Significance
The exploration of protease substrate specificity is generally restricted to naturally occurring amino acids, limiting the degree of conformational space that can be surveyed. We substantially enhanced this by incorporating 102 unnatural amino acids to explore the S1–S4 pockets of human neutrophil elastase. This approach provides hybrid natural and unnatural amino acid sequences, and thus we termed it the Hybrid Combinatorial Substrate Library. Using this approach, we have designed an extremely active substrate of NE and subsequently converted it into an ultrasensitive activity-based probe for imaging active elastase during the process of neutrophil extracellular trap formation. Our study could have a substantial effect on the design of substrates, inhibitors, and probes for any endopeptidase.
Abstract
The exploration of protease substrate specificity is generally restricted to naturally occurring amino acids, limiting the degree of conformational space that can be surveyed. We substantially enhanced this by incorporating 102 unnatural amino acids to explore the S1–S4 pockets of human neutrophil elastase. This approach provides hybrid natural and unnatural amino acid sequences, and thus we termed it the Hybrid Combinatorial Substrate Library. Library results were validated by the synthesis of individual tetrapeptide substrates, with the optimal substrate demonstrating more than three orders of magnitude higher catalytic efficiency than commonly used substrates of elastase. This optimal substrate was converted to an activity-based probe that demonstrated high selectivity and revealed the specific presence of active elastase during the process of neutrophil extracellular trap formation. We propose that this approach can be successfully used for any type of endopeptidase to deliver high activity and selectivity in substrates and probes.
Proteases play key roles in essentially all signaling pathways, with infection and inflammation, apoptosis, blood clotting, and cell cycle control being classic examples (1). Thus, misregulation of proteolysis can be deleterious and accompanies many human pathologies (2). The substrate specificity of proteolytic enzymes is dictated by the sequence of their target proteins, with proteinogenic (more frequently called natural) amino acid sequences directing selectivity. Several methods have been devised to define the optimal substrate specificity of proteases; one of the most commonly used is the Positional Scanning Substrate Combinatorial Library (PS-SCL) approach, in which tetrapeptides coupled to fluorogenic leaving groups are used to ascertain preferences (3–5). In this approach, only natural amino acids have previously been used, with the exception of norleucine (used instead of methionine) (6, 7). Data obtained using PS-SCL approaches have been used to design substrates, inhibitors, or activity-based probes for several families of proteases (3, 8, 9). However, restricting library design to natural amino acids narrows the amount of chemical space that can be explored to distinguish between closely related proteases of the same family.
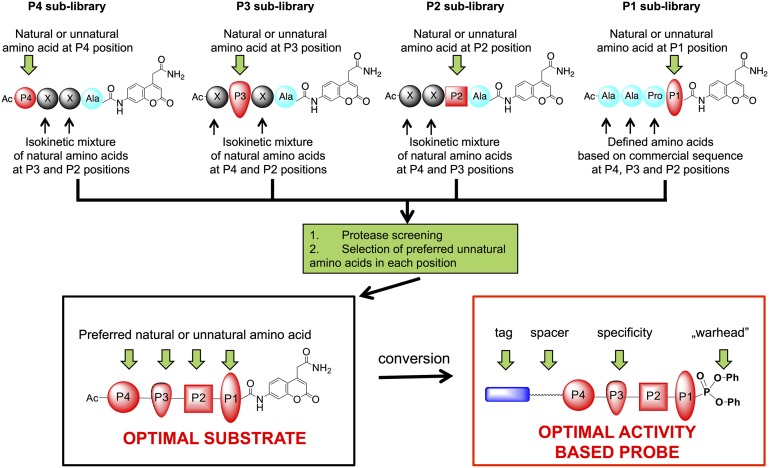
To overcome these limitations, we designed a combinatorial library of fluorogenic tetrapeptide substrates, exploring the primary specificity pockets of a protease [S1–S4 in the nomenclature of Schechter and Berger (10) (Fig. 1)] by applying a pool of 102 unnatural amino acids (defined as those amino acids not encoded in proteins) that are available in structurally different forms. The power of this approach was demonstrated initially for an individual fluorogenic substrates library screening of a family of aminopeptidases (11). Using this approach, unnatural amino acids were shown to be much better substrates in terms of specificity and selectivity compared with natural ones (12, 13). Demonstrating this in exopeptidases is relatively simple, as it only includes screening of a single position. Endopeptidases provide a greater challenge, as tetrapeptides are commonly used for substrate specificity profiling, severely complicating the combinatorial possibilities. Here we demonstrate a general approach for the synthesis of combinatorial libraries containing unnatural amino acids, with subsequent screening and analysis of large sublibraries. We term this approach the Hybrid Combinatorial Substrate Library (HyCoSuL). We demonstrate the utility of this approach in the design of a highly selective substrate and activity-based probe.
Fig. 1.
General scheme for HyCoSuL design, screening, optimal substrate selection, and activity-based probe design. The substrate specificity of NE, similar to that of many serine proteases, is dominated by surface enzyme pockets (subsites S4–S1) that occupy amino acid side chains P4–P1 (34). Preferred occupancy can be determined by positional scanning of P4–P1 residues.
As a target protease, we selected human neutrophil elastase (EC 3.4.21.37) (NE), a serine protease restricted to neutrophil azurophil granules (14). NE is released by neutrophils during inflammation, and its function is generally thought to be to degrade host tissue and destroy bacteria. Extended tissue destruction is deleterious, and NE is also involved in the development of chronic obstructive pulmonary diseases and in nonsmall-cell lung cancer progression (15). NE belongs to one of the most investigated protease families and has very broad substrate specificity (16). This enzyme does not efficiently process substrates based on natural amino acids, and there are no selective activity-based probes for investigating NE in situ or ex vivo. Using HyCoSuL, we set out to obtain highly efficient and selective substrates for NE and convert these to activity-based probes for microscopic imaging.
Results
HyCoSuL Libraries Synthesis Strategy.
Our objective was to develop selective substrates and probes based on the core tetrapeptide scaffold sequence Ala-Ala-Pro-Val commonly used for NE (17). Strategically, we explored the P2, P3, and P4 subsites preferences and followed-up by defining optimal occupancy in the key P1 position. Because of its convenience in solid-phase synthesis, 7-amino-4-carbamoylmethyl coumarin (ACC) was selected as the reporting fluorophore (18). To explore the S2, S3, and S4 pockets, we designed three sublibraries and fixed the P1 position as Ala (substituting Val, the preferred natural amino acid in this position, because of a problem with complete substitution to ACC fluorophore as a result of steric hindrance). Each library contained fixed P2, P3, or P4 components with a natural or unnatural amino acid and two components with isokinetic mixtures of natural amino acids (cysteine was omitted, Met was substituted with Nle). This approach guaranteed selection of the optimal residue occupancy (Fig. 1).
Each sublibrary contained 120 wells, with each well corresponding to the amino acid being profiled. In each well, there was a mixture of tetrapeptides composed of Ala fixed at P1, with one amino acid fixed in a defined position (P4, P3, or P2) and the remaining two positions composed of an equimolar mixture of 19 × 19 (361) tetrapeptidic substrates (Fig. 1). Unnatural amino acids were selected to cover a broad range of chemical space. They differed by side chain character (alkaline, acidic, neutral, hydrophilic, hydrophobic), size (small, big), shape (bulky, branched, unbranched), or stereoselectivity (d-amino acids). The free N terminus was capped with an acetyl group. All sublibraries were synthesized using standard solid-phase peptide synthesis protocols on a semiautomatic FlexChem synthesizer (SciGene).
Substrate Specificity Determination.
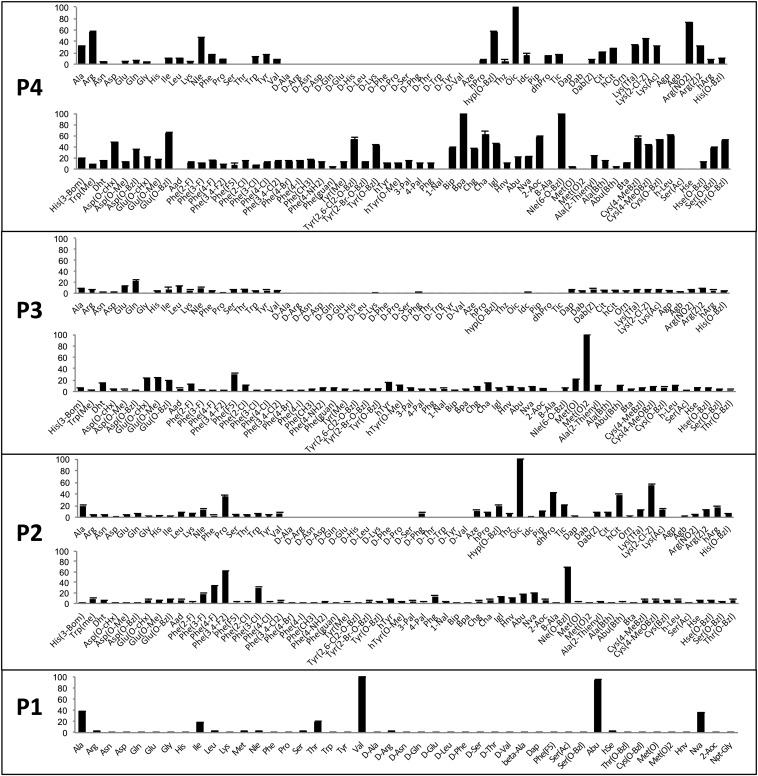
Each of the P2, P3, and P4 sublibraries was screened at 50 μM final total substrate mixture and 43 nM NE, and initial rates of substrate hydrolysis were recorded as relative fluorescent units over time. The signals from the two screens were combined and normalized by inclusion of at least 3 reference substrate mixtures.
The preferred P2 substrate substituent is octahydro-1H-indole-2-carboxylic acid (Oic), which is a Pro derivative with an additional cyclohexyl ring (Fig. 2). The S2 subsite displays a noticeable preference for bulky amino acid residues such as Nle(O-Bzl), Lys(2-Cl-Z), Phe(3,4-F), dhPro, and hCit. Pro was the best-recognized natural amino acid in P2, confirming a previous report (19).
Fig. 2.
Preferences at the P4–P1 subsites of NE. The P4, subsite preference of NE was determined using combinatorial substrate libraries of the general structure Ac-P4-X-X-Ala-ACC, where P4 represents a natural or unnatural amino acid and X represents an isokinetic mixture of natural amino acids (Cys and Met were omitted because of a problem with oxidation; Nle was used instead of Met). The P3 and P2 preferences were determined in a similar way. The P1 subsite preference was determined using individual substrate libraries of the general structure Ac-Ala-Ala-Pro-P1-ACC, where P1 is a natural or unnatural amino acid. Abbreviated amino acid names (SI Appendix, Fig. S2) are shown on the x axis. The y axis displays the average relative activity expressed as a percentage of the best amino acid. Error bars represent the SD (n = 3).
In the P3 position, NE favored methionine dioxide [Met(O)2] around five times more than Gln, which was the best-recognized natural amino acid (Fig. 2). Methionine oxide was around five times more weakly recognized compared with Met(O)2. Other unnatural amino acids well-tolerated in the S3 pocket were pentafluorinated phenylalanine and methyl, benzyl [Glu(O-Bzl)], and cyclohexyl esters of glutamic acid. Several other natural and unnatural amino acids were also recognized at P3.
Analysis of substrate specificity at the P4 position yields very interesting observations. Hydrophobic unnatural amino acids with very bulky side chains such as Bpa, Nle(O-Bzl), Oic, Glu(O-Bzl), or Cha (Fig. 2) are preferred. However, hydrophilic (but also bulky) Arg is the most preferred natural amino acid [around 60% activity of Glu(O-Bzl)]. Even better recognized is its derivative ArgNO2. With the exception of some hydrolysis of d-Phg at P2, d-amino acids were not selected at any position.
Preference in the P1 Position.
To complete the definition of optimal activity, we designed and screened a P1 library on the scaffold of the commonly used tetrapeptidic sequence Ala-Ala-Pro-Val (Fig. 1). In total, 45 individual substrate sequences with the general formula Ac-Ala-Ala-Pro-P1-ACC (P1 is a natural or unnatural amino acid) were synthesized using standard solid-phase peptide synthesis, and each substrate was purified using preparative HPLC.
P1 substrate specificity analysis revealed that NE demonstrates essentially exclusive selectivity for cleavage after small aliphatic residues, primarily Val and Abu, with tolerance also for Ala, Nva, Thr, and Ile (Fig. 2). Determination of second-order rate constants (Table 1) demonstrates that the reference substrate with Val in P1 is almost equal to Abu in terms of catalytic efficiency and is around 5 times more active than with Ala.
Table 1.
Kinetic analysis of best tetrapeptide substrates of the general formula Ac-Ala-Ala-Pro-P1-ACC, where P1 is a natural or unnatural amino acid selected for validation after library screening
| kcat/Km, M−1·s−1 | |
| Ac-Ala-Ala-ProVal-ACC | 4920 ± 278 |
| Ac-Ala-Ala-ProAbu-ACC | 4892 ± 56 |
| Ac-Ala-Ala-ProAla-ACC | 989 ± 36 |
Data represent the mean ± SD of three or more experiments.
Validation with Individual Tetrapeptide Sequences and Design of an Optimal Substrate.
To validate library screening data for each sublibrary, we synthesized a selection of “good” and “bad” representatives at each P2, P3, or P4 position and incorporated them into the reference sequence Ac-Ala-Ala-Pro-Ala-ACC. All individual sequences were synthesized by solid-phase methods, and their experimental catalytic efficiency (kcat/Km) was determined (SI Appendix, Tables S1, S2, and S3). The catalytic rates confirmed the order of selectivity predicted in the library screens, thereby validating the approach.
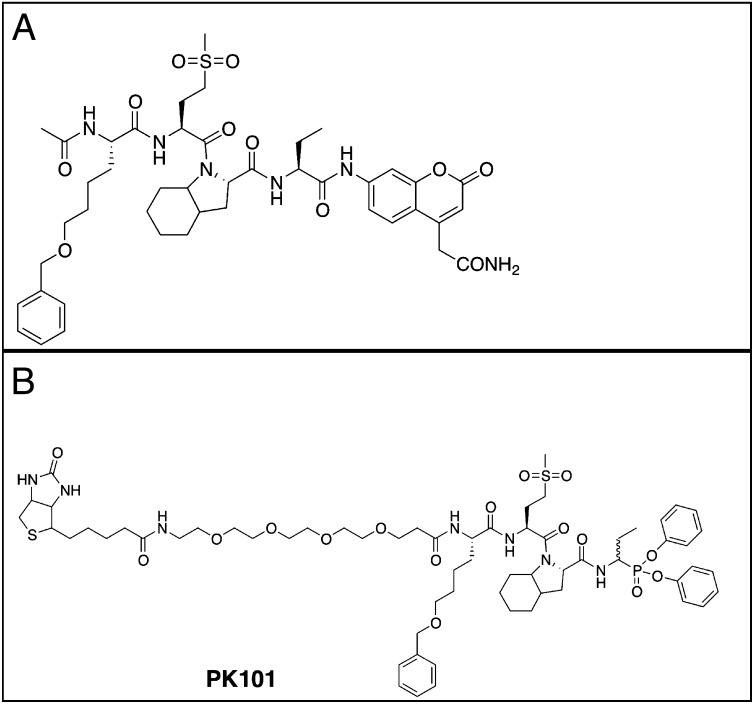
Finally, we selected the preferred P2, P3, and P4 substituents recognized by NE and synthesized the optimal fluorogenic substrate Ac-Nle(O-Bzl)-Met(O)2-Oic-Abu-ACC, containing the best unnatural amino acid Abu in the P1 position (Fig. 3A). Determination of catalytic efficiency revealed that this substrate is more than 7,000 times better hydrolyzed (kcat/Km = 4.79 × 107 M−1·s−1) by NE than the commonly used commercial peptide sequence based on the Ala-Ala-Pro-Val specificity sequence (Ac-Ala-Ala-Pro-Val-ACC, in our hands; kcat/Km = 5.81 × 103 M−1·s−1) see also ref. 20.
Fig. 3.
Structure of the optimized NE substrate and activity-based probe PK101. (A) Optimal substrate. (B) Activity-based probe derived from the optimal substrate. In the case of the probe PK101, enzyme inhibition is likely via attack of the serine nucleophile on the phosphorous of the probe, with release of a phenoxy group and formation of a highly stable, covalent intermediate complex, as previously discussed for this type of electrophilic warhead (35).
Selectivity of NE versus Proteinase 3.
The most closely related protease to NE is neutrophil proteinase 3 (PR3, also known as myeloblastin), which is 54% identical, located primarily in neutrophils, and shares with NE a preference for Val in the P1 (21). As a consequence, any attempt to devise specific substrates and probes for the analysis of NE must take into account PR3. To compare the two proteases, we obtained protease samples from the same supplier, purified from normal human neutrophils, determined their active site concentration by titration with 3,4-dichloroisocoumarin to ensure equal and optimal protease activity, and performed analysis with the optimal substrate Ac-Nle(O-Bzl)-Met(O)2-Oic-Abu-ACC to determine kinetic parameters. Because this substrate has a solubility limit of around 10 µM in aqueous solution, we were limited to this concentration. In addition, we shifted our assay buffer from a high-salt buffer (0.1 M Hepes, 0.5 M NaCl at pH 7.5) used in library screening to a low-salt buffer [50 mM Hepes, 0.1 M NaCl, 0.1% (vol/vol) Igepal CA-630 (Sigma-Aldrich) at pH 7.4] in which we found PR3 and NE to be optimally stable at 37 °C.
We readily observed substrate saturation with NE, but not with PR3, and thus we were able to determine individual kcat and Km values for NE, but only the ratio kcat/Km for PR3 (SI Appendix, Fig. S1). The kcat/Km value obtained for NE in low-salt buffer was about fourfold higher than that obtained in high-salt buffer, which is still in the same order of magnitude, More important, comparison of the kcat/Km values (Table 2) demonstrates that NE is 900-fold more efficient than PR3 in cleaving this substrate. Moreover, the Km for NE is about 0.28 µM, whereas for PR3 it must be above 10 µM, as we were unable to observe deviation from linearity in the PR3 substrate velocity plot.
Table 2.
Comparison of specificity constants for NE and PR3, using Ac-Nle(O-Bzl)-Met(O)2-Oic-Abu-ACC
| kcat, s−1 | Km, µM | kcat/Km, M−1·s−1 | |
| NE | 13.4 (±0.15) | 0.28 (±0.08) | 4.79 (±0.12) × 107 |
| PR3 | ND | ND | 5.4 (±0.1) × 104 |
Data represent the mean and SD of three or more experiments. ND, not able to determine.
By way of validating the approach for developing optimal substrates through HyCoSuL, we carried out a preliminary screen using PR3 (SI Appendix, Fig. S3), which revealed an optimal substrate Ac-Glu(O-Bzl)-Lys(Ac)-Hyp(Bzl)-Abu-ACC. This substrate had a kcat/Km of 7.9 × 104 M−1·s−1 and was 13-fold selective for PR3 over NE. Further experiments would be required to enhance the selectivity factor of substrates for PR3 compared with NE.
Design and Characteristics of Activity-Based Probes.
The high degree of selectivity of the optimal substrate Ac-Nle(O-Bzl)-Met(O)2-Oic-Abu-ACC provides proof of concept that the peptidic sequence recognition elements for NE can be used to convert the optimal substrate to an activity-based probe for NE. This compound was obtained using a mixed solid- and solution-phase approach. First, we synthesized the tripeptidic sequence Nle(O-Bzl)-Met(O)2-Oic, using chlorotrityl resin. The N terminus of the peptide was first equipped with a polyethylene glycol linker PEG (4), which was designed to improve solubility of the compound and allow for separation of a biotin tag from the recognition epitope. As an electrophilic warhead for the probe, we selected a phosphonate, +H3N-Abu-PO3Ph2, which was obtained according to the protocol described by Soroka and Goldeman (22). Finally, biotin-PEG (4)-Nle(O-Bzl)-Met(O)2-Oic-COOH was coupled to +H3N-Abu-PO3Ph2. After synthesis, Biotin-PEG (4)-Nle(O-Bzl)-Met(O)2-Oic-Abu-PO3Ph2 (Fig. 3B) was purified using HPLC, and its identity was confirmed by mass spectrometry.
We determined the selectivity of PK101 for NE by calculating apparent kobs(app)/I (second-order rate constants for inhibition) under pseudo first-order conditions by varying probe concentrations at constant 10 µM optimal substrate concentration (23). Because we had determined the Km of the optimal substrate for NE, we were able to calculate the exact kobs/I value for NE but were only able to place an upper limit of the kobs/I value for PR3. Comparison of the kobs/I values (Table 3) demonstrates that PK101 inhibits NE 150-fold more rapidly than PR3, mirroring the results with the optimal substrate.
Table 3.
Comparison of inhibition rate constants for NE and PR3, using 10 µM Ac-Nle(O-Bzl)-Met(O)2-Oic-Abu-ACC as substrate
| kobs(app)/I [M−1·s−1] | Km, µM | kobs/I, M−1·s−1 | |
| NE | 3.9 (±0.1) x 105 | 0.28 (±0.08) | 1.4 (±0.1) × 107 |
| PR3 | 9.3 × 104 | ND | 9.3 × 104 |
Data represent the mean and SD of three or more experiments with the exception of PR3, which was analyzed once.
Activity-Based Probe Labeling of NE.
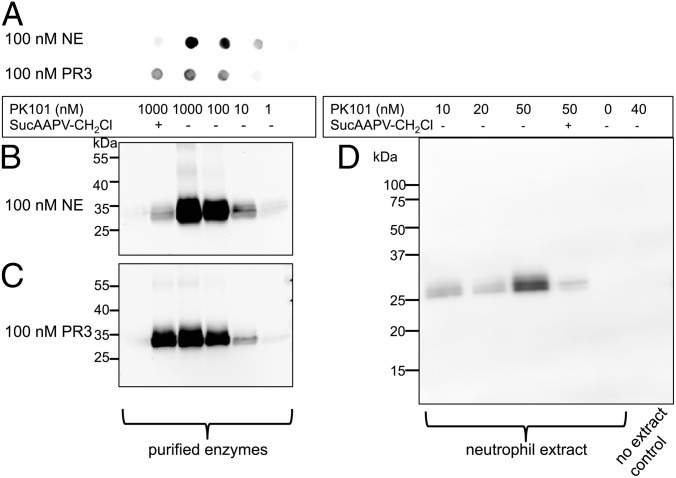
Armed with the information that the probe-optimized PK101 inhibits NE at least 150-fold more rapidly than PR3, we characterized the labeling by incubating the probe with either NE or PR3 for 20 min at 37 °C in low-salt buffer. We performed SDS/PAGE and dot blot analysis, followed by transfer to nitrocellulose, visualizing protein with fluorescent streptavidin (Fig. 4).
Fig. 4.
Reactivity of PK101 with NE and PR3. Purified NE or PR3 was treated with a range of PK101 concentrations, denatured in SDS sample buffer, and analyzed by dot blot (A) or SDS/PAGE followed by transfer (B and C). One sample was preincubated under the same conditions with 10 mM MeOSuc-Ala-Ala-Pro-Val-CH2Cl for 30 min to decrease probe binding (first samples in each column). (D) Supernatants from formyl-Met-Leu-Phe neutrophils were collected by centrifugation and treated with the indicated concentrations of PK101. Blots were developed with labeled streptavidin and imaged by fluorescence or bioluminescence scanning, as described in SI Appendix, Materials and Methods.
Each analysis revealed that the probe labels both NE and PR3, with labeling of NE being more efficient. This is to be expected from the calculated inhibition rates. We could have decreased the incubation time to decrease PR3 binding, but this was not practical, given the planned experiments with live neutrophils, so we settled on 20 min incubation. Importantly, the competing inhibitor MeOSuc-Ala-Ala-Pro-Val-CH2Cl almost completely abrogated probe binding to NE, with almost no effect on PR3 binding under the conditions used. PK101 showed binding to a pair of bands of ∼30 kDa in supernatants from neutrophils treated to secrete granule contents, which would include NE (Fig. 4D), indicating lack of off-target reactivity of the probe under the conditions analyzed. These bands are typical of carbohydrate microheterogeneity of natural NE (24), and PK101 reactivity was largely competed by MeOSuc-Ala-Ala-Pro-Val-CH2Cl, suggesting that the bands represent endogenous NE.
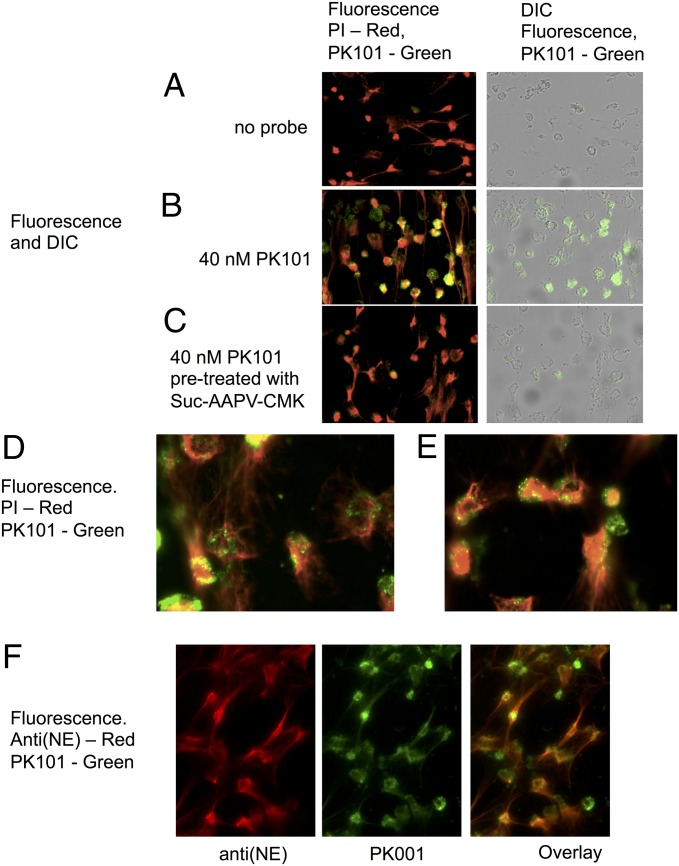
To test the specificity and utility of the biotinylated probe PK101, we chose to examine the location and activity of NE in neutrophil extracellular traps (NETs). These structures are reported to neutralize pathogens via the action of antimicrobial proteins bound to DNA that is extruded from neutrophils after stimulation or pharmacologic treatment in a manner proposed to be dependent on the activity of NE (25). NETs were formed from freshly isolated human neutrophils on glass coverslips after treatment with phorbol ester for 2.5 h at 37 °C in 5% (vol/vol) CO2. Probe binding was detected by fluorescent streptavidin, and NET formation was detected by propidium iodide (PI) staining of extruded DNA (Fig. 5).
Fig. 5.
Imaging of neutrophils and NETs treated with PK101. NET formation was triggered with phorbol ester, followed by 20 min of treatment with 40 nM PK101. It was briefly washed, fixed, and incubated with fluorescent streptavidin to visualize the probe, and PI was used to visualize DNA. (A) No probe. (B) Probe added. (C) Samples treated with 10 mM MeOSuc-Ala-Ala-Pro-Val-CH2Cl for 30 min before addition of probe. (A–C) Imaged using a 20× objective; two overlaid images from the same experiment are seen in D and E with a 40× objective. Representative images of 4 experiments using 2 different donors. NET formation is visible as PI-stained strings and dispersed nuclear material, and PK101 labeling is visible as punctate staining associated with cellular material, but not with NETs. NETs treated with PK101 followed by anti(NE) antiserum and imaged using a 40× objective are shown in F. The total NE antigen parallels PI binding, demonstrating that NETS are decorate with NE, but the overlaid image demonstrates than most of the NE on NETs is not active but is concentrated in the cell-associated punctate structures.
PK101 labeled material that was noticeably competed away with MeOSuc-Ala-Ala-Pro-Val-CH2Cl, indicating that proteolytic activity was present in punctate areas associated primarily in a perinuclear location. These are likely to be azurophil granules, the primary storage location of NE and PR3. NETs also contain PR3, but in lower abundance than NE (26). Because staining was substantially competed by MeOSuc-Ala-Ala-Pro-Val-CH2Cl, the majority of staining was caused by NE. Under the conditions used, although NET formation (revealed by string-like PI staining) is readily apparent, very little active NE is associated with the NETs. At first glance, this seems surprising, as it has been shown that NE is extruded together with nuclear material during NET formation (27). However, the advantage of using an activity-based probe is that it reveals the location of active, not just total, enzyme. Therefore, it appears NE associated with NETS is either not active in this location or is present only in a very low amount in the two individuals examined.
Discussion
Proteases define one of the largest groups of enzymes, and it has proven difficult to selectively measure their activity in biological samples because of the presence of multiple family members with overlapping specificity (1, 19, 28). To overcome this problem, substantial effort has been exerted by many groups over the years to understand the principles of recognition of natural and artificial substrates and to leverage this information to produce definitive diagnostic tools for in vivo and in vitro work. HyCoSuL provides a powerful tool in this quest, as it is based on classic and groundbreaking conventional positional scanning substrate library principles (6) but explores far more chemical space. Our objective was to create the very best NE substrate, not the most selective one. In doing so, we achieved the most sensitive substrate yet reported for NE, with a specificity constant of 4.79 × 107 M−1·s−1, about 100-fold more sensitive than the previous champion NE substrate Abz-Ala-Pro-Glu-Glu-Ile/Met-Arg-Arg-Gln-EDDnp [a fluorescence-quenched peptide where Abz is ortho-aminobenzoic acid and EDDnp is N-(2, 4-dinitrophenyl)ethylenediamine] (17). Interestingly, our optimal peptide has about the same degree of selectivity for human NE as Abz-APEEI/MRRQ-EDDnp. Importantly, our substrate can be converted to an activity-based probe, whereas the former champion cannot. One could imagine gaining even more selectivity for NE over PR3 by screening for subsite preferences of PR3, using HyCoSuL, and selecting ratio preferences that would substantially enhance the current selectivity ratio of 150 described by our PK101 probe, although this may take a hit in terms of absolute sensitivity for NE. This strategy seems not to be needed in the case of NE and PR3 but could become useful in situations in which more proteases with overlapping specificities need to be deconvoluted (e.g., caspases and kallikreins).
The conversion from a HyCoSuL-derived optimal substrate to an activity-based probe that preserves a similar ability to discriminate between NE and PR3 as the parent substrate has provided us with an important tool. With it we defined the location of active NE during NET formation, revealing the utility of this approach in dissecting biological events and as a potential biomarker. As pointed out earlier, in the case of proteases, which are usually stored in inactive forms until required, it is not the presence of protein that defines a biological event, it is the presence of active protein (29, 30). Thus, it comes as somewhat of a surprise that active NE revealed by PK101 is largely absent from NETs and is highly enriched in granular structures (Fig. 5 F and G), whereas NE antigen itself is readily found in NETs (Fig. 5H; ref. 31) and is reported to be the most abundant nonhistone protein present in NETs (26). Our data imply that NETs may be decorated with inactive NE. A recent report suggests that NE activity in NETs may vary considerably between donors (32), and at this time, we have only imaged NETs from three donors, with equivalent results. Not only is NE present on NETs but its activity is required for chromatin decondensation, leading to NET formation in response to phorbol ester and microbial stimuli (33). Signaling leading to NET formation, however, is dependent on the stimulus (25). PK101 provides an ideal tool to test the hypothesis that NE is required for NET formation by these and other stimuli in vitro and in vivo (33) and may form the basis of a sensitive and selective biomarker for defining the role of NE and neutrophils in innate immunity pathology.
Methods
Synthesis of the HyCoSuL library and individual substrates was carried out using solid phase strategies. The activity-based probe was synthesized using combined solid phase and solution phase strategies. Biochemical assays were performed with purified natural enzymes, and in situ activity-based probe labeling was carried out using freshly-isolated human donor neutrophils. For additional information please see SI Appendix, Materials and Methods.
Supplementary Material
Acknowledgments
The M.D. laboratory is supported by the Foundation for Polish Science. The C.C.W. laboratory is supported by the Health Research Council of New Zealand. The G.S.S. laboratory is supported by National Institutes of Health Grants R01GM09040 and R01CA163743.
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1318548111/-/DCSupplemental.
References
- 1.Drag M, Salvesen GS. Emerging principles in protease-based drug discovery. Nat Rev Drug Discov. 2010;9(9):690–701. doi: 10.1038/nrd3053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Turk B. Targeting proteases: Successes, failures and future prospects. Nat Rev Drug Discov. 2006;5(9):785–799. doi: 10.1038/nrd2092. [DOI] [PubMed] [Google Scholar]
- 3.Poreba M, Drag M. Current strategies for probing substrate specificity of proteases. Curr Med Chem. 2010;17(33):3968–3995. doi: 10.2174/092986710793205381. [DOI] [PubMed] [Google Scholar]
- 4.Lim MD, Craik CS. Using specificity to strategically target proteases. Bioorg Med Chem. 2009;17(3):1094–1100. doi: 10.1016/j.bmc.2008.03.068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Choe Y, et al. Substrate profiling of cysteine proteases using a combinatorial peptide library identifies functionally unique specificities. J Biol Chem. 2006;281(18):12824–12832. doi: 10.1074/jbc.M513331200. [DOI] [PubMed] [Google Scholar]
- 6.Harris JL, et al. Rapid and general profiling of protease specificity by using combinatorial fluorogenic substrate libraries. Proc Natl Acad Sci USA. 2000;97(14):7754–7759. doi: 10.1073/pnas.140132697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kasperkiewicz P, Gajda AD, Drąg M. Current and prospective applications of non-proteinogenic amino acids in profiling of proteases substrate specificity. Biol Chem. 2012;393(9):843–851. doi: 10.1515/hsz-2012-0167. [DOI] [PubMed] [Google Scholar]
- 8.Deu E, Verdoes M, Bogyo M. New approaches for dissecting protease functions to improve probe development and drug discovery. Nat Struct Mol Biol. 2012;19(1):9–16. doi: 10.1038/nsmb.2203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rano TA, et al. A combinatorial approach for determining protease specificities: Application to interleukin-1beta converting enzyme (ICE) Chem Biol. 1997;4(2):149–155. doi: 10.1016/s1074-5521(97)90258-1. [DOI] [PubMed] [Google Scholar]
- 10.Schechter I, Berger A. On the size of the active site in proteases. I. Papain. Biochem Biophys Res Commun. 1967;27(2):157–162. doi: 10.1016/s0006-291x(67)80055-x. [DOI] [PubMed] [Google Scholar]
- 11.Drag M, Bogyo M, Ellman JA, Salvesen GS. Aminopeptidase fingerprints, an integrated approach for identification of good substrates and optimal inhibitors. J Biol Chem. 2010;285(5):3310–3318. doi: 10.1074/jbc.M109.060418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zervoudi E, et al. Probing the S1 specificity pocket of the aminopeptidases that generate antigenic peptides. Biochem J. 2011;435(2):411–420. doi: 10.1042/BJ20102049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Poreba M, et al. Fingerprinting the substrate specificity of M1 and M17 aminopeptidases of human malaria, Plasmodium falciparum. PLoS ONE. 2012;7(2):e31938. doi: 10.1371/journal.pone.0031938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Korkmaz B, Horwitz MS, Jenne DE, Gauthier F. Neutrophil elastase, proteinase 3, and cathepsin G as therapeutic targets in human diseases. Pharmacol Rev. 2010;62(4):726–759. doi: 10.1124/pr.110.002733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Moroy G, Alix AJ, Sapi J, Hornebeck W, Bourguet E. Neutrophil elastase as a target in lung cancer. Anticancer Agents Med Chem. 2012;12(6):565–579. doi: 10.2174/187152012800617696. [DOI] [PubMed] [Google Scholar]
- 16.Rawlings ND, Barrett AJ, Bateman A. MEROPS: The database of proteolytic enzymes, their substrates and inhibitors. Nucleic Acids Res. 2012;40(Database issue):D343–D350. doi: 10.1093/nar/gkr987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Korkmaz B, et al. Measurement of neutrophil elastase, proteinase 3, and cathepsin G activities using intramolecularly quenched fluorogenic substrates. Methods Mol Biol. 2012;844:125–138. doi: 10.1007/978-1-61779-527-5_9. [DOI] [PubMed] [Google Scholar]
- 18.Maly DJ, et al. Expedient solid-phase synthesis of fluorogenic protease substrates using the 7-amino-4-carbamoylmethylcoumarin (ACC) fluorophore. J Org Chem. 2002;67(3):910–915. doi: 10.1021/jo016140o. [DOI] [PubMed] [Google Scholar]
- 19.Schilling O, Overall CM. Proteome-derived, database-searchable peptide libraries for identifying protease cleavage sites. Nat Biotechnol. 2008;26(6):685–694. doi: 10.1038/nbt1408. [DOI] [PubMed] [Google Scholar]
- 20.Castillo MJ, Nakajima K, Zimmerman M, Powers JC. Sensitive substrates for human leukocyte and porcine pancreatic elastase: A study of the merits of various chromophoric and fluorogenic leaving groups in assays for serine proteases. Anal Biochem. 1979;99(1):53–64. doi: 10.1016/0003-2697(79)90043-5. [DOI] [PubMed] [Google Scholar]
- 21. Rao NV, Hoidal JR (2013) Myeloblastin. Handbook of Proteolytic Enzymes, ed Rawlings ND, Salvesen G (Academic Press, London), 3rd Ed, pp 2666–2675.
- 22.Soroka M, Goldeman W. 2007. Polish Patent PL 196222.
- 23.Salvesen GS, Nagase H. 2001. Inhibition of proteolytic enzymes. Proteolytic Enzymes: A Practical Approach, ed Bond JS, Beynon RJ (Oxford University Press, Oxford), 2nd Ed, pp 105–130.
- 24.Watorek W, Farley D, Salvesen G, Travis J. Neutrophil elastase and cathepsin G: Structure, function, and biological control. Adv Exp Med Biol. 1988;240:23–31. doi: 10.1007/978-1-4613-1057-0_3. [DOI] [PubMed] [Google Scholar]
- 25.Parker H, Dragunow M, Hampton MB, Kettle AJ, Winterbourn CC. Requirements for NADPH oxidase and myeloperoxidase in neutrophil extracellular trap formation differ depending on the stimulus. J Leukoc Biol. 2012;92(4):841–849. doi: 10.1189/jlb.1211601. [DOI] [PubMed] [Google Scholar]
- 26.Urban CF, et al. Neutrophil extracellular traps contain calprotectin, a cytosolic protein complex involved in host defense against Candida albicans. PLoS Pathog. 2009;5(10):e1000639. doi: 10.1371/journal.ppat.1000639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fuchs TA, et al. Novel cell death program leads to neutrophil extracellular traps. J Cell Biol. 2007;176(2):231–241. doi: 10.1083/jcb.200606027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sexton KB, Witte MD, Blum G, Bogyo M. Design of cell-permeable, fluorescent activity-based probes for the lysosomal cysteine protease asparaginyl endopeptidase (AEP)/legumain. Bioorg Med Chem Lett. 2007;17(3):649–653. doi: 10.1016/j.bmcl.2006.10.100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bogyo M, et al. 2004. Applications for chemical probes of proteolytic activity. Curr Protoc Protein Sci Chapter 21:Unit 21.17.
- 30.Liu Y, Patricelli MP, Cravatt BF. Activity-based protein profiling: The serine hydrolases. Proc Natl Acad Sci USA. 1999;96(26):14694–14699. doi: 10.1073/pnas.96.26.14694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Brinkmann V, Zychlinsky A. Neutrophil extracellular traps: Is immunity the second function of chromatin? J Cell Biol. 2012;198(5):773–783. doi: 10.1083/jcb.201203170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Barrientos L, et al. An improved strategy to recover large fragments of functional human neutrophil extracellular traps. Front Immunol. 2013 doi: 10.3389/fimmu.2013.00166. 24;4:166–175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Papayannopoulos V, Metzler KD, Hakkim A, Zychlinsky A. Neutrophil elastase and myeloperoxidase regulate the formation of neutrophil extracellular traps. J Cell Biol. 2010;191(3):677–691. doi: 10.1083/jcb.201006052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bode W, et al. X-ray crystal structure of the complex of human leukocyte elastase (PMN elastase) and the third domain of the turkey ovomucoid inhibitor. EMBO J. 1986;5(10):2453–2458. doi: 10.1002/j.1460-2075.1986.tb04521.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sieńczyk M, Oleksyszyn J. Irreversible inhibition of serine proteases - design and in vivo activity of diaryl alpha-aminophosphonate derivatives. Curr Med Chem. 2009;16(13):1673–1687. doi: 10.2174/092986709788186246. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.