Significance
Nuclear pore complexes (NPCs) are proteinaceous transport channels gating transport of macromolecules across the nuclear envelope. Nup192 is one of five members of an adaptor nucleoporin complex (ANC) that is believed to be integral for linking the NPC subcomplexes and accommodating the dilations of the central transport channel. We present the structure of the N-terminal half of Nup192 that uncovers similarities with karyopherins, suggesting an evolutionary relationship. Using biochemical and in vivo data, we generated an interaction map of the ANC, providing a starting point for its mechanistic dissection.
Keywords: X-ray crystallography, site-directed mutagenesis, mega assembly, structure-function analysis
Abstract
Nucleocytoplasmic transport is facilitated by nuclear pore complexes (NPCs), which are massive proteinaceous transport channels embedded in the nuclear envelope. Nup192 is a major component of an adaptor nucleoporin subcomplex proposed to link the NPC coat with the central transport channel. Here, we present the structure of the ∼110-kDa N-terminal domain (NTD) of Nup192 at 2.7-Å resolution. The structure reveals an open ring-shaped architecture composed of Huntingtin, EF3, PP2A, and TOR1 (HEAT) and Armadillo (ARM) repeats. A comparison of different conformations indicates that the NTD consists of two rigid halves connected by a flexible hinge. Unexpectedly, the two halves of the ring are structurally related to karyopherin-α (Kap-α) and β-karyopherin family members. Biochemically, we identify a conserved patch that binds an unstructured segment in Nup53 and show that a C-terminal tail region binds to a putative helical fragment in Nic96. The Nup53 segment that binds Nup192 is a classical nuclear localization-like sequence that interacts with Kap-α in a mutually exclusive and mechanistically distinct manner. The disruption of the Nup53 and Nic96 binding sites in vivo yields growth and mRNA export defects, revealing their critical role in proper NPC function. Surprisingly, both interactions are dispensable for NPC localization, suggesting that Nup192 possesses another nucleoporin interaction partner. These data indicate that the structured domains in the adaptor nucleoporin complex are held together by peptide interactions that resemble those found in karyopherin•cargo complexes and support the proposal that the adaptor nucleoporins arose from ancestral karyopherins.
Nucleocytoplasmic transport is an essential process in which transport factors, called karyopherins, mediate nuclear transport of macromolecules larger than ∼40 kDa (1–3). Karyopherins are classified into two families, α-karyopherins and β-karyopherins, which are composed of Armadillo (ARM) and Huntingtin, EF3, PP2A, and TOR1 (HEAT) repeats, respectively (1, 3). Karyopherins facilitate nucleocytoplasmic transport of proteins by recognizing linear nuclear localization sequences (NLSs) or nuclear export sequences (NESs) in their cargo proteins. α-Karyopherins are import adaptors that interact with β-karyopherins through an N-terminal importin-β binding domain to coordinate cargo import through the nuclear pore complex (NPC) (1–4). Whereas α-karyopherins are structurally rigid, β-karyopherins display remarkable conformational flexibility, which enables them to regulate cargo recognition (1, 3, 5).
The NPC is the sole gateway for bidirectional nucleocytoplasmic transport (6). Its transport channel is lined with intrinsically disordered phenylalanine-glycine repeats, which form a diffusion barrier and bind karyopherin•cargo complexes (1, 6). In yeast, NPCs are composed of 34 different proteins, termed nucleoporins (nups), that assemble in multiple copies into an ∼60-MDa complex (6). EM revealed that the NPC consists of a doughnut-shaped symmetrical core, which is embedded in the nuclear envelope and decorated with filamentous structures on its cytoplasmic and nucleoplasmic faces. Cytoplasmic filaments extend into the cytoplasm and provide docking sites for karyopherins, the GTPase Ran, and the mRNA export machinery (1, 5, 6). On the nucleoplasmic face, a nuclear basket structure binds Ran and the transcription machinery and participates in chromatin organization (6).
The symmetrical NPC core can be considered schematically as a series of concentrical cylindrical layers, composed of integral membrane proteins of the pore membrane, the coat-forming Nup84 complex, the adaptor nucleoporin complex (ANC), and the central channel nucleoporins (6, 7). Structural studies provided key insights into the architecture and function of the coat and channel nucleoporins, as well as the interactions of the cytoplasmic filament nucleoporins with the mRNA export machinery (7–16).
An EM analysis of the coat-forming, heptameric Nup84 complex from Saccharomyces cerevisiae revealed an ∼400-Å long, Y-shaped architecture (17). Crystallographic studies established that the heptamer is composed of α-helical solenoids and β-propellers interacting through extensive hydrophobic interfaces (7–9, 12, 13, 15). The heptamer is architecturally similar to the membrane bending COP-I, COP-II, and clathrin coats, suggesting that the Nup84 complex forms a coat for the nuclear envelope (7–9, 18).
In contrast to the Nup84 complex, the interactions between the channel nucleoporins are mediated by ∼250-residue α-helical regions, which adopt a range of alternative conformations and interactions, including sliding α-helical domains, alternative assembly states, and changes in interaction partners (19, 20). The plasticity of these nucleoporins is believed to facilitate the transport of differently sized cargoes by dilating and contracting the transport channel (19, 20).
Unlike the coat and channel nucleoporins, the interactions within the ANC are poorly understood, largely due to the poor behavior of the large adaptor nucleoporins in solution. However, the adaptor nucleoporins of the eukaryotic thermophile Chaetomium thermophilum possess improved biochemical robustness (21). The C. thermophilum ANC is composed of Nup192, Nup188, Nup170, Nic96, and Nup53, which are conserved throughout the eukaryotic kingdom (6, 21). Nic96 and Nup192 are the only essential members of the ANC in S. cerevisiae and are thought to function as the scaffolds onto which the adaptor layer assembles (21, 22). Negative-stain EM of Nup192 revealed a question mark-shaped architecture (21).
To gain structural and functional insight into the ANC, we determined the crystal structure of the N-terminal domain (NTD) of C. thermophilum Nup192 (ctNup192NTD) at 2.7-Å resolution. Our structural analysis reveals that ctNup192NTD forms an α-helical solenoid with an ∼110-Å wide ring-shaped architecture with similarities to the β-karyopherin Cse1p and import adaptor karyopherin-α (Kap-α). Furthermore, we show that ctNup192NTD possesses a binding site on its convex surface for an unstructured fragment of ctNup53, whereas a C-terminal tail fragment of ctNup192 binds to an α-helical region of ctNic96. Hence, ctNup192 is an interaction platform in the ANC with distinct binding sites. Disruption of either or both of these interactions results in growth and mRNA export defects in vivo, establishing their physiological importance. However, both interactions are dispensable for NPC localization, suggesting that Nup192 possesses at least one additional nucleoporin binding site sufficient to anchor Nup192 in the NPC. Based on our data, we propose that the interactions between the adaptor nucleoporins are governed by short linear motifs resembling those found in classical karyopherin•cargo complexes.
Results
Structure Determination.
We identified a fragment of C. thermophilum Nup192 encompassing residues 1–958 that yielded well-behaved, soluble protein (ctNup192NTD) (Fig. 1A). ctNup192NTD crystallized in space group P43212 with two molecules in the asymmetric unit. The structure was solved by single-wavelength anomalous dispersion using X-ray diffraction data from a seleno–l-methionine derivative (Fig. S1 A and B). The final model was refined to 2.7-Å resolution with Rwork and Rfree values of 19.1% and 23.1%, respectively. Data collection and refinement statistics are provided in Table S1. Because there was no dimerization in solution, we focused on the ctNup192NTD monomer (Fig. S1C).
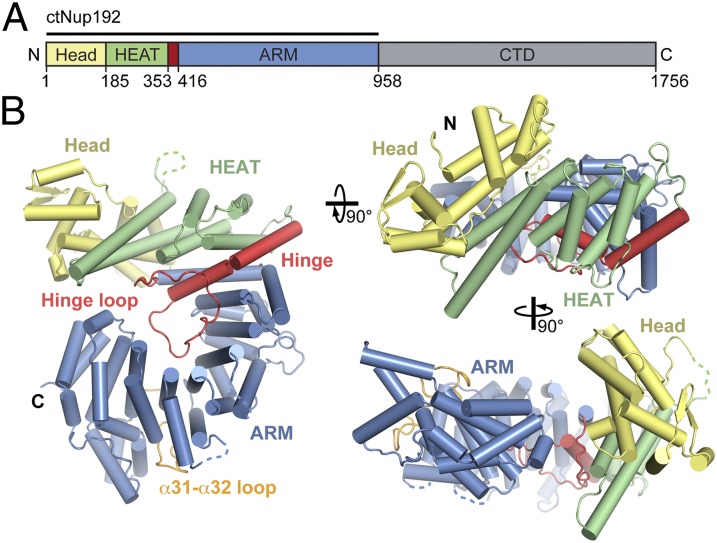
Fig. 1.
Structural overview of the NTD of ctNup192. (A) Domain structure of ctNup192 (crystallized fragment is indicated by a black bar). The N-terminal Head module (yellow), HEAT module (green), hinge module (red), ARM module (blue), and CTD (gray) are indicated. (B) Structure of ctNup192NTD shown in a cartoon representation using the same coloring scheme as in A. The hinge and α31–α32 loops, which line the inner arch of the ring, are colored in red and orange, respectively. Dashed lines indicate disordered loops.
Structural Overview.
ctNup192NTD folds into a ring-shaped solenoid composed of 42 α-helices (α1–α42) and a single β-hairpin (β1–β2) with a right-handed superhelical twist and dimensions of 110 Å × 90 Å × 40 Å (Fig. 1). The N- and C-terminal halves of the solenoid do not form direct contacts, resulting in an ∼10-Å gap in the ring and an overall lock washer-like shape. ctNup192NTD is divided into four structural segments: the N-terminal Head (residues 1–184), HEAT repeat (residues 185–352), small hinge (residues 353–415), and C-terminal ARM repeat modules (residues 416–958) (Fig. 1).
The Head module consists of two helical pairs, α1–α2 and α7–α8, arranged in a HEAT-like topology interrupted by an α-helical insertion (α3–α6) containing the β-hairpin (β1–β2). The Head module is connected to the HEAT module by a long, disordered loop between helices α8 and α9 and makes hydrophobic contacts with HEAT repeat 1 (α9–α10) (Fig. 1B and Fig. S1D).
The HEAT module is composed of three HEAT repeats that each form one turn of a right-handed superhelix. The HEAT and ARM modules are bridged by a short hinge module composed of the hinge loop and two helices, α15 and α16. These helices are connected by a single glycine residue that is invariant across fungi, suggesting that this feature is evolutionarily conserved (Fig. S2). Helix α15 caps the end of the HEAT module superhelix, and helix α16 initiates the ARM module superhelix. The hinge loop is an ordered, 31-residue connector, which forms contacts along the entire concave surface of the ring. The ARM module forms a right-handed superhelix composed of noncanonical ARM repeats. ARM repeat 6 (α32–α35) is a degenerate ARM repeat in which the α31–α32 loop replaces the first helix (Fig. 1 and Fig. S1D).
Structural Similarity to Karyopherins.
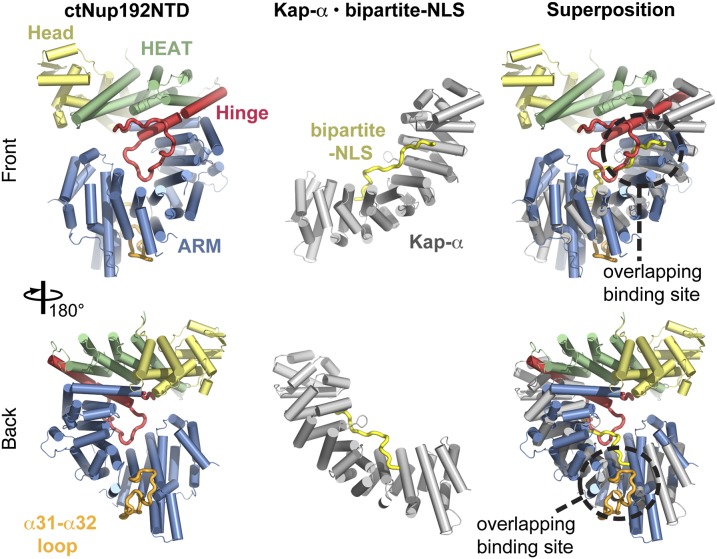
The unusual combination of HEAT and ARM repeats in ctNup192NTD prompted us to look for proteins with a similar architecture. In structural homology searches with the Dali server, we found that yeast Kap-α was most similar to the ARM module of ctNup192NTD (23). The structure of yeast Kap-α superposes with the ARM module with an rmsd of 5.7 Å over 303 Cα atoms (Fig. 2). Both proteins have an identical topology, but the α31–α32 loop perturbs the curvature of the ARM module superhelix, generating a greater curvature in ctNup192NTD. Intriguingly, the hinge and α31–α32 loops overlap with the Kap-α recognition sites for a bipartite NLS (Fig. 2). However, the conserved tryptophans that line the concave surface of Kap-α and bind the NLS peptide are absent in ctNup192NTD. Likewise, a Dali search on the HEAT module of ctNup192NTD found the export β-karyopherin Cse1p to be structurally similar. The HEAT module superposed with three N-terminal HEAT repeats of Cse1p with an rmsd of 5.2 Å over 117 Cα atoms (Fig. S3A). Thus, ctNup192NTD possesses a surprising architectural similarity to members of both karyopherin families, wherein the C-terminal ARM module is structurally analogous to Kap-α and the N-terminal HEAT module is similar to β-karyopherins.
Fig. 2.
Structural similarity of the ctNup192NTD ARM module to Kap-α. Overview of ctNup192NTD (Left) colored as in Fig. 1B, the Kap-α•bipartite-NLS complex (Center, gray and yellow; Protein Data Bank ID code 1EE5) (28), and their superposition (Right).
Structural Analysis.
Given the unusual topology of ctNup192NTD, we further analyzed the individual ARM and HEAT repeats. Sequence and structural alignments revealed that whereas the ARM and HEAT repeats of ctNup192NTD possess the hallmark sequence characteristics for their respective motifs, they are also more divergent in helical length and position (Fig. S3 B and C). Superposition of the two molecules in the asymmetric unit and the structure of S. cerevisiae Nup192NTD (scNup192NTD) revealed conformational changes in the entire ring that open and close the gap between the two ends of the ring by ∼10 Å (24) (Fig. S4A). These conformational changes are mostly the result of rigid body rearrangements around the hinge module (Fig. S4 B and C). A detailed analysis is provided in SI Text.
Surface Properties.
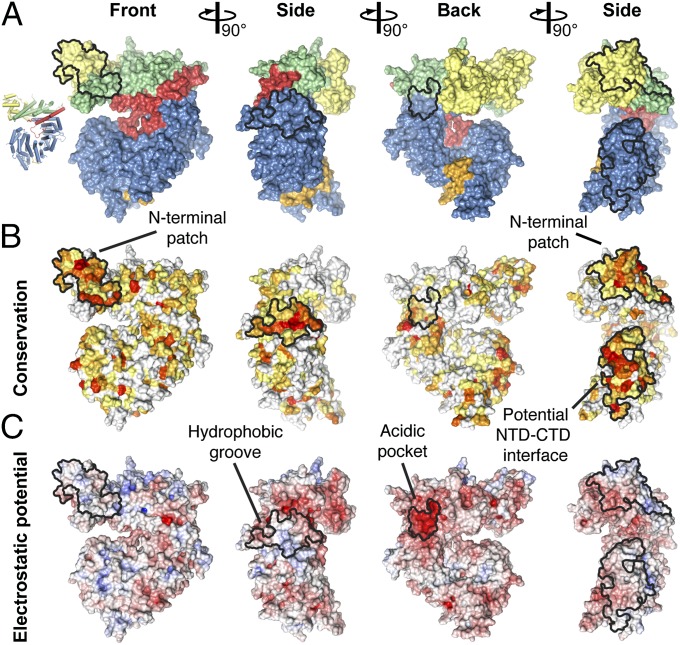
To identify functionally important surfaces in ctNup192NTD, we created surface maps depicting evolutionary conservation and electrostatic potential (Fig. 3 and Fig. S2). An immediately apparent feature is a conserved hydrophobic groove on the convex surface of the ring, next to the hinge module (Fig. 3 A and B). In CRM1, the availability of the hydrophobic NES binding cleft is modulated by a RanGTP-dependent rearrangement of an acidic loop (25). The hydrophobic groove observed in ctNup192NTD is located in a similar position on the outside of the ring, suggesting that the interaction with another nucleoporin at this site is regulated similarly.
Fig. 3.
Surface properties of ctNup192NTD. (A) Surface representation of ctNup192NTD in four different orientations colored as in Fig. 1B. (B) Surface representation of ctNup192NTD colored according to conservation within seven fungal Nup192 sequences (Fig. S2). Sequence conservation is shaded from white (<45% similarity), to yellow (45% similarity), to red (100% identity). (C) Surface representation is colored according to electrostatic potential from −10 kBT/e (red) to +10 kBT/e (blue).
Additionally, we could identify a conserved, charged surface patch that spans the α-helical insertion of the Head module and the front face of the HEAT module (Fig. 3). A third, C-terminal surface also displays a high degree of conservation, but it is formed by the last α-helix in the truncated construct, suggesting that it is normally buried in the interface between the NTD and C-terminal domain (CTD) of full-length ctNup192 (Fig. 3B). Finally, we identified a surface pocket with a highly negative electrostatic potential on the back face of ctNup192NTD (Fig. 3C). Although this pocket is not strictly conserved in sequence, its negative character is maintained in the structure of scNup192NTD (24). Furthermore, this acidic pocket is immediately adjacent to the hydrophobic groove, suggesting that these two features form a composite binding site for another nucleoporin. Given their distinct chemical natures, these identified surfaces represent likely protein–protein interaction sites.
Biochemical Analysis.
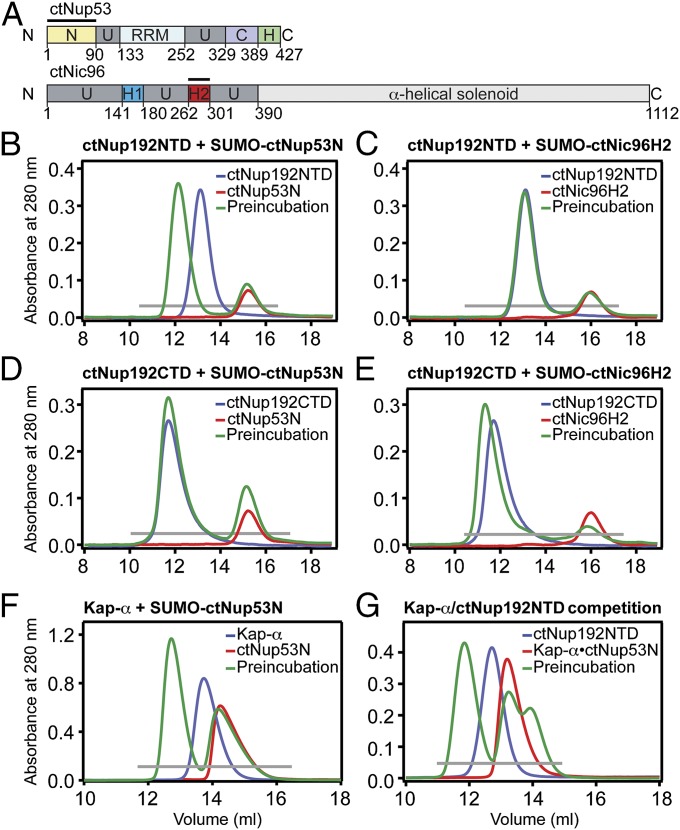
Nup192 was originally identified as the S. cerevisiae homolog of vertebrate Nup205 and, subsequently, as an interaction partner of Nic96 (22, 26). Recently, ctNup192 was found to interact with both a fragment of ctNic96 and a region of ctNup53 (21). Based on these results, we tested whether an N-terminal fragment of ctNup53 (ctNup53N, residues 1–90) and an N-terminal, α-helical segment of ctNic96 (ctNic96H2, residues 262–301) form a complex with ctNup192NTD (Fig. 4A). In size-exclusion chromatography (SEC) interaction experiments, ctNup192NTD formed a stoichiometric complex with ctNup53N but failed to interact with ctNic96H2 (Fig. 4 B and C and Fig. S5 A–C). In contrast, the ctNup192 CTD (ctNup192CTD, residues 976–1,756) is capable of forming a stoichiometric complex with ctNic96H2 but fails to form a complex with ctNup53N (Fig. 4 D and E and Fig. S5 D and E). Further mapping revealed that only the C-terminal 340 residues of ctNup192, ctNup192TAIL, mediate the interaction with ctNic96H2 (Fig. S5F).
Fig. 4.
Biochemical interaction analysis. (A) Domain organization of ctNup53 and ctNic96. C, C-terminal segment; H, amphipathic helix; H1, Helix 1; H2, Helix 2; N, N-terminal segment; RRM, RNA-recognition motif domain; U, unstructured segment. Black bars highlight fragments used for interaction analysis. SEC analysis of ctNup192NTD with ctNup53N (B) or ctNic96H2 (C). SEC analysis of ctNup192CTD with ctNup53N (D) or ctNic96H2 (E). (F) SEC analysis of Kap-α with ctNup53N. (G) SEC analysis of competitive binding between Kap-α•ctNup53N and ctNup192NTD. Gray bars and colored lines designate the analyzed fractions in the respective SDS/PAGE gels stained with Coomassie brilliant blue (Fig. S5 B–E, G, and H).
ctNup53N contains a 37-residue region (residues 31–67) that resembles a classical bipartite NLS with the consensus sequence KR(X)10–12K(K/R)X(K/R) but with deviations wherein lysine and arginine residues are interchanged (Fig. 5A). Given the structural similarity of ctNup192NTD to Kap-α, we tested whether ctNup53N is also capable of interacting with Kap-α. Indeed, ctNup53N and Kap-α form a stoichiometric complex on a gel filtration column (Fig. 4F and Fig. S5G). We next tested whether ctNup53N could simultaneously interact with both ctNup192NTD and Kap-α by incubating the preformed Kap-α•ctNup53N pair with ctNup192NTD. However, in our SEC interaction experiments, ctNup53N binding to ctNup192NTD and Kap-α is mutually exclusive (Fig. 4G and Fig. S5H). The unexpected finding that ctNup192NTD and Kap-α both interact with the same ctNup53 fragment further supports an evolutionary connection between karyopherins and adaptor nucleoporins.
Fig. 5.
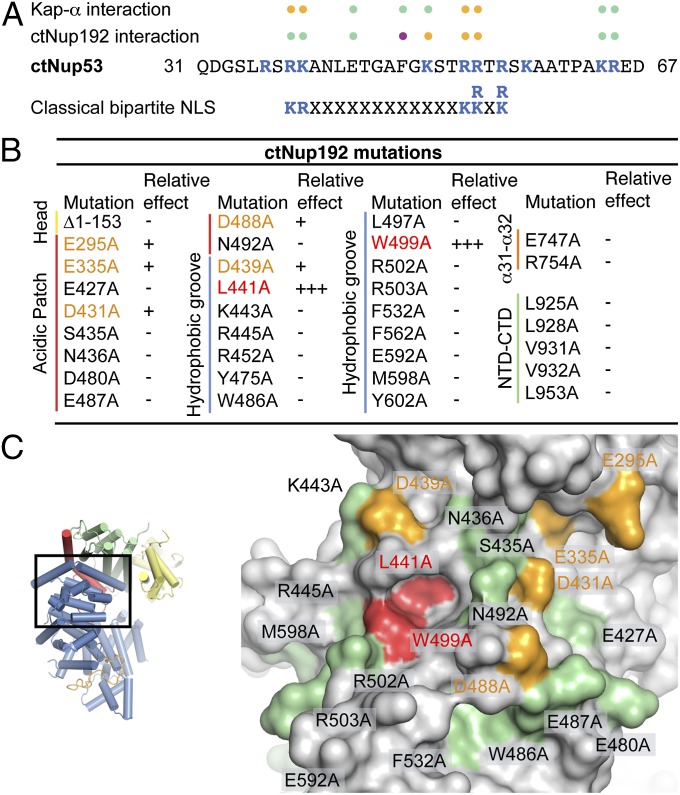
Mutational analysis of the ctNup53 interactions with ctNup192 and Kap-α. (A) Sequence comparison of ctNup5331–67 with a consensus classical bipartite NLS and summary of the effects of ctNup5331–67 mutations on ctNup192NTD and Kap-α binding. Basic residues are highlighted in blue, and dots indicate mutations that cause no effect (green), reduced binding (orange), or complete disruption (purple) (Fig. S6 A and B). (B) Mutational analysis of the ctNup192NTD–ctNup5331–67 interaction. Mutations that have no effect (−, black), reduce binding (+, orange), or abolish the interaction (+++, red) are indicated (Fig. S7A). (C, Right) Surface mapping of ctNup192NTD mutagenesis results. (C, Left) As a reference, a cartoon representation of ctNup192NTD, colored as in Fig. 1B, is shown. The locations of mutations on the ctNup192NTD surface are labeled and colored in green, orange, and red to indicate no effect, reduced binding, or complete disruption of ctNup5331–67 binding, respectively.
To gain additional insight into the molecular details of the ctNup53N interactions, we generated a minimal fragment comprising residues 31–67, ctNup5331–67, and purified nine ctNup5331–67 alanine mutants of conserved residues. In SEC interaction experiments, only mutations of basic residues at consensus bipartite NLS positions (R39, K40, R53, and R54) impaired the interaction with Kap-α (Fig. 5A and Fig. S6A). Additionally, two of the mutations affecting Kap-α binding, R53A and R54A, also weakened the interaction with ctNup192NTD (Fig. 5A and Fig. S6B), providing a molecular explanation for the exclusivity of these interactions. Strikingly, a single mutation, F48A, completely abolished the ctNup5331–67–ctNup192NTD interaction, with no effect on Kap-α binding (Fig. 5 A and B and Fig. S6B). Thus, the ctNup5331–67–Kap-α interaction appears to be mechanistically similar to classical NLS binding, whereas the ctNup5331–67–ctNup192NTD interaction is mediated by a distinct binding mode.
We next determined whether any of the potential protein–protein interaction sites on the ctNup192NTD surface mediate binding to ctNup5331–67. We purified 33 single-alanine mutants of conserved residues distributed throughout the identified surfaces, as well as a variant of ctNup192NTD that lacked the Head module (Fig. 5). All tested mutants were properly folded, because they were indistinguishable in their behavior from WT ctNup192NTD. Whereas deletion of the Head module and mutations in the potential NTD–CTD interface and α31–α32 loop had no effect on ctNup5331–67 binding, there was a strong clustering of mutations in the acidic pocket and hydrophobic groove that reduced (E295A, E335A, D431A, D488A, and D439A) or completely abolished (L441A and W499A) ctNup5331–67 binding (Fig. 5B and Fig. S7A). These mutations are on the convex surface of ARM1 and ARM2 or immediately adjacent to it (Fig. 5C). L441 and W499, which are essential for ctNup5331–67 binding, form a deep hydrophobic pocket in the conserved hydrophobic groove (Fig. 5C). These results establish that ctNup192NTD binds to ctNup5331–67 with a combined surface that stretches from the hydrophobic groove formed by the convex surfaces of ARM1 and ARM2 to the immediately adjacent acidic pocket. The mechanism of the Nup192–Nup53 interaction is evolutionarily conserved, because WT scNup192NTD and scNup53N form stoichiometric complexes that are abolished by the corresponding point mutations, scNup192NTD, W513A and scNup53N, F124A (Figs. S6C and S7B).
Using isothermal titration calorimetry (ITC), we found that the ctNup192NTD–ctNup5331–67 interaction has a dissociation constant of ∼0.1 μM and that single-alanine mutations, ctNup5331–67, F48A and ctNup192NTD, W499A, reduce binding to undetectable levels, consistent with our SEC interaction experiments (Fig. S7 C–E). Surprisingly, the interaction between ctNup5331–67 and Kap-α is 250-fold weaker (∼25 μM) than the interaction between ctNup5331–67 and ctNup192NTD (Fig. S7F), likely as a result of a deviating NLS sequence observed in ctNup5331–67. The ITC results are summarized in Fig. S7G.
Given the similarities in overall shape and architecture between Nup192 and the other large adaptor nucleoporin, Nup188 (6), we tested whether ctNup188NTD also interacts with ctNup5331–67 but detected no binding in an SEC interaction experiment (Fig. S8A). A comparison of the ctNup192NTD and Myceliophthora thermophila Nup188NTD (mtNup188NTD) structures provides a structural explanation for this result (27) (Fig. S8 B and C). Although the two proteins share the same topology and organization, two features are unique to mtNup188NTD: an N-terminal clamp module that binds the C terminus of mtNup188NTD and an SH3-like insertion in ARM repeat 3 between helices α23 and α24, protruding out of the convex surface (Fig. S8B). Furthermore, the ctNup5331–67 binding site is not conserved in mtNup188NTD and possesses the following significant alterations: (i) The bulky hydrophobic residues that form the deep pocket in ctNup192NTD are absent in mtNup188NTD; instead, the helices pack more closely together, and (ii) helix α18, which forms a large part of the conserved hydrophobic groove, is absent in mtNup188NTD; this part of the surface is instead occupied by the SH3-like insertion, thus restricting access to the convex surface (Fig. S8C).
Altogether, these results show that Nup192 interacts with two other adaptor nucleoporins, Nup53 and Nic96, via spatially separated binding sites at opposite ends of the question mark-shaped molecule, consistent with its ANC-scaffolding role. Despite their structural and evolutionary relationship, the large adaptor nucleoporins Nup192 and Nup188 fulfill functionally distinct roles in the NPC, because the interaction with Nup53 is not replicated in Nup188.
Functional Analysis.
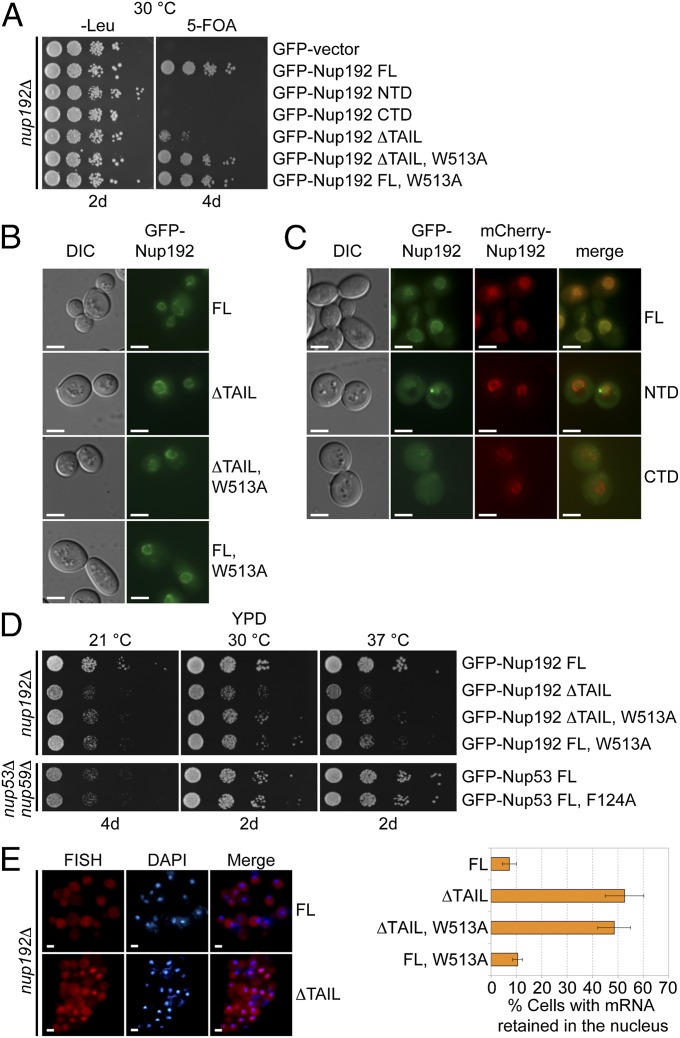
Nup192 is essential for viability in S. cerevisiae, but its roles in NPC structure and function are poorly understood (22). To identify the roles of Nup192 and its binding partners in vivo, we determined the viability of S. cerevisiae nup192Δ deletion strains complemented by various GFP-tagged Nup192 constructs. Neither Nup192NTD nor Nup192CTD alone was sufficient to overcome the lethality of the Nup192 KO (Fig. 6A). In contrast, Nup192 variants that abolish the interaction with Nup53 (Nup192FL, W513A) or Nic96 (Nup192∆TAIL) were sufficient to restore viability (Fig. 6A). Viability correlated with targeting of the GFP-Nup192 fusion constructs to the nuclear envelope. Whereas GFP-Nup192NTD and GFP-Nup192CTD were diffusely localized throughout the entire cell, GFP-Nup192FL, W513A and GFP-Nup192∆TAIL variants displayed robust nuclear rim staining consistent with NPC incorporation (Fig. 6 B and C). Despite rescuing lethality, individual disruption of the Nup53 or Nic96 interactions yielded mild and severe growth defects, respectively (Fig. 6D). Consistent with these results, complementation of a nup53Δnup59Δ strain with the GFP-Nup53F124A mutant, which is defective in Nup192 binding, exhibited no observable growth defect (Fig. 6D).
Fig. 6.
In vivo analyses of Nup192 and Nup53 mutants in S. cerevisiae. (A) The nup192Δ shuffle strain containing an mCherry-Nup192 cover plasmid (URA3) was transformed with control or GFP-Nup192 variants [leucine (Leu)]. Growth was followed on synthetic dextrose complete (SDC)-Leu and 5-fluoroorotic acid (5-FOA)/SDC plates for the indicated times and temperatures. FL, full-length. (B and C) In vivo localization of GFP-Nup192 variants in the nup192Δ strain visualized by fluorescence and differential interference contrast (DIC) microscopy. (C) Cells still carry the mCherry-Nup192 full-length plasmid. (D) Growth analysis of nup192Δ and nup53Δnup59Δ strains. Cells were spotted on yeast extract peptone dextrose (YPD) plates and grown for 2–4 d at the indicated temperatures. (E) mRNA export assay of GFP-Nup192 variants. (Left) Representative images of WT GFP-Nup192– (Upper) and GFP-Nup192∆TAIL– (Lower) complemented nup192Δ cells are shown. (Right) Quantification of nuclear poly(A) mRNA retention is shown. Error bars indicate SDs derived from six independent images, each containing ∼100 cells. (Scale bars: 5 μm.)
We next assayed mRNA export by FISH of an Alexa647-labeled dT50 oligonucleotide as a probe for NPC function. In agreement with the observed growth phenotypes, disruption of Nic96 binding (Nup192∆TAIL) caused a substantial mRNA export defect, because ∼53% of cells displayed nuclear mRNA retention compared with ∼7% of cells containing full-length Nup192. Loss of Nup53 binding (Nup192FL, W513A) had only a mild effect on mRNA export, with ∼10% of cells displaying a defect (Fig. 6E). Interestingly, the loss of both the Nic96 and Nup53 interactions failed to have an additive effect, because the Nup192∆TAIL, W513A variant exhibited a less pronounced growth phenotype than Nup192∆TAIL, despite still possessing a major mRNA export defect with ∼49% of cells displaying nuclear mRNA retention (Fig. 6 D and E). Unexpectedly, GFP-Nup192∆TAIL, W513A still demonstrated robust rim staining, indicating that both interactions are dispensable for NPC incorporation. These results confirm the physiological importance of the interactions within the ANC interaction network that we characterized biochemically, and further suggest a yet to be identified nucleoporin interaction that is sufficient to anchor Nup192 in the NPC.
Discussion
We determined the structure of the NTD of the large adaptor nucleoporin Nup192 from C. thermophilum. The structure revealed a ring-shaped architecture composed of an unusual combination of HEAT and ARM repeats that display unexpected structural similarities with karyopherins. Comparison of distinct conformational states of ctNup192NTD revealed a hinge module that bridges the two halves of the ring and confers conformational plasticity. Furthermore, ctNup192 binds to linear sequence motifs in two other adaptor nucleoporins, ctNup53 and ctNic96, in a manner that resembles known karyopherin•cargo complex interactions (1–3, 5). We also found that the same segment in ctNup53 interacts with both ctNup192NTD and Kap-α in a mutually exclusive fashion, using partially overlapping, interdigitated binding motifs. Together, these data suggest an evolutionary connection between the karyopherins and the large, all-helical adaptor nucleoporin Nup192.
Our interaction analyses facilitate the construction of a high-resolution biochemical map of the adaptor nucleoporin interaction network. Mutational analysis of the ctNup192NTD surface revealed that ctNup53 binds at the midpoint of the convex surface of the ring in a combined interface that includes a conserved hydrophobic groove and an acidic pocket. These findings suggest that ctNup53 binding to ctNup192NTD is mechanistically distinct from the classical bipartite NLS–Kap-α interaction, which is mediated by an array of tryptophans and acidic residues lining the concave Kap-α surface (28). Our finding that a single hydrophobic residue in ctNup53 is essential for ctNup192NTD binding but dispensable for the Kap-α interaction further supports this conclusion. Whether ctNup53 indeed binds to Kap-α in a classical fashion or possesses a distinct interaction mode, as previously shown for other nucleoporins, awaits further structural characterization (29).
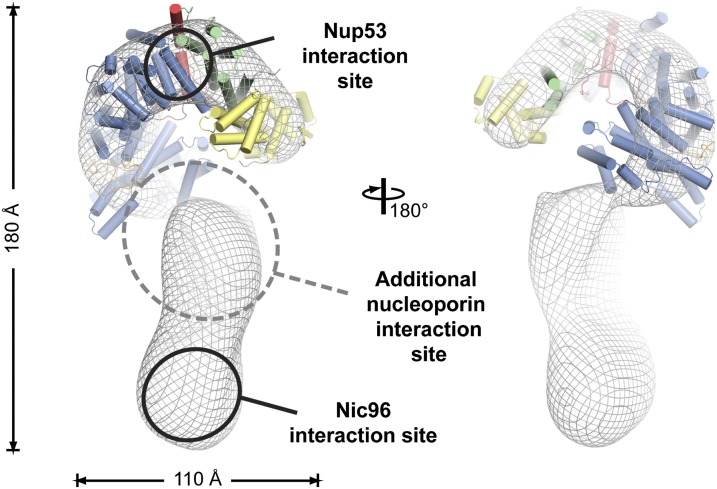
We also found that the ctNic96H2 interaction site maps to the C-terminal tail of ctNup192. By docking the structure of ctNup192NTD into a negative-stain EM envelope of full-length ctNup192 (21), we establish that the ctNup53 binding site is located at the top of the question mark-shaped molecule, which is the furthest possible point from the C-terminal tail that binds ctNic96, a distance of ∼150 Å (Fig. 7). Our in vivo localization analysis further suggests that there is at least one additional nucleoporin interaction that is sufficient to anchor Nup192 in the NPC, because the interactions with Nup53 and Nic96 are dispensable for its localization. Nevertheless, the observed growth and mRNA export defects establish both identified interactions as important for proper NPC function.
Fig. 7.
Model for the ctNup192 interaction network. The structure of ctNup192NTD is docked into the EM envelope of full-length ctNup192 (21). The general locations of the interaction sites for Nup5331–67 (Upper) and Nic96H2 (Lower) on Nup192 are shown. The location of a yet to be identified nucleoporin interaction site is indicated.
We propose that the interactions between the structured domains in the ANC are primarily mediated by short linear motifs, which we suggest terming “nucleoporin anchor sequences” (NASs). Such a peptide interaction network would be in stark contrast to the 3D interfaces previously identified in the Nup84 complex (7–9, 12, 15). By tethering large, distortable adaptor nucleoporin solenoids with predominantly unstructured “linker” nucleoporins, the adaptor layer would be able to cushion the proposed dilations of the central transport channel dynamically during cargo translocation. Furthermore, the interactions between the linker nucleoporins and the large structural nucleoporins may allow for a hierarchical NPC assembly pathway resembling the assembly and disassembly of transport factor•cargo complexes. An intriguing possibility is that the mutually exclusive interactions of Nup53 with Kap-α and Nup192 may play a role in the regulation of NPC assembly or cargo transport, possibly in a manner similar to the competitive interactions in the Nup53-Nup170-Kap121 network (30, 31). Finally, such a NAS interaction network is also likely to facilitate the oligomerization of the NPC subcomplexes in the assembled NPC, a possibility exemplified by our previous observation that an unstructured segment in Nup133 mediates the head-to-tail arrangement of the Nup84 complex (13). Future work is necessary to identify all interactions within the ANC, understand its overall organization, and determine its roles in NPC function.
Methods
The details of X-ray diffraction data collection and structure data are described in SI Text and Table S1. The details of molecular cloning, expression, purification, crystallization, protein interaction and multiangle light scattering analyses, biochemistry experiments, and yeast experiments are described in SI Text and Tables S2 and S3. ctNup192NTD was expressed using a modified pET28a vector (32).
Supplementary Material
Acknowledgments
We thank members of the A.H. laboratory, Alina Patke, and Yunji Wu for critical reading of the manuscript; David King for MS analysis; Jens Kaiser and the scientific staff of Stanford Synchrotron Radiation Lightsource (SSRL) beam line 12-2 for their support with X-ray diffraction measurements; the University of Colorado Biophysics Core for assistance with ITC measurements; and Elena Conti for material. We acknowledge the Gordon and Betty Moore Foundation, the Beckman Institute, and the Sanofi–Aventis Bioengineering Research Program for their support of the Molecular Observatory at the California Institute of Technology. The SSRL is supported by the Department of Energy and by the National Institutes of Health (NIH). T.S. is supported by a Deutsche Forschungsgemeinschaft (DFG) postdoctoral fellowship. D.H.L. is supported by an NIH Research Service Award (Grant 5 T32 GM07616). A.H. was supported by the Albert Wyrick V Scholar Award of the V Foundation for Cancer Research, the 54th Mallinckrodt Scholar Award of the Edward Mallinckrodt, Jr. Foundation, and a Kimmel Scholar Award of the Sidney Kimmel Foundation for Cancer Research.
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
Data deposition: The structure factors and atomic coordinates of the Nup192 N-terminal domain have been deposited in the Protein Data Bank, www.pdb.org (PDB ID code 4KNH).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1311081111/-/DCSupplemental.
References
- 1.Cook A, Bono F, Jinek M, Conti E. Structural biology of nucleocytoplasmic transport. Annu Rev Biochem. 2007;76:647–671. doi: 10.1146/annurev.biochem.76.052705.161529. [DOI] [PubMed] [Google Scholar]
- 2.Hoelz A, Blobel G. Cell biology: Popping out of the nucleus. Nature. 2004;432(7019):815–816. doi: 10.1038/432815a. [DOI] [PubMed] [Google Scholar]
- 3.Xu D, Farmer A, Chook YM. Recognition of nuclear targeting signals by Karyopherin-β proteins. Curr Opin Struct Biol. 2010;20(6):782–790. doi: 10.1016/j.sbi.2010.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lott K, Cingolani G. The importin β binding domain as a master regulator of nucleocytoplasmic transport. Biochim Biophys Acta. 2011;1813(9):1578–1592. doi: 10.1016/j.bbamcr.2010.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Debler EW, Blobel G, Hoelz A. Nuclear transport comes full circle. Nat Struct Mol Biol. 2009;16(5):457–459. doi: 10.1038/nsmb0509-457. [DOI] [PubMed] [Google Scholar]
- 6.Hoelz A, Debler EW, Blobel G. The structure of the nuclear pore complex. Annu Rev Biochem. 2011;80:613–643. doi: 10.1146/annurev-biochem-060109-151030. [DOI] [PubMed] [Google Scholar]
- 7.Hsia KC, Stavropoulos P, Blobel G, Hoelz A. Architecture of a coat for the nuclear pore membrane. Cell. 2007;131(7):1313–1326. doi: 10.1016/j.cell.2007.11.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brohawn SG, Leksa NC, Spear ED, Rajashankar KR, Schwartz TU. Structural evidence for common ancestry of the nuclear pore complex and vesicle coats. Science. 2008;322(5906):1369–1373. doi: 10.1126/science.1165886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Debler EW, et al. A fence-like coat for the nuclear pore membrane. Mol Cell. 2008;32(6):815–826. doi: 10.1016/j.molcel.2008.12.001. [DOI] [PubMed] [Google Scholar]
- 10.Kassube SA, et al. Crystal structure of the N-terminal domain of Nup358/RanBP2. J Mol Biol. 2012;423(5):752–765. doi: 10.1016/j.jmb.2012.08.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lin DH, Zimmermann S, Stuwe T, Stuwe E, Hoelz A. Structural and functional analysis of the C-terminal domain of Nup358/RanBP2. J Mol Biol. 2013;425(8):1318–1329. doi: 10.1016/j.jmb.2013.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nagy V, et al. Structure of a trimeric nucleoporin complex reveals alternate oligomerization states. Proc Natl Acad Sci USA. 2009;106(42):17693–17698. doi: 10.1073/pnas.0909373106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Seo HS, et al. Structural and functional analysis of Nup120 suggests ring formation of the Nup84 complex. Proc Natl Acad Sci USA. 2009;106(34):14281–14286. doi: 10.1073/pnas.0907453106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Stuwe T, von Borzyskowski LS, Davenport AM, Hoelz A. Molecular basis for the anchoring of proto-oncoprotein Nup98 to the cytoplasmic face of the nuclear pore complex. J Mol Biol. 2012;419(5):330–346. doi: 10.1016/j.jmb.2012.03.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Whittle JR, Schwartz TU. Architectural nucleoporins Nup157/170 and Nup133 are structurally related and descend from a second ancestral element. J Biol Chem. 2009;284(41):28442–28452. doi: 10.1074/jbc.M109.023580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yoshida K, Seo HS, Debler EW, Blobel G, Hoelz A. Structural and functional analysis of an essential nucleoporin heterotrimer on the cytoplasmic face of the nuclear pore complex. Proc Natl Acad Sci USA. 2011;108(40):16571–16576. doi: 10.1073/pnas.1112846108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lutzmann M, Kunze R, Buerer A, Aebi U, Hurt E. Modular self-assembly of a Y-shaped multiprotein complex from seven nucleoporins. EMBO J. 2002;21(3):387–397. doi: 10.1093/emboj/21.3.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Debler EW, Hsia KC, Nagy V, Seo HS, Hoelz A. Characterization of the membrane-coating Nup84 complex: Paradigm for the nuclear pore complex structure. Nucleus. 2010;1(2):150–157. doi: 10.4161/nucl.1.2.11120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Melcák I, Hoelz A, Blobel G. Structure of Nup58/45 suggests flexible nuclear pore diameter by intermolecular sliding. Science. 2007;315(5819):1729–1732. doi: 10.1126/science.1135730. [DOI] [PubMed] [Google Scholar]
- 20.Solmaz SR, Blobel G, Melcák I. Ring cycle for dilating and constricting the nuclear pore. Proc Natl Acad Sci USA. 2013;110(15):5858–5863. doi: 10.1073/pnas.1302655110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Amlacher S, et al. Insight into structure and assembly of the nuclear pore complex by utilizing the genome of a eukaryotic thermophile. Cell. 2011;146(2):277–289. doi: 10.1016/j.cell.2011.06.039. [DOI] [PubMed] [Google Scholar]
- 22.Kosova B, Panté N, Rollenhagen C, Hurt E. Nup192p is a conserved nucleoporin with a preferential location at the inner site of the nuclear membrane. J Biol Chem. 1999;274(32):22646–22651. doi: 10.1074/jbc.274.32.22646. [DOI] [PubMed] [Google Scholar]
- 23.Holm L, Rosenström P. Dali server: Conservation mapping in 3D. Nucleic Acids Res. 2010;38(Web Server issue):W545–W459. doi: 10.1093/nar/gkq366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sampathkumar P, et al. Structure, dynamics, evolution, and function of a major scaffold component in the nuclear pore complex. Structure. 2013;21(4):560–571. doi: 10.1016/j.str.2013.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Monecke T, et al. Structural basis for cooperativity of CRM1 export complex formation. Proc Natl Acad Sci USA. 2013;110(3):960–965. doi: 10.1073/pnas.1215214110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Grandi P, et al. Nup93, a vertebrate homologue of yeast Nic96p, forms a complex with a novel 205-kDa protein and is required for correct nuclear pore assembly. Mol Biol Cell. 1997;8(10):2017–2038. doi: 10.1091/mbc.8.10.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Andersen KR, et al. Scaffold nucleoporins Nup188 and Nup192 share structural and functional properties with nuclear transport receptors. Elife. 2013;2:e00745. doi: 10.7554/eLife.00745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Conti E, Kuriyan J. Crystallographic analysis of the specific yet versatile recognition of distinct nuclear localization signals by karyopherin alpha. Structure. 2000;8(3):329–338. doi: 10.1016/s0969-2126(00)00107-6. [DOI] [PubMed] [Google Scholar]
- 29.Matsuura Y, Stewart M. Nup50/Npap60 function in nuclear protein import complex disassembly and importin recycling. EMBO J. 2005;24(21):3681–3689. doi: 10.1038/sj.emboj.7600843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lusk CP, Makhnevych T, Marelli M, Aitchison JD, Wozniak RW. Karyopherins in nuclear pore biogenesis: A role for Kap121p in the assembly of Nup53p into nuclear pore complexes. J Cell Biol. 2002;159(2):267–278. doi: 10.1083/jcb.200203079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kobayashi J, Matsuura Y. Structural basis for cell-cycle-dependent nuclear import mediated by the karyopherin Kap121p. J Mol Biol. 2013;425(11):1852–1868. doi: 10.1016/j.jmb.2013.02.035. [DOI] [PubMed] [Google Scholar]
- 32.Hoelz A, Nairn AC, Kuriyan J. Crystal structure of a tetradecameric assembly of the association domain of Ca2+/calmodulin-dependent kinase II. Mol Cell. 2003;11(5):1241–1251. doi: 10.1016/s1097-2765(03)00171-0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.