, Choi WC, Lee S, Hwang J, Hwang E, Guchhait K, Haas J, et al. Bilateral inhibition of HAUSP deubiquitinase by a viral interferon regulatory factor protein. Nat Struct Mol Biol. 2011;18:1336–1344. doi: 10.1038/nsmb.2142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Yamaguchi M, Miyazaki M, Kodrasov MP, Rotinsulu H, Losung F, Mangindaan RE, de Voogd NJ, et al. Spongiacidin C, a pyrrole alkaloid from the marine sponge Stylissa massa, functions as a USP7 inhibitor. Bioorg Med Chem Lett. 2013;23:3884–3886. doi: 10.1016/j.bmcl.2013.04.066. [DOI] [PubMed] [Google Scholar]
- 64.Popovic R, Shah MY, Licht JD. Epigenetic therapy of hematological malignancies: where are we now? Ther Adv Hematol. 2013;4:81–91. doi: 10.1177/2040620712466864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Brueckner B, Lyko F. DNA methyltransferase inhibitors: old and new drugs for an epigenetic cancer therapy. Trends Pharmacol Sci. 2004;25:551–554. doi: 10.1016/j.tips.2004.09.004. [DOI] [PubMed] [Google Scholar]
- 66.Brueckner B, Garcia Boy R, Siedlecki P, Musch T, Kliem HC, Zielenkiewicz P, Suhai S, et al. Epigenetic reactivation of tumor suppressor genes by a novel small-molecule inhibitor of human DNA methyltransferases. Cancer Res. 2005;65:6305–6311. doi: 10.1158/0008-5472.CAN-04-2957. [DOI] [PubMed] [Google Scholar]
- 67.Mohammad F, Pandey GK, Mondal T, Enroth S, Redrup L, Gyllensten U, Kanduri C. Long noncoding RNA-mediated maintenance of DNA methylation and transcriptional gene silencing. Development. 2012;139:2792–2803. doi: 10.1242/dev.079566. [DOI] [PubMed] [Google Scholar]
- 68.Mohammad F, Mondal T, Guseva N, Pandey GK, Kanduri C. Kcnq1ot1 noncoding RNA mediates transcriptional gene silencing by interacting with Dnmt1. Development. 2010;137:2493–2499. doi: 10.1242/dev.048181. [DOI] [PubMed] [Google Scholar]
- 69.Di Ruscio A, Ebralidze AK, Benoukraf T, Amabile G, Goff LA, Terragni J, Figueroa ME, et al. DNMT1-interacting RNAs block gene-specific DNA methylation. Nature. 2013;503:371–376. doi: 10.1038/nature12598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Rothbart SB, Dickson BM, Ong MS, Krajewski K, Houliston S, Kireev DB, Arrowsmith CH, et al. Multivalent histone engagement by the linked tandem Tudor and PHD domains of UHRF1 is required for the epigenetic inheritance of DNA methylation. Genes Dev. 2013;27:1288–1298. doi: 10.1101/gad.220467.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Hayashi N, Kobayashi M, Shamma A, Morimura Y, Takahashi C, Yamamoto KI. Regulatory interaction between NBS1 and DNMT1 responding to DNA damage. J Biochem. 2013;154:429–435. doi: 10.1093/jb/mvt071. [DOI] [PubMed] [Google Scholar]
- 72.Espada J. Non-catalytic functions of DNMT1. Epigenetics. 2012;7:115–118. doi: 10.4161/epi.7.2.18756. [DOI] [PubMed] [Google Scholar]


