Significance
This paper uses NMR methods to compare the dynamics of a protein kinase in its active and inactive states. The results show that domain movements in the MAP kinase ERK2 are inherently constrained until the enzyme is activated by phosphorylation, with the constraint located at the hinge region. This represents an important mode for dynamical regulation in ERK2, not anticipated from previous X-ray structural analyses.
Abstract
Protein motions control enzyme catalysis through mechanisms that are incompletely understood. Here NMR 13C relaxation dispersion experiments were used to monitor changes in side-chain motions that occur in response to activation by phosphorylation of the MAP kinase ERK2. NMR data for the methyl side chains on Ile, Leu, and Val residues showed changes in conformational exchange dynamics in the microsecond-to-millisecond time regime between the different activity states of ERK2. In inactive, unphosphorylated ERK2, localized conformational exchange was observed among methyl side chains, with little evidence for coupling between residues. Upon dual phosphorylation by MAP kinase kinase 1, the dynamics of assigned methyls in ERK2 were altered throughout the conserved kinase core, including many residues in the catalytic pocket. The majority of residues in active ERK2 fit to a single conformational exchange process, with kex ≈ 300 s−1 (kAB ≈ 240 s−1/kBA ≈ 60 s−1) and pA/pB ≈ 20%/80%, suggesting global domain motions involving interconversion between two states. A mutant of ERK2, engineered to enhance conformational mobility at the hinge region linking the N- and C-terminal domains, also induced two-state conformational exchange throughout the kinase core, with exchange properties of kex ≈ 500 s−1 (kAB ≈ 15 s−1/kBA ≈ 485 s−1) and pA/pB ≈ 97%/3%. Thus, phosphorylation and activation of ERK2 lead to a dramatic shift in conformational exchange dynamics, likely through release of constraints at the hinge.
The MAP kinase, extracellular signal-regulated kinase 2 (ERK2), is a key regulator of cell signaling and a model for protein kinase activation mechanisms (1). ERK2 can be activated by MAP kinase kinases 1 and 2 (MKK1 and 2) through dual phosphorylation of Thr and Tyr residues located at the activation loop (Thr183 and Tyr185, numbered in rat ERK2) (1, 2). Phosphorylation at both sites is required for kinase activation, resulting in increased phosphoryl transfer rate and enhanced affinity for ATP and substrate (3).
Conformational changes accompanying the activation of ERK2 have been documented by X-ray structures of the inactive, unphosphorylated (0P-ERK2) and the active, dual-phosphorylated (2P-ERK2) forms (4, 5). Phosphorylation rearranges the activation loop, leading to new ion-pair interactions between phospho-Thr and phospho-Tyr residues and basic residues in the N- and C-terminal domains of the kinase core structure. This leads to a repositioning of active site residues surrounding the catalytic base, enabling recognition of the Ser/Thr-Pro sequence motif at phosphorylation sites and exposing a recognition site for interactions with docking sequences in substrates and scaffolds (6).
Less is known about how changes in internal motions contribute to kinase activation. Previous studies using hydrogen-exchange mass spectrometry (HX-MS) and electron paramagnetic resonance spectroscopy (7–9) led to a model where conformational mobility at the hinge linking the N- and C-terminal domains is increased by phosphorylation, therefore releasing constraints needed for activation. Such a model differs from other types of autoinhibitory mechanisms in protein kinases, which involve interactions with domains outside the kinase core (10, 11). However, how hinge flexibility regulates ERK2 is unknown.
NMR relaxation dispersion methods enable protein dynamics to be monitored by measuring exchange between conformational states (12). In particular, Carr–Purcell–Meiboom–Gill (CPMG) relaxation dispersion experiments report on motions on slow (100–2,000 s−1) timescales (13), which are often important for enzymatic function (13–16). In the CPMG experiment, exchange between different conformational states is probed with varying times between “refocusing” pulses. Conformational exchange leads to imperfect refocusing, thus decreasing the intensity of the NMR signal. Increasing the pulse frequency allows less chance for conformational exchange, and therefore increased NMR signal intensity. For a given pulse frequency, analysis of the signal intensity yields the effective relaxation rate for the resonance, R2,eff. This is typically plotted as a relaxation dispersion curve, which can be fit to a two-state conformational exchange process (e.g., A ⇌ B interconversion). Fitting extracts the populations and the exchange rates between states, thus reflecting the thermodynamics and kinetics of the system (17, 18).
Here we performed CPMG relaxation dispersion experiments at multiple field strengths to compare the dynamic properties of [13C]methyl-labeled ERK2 in its phosphorylated and unphosphorylated states. The results demonstrate that phosphorylation causes a significant change in exchange dynamics throughout the kinase core, consistent with a global domain motion. Increasing hinge mobility by introducing mutations at the hinge also promotes domain motion within the core but with differing kinetics and populations. Taken together, the results show that large changes in dynamics accompany ERK2 phosphorylation, which are influenced by conformational mobility at the hinge. We propose that the activation of ERK2 involves removing inhibitory constraints to domain motion, which are conferred by the internal architecture of the kinase.
Results
NMR Peak Assignments and Chemical Shift Behavior.
ERK2 was selectively labeled with [methyl-1H,13C]Ile, -Leu, and -Val (ILV) as described in Materials and Methods (19, 20) and activated by in vitro phosphorylation using active MKK1 (21). Fig. 1A overlays 2D (13C,1H) methyl transverse relaxation-optimized spectroscopy (TROSY) heteronuclear multiple-quantum coherence (HMQC) spectra for 0P- and 2P-ERK2 at 25 °C. In 0P-ERK2, 140 of 144 predicted ILV methyl resonance peaks were observed. Of these, 70 methyl peaks were assigned to 60 residues by combining data from site-directed mutagenesis, through-space 1H-1H nuclear Overhauser effects (NOEs) between 1H,13C-labeled methyls analyzed using the X-ray structure of 0P-ERK2, and through-bond intraresidue (HMCM[CG]CBCA) experiments (Fig. S1) (4, 5, 22). Most ILV residues in the hydrophobic core of the kinase were assigned, including those in the structurally conserved α-helices (αC–αH) and β-strands (β1–β5, β7, β8) as well as nonconserved helices in the MAP kinase insert (α1L14, α2L14) and C terminus (αL16). Assignments were limited in the activation loop and surface-accessible loops, due to the lack of methyl-methyl NOEs in these regions. In 2P-ERK2, 137 of 144 predicted methyl resonances were observed, of which 67 were assigned by transferring assignments from 0P-ERK2 and confirmed by methyl-methyl NOE measurements of 2P-ERK2.
Fig. 1.
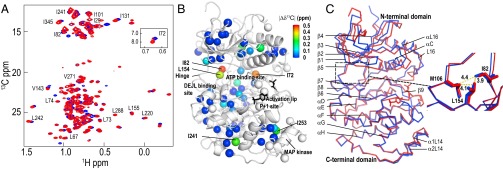
ERK2 phosphorylation induces chemical shift changes at the hinge region. (A) Two-dimensional (13C,1H) HMQC spectra of [methyl-13C]ILV-labeled 0P-ERK2 (blue) and 2P-ERK2 (red) recorded at 800 MHz and 25 °C. (B) X-ray structure of 0P-ERK2 (Protein Data Bank ID code 1ERK) showing spheres representing positions of ILV methyls, with 13C chemical shift differences (|Δδ13C|) between 0P-ERK2 and 2P-ERK2 indicated by the color scale (also see Fig. S2). Unassigned ILV methyls are represented by white spheres. DEJL is the docking site motif for ERK, JNK, and LXL. (C) Structural alignment of 0P-ERK2 (blue) and 2P-ERK2 (red). The expansion shows that the side chains of L154 and I82, which have two of the largest |Δδ13C| (B), form hydrophobic interactions with each other and with M106 at the hinge. Structure images were prepared using PyMOL (www.pymol.org).
The differences in [13C]methyl chemical shifts between 0P- and 2P-ERK2 (in ppm), |Δδ13C|, provide a sensitive probe of changes in the local environment. Eight residues showed significant chemical shift differences between the two forms of ERK2, ranging between 0.16 and 0.47 ppm, whereas 16 residues showed moderate changes, ranging between 0.1 and 0.15 ppm (Fig. 1B and Fig. S2). Two of the three residues with the largest |Δδ13C| were I82 and L154 (0.47 and 0.29 ppm, respectively), which interact with the hinge that links the N- and C-terminal domains. L154 is located in the β7-strand, and I82 is located in the αC-β4 loop. These form hydrophobic contacts with each other as well as with M106 at the hinge (Fig. 1C). Large chemical shift changes at these residues were surprising, given their distance from the site of phosphorylation. The X-ray structures showed no significant conformational differences between 0P- and 2P-ERK2 around these residues (Fig. 1C). Significant |Δδ13C| were also observed at I72 near the ATP binding site in helix αC, I241 in helix αG, and I253 in the MAP kinase insert (0.41, 0.26, and 0.25 ppm, respectively). Other residues with measurable |Δδ13C| are shown in Fig. S2. Based on the X-ray structures, chemical shift changes were expected in the activation loop and C-terminal L16 loop, but could not be determined due to incomplete assignments. Overall, the NMR chemical shift data reported significant changes in the chemical environment of ILV methyl side chains at the hinge, ATP binding site, and MAP kinase insert upon phosphorylation of ERK2.
Residues in 0P-ERK2 Show Local Conformational Dynamics.
Conformational dynamics of ILV methyl groups were examined in 0P-ERK2 using (13C,1H) multiple-quantum CPMG relaxation dispersion experiments (18). Methyl groups undergoing conformational exchange on the microsecond-to-millisecond timescale showed changes in their effective relaxation rate R2,eff, measured as a function of the frequency of the refocusing pulses (νCPMG). Relaxation dispersion curves, collected at 25 °C and at three field strengths (600, 800, and 900 MHz), are shown in Fig. 2 and Fig. S3. Dynamics were indicated when Rex, the contribution to R2,eff from exchange, was significant (>4 s−1), yielding curvature in the plot of R2,eff vs. νCPMG (Fig. 2 and Table S1). No evidence for dynamics was indicated when Rex ≈ 0 s−1. For individual ILV methyl groups, the dispersion curves at 600, 800, and 900 MHz were fit to a two-state exchange model (A ⇌ B), yielding exchange rate constants (kex = kAB + kBA) (17, 18). Under optimal conditions, the populations (pA and pB) and the 13C chemical shift differences between the two states, |Δω13C| (in Hz, or |ΔδCPMG13C| in ppm), were obtained for individual methyl groups (17, 18).
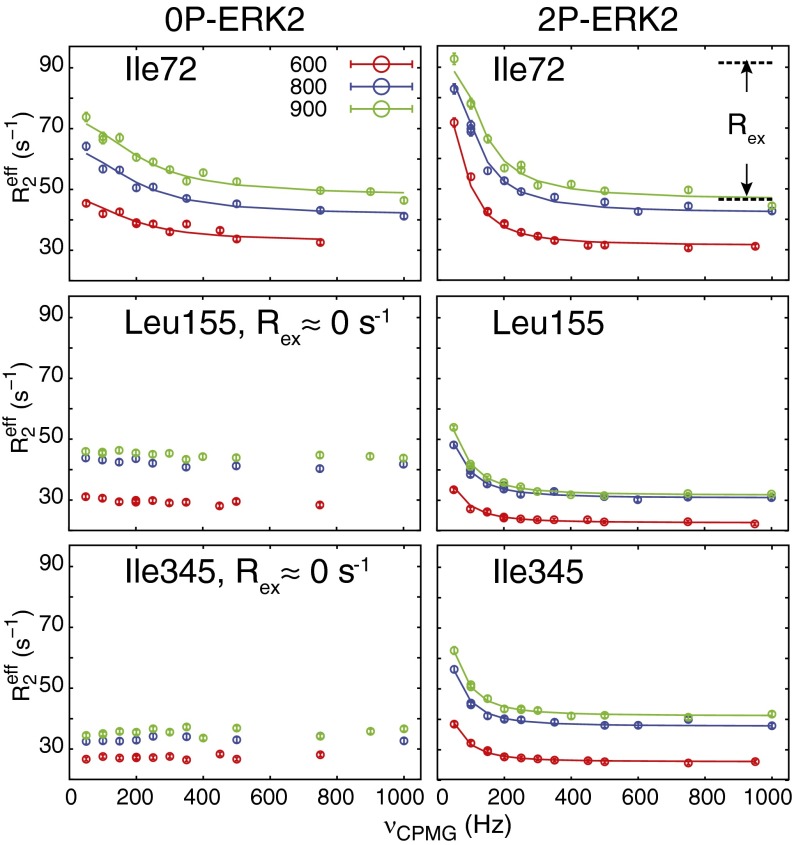
Fig. 2.
CPMG dispersion experiments show changes in methyl dynamics following ERK2 phosphorylation. The dispersion curves measured at 25 °C and 900 (green), 800 (blue), and 600 (red) MHz are shown for representative residues in 0P-ERK2 (Left) and 2P-ERK2 (Right). The Rex is shown for I72 in 2P-ERK2 at 800 MHz. Lines show individual fits to the Carver–Richards equation, and the error bars were estimated from peak intensities of duplicate measurements.
Methyl peaks from 13 residues in 0P-ERK2 showed significant conformational exchange dynamics (Fig. 3A and Table S1). These residues were clustered in three regions located in the cleft between the N- and C-terminal domains, including the αC-β4 loop, β7-β8 loop, and helix αE. Other regions with significant exchange were located near the activation loop and the P+1 loop, helix αG, and helices in the MAP kinase insert, α1L14/α2L14. Individual fits of the relaxation dispersion data yielded kex ranging between 900 and 2,200 s−1 (Table S1). These exchange processes were fast on the NMR chemical shift timescale (i.e., kex > |Δω13C|), and therefore it was not possible to confidently extract populations and chemical shift differences for individual methyls. Attempts to globally fit all or subsets of residues resulted in poorly defined populations. Thus, relaxation dispersion measurements on 0P-ERK2 were inconsistent with ILV residues undergoing a single exchange process. Instead, they reported fast conformational exchange processes in subdomains of the kinase, with little or no evidence for coupling between residues, including those with interacting methyl groups.
Fig. 3.
ERK2 phosphorylation induces global conformational exchange in the kinase core. (A) Assigned methyls in 0P-ERK2 are shown as spheres, with kex values indicated by the color scale for individual fits of methyls that have Rex > 4 s−1. The asterisks and double asterisks indicate methyls having errors in kex > 50% and kex > 100%, respectively, with the latter methyls shown as black spheres. Methyls with no observable dynamics (Rex ≈ 0 s−1) are shown as white spheres. Thr183 and Tyr185 are shown as black sticks. (B) Residues in 2P-ERK2 with observable dynamics each fitted individually, colored as in A. (C) In 2P-ERK2, 19 methyls could be fit globally to a single rate constant (kex = 300 ± 10 s−1) and population (pA/pB = 80/20 ± 0.6%), demonstrating global domain motion for residues throughout the kinase core. Residues in the MAP kinase insert subdomain could not be fit by a global process.
Phosphorylation of ERK2 Induces Large-Scale Conformational Exchange Dynamics.
Significant changes in conformational exchange dynamics were observed between 0P-ERK2 and 2P-ERK2, as reflected by differences in their dispersion curves (Fig. 2). Methyls from 22 residues in 2P-ERK2 showed significant Rex. For many ILV methyls, the values of kex, populations, and 13C chemical shift differences between the two states in ppm |ΔδCPMG13C| could be confidently fit for individual residues (Fig. 3B and Table S2). Nineteen residues throughout the kinase core and in the catalytic pocket could be globally fit (Fig. 3C), consistent with a single exchange process with kex = 300 ± 10 s−1 and pA and pB populations of 20% and 80% (± 0.6%). They included residues at the hinge, αC-β4 loop, and β5, which formed the hydrophobic cluster at the cleft between the N- and C-terminal domains; N-terminal domain residues located at the interface between αC and β3-β4 strands; C-terminal domain residues at the interface between αE and αF; and C-terminal domain residues between αF and αG (Fig. 3C). In contrast, residues in the MAP kinase insert could not be fit globally, but individual fits yielded kex values of 640–750 s−1, significantly lower than the corresponding kex of 1,200–2,200 s−1 in 0P-ERK2. The results revealed that phosphorylation of ERK2 induces a dramatic change in dynamics, where internal motions become dominated by a slow-exchange process characterized by interconversion between two major conformational states with populations of 20% and 80%.
Phosphorylation Shifts the Equilibrium Between Two Conformational States.
We next examined the temperature dependence of the HMQC spectra of 2P-ERK2. Several methyls appeared as two discrete peaks at 5 °C, indicating two conformational states in slow exchange on the NMR chemical shift timescale (Fig. 4A). These methyls showed similar populations of the two states as a function of temperature, varying from 50%:50% at 5 °C to 80%:20% at 25 °C. Importantly, the populations at 25 °C matched those obtained from the global fits of the relaxation dispersion experiments (Table S2). This demonstrates that the conformational exchange process reflects two discrete conformers, which shift their populations with temperature.
Fig. 4.
Methyl peaks reveal a slow conformational exchange process in 2P-ERK2. (A) Two-dimensional (13C,1H) methyl HMQC spectra of 2P-ERK2 from 5 to 25 °C show that many methyls have two distinct peaks that are undergoing slow conformational exchange on the NMR chemical shift timescale. The black lines on the 5-°C spectra highlight the two exchanging peaks, indicating two well-populated states. (B) Overlay of spectra from 0P-ERK2 (black) and 2P-ERK2 (red) at 25 °C shows that the higher-populated state in 0P-ERK2 corresponds to the lower-populated state in 2P-ERK2.
Thermodynamic parameters determined from van’t Hoff plots were most accurately measured for residue I72, yielding ΔH° = 9.7 ± 1.1 kcal/mol and ΔS° = 36 ± 2 cal⋅mol−1⋅K−1 (Fig. S4A). Thermodynamic parameters estimated for other residues (e.g., I72, I131, and L288) were consistent with these values, indicating that the same conformational exchange processes underlie dynamics throughout most of the molecule. The exception was I241, located within helix αG, which gave ΔH° = 5.8 ± 1.1 kcal/mol and ΔS° = 22 ± 3 cal⋅mol−1⋅K−1 (Fig. S4B), indicating a distinct process.
In 0P-ERK2, each methyl group appeared as one peak in the HMQC spectra. In each case, peaks from 0P-ERK2 overlapped well with those corresponding to the minor conformational state in 2P-ERK2 (i.e., with 20% population at 25 °C) (Fig. 4B). Thus, based on chemical shifts, the major state in 0P-ERK2 corresponds to the minor state in 2P-ERK2. In 0P-ERK2, the single observed conformer (population >99.5%) exchanges with an unobservable conformer (population <0.5%), from which ΔG° can be estimated as >+3.1 kcal/mol. In 2P-ERK2, interconverting conformers represent two states with populations of 80% and 20%, yielding ΔG° = −0.8 kcal/mol. Two-dimensional (13C,1H) HMQC experiments performed on 0P-ERK2 and 2P-ERK2 in the presence and absence of saturating Mg2+-adenosine-5′-(β,γ-imido)triphosphate (AMP-PNP) showed little or no effect on the populations in either kinase, indicating that the major state in 2P-ERK2 is not affected by nucleotide binding (Fig. S5).
The Conformational Switch Is Controlled by Hinge Residues.
We next asked what type of dynamics might be reflected by a large-scale two-state exchange process involving residues throughout the consensus kinase core. Prior HX-MS studies suggested that phosphorylation of ERK2 leads to enhanced backbone flexibility at the hinge, reducing the constraint for interdomain motion between the N- and C-terminal domains (9). We therefore asked whether increased hinge motion could affect the conformational exchange process, as measured by NMR.
To address this, mutations were engineered to increase conformational mobility at the hinge, by replacing residues M106-E107 with G-G to create an “ME/GG-ERK2” mutant. HMQC spectra were acquired on the [methyl-13C]ILV-labeled ME/GG-ERK2 and assigned as described in Materials and Methods. The chemical shifts of the methyls in ME/GG-ERK2 overlaid well with wild-type 0P-ERK2, except for residues near the mutation site, where structural changes would be expected (Fig. 5 A and B).
Fig. 5.
ME/GG hinge mutations induce conformational exchange in the kinase core but not the MAP kinase insert. (A) Overlays of several methyl regions of the 2D HMQC spectra showing similar chemical shifts for these methyls in 0P-ERK2 (blue) and ME/GG-ERK2 (red). (B) The |Δδ13C| between 0P-ERK2 and ME/GG-ERK2 are indicated by the color scale, and unassigned ILV methyls are represented by white spheres. The |Δδ13C| between 0P-ERK2 and ME/GG-ERK2 are small throughout the protein, except for residues close to the mutated M106 and E107 (shown in pink). (C) Sixteen methyls in ME/GG-ERK2 could be globally fit with kex (500 ± 60 s−1; shown as cyan spheres) and population (97 and 3 ± 0.2%), indicating a single exchange process throughout the kinase core, similar to 2P-ERK2. The assigned methyls in the MAP kinase insert are shown as spheres, with kex values indicated by the color scale.
Relaxation dispersion experiments on unphosphorylated ME/GG-ERK2 showed that methyls on 21 ILV residues had significant dynamics, located throughout the enzyme (Fig. 5C and Table S3). These corresponded to 12 of the 13 residues with significant Rex in 0P-ERK2, and 14 of 22 residues with significant Rex in 2P-ERK2. Importantly, 16 residues throughout the kinase core could be fit to a single exchange model, indicating a global motion with kex = 500 ± 60 s−1 and populations of 97 and 3 ± 0.2% (Fig. 5C). The similar chemical shifts for ME/GG-ERK2 and 0P-ERK2 mean that the dominant state is similar in both proteins. Within the MAP kinase insert of ME/GG-ERK2, kex values of 1,200–1,600 s−1 were observed, comparable to the exchange rate constants in 0P-ERK2. Therefore, ME/GG partially mimics the shift in conformational exchange dynamics induced by phosphorylation and activation of ERK2.
Discussion
Our study demonstrates that the catalytic activation of a eukaryotic protein kinase elicits significant changes in protein dynamics. In 0P-ERK2, side chains show fast, localized motions on a microsecond-to-millisecond timescale, with little evidence of coupling. In the phosphorylated enzyme, side-chain motions are dominated by a global exchange process involving residues throughout the kinase core. The NMR data demonstrate that large-scale interdomain motion, with an exchange rate constant of 300 s−1, accompanies phosphorylation and activation. Motions within the MAP kinase insert indicated a separate process, illustrating that independent subdomain motion occurs outside the consensus kinase core. The results support a model, illustrated in Fig. 6A, in which unphosphorylated ERK2 is stabilized in the inactive conformer by an inhibitory constraint. This is released upon phosphorylation, allowing a global shift in equilibrium between conformational states, illustrated here as a hypothetical domain movement.
Fig. 6.
Model for activation of ERK2, involving the release of constraints to domain movement and change in energy landscape induced by phosphorylation. (A and B) Before phosphorylation, 0P-ERK2 is in an inactive conformation that is constrained from domain motion involving rigidity at the hinge (illustrated by the straight thick line between hinge residues M and E). The CPMG dispersion data show no evidence for additional conformations, indicating they must have low populations (<0.5%), reflecting ΔG° > +3.1 kcal/mol. Phosphorylation enhances hinge mobility (illustrated by the wavy thin line) and shifts the equilibrium to favor the active conformer with ΔG° = −0.8 kcal/mol and rate constants of kAB = 240 s−1 and kBA = 60 s−1. The ME/GG mutation partially relieves the constraint to domain motion by increasing mobility at the hinge, sampling of the active-like conformer (3%), and reducing ΔG° to +2.1 kcal/mol. The activation energy is not known for 0P-ERK2, indicated by the dashed line in B. (C) 0P-ERK2 and 2P-ERK2 structures show how phosphorylation at the activation loop promotes interactions between the N- and C-terminal domains through ion pairing between pT183 and Arg65/68 in helix αC, which may stabilize the active form. The ERK2 backbone is colored white, phosphates are colored orange, Thr183 and Tyr185 are colored red, Arg side chains are represented as sticks, and surfaces are shown for the guanidinium groups.
Lateral or rotational movements between the N- and C-terminal domains provide one conceptual model to describe the global motions observed in active ERK2. Interconversions between “open” and “closed” domain conformations have been noted in other enzymes, including protein kinases (23, 24). In the cAMP-dependent protein kinase catalytic subunit (PKA-C), the X-ray structure of the apo form shows an open conformation, whereas a binary complex with Mg2+-AMP-PNP shows a closed conformation formed by rotation of the N-terminal domain and closure of the catalytic cleft around the nucleotide (25, 26). NMR relaxation measurements of backbone amides in the nucleotide–PKA-C binary complex show global exchange behavior in residues lining the catalytic core (27, 28), which are absent in apo PKA-C, and have been interpreted as an equilibrium shift to a closed conformation. Thus, PKA-C and active ERK2 share characteristics consistent with interconversion between open and closed conformers, with comparable rates of conformational exchange (kex ≈ 200 s−1 in PKA-C and 300 s−1 in 2P-ERK2). However, in PKA-C, the closed conformation is stabilized by nucleotide binding (28), whereas Mg2+-AMP-PNP has little or no effect on the equilibrium between conformers in ERK2 (Fig. S5). This suggests that the simple domain closure observed in PKA-C may not adequately describe the dynamics in ERK2, where global exchange is allosterically stabilized by phosphorylation but not by ligand binding.
Other models are possible, for example, one involving rotation of secondary structures within the N-terminal domain. In X-ray structures of ERK2, phosphorylation induces rearrangement of the activation loop, which in turn directs movement of helices αC and αL16 and refolding of the C-terminal L16 extension (4, 5). A network of hydrogen bond and hydrophobic residue interactions throughout the N-terminal domain enables communication between these structures as well as connectivity with residues in the β3–β5 strands. It is possible that this network extends to the hinge through connections with the αC-β4 loop, and that the large chemical shift changes induced by phosphorylation within this region (Fig. 1B) reflect environmental changes due to rotational movements in the N-terminal domain.
An important finding from our study was that the unphosphorylated ME/GG hinge mutant induced a global exchange process similar to that of 2P-ERK2. This involved residues as widely spread across the kinase core as in 2P-ERK2, strongly implying that the constraints to global exchange in 0P-ERK2 can be released by enhancing hinge flexibility. Mechanically, ERK2 might be conceptualized as having a strong spring constant at the hinge, which is weakened upon either phosphorylation or mutation, thus leading to a change in population of the conformational states. Fig. 6A illustrates this model, where the ME/GG mutation bypasses the constraint to domain movement in 0P-ERK2 by mimicking the effect of phosphorylation on conformational mobility at the hinge. This increased hinge flexibility lowers the barrier to domain movement relative to 0P-ERK2. However, unlike 2P-ERK2, ME/GG-ERK2 showed a smaller population (3%, instead of 80%) of the “active-like” conformer (Fig. 6).
The specific activity of ME/GG-ERK2 (0.06 nmol⋅min−1⋅mg−1) was comparable to that of 0P-ERK2 (0.08 nmol⋅min−1⋅mg−1), both significantly lower than 2P-ERK2 (264 nmol⋅min−1⋅mg−1). The NMR relaxation dispersion data summarized in Fig. 6A show no correlation of the kinase activity with the kinetics or populations for any of the studied forms of ERK2. Instead, the data support a model where multiple events take place following the phosphorylation of ERK2. One event involves removal of a constraint to global exchange dynamics, illustrated by domain movement, which can be induced by either ME/GG or phosphorylation. A second event involves stabilization of the “active” conformation in 2P-ERK2, illustrated by ionic interactions that promote interactions between the N- and C-terminal domains (Fig. 6C). Further work is needed to understand the details of the conformational transitions, as well as the kinetic and thermodynamic contributions, that occur upon activation of ERK2.
Compared with the large amount of structural information on protein kinases, the understanding of how kinases are regulated at the level of internal protein motions is much less well developed. A major conclusion of this study is that, before phosphorylation, ERK2 is maintained in its inactive form by a mechanism that imposes constraints on protein dynamics. Unlike autoinhibitory mechanisms that involve intra- or intermolecular occlusion at the catalytic site in other kinases, the constraints in ERK2 involve the hinge region, distal from the site of phosphorylation. The importance of hinge mobility has been suggested in other protein kinases. For example, allosteric communication between the activation loop and hinge was reported in fibroblast growth factor receptor 2 (FGFR2), which forms an autoinhibitory “molecular brake” involving a triad of interacting residues at the hinge, αC-β4 loop, and β8 strand (29). Structural studies also showed an extensive hydrogen-bond network accompanied by reduced hinge flexibility in zeta-chain–associated protein kinase 70 (ZAP-70), which was proposed to maintain the kinase-inactive form (30). Finally, molecular dynamics calculations suggested local unfolding of the hinge in the catalytic domain of epidermal growth factor receptor (EGFR), which was proposed as an intermediate step toward the transition from the inactive to the active state (31, 32). However, in contrast to FGFR2, ZAP-70, and EGFR, the X-ray structures of ERK2 show no significant conformational changes around the hinge upon phosphorylation (4, 5). Likewise, both inactive and active X-ray structures of ERK2 show a properly formed catalytic site and an intact “hydrophobic spine” network, whose misalignment underlies the mechanism of inactivation in many protein kinases (33, 34). Thus, the mechanism of communication from the activation loop to the hinge most likely involves subtle reorganization of residues, suggesting that the autoinhibitory regulation in ERK2 involves architectural features that may be unique to this kinase. Our findings now set the stage for future studies to define the details of this architecture and understand its prevalence across the protein kinase superfamily.
Materials and Methods
Protein Preparation and Enzyme Assays.
Wild-type or mutant rat His6-ERK2 was expressed from the pET23a plasmid in Escherichia coli BL21(DE3) cells (35). [methyl-13C]ILV-labeled samples of 0P-ERK2 were prepared and purified as described in SI Materials and Methods. 2P-ERK2 was prepared from 0P-ERK2 in vitro using constitutively active mutant MKK1 as described (21). The buffer used for ERK2 in NMR experiments contained 50 mM Tris (pH 7.4), 150 mM NaCl, 5 mM MgSO4, 0.1 mM EDTA, 5 mM DTT, 100% D2O, and 2.5% (vol/vol) glycerol. Kinase activities of 0P-ERK2, ME/GG-ERK2, and 2P-ERK2 were measured as described in a previous study (36). Nucleotide binding experiments were performed by titrating AMP-PNP (Roche Applied Science) into the NMR samples of 0P-ERK2 and 2P-ERK2, to final AMP-PNP concentrations of 1.35 mM and 1.15 mM, respectively.
NMR Spectroscopy.
De novo nonstereospecific Ile, Leu, and Val methyl assignments ([1Hm,13Cm]) were made for methyl peaks of 0P-ERK2 using 3D (13C,13C,1H) HMQC-NOESY, 2D (13C,1H) methyl TROSY HMQC, and 3D (13C,13C,1H) HMCM[CG]CBCA experiments (SI Materials and Methods), carried out at 25 °C on a Varian 800-MHz NMR spectrometer with a cryoprobe. Leu and Val methyls were distinguished using HMCM[CG]CBCA experiments to facilitate accurate identification of ILV clusters (Fig. S1A). Spatially adjacent ILV methyls were identified in 3D (13C,13C,1H) HMQC-NOESY spectra, and assigned by mapping onto the X-ray structure of 0P-ERK2 (Fig. S1 B and C). Two-dimensional (13C,1H) methyl TROSY HMQC spectra of 10 ERK2 I, L, or V to A mutants were used to help assign individual methyls (Fig. S1D). Methyl assignments for ME/GG-ERK2 were made by transferring assignments from 0P-ERK2.
Methyl side-chain dynamics were measured using 2D (13C,1H) multiple-quantum (MQ) CPMG relaxation dispersion experiments (17, 18) on 0P- and 2P-ERK2 (SI Materials and Methods), performed at 25 °C on Varian 600-, 800-, and 900-MHz NMR spectrometers with cryoprobes. The CPMG dispersion profiles were fit on a per-methyl basis to a two-state exchange model using the Carver–Richards equation (18, 37), obtaining the exchange rate constant (kex), populations (pA, pB), and 13C chemical shift changes (in ppm) between states A and B (|ΔδCPMG13C|, assuming |ΔδCPMG1H| = 0). Fittings were performed using the program CATIA (http://pound.med.utoronto.ca/software). The estimated errors of the individual fittings (Tables S1–S3) assumed a two-state exchange process, and could be underestimated due to the existence of more than two states.
Slow exchange in 2P-ERK2 was probed from analysis of methyl peaks in HMQC spectra, collected on a Varian 800-MHz spectrometer with a cryoprobe at 5, 10, 15, 20, and 25 °C. The peak volumes of slow-exchanging peaks were estimated using CcpNmr Analysis software (38). Equilibrium constants of two-state exchange (Keq) were calculated from the volume ratios of the two slow-exchanging peaks.
Two-dimensional (13C,1H) single-quantum CPMG measurements (39) on 0P- and 2P-ERK2 were carried out at 900 MHz, in parallel with 2D (13C,1H) MQ CPMG experiments. The results showed dispersion curves (Fig. S6) comparable to MQ CPMG experiments, validating the assumption that |ΔδCPMG1H| is ≈ 0.
Supplementary Material
Acknowledgments
We are indebted to Lewis Kay (University of Toronto) for NMR pulse sequences, analysis software, and insightful discussions. We also thank Geoffrey Armstrong and Richard Shoemaker for help with NMR data acquisition, and Rebecca Page and Wolfgang Peti (Brown University) for their gift of the ERK2 expression plasmid. These studies were supported by National Institutes of Health Grants R01GM074134 (to N.G.A.), T32GM008759 (to L.R.W.), and T32GM065103 (to M.P.L.). NMR instrumentation was purchased with partial support from NIH grants GM068928, RR11969, and RR16649; National Science Foundation Grants 9602941 and 0230966; and the W. M. Keck Foundation.
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1318899111/-/DCSupplemental.
References
- 1.Roskoski R., Jr ERK1/2 MAP kinases: Structure, function, and regulation. Pharmacol Res. 2012;66(2):105–143. doi: 10.1016/j.phrs.2012.04.005. [DOI] [PubMed] [Google Scholar]
- 2.Pearson G, et al. Mitogen-activated protein (MAP) kinase pathways: Regulation and physiological functions. Endocr Rev. 2001;22(2):153–183. doi: 10.1210/edrv.22.2.0428. [DOI] [PubMed] [Google Scholar]
- 3.Prowse CN, Lew J. Mechanism of activation of ERK2 by dual phosphorylation. J Biol Chem. 2001;276(1):99–103. doi: 10.1074/jbc.M008137200. [DOI] [PubMed] [Google Scholar]
- 4.Canagarajah BJ, Khokhlatchev A, Cobb MH, Goldsmith EJ. Activation mechanism of the MAP kinase ERK2 by dual phosphorylation. Cell. 1997;90(5):859–869. doi: 10.1016/s0092-8674(00)80351-7. [DOI] [PubMed] [Google Scholar]
- 5.Zhang F, Strand A, Robbins D, Cobb MH, Goldsmith EJ. Atomic structure of the MAP kinase ERK2 at 2.3 Å resolution. Nature. 1994;367(6465):704–711. doi: 10.1038/367704a0. [DOI] [PubMed] [Google Scholar]
- 6.Lee T, et al. Docking motif interactions in MAP kinases revealed by hydrogen exchange mass spectrometry. Mol Cell. 2004;14(1):43–55. doi: 10.1016/s1097-2765(04)00161-3. [DOI] [PubMed] [Google Scholar]
- 7.Lee T, Hoofnagle AN, Resing KA, Ahn NG. Hydrogen exchange solvent protection by an ATP analogue reveals conformational changes in ERK2 upon activation. J Mol Biol. 2005;353(3):600–612. doi: 10.1016/j.jmb.2005.08.029. [DOI] [PubMed] [Google Scholar]
- 8.Hoofnagle AN, Stoner JW, Lee T, Eaton SS, Ahn NG. Phosphorylation-dependent changes in structure and dynamics in ERK2 detected by SDSL and EPR. Biophys J. 2004;86(1 Pt 1):395–403. doi: 10.1016/S0006-3495(04)74115-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hoofnagle AN, Resing KA, Goldsmith EJ, Ahn NG. Changes in protein conformational mobility upon activation of extracellular regulated protein kinase-2 as detected by hydrogen exchange. Proc Natl Acad Sci USA. 2001;98(3):956–961. doi: 10.1073/pnas.98.3.956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Smock RG, Gierasch LM. Sending signals dynamically. Science. 2009;324(5924):198–203. doi: 10.1126/science.1169377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nagar B, et al. Structural basis for the autoinhibition of c-Abl tyrosine kinase. Cell. 2003;112(6):859–871. doi: 10.1016/s0092-8674(03)00194-6. [DOI] [PubMed] [Google Scholar]
- 12.Baldwin AJ, Kay LE. NMR spectroscopy brings invisible protein states into focus. Nat Chem Biol. 2009;5(11):808–814. doi: 10.1038/nchembio.238. [DOI] [PubMed] [Google Scholar]
- 13.Mittermaier AK, Kay LE. Observing biological dynamics at atomic resolution using NMR. Trends Biochem Sci. 2009;34(12):601–611. doi: 10.1016/j.tibs.2009.07.004. [DOI] [PubMed] [Google Scholar]
- 14.Bhabha G, et al. A dynamic knockout reveals that conformational fluctuations influence the chemical step of enzyme catalysis. Science. 2011;332(6026):234–238. doi: 10.1126/science.1198542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Eisenmesser EZ, et al. Intrinsic dynamics of an enzyme underlies catalysis. Nature. 2005;438(7064):117–121. doi: 10.1038/nature04105. [DOI] [PubMed] [Google Scholar]
- 16.Fraser JS, et al. Hidden alternative structures of proline isomerase essential for catalysis. Nature. 2009;462(7273):669–673. doi: 10.1038/nature08615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Korzhnev DM, Kloiber K, Kanelis V, Tugarinov V, Kay LE. Probing slow dynamics in high molecular weight proteins by methyl-TROSY NMR spectroscopy: Application to a 723-residue enzyme. J Am Chem Soc. 2004;126(12):3964–3973. doi: 10.1021/ja039587i. [DOI] [PubMed] [Google Scholar]
- 18.Korzhnev DM, Kloiber K, Kay LE. Multiple-quantum relaxation dispersion NMR spectroscopy probing millisecond time-scale dynamics in proteins: Theory and application. J Am Chem Soc. 2004;126(23):7320–7329. doi: 10.1021/ja049968b. [DOI] [PubMed] [Google Scholar]
- 19.Gardner KH, Kay LE. Production and incorporation of N-15, C-13, H-2 (H-1-delta 1 methyl) isoleucine into proteins for multidimensional NMR studies. J Am Chem Soc. 1997;119(32):7599–7600. [Google Scholar]
- 20.Goto NK, Gardner KH, Mueller GA, Willis RC, Kay LE. A robust and cost-effective method for the production of Val, Leu, Ile (delta 1) methyl-protonated 15N-, 13C-, 2H-labeled proteins. J Biomol NMR. 1999;13(4):369–374. doi: 10.1023/a:1008393201236. [DOI] [PubMed] [Google Scholar]
- 21.Shapiro PS, et al. Activation of the MKK/ERK pathway during somatic cell mitosis: Direct interactions of active ERK with kinetochores and regulation of the mitotic 3F3/2 phosphoantigen. J Cell Biol. 1998;142(6):1533–1545. doi: 10.1083/jcb.142.6.1533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tugarinov V, Kay LE. Ile, Leu, and Val methyl assignments of the 723-residue malate synthase G using a new labeling strategy and novel NMR methods. J Am Chem Soc. 2003;125(45):13868–13878. doi: 10.1021/ja030345s. [DOI] [PubMed] [Google Scholar]
- 23.Sinha N, Kumar S, Nussinov R. Interdomain interactions in hinge-bending transitions. Structure. 2001;9(12):1165–1181. doi: 10.1016/s0969-2126(01)00687-6. [DOI] [PubMed] [Google Scholar]
- 24.Johnson DA, Akamine P, Radzio-Andzelm E, Madhusudan M, Taylor SS. Dynamics of cAMP-dependent protein kinase. Chem Rev. 2001;101(8):2243–2270. doi: 10.1021/cr000226k. [DOI] [PubMed] [Google Scholar]
- 25.Robbins DJ, et al. Regulation and properties of extracellular signal-regulated protein kinases 1 and 2 in vitro. J Biol Chem. 1993;268(7):5097–5106. [PubMed] [Google Scholar]
- 26.Akamine P, et al. Dynamic features of cAMP-dependent protein kinase revealed by apoenzyme crystal structure. J Mol Biol. 2003;327(1):159–171. doi: 10.1016/s0022-2836(02)01446-8. [DOI] [PubMed] [Google Scholar]
- 27.Masterson LR, et al. Dynamically committed, uncommitted, and quenched states encoded in protein kinase A revealed by NMR spectroscopy. Proc Natl Acad Sci USA. 2011;108(17):6969–6974. doi: 10.1073/pnas.1102701108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Masterson LR, et al. Dynamics connect substrate recognition to catalysis in protein kinase A. Nat Chem Biol. 2010;6(11):821–828. doi: 10.1038/nchembio.452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chen H, et al. A molecular brake in the kinase hinge region regulates the activity of receptor tyrosine kinases. Mol Cell. 2007;27(5):717–730. doi: 10.1016/j.molcel.2007.06.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Deindl S, et al. Structural basis for the inhibition of tyrosine kinase activity of ZAP-70. Cell. 2007;129(4):735–746. doi: 10.1016/j.cell.2007.03.039. [DOI] [PubMed] [Google Scholar]
- 31.Shan Y, et al. Oncogenic mutations counteract intrinsic disorder in the EGFR kinase and promote receptor dimerization. Cell. 2012;149(4):860–870. doi: 10.1016/j.cell.2012.02.063. [DOI] [PubMed] [Google Scholar]
- 32.Shan Y, Arkhipov A, Kim ET, Pan AC, Shaw DE. Transitions to catalytically inactive conformations in EGFR kinase. Proc Natl Acad Sci USA. 2013;110(18):7270–7275. doi: 10.1073/pnas.1220843110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kornev AP, Haste NM, Taylor SS, Eyck LF. Surface comparison of active and inactive protein kinases identifies a conserved activation mechanism. Proc Natl Acad Sci USA. 2006;103(47):17783–17788. doi: 10.1073/pnas.0607656103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Taylor SS, Kornev AP. Protein kinases: Evolution of dynamic regulatory proteins. Trends Biochem Sci. 2011;36(2):65–77. doi: 10.1016/j.tibs.2010.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Francis DM, et al. Resting and active states of the ERK2:HePTP complex. J Am Chem Soc. 2011;133(43):17138–17141. doi: 10.1021/ja2075136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Emrick MA, et al. The gatekeeper residue controls autoactivation of ERK2 via a pathway of intramolecular connectivity. Proc Natl Acad Sci USA. 2006;103(48):18101–18106. doi: 10.1073/pnas.0608849103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Palmer AG., III NMR characterization of the dynamics of biomacromolecules. Chem Rev. 2004;104(8):3623–3640. doi: 10.1021/cr030413t. [DOI] [PubMed] [Google Scholar]
- 38.Vranken WF, et al. The CCPN data model for NMR spectroscopy: Development of a software pipeline. Proteins. 2005;59(4):687–696. doi: 10.1002/prot.20449. [DOI] [PubMed] [Google Scholar]
- 39.Skrynnikov NR, Mulder FA, Hon B, Dahlquist FW, Kay LE. Probing slow time scale dynamics at methyl-containing side chains in proteins by relaxation dispersion NMR measurements: Application to methionine residues in a cavity mutant of T4 lysozyme. J Am Chem Soc. 2001;123(19):4556–4566. doi: 10.1021/ja004179p. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.