Significance
During embryonic development of the central nervous system (CNS), the neural progenitor cells (NPC) not only produce diverse types of neurons and glial cells but also self-renew to maintain a pool of undifferentiated cells. The mechanism of the self-renewal is not well understood. We show that a positive feedback loop exists between two transcription factors, Sox2 and Sox6, which are expressed in the NPC. It appears that the mutual stimulation of expression between the two genes is important for maintenance of the NPC pool as its disruption leads to overt and precocious neuronal differentiation. Our results provide a mechanistic insight into the regulation of CNS development.
Keywords: CNS, neural development, neural stem cell, SoxB1, SoxD
Abstract
How a pool of undifferentiated neural progenitor cells is maintained in the developing nervous system is an issue that remains unresolved. One of the key transcription factors for self-renewal of these cells is Sox2, the forced expression of which has been shown to inhibit neuronal differentiation in vivo. To dissect the molecular mechanisms of Sox2 activity, a ChIP-on-chip assay has been carried out for Sox2, and multiple candidate direct target genes have been isolated. In this report, we provide evidence indicating that Sox6, which like Sox2 belongs to the SRY-related HMG box transcription factor family, is a bona-fide direct regulatory target of Sox2. In vivo, Sox6 expression is seen with a temporal lag in Sox2-positive neural precursor cells in the ventricular zone, and Sox2 promotes expression of Sox6 as a transcriptional activator. Interestingly, gain- and loss-of-function assays indicate that Sox6 in turn is required for the maintenance of Sox2 expression, suggesting that a positive feedback loop, which functions to inhibit premature neuronal differentiation, exists between the two transcription factors.
Embryonic development of the central nervous system (CNS) features multiple types of differentiated cells generated from neural progenitor cells (NPC) present in the ventricular zone (VZ) (1). Compared with spatial and temporal control of the differentiation, relatively little is known about the mechanism of self-renewal or the maintenance of the progenitor population that must be robust during embryogenesis and at the same time coordinated with differentiation. Central among the genes implicated in the control of the process is Sox2, a member of the SRY-related HMG box transcription factor family, which has been shown to be functional in the maintenance of a wide variety of stem cells including embryonic stem cells (ESC), embryonic and adult NPC, and adult stem cells of multiple endodermal and ectodermal organs (2–4). Sox2, along with other members of the SoxB1 subfamily, Sox1 and Sox3, is expressed in the progenitor cells in the VZ throughout the course of embryonic CNS development (5, 6). It has been shown that forced expression of SoxB1 subfamily members or their constitutive active derivatives inhibits neuronal differentiation whereas forced expression of dominant negative derivatives induces premature neuronal differentiation (5, 7). This implies that SoxB1 genes function as the so-called stemness factors of NPC, and their direct target genes should at least partially compose the regulatory network for self-renewal of NPC. Notably, Notch1 and Sonic Hedgehog have been shown to be direct regulatory targets of Sox2 in NPC (8, 9). The identification of the two genes was in large part based on the knowledge of their functions in neural development and/or in NPC culture in vitro. More recently, two genome-wide studies based on chromatin immunoprecipitation (ChIP) techniques led to the isolation of multiple candidate direct target genes of Sox2. Engelen and coworkers used mouse ESC-derived NPC stably expressing Flag-epitope–tagged Sox2 to find target genes of Sox2 (10). Notable among the targets are Delta-Notch pathway genes such as Jag1, Rbpj, and Hes5 and SHH pathway genes such as Gli2, Gli3, Mycn, and Tulip. Furthermore, expression of these genes was shown to be down-regulated upon Sox2 knockdown, providing supporting evidence for earlier studies on the involvement of Sox2 in regulation of these pathways in NPC (8, 9). In another study by Bergsland and coworkers, again using a mouse ESC-derived NPC and neurons, Sox2 was shown to target a set of genes in common with Sox3 and Sox11 that act in sequence on these genes during the course of neurogenesis (11). This study thus confirmed the role of Sox2 as a prebinding factor of neural lineage-specific genes, including those later expressed in differentiating neurons.
Here, we report the identification and characterization of Sox6, a member of the SoxD subfamily, as a direct target of Sox2 in the developing CNS. Expression pattern analyses and gene function studies in vivo confirm that Sox6 is indeed a regulatory target of Sox2 and forms a positive feedback network with Sox2 that inhibits neuronal differentiation. Our study adds Sox6 as an important regulator of NPC and provides an insight into the mechanism of NPC maintenance during neurogenesis.
Results
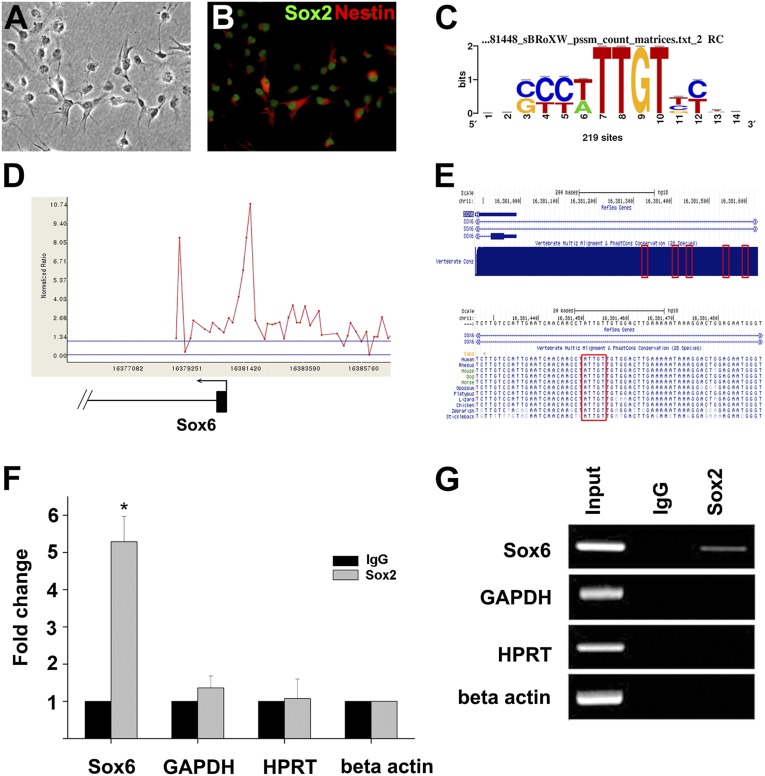
We used ReNcell CX cells for the ChIP-on-chip assay. Derived from human fetal cortical brain tissue and immortalized by c-myc expression, these cells express high levels of nestin and Sox2, two neural stem-cell marker proteins (Fig. 1 A and B). We used a commercial microarray chip on which genomic regions near the transcription initiation site (typically, −5.5 to +2.5 kb) of >17,000 genes are represented. Of these, we propose 811 genes as potential Sox2 target genes (Dataset S1) based on the significance of the ChIP signal. The result of ChIP-on-chip assay has been deposited in the Gene Expression Omnibus database (GSE34395). There was a clear overrepresentation of the SOX-binding element, 5′-ACAAT/A-3′, or the complementary sequence, 5′-A/TTTGT-3′, near the peaks of ChIP signals (P value 2.84E-65) (Fig. 1C and SI Materials and Methods). Interestingly, the candidate target gene set from this study showed a limited overlap with those from two other studies using mouse NPC derived from ESC although multiple common targets (ETV1, HMGB1, and PIPOX) as well as targets unique to our study (ARID2 and CBY1) have been confirmed by conventional ChIP assay (Discussion and Fig. S1).
Fig. 1.
Sox2 directly targets Sox6. Phase-contrast (A) and epifluorescence images (B) of ReN cells immunostained for Sox2 and Nestin. (C) Sequence logo obtained using WebLogo (http://weblogo/berkeley.edu/loco/cgi) based on RSAT analysis. Enrichment of the consensus SOX-binding DNA motif is seen. (D) Sox2 occupancy of Sox6 promoter from ChIP-on-chip assay. The plot shows ChIP-enrichment ratios (y axis) for probes within the genomic region indicated. The numbers on the x axis indicate the genomic location (NCBI36/hg18). Sox6 promoter is shown in scale below the plot (exon in thick line and intron in thin line), and the arrow denotes the start site and direction of the transcription. (E) Sox6 promoter on human chromosome 11 (adapted from the University of California at Santa Cruz Genome Browser). (Upper) The locations of the five potential Sox2-binding sites near the Sox6 promoter are indicated by red boxes. (Lower) The nucleotide sequences surrounding the third binding site are shown in detail. Within the red box is the conserved core SOX-binding sequence, 5′-ATTGT-3′. (F) Interaction of Sox2 with potential binding sites of the Sox6 promoter was analyzed by ChIP assay using anti-Sox2 antibody and control IgG antibody. Genomic sites near GAPDH, hypoxanthine phosphoribosyltransferase 1 (HPRT), and β-actin loci were used as controls, and normalization was carried out using β-actin. Data are the average of three independent experiments, each with two measurements, and error bars represent SDs (*P < 0.01). (G) Gel electrophoresis analysis of the ChIP assay. PCR product is seen only with input DNA control or with the combination of anti-Sox2 antibody and the Sox6 locus.
Among the candidate target genes was Sox6, another member of the SRY-related HMG box transcription factor family. Sox6, a gene belonging to the SoxD subfamily, has been ascribed with diverse functions during embryogenesis in multiple tissues, including regulation of oligodendrocyte and interneuron differentiation in CNS (12). However, Sox6 is also expressed in the VZ before oligodendrocyte formation, but its function is currently unknown (13). Isolation of Sox6 as a Sox2 target therefore represents an unexplored context to study its role in NPC. The ChIP signal indicated that the Sox2-binding site is located ∼400–600 bases 5′ to the transcription initiation site of two of the splice variants with an identical first exon (RefSeq Ids, NM_017508 and NM_001145819) and within the first intron of other splice variants (RefSeq IDs: NM_033326 and NM_001145811). We found five potential SOX-binding elements within a 250-bp stretch (Fig. 1 D–E and Fig. S2) in the peak signal region. Comparison with genomic DNA sequences of other species indicated that the cluster was highly conserved among vertebrate species. Importantly, the homologous DNA cluster for mice, located 5′ to the transcription initiation site of several Sox6 splice variants (RefSeq IDs: NM_001277326, NM_001277327, and NM_001277328) and in the second intron of other splice variants (RefSeq IDs: NM_011445, NM_001025559, and NM_001025560), also has been designated as a Sox2-binding site in one of the previous studies (11). We confirmed the binding of Sox2 to this cluster using conventional ChIP assays (Fig. 1 F and G). Also, several luciferase reporter constructs have been generated and tested for Sox2-dependent activation in 293T cells, and at least three of the five SOX-binding sites appeared to be functional (Fig. S3).
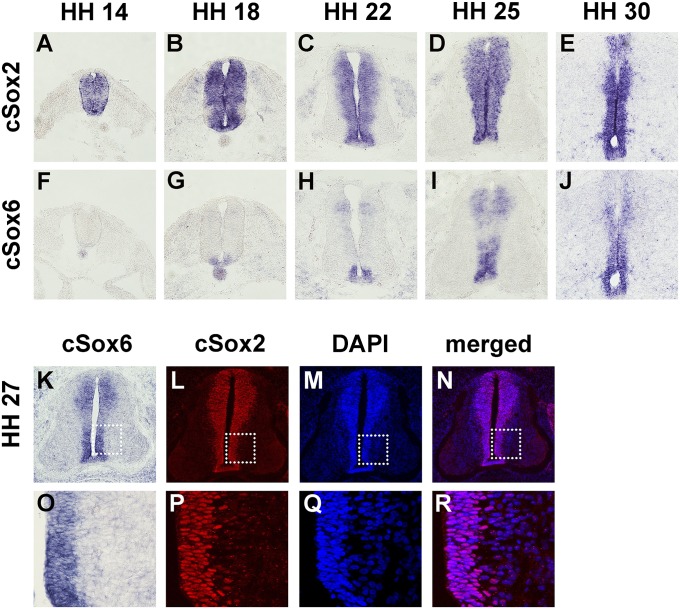
We chose chicken as the in vivo model to dissect the nature of interaction between Sox2 and Sox6. First, the spatial and temporal expression patterns of the two genes were examined by RNA in situ hybridization. As has been previously reported, Sox2 expression is seen in the VZ of the neural tube throughout embryogenesis (Fig. 2 A–E). In contrast, Sox6 expression was first seen in the floor plate region around the Hamburger–Hamilton (HH) stage 18 and later in more dorsal regions of the neural tube (Fig. 2 F–J). Importantly, the extent of Sox6 expression was limited to regions of Sox2 expression in all stages examined. A similar expression pattern of the two genes was also seen in the mouse neural tube (Fig. S4). We sought to determine if the expression of two genes overlapped at the cellular level. Due to the lack of an appropriate antibody for immunohistochemical staining of chicken Sox6, we resorted to comparing Sox6 RNA in situ hybridization staining with Sox2 immunostaining. By HH stage 27, Sox6 expression is seen throughout the VZ in a majority of the cells (Fig. 2 K and O). That Sox2 also is expressed in an overwhelming majority of cells in VZ indicates that these two genes are expressed together in a large majority of NPC (Fig. 2 L–N and P–R).
Fig. 2.
Expression of Sox2 and Sox6 in the developing chick neural tube. RNA in situ hybridization was performed for Sox2 (A–E) or Sox6 (F–J) on adjacent transverse sections at the forelimb levels of the chicken embryos at HH stages 14 (A and F), 18 (B and G), 22 (C and H), 25 (D and I), and 30 (E and J). Coexpression of Sox6 and Sox2 at the cellular level was examined by staining for Sox6 by RNA in situ hybridization (K and O) and for Sox2 by immunohistochemistry (L and P). Sox2 immunostained sections were DAPI-counterstained (M and Q), and merged images (N and R) show that virtually all cells in the ventricular zone are positive for Sox2. Boxed areas in K, L, M, and N are presented in enlarged forms in O, P, Q, and R, respectively.
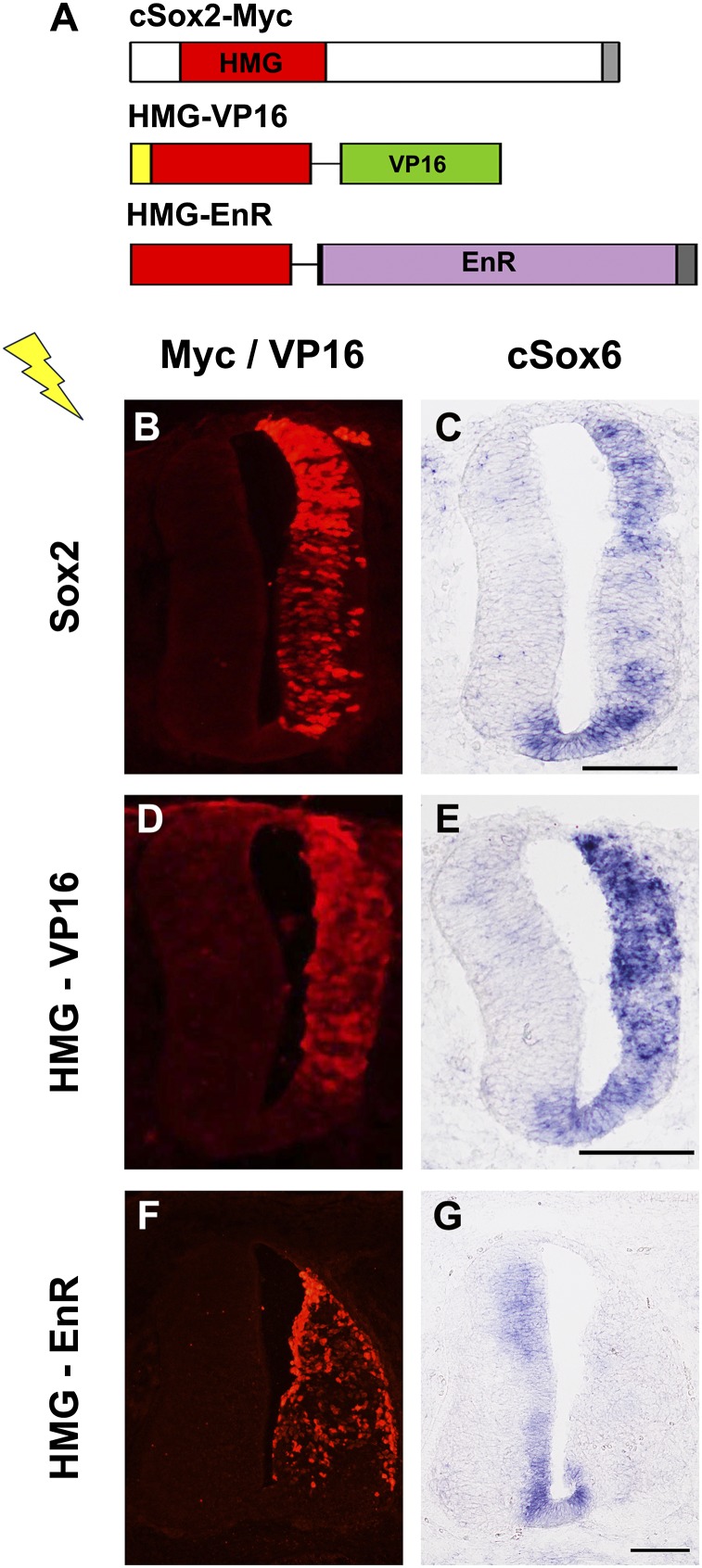
To determine if Sox2 promotes the expression of Sox6 in the developing neural tube, a series of in ovo electroporation experiments was carried out. First, an expression construct of the full-length Sox2 was introduced into one side of the neural tube, and the change in Sox6 expression was examined. At HH stage 15, when Sox6 expression was not yet strongly seen in the control side, Sox2 overexpression induced Sox6 expression (Fig. 3 A–C). We next used obligate activator and repressor versions of Sox2, which, respectively, contain the activation domain of viral protein VP16 and the repression domain of Drosophila Engrailed protein fused to the HMG domain of Sox2 (Fig. 3A). These constructs, HMG-VP16 and HMG-EnR, have been used to demonstrate that Sox2 inhibits neuronal differentiation and maintains NPC in the progenitor state by functioning as a transcriptional activator (5). Upon forced expression of HMG-VP16, Sox6 expression was strongly induced as seen in the case of full-length Sox2 expression (Fig. 3 D and E). In contrast, upon forced expression of HMG-EnR, the robust expression of Sox6 at HH stage 25 was strongly inhibited (Fig. 3 F and G). Taken together, these data indicate that Sox2 functions as a cell-autonomous direct activator of Sox6 in the developing CNS.
Fig. 3.
Activation of Sox6 expression by Sox2 in vivo. (A) Expression constructs are schematically illustrated. cSox2-myc: Sox2 with a myc-epitope tag (dark gray) at the C terminus. HMG-VP16: The HMG domain of Sox2 fused to the transactivation domain of VP16 (green) and nuclear localization signal (yellow) at the N terminus. HMG-EnR: The HMG domain of Sox2 fused to the repressor domain of Engrailed (purple) with a myc-epitope tag (dark gray) at the C terminus. (B–G) Electroporated chicken embryos were harvested 30–48 h after electroporation and processed for immunohistochemical staining and RNA in situ hybridization on serial sections. Expression of constructs was confirmed by immunostaining for the myc epitope or VP16 (B, D, and F). Forced expression of Sox2 (B and C) or HMG-VP16 (D and E) promoted the expression of Sox6, but forced expression of HMG-EnR (F and G) suppressed the expression of Sox6. Adjacent sections at the forelimb level were used.
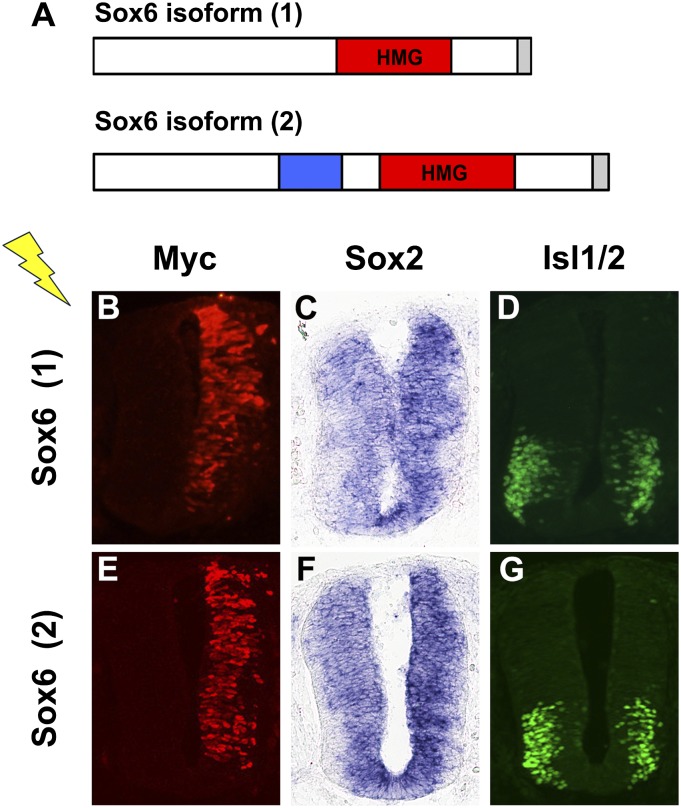
The fact that Sox6 is induced by Sox2 in precursor cells raised the issue of its role in these cells. To address this, we attempted overexpression of Sox6 in the developing spinal cord. First, we isolated the full-length Sox6 cDNA from chicken spinal cord at HH27. We found two splice variants (GenBank accession nos. KC832418 and KC832419), one of which matches the predicted chicken Sox6 (National Center for Biotechnology Information reference sequence XM_421000.3). KC832419 differs from KC832418 by inclusion of a 60-nucleotide-long exon resulting in an extra 20 amino acids (Fig. 4A). Upon introduction into the spinal cord, both splice variants of Sox6 led to a slight decrease in the size of the marginal zone on the electroporated side, which was reminiscent of the overexpression of Sox2 (Fig. 4 B–G). We thus examined the expression of Sox2 by RNA in situ hybridization and found a detectable increase on the Sox6-electroporated side compared with the control side (Fig. 4 C and F). The effect of Sox2 induction was similar for both splice variants. We also examined neuronal differentiation by immunostaining for the transcription factor Islet-1/2 (Isl1/2), an early neuronal marker for terminally differentiated motor neurons. A significant decrease (35 ± 9% and 32 ± 9% for splice variants 1 and 2, respectively; P < 0.05) was noted in the number of Isl1/2-positive cells, consistent with the increased Sox2 expression (Fig. 4 D and G). A similar decrease of Isl1/2-positive cells was also seen by ectopic expression of Sox2, consistent with its inhibitory effect on neurogenesis (Fig. S5).
Fig. 4.
Ectopic expression of Sox6. (A) Schematic illustration of the two isoforms of chick Sox6 with a myc-epitope tag (gray) at the C terminus. (Lower) The blue box represents a 60-base-long exon found in isoform 2. (B–G) Chicken embryos at HH stage 14–16 were used for transfection by electroporation. Expression was confirmed by immunostaining for the myc epitope (B and E). Embryos were harvested 24 h later, and RNA in situ hybridization and immunostaining were carried out to examine the expression of Sox2 and Isl1/2, respectively. Forced expression of Sox6 led to up-regulation of Sox2 (C and F) and down-regulation of Isl1/2 (D and G). Serial sections at the forelimb level were used.
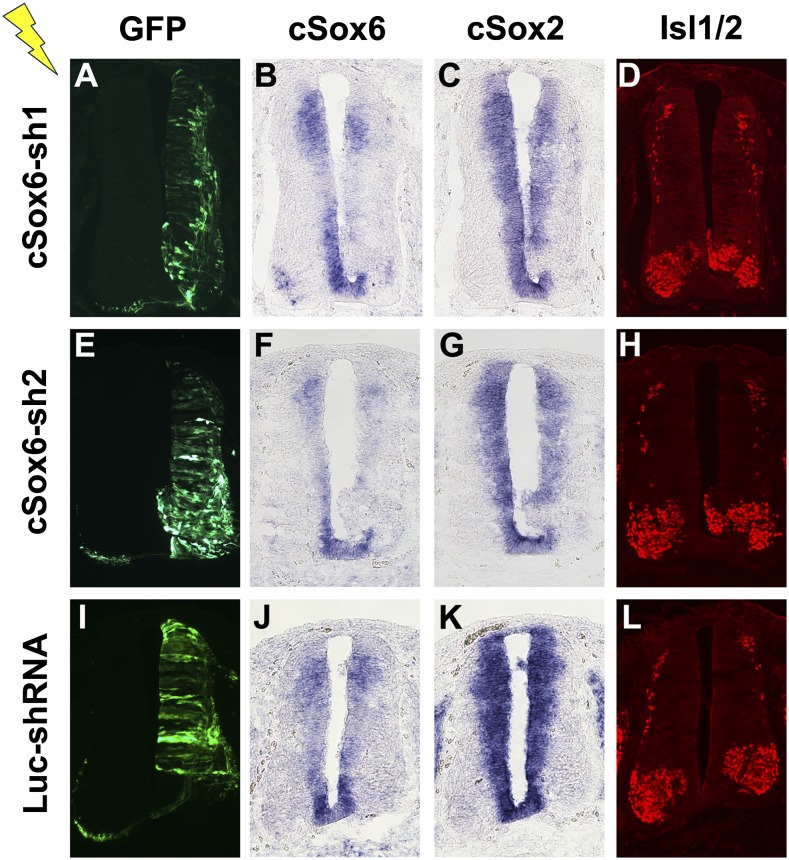
We next sought to perform Sox6 loss-of-function experiments. To this end, we generated a battery of pSuperior plasmid constructs, each designed to express an shRNA specific to Sox6 and the green fluorescent protein (GFP) (Fig. 5 A, E, and I). At least two distinct shRNAs targeting different portions of Sox6 mRNA led to RNA interference as determined by RNA in situ hybridization and comparison with the contralateral side (Fig. 5 B, F, and J). Importantly, where Sox6 expression decreased, Sox2 expression also decreased to a detectable level (Fig. 5 C, G, and K). A similar decrease was also seen for NK6 homeobox 1 (Nkx6.1)-positive progenitor cells in the VZ (Fig. S6). Furthermore, motor neuron differentiation as determined by immunostaining for Isl1/2 occurred in a spatially premature manner. Specifically, Isl1/2-positive cells were found in what should be the ventricular zone where only undifferentiated and Isl1/2-negative cells are present as seen in the control side (Fig. 5 D, H, and L). Similar precocious neuronal differentiation is induced by expression of the obligate repressor derivative of Sox2, HMG-EnR (Fig. S5). Taken together with results from Sox6 overexpression, these data indicate that Sox6 in fact is necessary for the maintenance of Sox2 expression and in turn of the NPC population, including precursors of motor neurons.
Fig. 5.
Knockdown of Sox6. Chicken embryos at HH stage 14–16 were used for transfection by electroporation of pSuperior shRNA-expressing plasmids. Embryos were harvested 48 h later, and electroporation was confirmed by immunostaining for GFP (A, E, and I). cSox6-sh1 and cSox6-sh2 express shRNAs specifically targeting cSox6 mRNA whereas the control plasmid Luc-shRNA is designed to target luciferase mRNA. RNA in situ hybridization was used to examine the expression of Sox6 (B, F, and J) and Sox2 (C, G, and K). Down-regulation of Sox6 is seen only in the cases of specific targeting (B and F) but not in the control case (J). Down-regulation of Sox2 was seen only in the cases of specific targeting of Sox6 (C and G) but not in the control case (K). Mis-expression of Isl1/2 in the ventricular zone was seen only in the cases of specific targeting of Sox6 (D and H) but not in the control case (L). Serial sections at the forelimb level were used.
Discussion
The advent of high-throughput techniques such as microarray and deep sequencing combined with a ChIP assay opened the way to identifying direct regulatory targets of transcription factors (14). Sox2 was one of the earliest to be analyzed mainly due to its key role in ES cells as a component of the network that maintains the self-renewal capacity and multipotency of these cells (3). Function of Sox2 as a stemness factor is not limited to ES cells as loss-of-function and lineage tracing experiments indicate that Sox2 is a stemness factor in multiple types of stem cells (2, 6). In particular, maintenance of the NPC pool in the ventricular zone of the developing CNS has been a well-established role of Sox2 (5, 7).
Two studies recently published describe isolation of direct targets of Sox2 in NPC (10, 11). We provide another set of potential target genes in this report. Interestingly, the gene sets from the three studies show a limited overlap, and only 39 genes are found to be targeted by Sox2 in all three studies (Fig. S1). Clearly, cells cultured under different conditions most likely end up expressing at least a partly different set of genes. Also, because the analysis in this study was limited to promoter regions of ∼17,000 genes, some of the genes not represented on the microarray used or the genes regulated from a distance may have been missed. In addition, technical and statistical issues such as determining significant signal outputs in microarray-based screening and read counts in deep-sequencing–based screening may generate disparity. Another source of difference between this study and others could be that human cells were used as opposed to mouse cells in the other two studies. Odom and coworkers examined the binding pattern of four liver-specific transcription factors, FOXA2, HNF1A, HNF4A and HNF6, from mice and humans on 4,000 unambiguously orthologous promoters and found, surprisingly, that 41–89% of the promoters bound by a protein in one species were not bound by the orthologous transcription factor in the other (15). It remains to be determined if such species-specific patterns are seen in the case of other transcription factors or cell types, but species-specific targeting is likely at least partly responsible for the limited overlap seen between this study and the two others. Another source of disparity may be the ever-changing chromatin environment within the differentiating cells. Bergsland and coworkers showed the dynamic nature of binding-site recognition by a series of Sox proteins including Sox2, suggesting that, even before overt differentiation of NPC, the nature of interaction between Sox2 and target sites can evolve (11). Thus, the developmental stages that various NPC lines represent may substantially differ from one another. At any rate, the utility of high-throughput assays notwithstanding, the current situation points to the need for a careful evaluation of the proposed interactions between transcription factors and target sites or target genes derived from high-throughput assays.
In this regard, Sox6 is likely to be a bona-fide direct target of Sox2. First, all three studies find Sox6 as the most proximal gene to one of the Sox2-binding sites. Second, the cluster we found as a Sox2-binding site not only is conserved across the vertebrate species, but also has been identified in another study using mouse NPC as a Sox2-binding site (11). Finally, expression and functional analyses in vivo provide evidence that Sox2 is a cell-autonomous activator of Sox6. Although Sox6 has been shown to function downstream from Sox9 in chondrocyte differentiation (16), this is a previously undescribed study of Sox6 as a direct target of another Sox transcription factor and a mediator of positive feedback to its upstream regulator.
Sox6 is expressed in diverse cell types during embryogenesis. The lack of the activation or repression domain and the association with cofactors with diverse biochemical functions indicate that Sox6 plays the role of organizing transcriptional or epigenetic regulatory complexes (12). In the CNS, Sox6 has been shown to regulate oligodendrocyte precursor cells by inhibiting premature exit from the cell cycle (13). In addition, Sox6 appears to have a role in specifying dorsal interneurons in telencephalon and promoting their differentiation (17). Our results endow an additional role to Sox6 as a regulator of NPC in the developing CNS. Specifically, gain- and loss-of-function assays of Sox6 in NPC indicate that Sox6 functions to maintain a precursor state of these cells. In fact, that maintenance of Sox2 expression is dependent on Sox6 expression indicates that a positive feedback loop has been established between these two Sox genes. It can be envisioned then that, whereas Sox2-positive, Sox6-negative cells are subject to neuronal differentiation, Sox2 and Sox6 double-positive cells continue to stay as NPC through the later stages of CNS development. How generally Sox6 functions in the CNS or neural tube development remains to be examined further. At this point, the clear effect is seen only for motor neurons and their precursors (Fig. 5 and Fig. S6). This could be because spatiotemporally confined expression of Sox6 (Fig. 2) and the inherent neurogenesis program make motor neurons most sensitive to perturbation of Sox6 at least via in ovo electroporation of chicken embryos. How Sox6 exactly performs its function also is an issue to be resolved. A recent genome-wide mapping of Sox6 targets by ChIP-seq analysis in skeletal muscles revealed that Sox6 binds and represses a group of genes to be specifically expressed in slow-fiber muscle cells, indicating that Sox6 regulates the balance between slow-twitch and fast-twitch muscle fiber groups (18). It would be interesting to see if a parallel mechanism is in place for the regulation of CNS development, and genome-wide mapping studies for Sox6 binding and Sox6-dependent expression profiling assays in NPC should add to our understanding of the self-renewal of NPC in the developing CNS.
Materials and Methods
Cell Culture and ChIP-on-Chip Assay.
ReNcell CX cell line was purchased from Chemicon Inc. Cells were grown in ReN maintenance medium (Chemicon Inc.) supplemented with 20 ng/mL of EGF (Invitrogen Inc.) and basic fibroblast growth factor (R&D Systems) on laminin-coated dishes. ChIP-on-chip assay was performed as previously described with minor modifications (3). Anti-Sox2 antibody (AF2018) was purchased from R&D Systems. Detailed protocol is available upon request. Isolated and labeled DNA was applied to the Agilent human promoter 2 array set, which covers regions surrounding the transcription initiation site (from −5.5 to +2.5 kb) for ∼17,000 genes. We used DNA Analytics software to select from the raw output data statistically significant (P value cutoff of <0.01) and consecutive (at least two) probes leading to 811 Sox2-binding regions (Dataset S1). The sequence motif search was carried out using the oligo-analysis program from Regulatory Sequence Analysis Tools (RSAT; http://rsat.bigre.ulb.ac.be/rsat/) on the 811 Sox2-binding regions (Fig. 1C). Details of in silico analysis are described in the Supporting Information.
ChIP Assay.
ChIP assays were performed using EZ ChIP Chromatin immunoprecipitation kits (Upstate Inc.) following the manufacturer’s protocols with some minor modifications. Oligonucleotide primers used for PCR are listed in Table S1.
Immunohistochemistry.
Embryos were typically fixed in 4% (wt/vol) paraformaldehyde in PBS for 2 h, cryo-preserved by sinking in 30% (wt/vol) sucrose in PBS, embedded in optimal cutting temperature compound (Leica), and sectioned on a Microtome cryostat. Frozen sections were incubated overnight at 4 °C with the following primary antibodies at the indicated dilution: anti-Isl1/2 antibody (1:20; 39.4D5, Developmental Studies Hybridoma Bank), anti-Nkx6.1 (1:20; F55A10, Developmental Studies Hybridoma Bank), anti-Tuj1 (1:400; MMS-435P, Covance), anti-Nestin (1:500; MAB5326, Chemicon), anti-Lim1/2 (1:2; 42F, Developmental Studies Hybridoma Bank), anti-Pax6 (1:20; PAX6, Developmental Studies Hybridoma Bank), anti-myc antibody (1:50; sc-40, Santa Cruz Biotechnology Inc.), anti-VP16 antibody (1:50; sc-7545, Santa Cruz Biotechnology Inc.), anti-Sox2 antibody (1:500; AB5603, Chemicon Inc.), anti-GFP Alexa Fluor 488 conjugate (1:400; A21311, Life Technologies). Appropriate secondary antibodies conjugated with Alexa Fluor 488, Alexa Fluor 594, and Alexa Fluor 633 (Life Technologies) were used for detection by epifluorescence microscopy.
RNA in Situ Hybridization.
RNA in situ hybridization was performed on frozen sections essentially as described with minor modifications (19). Detailed protocol is available upon request. The antisense chick Sox2 probe was derived from a cDNA clone encompassing the ORF region (NM_205188). The template for chick Sox6 probe was amplified from the chick cDNA (GenBank accession no. KC832418) and corresponds to the nucleotide 1–1361 region. Linearized templates were transcribed with SP6 RNA polymerase and digoxygenin-labeling mix (Roche). The probe was detected with alkaline phosphatase-conjugated anti-digoxigenin antibody and developed with nitro blue tetrazolium and 5-bromo-4-chloro-3-indoly phosphate, which yield purple precipitates.
Isolation of Chicken Sox6 cDNA Clones.
Neural tubes of HH stage 27 chicken embryos were dissociated using collagenase (Worthington) and dispase (Gibco) in HBSS by trituration. This digestion solution was quenched with FBS, the total RNA was extracted with TriZol reagent (Gibco), and cDNA synthesis was performed with the SuperScript system (Invitrogen) according to the manufacturer’s instructions using oligo(dT) primers. The two chick Sox6 cDNA variants encompassing the full-length ORF were amplified by PCR using oligonucleotide primers derived from the predicted chick Sox6 (XM_421000.3).
Expression Constructs and Chick in ovo Electroporation.
pCAG constructs for chick Sox1, Sox2, Sox3, HMG-VP16, and HMG-EnR have been described (5). Full-length cDNAs encoding the two isoforms of Sox6 were myc-tagged and subcloned into the pCAGGS vector. Details are available upon request. RNA interference was performed using the pSUPERIOR vector system (OligoEngine), which coexpresses the cloned shRNA under the control H1 promoter along with GFP under the control mouse phosphoglycerate kinase 1 promoter. The targeted sequences for Sox6 were 5′-TCGAAATGTACGAGGACTA-3′ (cSox6-sh1) and 5′-TCTAAAGACTGGAAGGAGA-3′ (cSox6-sh2). The control sequence targets the firefly luciferase gene, 5′-TGCTGGTGCCAACCCTATT-3′ (Luc-shRNA). Chicken embryos at HH stage 14–16 were used for in ovo electroporation assays. Approximately 0.1–2 μg/μL of expression vectors was injected into the neural-tube lumen along with 1 μL of 0.4% Trypan blue, and embryos were electroporated with five pulses of 22 V for 50 ms using a BTX electroporator. Eggs were incubated at 38 °C in an atmosphere of 70% humidity. After 24–48 h, embryos were fixed and processed for immnohistochemistry and RNA in situ hybridization.
Supplementary Material
Acknowledgments
This research was supported by Grant 2010K000803 from the Brain Research Center of the 21st Century Frontier Research Program and by the “Systems biology infrastructure establishment grant” provided by the Gwangju Institute of Science and Technology in 2008 through the Ewha Research Center for Systems Biology. Supports also came from Ministry of Science, ICT and Future Planning via National Research Foundation Grants NRF-2013M3C7A1056563 and NRF-2012M3A9B4028766 and Redoxomics Grant 2012M3A9C5048708.
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession nos. KC832418 and KC832419) and in the Gene Expression Omnibus database (accession no. GSE34395).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1308758111/-/DCSupplemental.
References
- 1.Kintner C. Neurogenesis in embryos and in adult neural stem cells. J Neurosci. 2002;22(3):639–643. doi: 10.1523/JNEUROSCI.22-03-00639.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arnold K, et al. Sox2(+) adult stem and progenitor cells are important for tissue regeneration and survival of mice. Cell Stem Cell. 2011;9(4):317–329. doi: 10.1016/j.stem.2011.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Boyer LA, et al. Core transcriptional regulatory circuitry in human embryonic stem cells. Cell. 2005;122(6):947–956. doi: 10.1016/j.cell.2005.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Suh H, et al. In vivo fate analysis reveals the multipotent and self-renewal capacities of Sox2+ neural stem cells in the adult hippocampus. Cell Stem Cell. 2007;1(5):515–528. doi: 10.1016/j.stem.2007.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bylund M, Andersson E, Novitch BG, Muhr J. Vertebrate neurogenesis is counteracted by Sox1-3 activity. Nat Neurosci. 2003;6(11):1162–1168. doi: 10.1038/nn1131. [DOI] [PubMed] [Google Scholar]
- 6.Sarkar A, Hochedlinger K. The Sox family of transcription factors: Versatile regulators of stem and progenitor cell fate. Cell Stem Cell. 2013;12(1):15–30. doi: 10.1016/j.stem.2012.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Graham V, Khudyakov J, Ellis P, Pevny L. SOX2 functions to maintain neural progenitor identity. Neuron. 2003;39(5):749–765. doi: 10.1016/s0896-6273(03)00497-5. [DOI] [PubMed] [Google Scholar]
- 8.Favaro R, et al. Hippocampal development and neural stem cell maintenance require Sox2-dependent regulation of Shh. Nat Neurosci. 2009;12(10):1248–1256. doi: 10.1038/nn.2397. [DOI] [PubMed] [Google Scholar]
- 9.Taranova OV, et al. SOX2 is a dose-dependent regulator of retinal neural progenitor competence. Genes Dev. 2006;20(9):1187–1202. doi: 10.1101/gad.1407906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Engelen E, et al. Sox2 cooperates with Chd7 to regulate genes that are mutated in human syndromes. Nat Genet. 2011;43(6):607–611. doi: 10.1038/ng.825. [DOI] [PubMed] [Google Scholar]
- 11.Bergsland M, et al. Sequentially acting Sox transcription factors in neural lineage development. Genes Dev. 2011;25(23):2453–2464. doi: 10.1101/gad.176008.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hagiwara N. Sox6, jack of all trades: A versatile regulatory protein in vertebrate development. Dev Dyn. 2011;240(6):1311–1321. doi: 10.1002/dvdy.22639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Stolt CC, et al. SoxD proteins influence multiple stages of oligodendrocyte development and modulate SoxE protein function. Dev Cell. 2006;11(5):697–709. doi: 10.1016/j.devcel.2006.08.011. [DOI] [PubMed] [Google Scholar]
- 14.Park PJ. ChIP-seq: Advantages and challenges of a maturing technology. Nat Rev Genet. 2009;10(10):669–680. doi: 10.1038/nrg2641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Odom DT, et al. Tissue-specific transcriptional regulation has diverged significantly between human and mouse. Nat Genet. 2007;39(6):730–732. doi: 10.1038/ng2047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Akiyama H, Chaboissier MC, Martin JF, Schedl A, de Crombrugghe B. The transcription factor Sox9 has essential roles in successive steps of the chondrocyte differentiation pathway and is required for expression of Sox5 and Sox6. Genes Dev. 2002;16(21):2813–2828. doi: 10.1101/gad.1017802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Batista-Brito R, et al. The cell-intrinsic requirement of Sox6 for cortical interneuron development. Neuron. 2009;63(4):466–481. doi: 10.1016/j.neuron.2009.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.An CI, Dong Y, Hagiwara N. Genome-wide mapping of Sox6 binding sites in skeletal muscle reveals both direct and indirect regulation of muscle terminal differentiation by Sox6. BMC Dev Biol. 2011;11:59. doi: 10.1186/1471-213X-11-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Birren SJ, Lo L, Anderson DJ. Sympathetic neuroblasts undergo a developmental switch in trophic dependence. Development. 1993;119(3):597–610. doi: 10.1242/dev.119.3.597. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.