Significance
Trimethylamine N-oxide (TMAO) is a nitrogen-containing osmolyte found in a wide variety of marine biota and has been detected at nanomolar concentrations in surface seawaters. This study provides the first genetic and biochemical evidence for uptake and catabolism of TMAO by marine heterotrophic bacteria that are abundant in the oceans. The genes conferring the ability of bacteria to catabolize TMAO we identified in this study are highly expressed in the marine environment and can be used as functional biomarkers to better understand oceanic microbial-mediated carbon and nitrogen cycles. Our data suggest that TMAO represents a significant, yet overlooked, nutrient for marine bacteria in the surface oceans.
Keywords: TMAO transporter, nitrogenous osmolyte, methylated amine metabolism, marine nitrogen cycle
Abstract
Trimethylamine N-oxide (TMAO) is a common osmolyte found in a variety of marine biota and has been detected at nanomolar concentrations in oceanic surface waters. TMAO can serve as an important nutrient for ecologically important marine heterotrophic bacteria, particularly the SAR11 clade and marine Roseobacter clade (MRC). However, the enzymes responsible for TMAO catabolism and the membrane transporter required for TMAO uptake into microbial cells have yet to be identified. We show here that the enzyme TMAO demethylase (Tdm) catalyzes the first step in TMAO degradation. This enzyme represents a large group of proteins with an uncharacterized domain (DUF1989). The function of TMAO demethylase in a representative from the SAR11 clade (strain HIMB59) and in a representative of the MRC (Ruegeria pomeroyi DSS-3) was confirmed by heterologous expression of tdm (the gene encoding Tdm) in Escherichia coli. In R. pomeroyi, mutagenesis experiments confirmed that tdm is essential for growth on TMAO. We also identified a unique ATP-binding cassette transporter (TmoXWV) found in a variety of marine bacteria and experimentally confirmed its specificity for TMAO through marker exchange mutagenesis and lacZ reporter assays of the promoter for genes encoding this transporter. Both Tdm and TmoXWV are particularly abundant in natural seawater assemblages and actively expressed, as indicated by a number of recent metatranscriptomic and metaproteomic studies. These data suggest that TMAO represents a significant, yet overlooked, nutrient for marine bacteria.
Trimethylamine N-oxide (TMAO) frequently occurs in the tissues of a variety of marine biota (1) and is predicted to have a number of important physiological roles (2). In marine elasmobranchs (sharks and rays), TMAO accumulates at high concentrations (up to 500 mM), helping to offset the destabilizing effects of urea on cellular proteins (1, 3, 4). TMAO can be metabolized to small methylated amines, for example, tri-, di-, and monomethylamine (TMA, DMA, and MMA, respectively). These volatile organic N compounds are precursors of marine aerosols and the potent greenhouse gas nitrous oxide in the marine atmosphere (5). In anoxic sediments or pockets of hypoxic conditions, such as in marine snow, they are precursors for the potent greenhouse gas methane (6). In marine surface waters, TMAO concentrations can reach up to 79 nM; however, owing to the technical difficulties associated with quantifying TMAO in seawater, reports of in situ concentrations of TMAO are limited (7, 8). In a previously published study in which TMAO and TMA were quantified in the marine environment, TMAO had a higher average concentration throughout the water column and over a seasonal cycle (8).
TMAO is a well-studied terminal electron acceptor for anaerobic microbial respiration (9, 10), but its catabolism in aerobic surface seawater is not well understood. Recent studies have shown that TMAO in the Sargasso Sea is predominantly oxidized by bacterioplankton as an energy source (11) and that the marine methylotrophic bacterium Methylophilales sp. HTCC2181 oxidizes TMAO to CO2 to generate energy (12). However, the genes and enzymes responsible for the metabolism and uptake of TMAO by marine bacteria are not known. It has previously been suggested that in Methylocella silvestris, a TMA-degrading soil bacterium, an aminotransferase protein containing a conserved C-terminal tetrahydrofolate (THF)-binding domain (Msil_3603) is probably involved in the metabolism of TMAO, because this polypeptide was highly enriched in TMA-grown cells and TMAO is a known intermediate of TMA metabolism by TMA monooxygenase, Tmm, in this bacterium (TMA + NADPH + O2 + H+ → TMAO + H2O + NADP+) (13). It is hypothesized that TMAO is further metabolized to ammonium and formaldehyde, which serve as N and C/energy sources, respectively, for this bacterium (13).
ATP-binding cassette (ABC) transporters form one of the largest gene superfamilies found within many bacterial genomes (14), and their expression is frequently detected in the marine environment (15–17). ABC transporters are essential for bacteria because they are responsible for the uptake of a wide range of compounds, such as sugars, amino acids, metals, and vitamins, at the expense of ATP (18). They usually consist of three subunits: a transmembrane domain that is bound to an inner membrane-bound ATP-binding domain and a periplasmic substrate-binding protein (SBP), which binds a given ligand. SBPs confer substrate specificity and can bind their ligands with very high affinity (19, 20). One group of ABC transporters specialize in the uptake of compatible osmolytes and structurally related compounds, such as glycine betaine (GBT), choline, carnitine, and proline betaine (21, 22). These transporters either function in osmoregulation (23) or play a role in substrate catabolism (19). A bacterial ATP-dependent TMAO transporter has been identified (24), but the genes encoding this transport system are unknown.
The SAR11 clade (Pelagibacteraceae) and the marine Roseobacter clade (MRC, Rhodobacteraceae) are two groups of marine bacteria that differ in their ecology, but both play important roles in marine C, S, and N cycles (25–27). Bacteria of the SAR11 clade bacteria dominate low-nutrient environments, have streamlined genomes, are generally slow-growing and have distinct auxotrophic requirements for certain compounds (28–30). In contrast, bacteria of the MRC have larger genomes, display high metabolic versatility, can live a particle-associated lifestyle, and often represent a large proportion of the metabolically active bacterial community in coastal oceans (25, 31–34). Ecologically relevant representatives of the MRC are readily cultivated and amenable to genetic manipulation, thereby making them good model organisms to investigate bacterial ecophysiology in the marine environment. Ruegeria pomeroyi DSS-3, isolated off the coast of Oregon in the United States (35), is the best characterized model marine organism in this clade (32, 36–39).
Here, we identify a TMAO-specific microbial ABC transporter and the TMAO demethylase, Tdm (TMAO → DMA + formaldehyde), from key marine heterotrophs, including bacteria from the SAR11 clade and the MRC. This transporter and Tdm are highly expressed in the marine environment, as indicated by a number of recent metatranscriptomic and metaproteomic studies. Therefore, our data suggest that TMAO is an important, yet overlooked, nutrient for marine bacteria.
Results
Identification and Confirmation of a Functional Tdm in R. pomeroyi.
We used R. pomeroyi DSS-3 as the model organism to study TMAO metabolism. This bacterium can grow on methylated amines, including TMAO, as a sole N source (Fig. 1A). In the genome sequence of R. pomeroyi we identified an ORF (SPO1562) that has high sequence similarity (54%) to Msil_3603, the ORF predicted to encode the Tdm in M. silvestris (13). Sequence analysis has shown that both proteins contain an uncharacterized domain (DUF1989) and a THF-binding domain, which is likely to be important in conjugating formaldehyde released from the demethylation of TMAO. In a representative of the SAR11 clade it has been suggested that TMAO demethylation through THF-mediated one-carbon oxidation provides cellular energy (11). To confirm that SPO1562 in R. pomeroyi encodes for a bona fide Tdm, this gene was cloned and overexpressed in Escherichia coli. In the presence of TMAO, E. coli cells expressing the putative Tdm from R. pomeroyi produced 984 ± 45 µM DMA in the culture medium (Fig. 2B), confirming that SPO1562 does indeed encode for a Tdm. E. coli cells transformed with vector, pET28a alone, did not produce DMA.
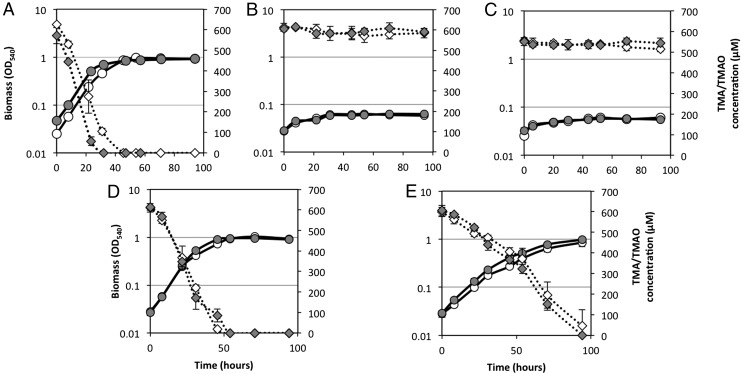
Fig. 1.
Growth of R. pomeroyi DSS-3 on TMA and TMAO as a sole N source. R. pomeroyi DSS-3 was grown on either TMA (white circles) or TMAO (gray circles) and concentrations of TMA (white diamonds) and TMAO (gray diamonds) were quantified throughout the growth. The function of tdm was determined by comparing growth of the wild type (A) against the Δtdm::Gm mutant (B). When the mutant was corrected with a native tdm from either R. pomeroyi DSS-3 (D) or Pelagibacteraceae strain HIMB59 (E) growth was restored, whereas the vector control (pBBR1MCS-km) did not restore the growth of the mutant on TMA or TMAO (C). All cultures were grown in triplicate and error bars denote SDs.
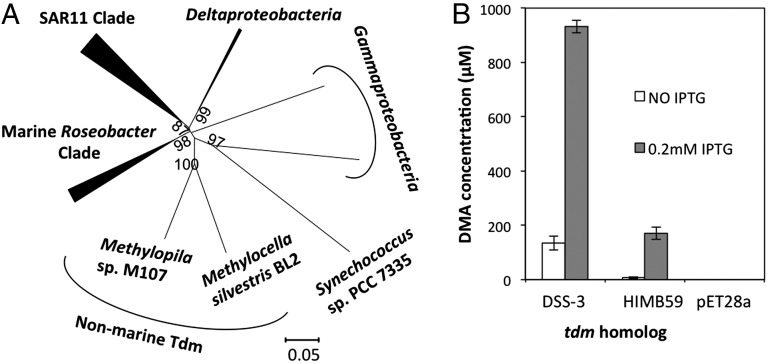
Fig. 2.
(A) Neighbor-joining phylogenetic analysis of Tdm retrieved from the genomes of sequenced marine bacteria. Bootstrap values (500 replicates) greater than 60% are shown. The scale bar denotes the number of amino acid differences per site. The analysis involved 49 Tdm sequences. There were a total of 468 amino acid residues in the alignment. Evolutionary analyses were conducted in MEGA5.1 (52). (B) Production of DMA from TMAO demethylation by recombinant Tdm of R. pomeroyi DSS-3 and Pelagibacteraceae strain HIMB59. pET28a represents the control empty vector with no insert. Error bars denote SDs of triplicate measurements. IPTG, isopropyl β-d-1-thiogalactopyranoside.
To determine whether SPO1562 is required for growth of R. pomeroyi on TMAO, this gene was mutated. As predicted, the mutant (Δtdm::Gm) could not grow on TMAO or its upstream precursor TMA (Fig. 1B), although it could grow on DMA and MMA (Table S1). To confirm whether tdm is essential in R. pomeroyi, tdm was cloned along with its promoter from R. pomeroyi into the broad-host-range plasmid pBBR1MCS-km (40), which was then mobilized into the Δtdm::Gm mutant via conjugation. Complementation of the mutant with the native tdm gene from R. pomeroyi reversed the phenotype, restoring growth on both TMAO and TMA as a sole N source (Fig. 1D). Complementation of this mutant with the vector pBBR1MCS-km alone did not result in growth on TMA and TMAO (Fig. 1C).
Distribution of Tdm Homologs in Other Marine Bacteria.
To test the importance of the tdm gene and to investigate its occurrence in the marine environment, we further investigated the distribution of Tdm in the genomes of isolated marine bacteria. The Tdm from R. pomeroyi was used as the query sequence to generate a BLASTP database using the Integrated Microbial Genomes system at the Joint Genome Institute. Closely related homologs (E value = 0.0) of Tdm were retrieved from representatives of the SAR11 clade and the MRC of the Alphaproteobacteria, the SAR324 cluster of Deltaproteobacteria, and some Gammaproteobacteria (Fig. 2A and Fig. S1). In general, the presence of tmm, the gene encoding TMA monooxygenase, coincides with the presence of tdm, but not vice versa. Those bacteria lacking tmm do, however, have the genes necessary for further downstream catabolism of MMA (13, 26). One example is Roseobacter sp. SK209-2-6, a representative of the MRC. This bacterium lacks tmm in its genome but does contain tdm and genes required for MMA catabolism (e.g., gmaS) (26). As predicted, Roseobacter sp. SK209-2-6 failed to grow on TMA but could grow on TMAO (Table S2).
We generated another BLASTP database using the Global Ocean Sampling (GOS) Expedition database (41) and we estimated that Tdm homologs are present in 21% of bacterial cells inhabiting surface seawater, comparable to estimates for Tmm (20%) and GmaS (23%) (13). Tdm sequences were present in both open ocean and coastal ocean surface waters (Fig. S2). Phylogenetic analysis indicated that the majority of Tdm homologs (92%) identified from the GOS dataset were related to the Tdm of the SAR11 clade, and the remaining were related to the MRC (5%), Gammaproteobacteria (2%), and Deltaproteobacteria (1%).
Tdm homologs from representatives of the SAR11 clade share ∼57% sequence similarity at the amino acid level to the Tdm from R. pomeroyi DSS-3. As yet, no genetic system has been established for SAR11 strains, so to confirm that these Tdm homologs are functional, a Tdm homolog from the SAR11 clade representative, Pelagibacteraceae strain HIMB59, was cloned and overexpressed in E. coli. In the presence of TMAO, E. coli cells expressing Tdm produced 171 ± 34 µM DMA (Fig. 2B). Complementation of the R. pomeroyi mutant (Δtdm::Gm) with the native tdm homolog from Pelagibacteraceae strain HIMB59 also reversed the phenotype (Fig. 1E). These experiments suggest that the SAR11 tdm homologs also encode a functional Tdm.
Identification and Characterization of a TMAO-Specific ABC Transporter.
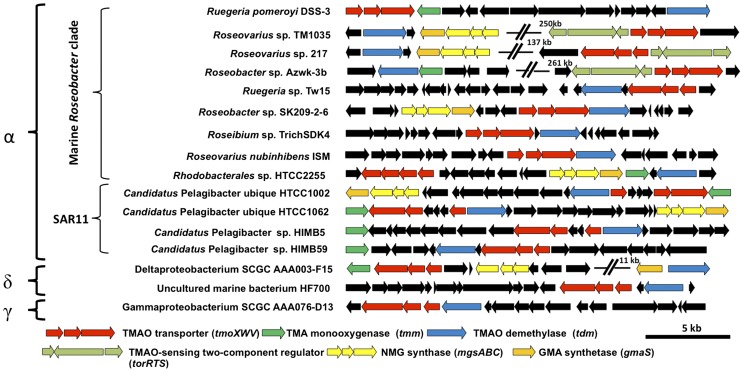
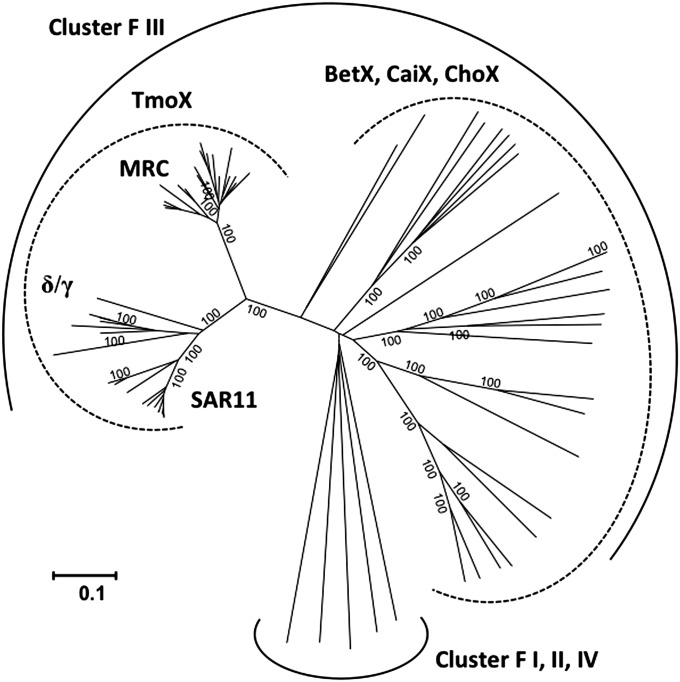
The fact that some bacteria, such as Roseobacter sp. SK209-2-6, can metabolize TMAO but not TMA suggests that TMAO transport into the cell can be independent of TMA metabolism. This led us to hypothesize that a specific transporter for TMAO is needed for such microorganisms. We therefore systematically investigated the presence of membrane transporter proteins in the genomes of marine bacteria possessing a Tdm and paid particular attention to the neighborhoods of genes known to be involved in methylated amine metabolism (e.g., tdm, tmm and gmaS). We found a conserved three-ORF gene cluster encoding a putative GBT/proline betaine ABC transporter present in the neighborhood of tdm in many marine bacterial genomes, including Roseobacter sp. SK209-2-6 (Fig. 3). These genes encode a periplasmic SBP, an ATP-binding domain protein, and a transmembrane permease protein and are hereafter designated as tmoX, tmoW, and tmoV, respectively. In some MRC bacteria (Roseovarius sp. 217, Roseovarius sp. TM1035, and Roseobacter sp. Azwk-3B) this tmoXWV gene cluster is located adjacent to genes encoding a two-component regulatory system, torRTS. These regulatory proteins are known to be involved in the regulation of the TMAO reductase in E. coli, which is required for anaerobic respiration of TMAO (10, 42). None of these three MRC bacteria possesses a TMAO reductase homolog, and we therefore conclude that these two gene clusters are involved in aerobic catabolism of TMAO. Our conclusion is further supported by phylogenetic analysis of the SBPs of the GBT/proline betaine-type ABC transporter family. TmoX is part of the cluster F III of the ABC transporter superfamily, containing SBPs specific for compatible osmolytes (22). However, TmoX forms a distinct subcluster within cluster F III that does not contain any previously characterized SBPs (Fig. 4). Other GBT/proline betaine-type SBPs from R. pomeroyi, Roseovarius sp. 217, Pelagibacteraceae strain HIMB59, and Candidatus Pelagibacter ubique sp. HTCC1002/ HTCC1062 fall within the traditional F III subcluster (Fig. S3).
Fig. 3.
Genetic neighborhoods of the genes (tmoXWV) that encode the TMAO transporter (red) among representative genome-sequenced marine bacteria. All genes colored black have no confirmed functional relationship with TMAO metabolism. α, Alphaproteobacteria; δ, Deltaproteobacteria; γ, Gammaproteobacteria; GMA, γ-glutamylmethylamide; NMG, N-methylglutamate.
Fig. 4.
Phylogenetic analysis of the SBP, TmoX, of the TMAO-specific transporter in relation to other characterized SBPs. Current known SBPs specific for osmolytes, such as choline, glycine betaine, and carnitine, fall into the cluster F of the ABC superfamily (22). The evolutionary history was inferred using the neighbor-joining method (53). Bootstrap values (500 replicates) greater than 99% are shown. The scale bar represents the number of amino acid differences per site. The analysis involved 69 SBP sequences. There were a total of 296 amino acids positions in the alignment. Evolutionary analyses were conducted in MEGA5.1 (52). δ, Deltaproteobacteria; γ, Gammaproteobacteria; BetX, glycine betaine/proline betaine SBP; CaiX, carnitine SBP; ChoX, choline SBP.
The tmoXWV gene cluster (SPO1548–SPO1550) was targeted for mutagenesis again using R. pomeroyi as a model bacterium. Two transporter mutants were generated, one targeting both tmoX and tmoW to mutate the entire membrane component of the transporter (ΔtmoXW::Gm) and the other targeting only the periplasmic SBP (ΔtmoX::Gm), leaving the core transporter domain intact. Growth on TMAO as a sole N source was significantly reduced for mutants ΔtmoX::Gm (Fig. 5) and ΔtmoXW::Gm (Fig. S4). Over 96 h, wild-type cells metabolized over 1 mM of TMAO whereas the two mutants only metabolized 87 ± 14 µM added TMAO (Fig. 5 A and C and Fig. S4). The growth of the mutants on TMA, however, was unaffected (Fig. 5B and Fig. S4), suggesting that this transporter is only involved in TMAO and not in TMA metabolism. Complementation of the ΔtmoX::Gm mutant with the native tmoX from R. pomeroyi reversed the phenotype (Fig. 5C).
Fig. 5.
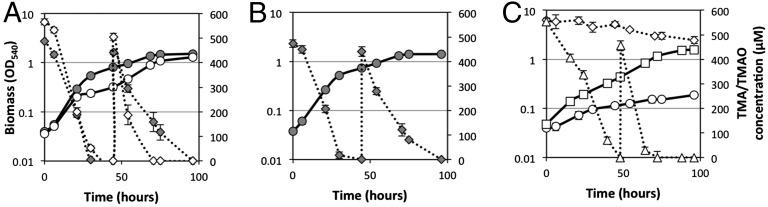
Growth of R. pomeroyi DSS-3 and the TMAO transporter mutants on TMA and TMAO as a sole N source. (A) R. pomeroyi wild type was grown on either TMA (gray circles) or TMAO (white circles) and concentrations of TMA (gray diamonds) and TMAO (white diamonds) were quantified during growth. (B) R. pomeroyi mutant ΔtmoX::Gm was grown on TMA (gray circles) and the concentration of TMA (gray diamonds) was quantified throughout growth. (C) R. pomeroyi mutant ΔtmoX::Gm was grown on TMAO (white circles) and the concentration of TMAO (white diamonds) was quantified throughout growth. The mutant was complemented with the native tmoX from R. pomeroyi, which was grown on TMAO as a sole N source (white squares), and the concentration of TMAO was quantified (white triangles). Once TMA/TMAO was depleted in the medium, a second dose (final concentration 0.5 mM) was added at 48 h. All cultures were grown in triplicate and error bars denote SDs.
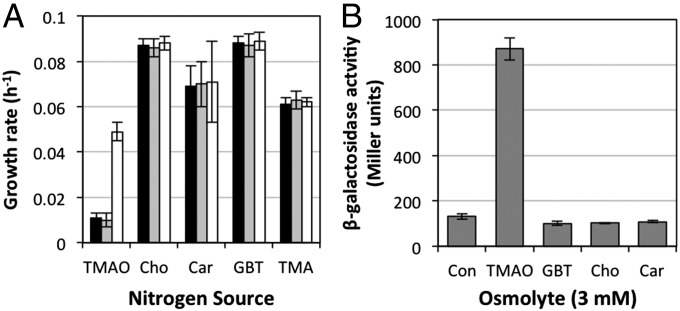
To better understand the specificity of this transporter, the transporters mutants (ΔtmoXW::Gm and ΔtmoX::Gm) were tested for their growth on structurally related compounds (GBT, choline, and carnitine) as a sole N source. Growth rates of the mutants (ΔtmoXW::Gm and ΔtmoX::Gm) were unaffected when grown on these three osmolytes and TMA (Fig. 6A). We probed the transcriptional specificity of the promoter of the tmoXWV gene cluster in R. pomeroyi. The promoter of tmoXWV (∼250 bp upstream region) was cloned into the broad-host-range promoter probe vector, pBIO1878 (36), upstream of its lacZ reporter region. The resulting plasmid pBIOIL101 was mobilized into R. pomeroyi DSS-3 and a transconjugant was grown overnight in minimal medium either lacking any osmolyte or containing GBT, choline, carnitine, or TMAO (3 mM) before assaying for β-galactosidase activity. The presence of TMAO led to a sixfold increase in induction of the tmoX–lacZ fusion, whereas no induction was observed with the other osmolytes tested (Fig. 6B). TMA also led to the induction of the transporter (Fig. S5); however, we hypothesized that intracellular production of TMAO through TMA oxidation was responsible for this phenomenon. To test this hypothesis, we mobilized the pBIOIL101 plasmid into the mutant Δtmm::Gm, which can no longer grow on TMA as a sole N source (Fig. S5). In this strain, TMAO still induced the transporter, but the sensitivity of the transporter to TMA was significantly reduced (Fig. S5). Together, these data suggest that the ABC transporter tmoXWV is specific for TMAO and is essential for TMAO metabolism in R. pomeroyi DSS-3.
Fig. 6.
Effects of different compatible osmolytes on the growth of R. pomeroyi DSS-3 and regulation of the TMAO transporter tmoXWV. The growth rates of R. pomeroyi wild type (white bars) and the two transporter mutants, ΔtmoX::Gm (gray bars) and ΔtmoXW::Gm (black bars), were determined for each osmolyte and TMA as a sole N source. (A) Cultures of R. pomeroyi DSS-3 containing the tmoX–lacZ fusion plasmid pBIL101 were grown in the presence of each compatible osmolyte (3 mM). (B) Cultures were grown and assayed in triplicate for β-galactosidase activity and error bars denote SDs. Car, carnitine; Cho, choline; Con, control.
Discussion
We report the identification of the genes encoding the Tdm and a TMAO-specific ABC transporter in a number of divergent marine bacteria, including MRC and SAR11 clade Alphaproteobacteria, SAR324 clade Deltaproteobacteria, and some Gammaproteobacteria (Figs. 2–4). The Tdm and the associated TMAO transporter and the genes encoding these proteins are widespread in both coastal and open ocean surface seawater, and we estimate using the GOS metagenome dataset that one in five bacterial cells is capable of TMAO catabolism (Fig. S2). It is noteworthy that Tdm and TmoXWV are found not only in cultivated representatives of abundant marine bacteria (e.g., SAR11 and MRC), but also in as-yet uncultivated marine bacteria inhabiting the surface oceans with streamlined genomes (Figs. S1 and S3). For example, these genes are found in single-cell amplified genomes of uncultivated Roseobacters that are prevalent in tropical and temperate regions of the oceans (AAA298-K06) as well as in polar oceans (AAA076-C03) (43).
The ability to use the potentially more abundant TMAO directly from the water column would provide an energetic and ecological benefit to marine bacteria. Conversely, the conversion of TMA to TMAO requires an extra enzyme and NADPH as a reducing equivalent, and production of TMA is reliant on the anaerobic conversion of quaternary amines, including TMAO, and may not be relevant to open ocean systems. Our study has shown that some bacteria do not have the genetic potential to metabolize TMA but are still able to metabolize TMAO (e.g., Roseobacter sp. SK209-2-6). In addition, all Tdm-containing marine bacteria have a TMAO-specific transporter, thereby strengthening the hypothesis that TMAO is an important nutrient in the marine environment and not simply an intermediate of intracellular TMA metabolism, as proposed previously (13). This hypothesis is supported by at least three key observations. First, TMAO is directly produced in a diverse range of marine biota and has been detected in marine surface seawater (2, 8). Second, TMAO added to surface seawater can be metabolized to CO2 by marine microorganisms to generate cellular energy (11). Third, reanalyses of a number of recent metatranscriptomic and metaproteomic datasets has indicated that Tdm and the newly identified TMAO-specific ABC transporter are highly expressed in situ (15, 17, 44–46). For example, analysis of metatranscriptomic data of bacterioplankton from the Monterey Bay of California showed that the TMAO transporter is one of the most highly expressed transporters in the MRC representative, Rhodobacterales sp. HTCC2255 (ZP_01447069), an abundant member of the microbial community (17), and off the coast of northern California tmoX from SAR11 bacteria (Cluster 686, YP_266709) is among the 10 most highly expressed genes (44). Metaproteomic data collected from the Sargasso Sea also revealed that a polypeptide identified as TmoX, closely related to TmoX of the SAR11 isolate Candidatus Pelagibacter sp. 7211 (PB7211_687), was among the 10 most highly expressed transporter proteins (15). During the summer and winter months in Antarctic surface seawater, a TmoX closely related to the TmoX of Candidatus Pelagibacter ubique HTCC1002 (PU1002_06741) was also highly expressed (46). Not only has expression of the TMAO transporter been frequently detected in natural seawater by metatranscriptomic and metaproteomic studies, but Tdm expression (Cluster 435, YP_266710) has also been found in bacterial plankton assemblages in the surface seawater (44). The high level of tmoX and tdm expression in SAR11 and MRC bacteria from natural bacterioplankton communities points toward TMAO serving as an important substrate for energy generation (11), and it may also be an important source of N for these heterotrophs in the marine environment.
Several lines of evidence further suggest that the metabolism of TMAO is important in the marine environment. For example, a tmm homolog is present in the genome of the marine N2 fixer Trichodesmium erythraeum IMS101. Although there are no data regarding the function of Tmm or whether TMAO has any physiological role in Trichodesmium, a MRC bacterium, Roseibium sp. TrichSDK4, isolated from Trichodesmium colonies, has the genes necessary for TMAO catabolism but lacks a Tmm. It is therefore tempting to speculate that this bacterium may benefit from TMAO released by Trichodesmium cells. We also found Tdm and the TMAO transporter in the genome of a SAR324 cluster bacterium, which is predominantly found in the deep ocean “twilight zone” where photosynthesis does not occur (47, 48). TMAO metabolism by SAR324 bacteria may help facilitate their chemoautotrophic lifestyle, supplementing energy predominantly derived from the oxidation of reduced S compounds (47). Genes required for the THF-linked oxidation of methyl groups cleaved off during the dissimilation of TMAO were indeed expressed among the SAR324 cluster bacteria inhabiting deep-sea marine plumes (48). The ability of SAR324 bacteria to use TMAO is in line with the recent discovery that they are capable of using a range of electron donors and acceptors, which helps explain their prevalence in the dark ocean (48).
We noticed that both transporter mutants (ΔtmoXW::Gm and ΔtmoX::Gm) can still deplete TMAO from the medium, albeit at much slower rates (Fig. 5 and Fig. S4), suggesting the presence of another yet-undiscovered membrane transporter for TMAO. Indeed, in the genome of M. silvestris (13) no homologs of tmoXWV were found, although it can use TMAO as a sole N source. It is also likely that in R. pomeroyi there is an SBP of broad specificity but lower affinity for TMAO, therefore contributing to the slower growth rates on TMAO observed in the mutants, and clearly this warrants further investigation. We cannot rule out the possibility that TmoXWV may also serve as a high-affinity TMA transporter, and further investigation is required to determine the affinity of this transporter for both TMA and TMAO. In Aminobacter aminovorans a high concentration of TMA (5 mM) only partially inhibited uptake of TMAO (at 10 µM), and it was proposed that there might be two different high-affinity transporters for these two compounds (24). Because we observed no difference in TMA metabolism in the mutant, ΔtmoX::Gm, we also propose that in R. pomeroyi another high-affinity transport system is necessary for TMA uptake. Alternative microbial pathways for TMAO catabolism in surface seawaters are also likely. For example, Methylophilales sp. HTCC2181 lacks the tdm gene required for TMAO metabolism, but it can oxidize TMAO to CO2, as demonstrated previously (12). Similarly, multiple enzymes responsible for the cleavage of the compatible osmolyte dimethylsulfoniopropionate into the climate-active gas dimethylsulfide have now been identified (36, 49).
In conclusion, our discovery of the genes encoding the TMAO demethylase and a TMAO-specific ABC transporter in abundant members of the bacterioplankton and the prevalence of these genes and their transcription and subsequent expression in natural surface seawaters implies that this compound is an important nutrient for different groups of heterotrophic bacteria in the marine environment.
Materials and Methods
Cultivation of MRC Bacteria on Methylamines.
MRC bacteria were grown at 30 °C in 125-mL serum vials in triplicate using a defined medium as previously described (13). Methylated amines (0.5 mM) were used as the sole N source. Succinate (5–10 mM) was used as the sole C source. Vitamins were added as described previously (13). To test whether the TMAO demethylase mutant (Δtdm::Gm) and the TMAO ABC transporter mutants (ΔtmoXW::Gm and ΔtmoX::Gm) could grow on methylated amines, growth experiments were set up in triplicate using 120-mL serum vials, containing 20 mL medium with an inoculum size of 10%.
Marker Exchange Mutagenesis and Complementation of R. pomeroyi Mutants.
All strains used for cloning are listed in Table S3. All primers used for PCR and sequencing are listed in Table S4. The method for marker exchange mutagenesis was modified from ref. 50. Detailed protocols for marker exchange mutagenesis and complementation of mutants in R. pomeroyi are described in SI Materials and Methods.
Overexpression of Tdm in E. coli.
The tdm gene from R. pomeroyi DSS-3 was amplified by PCR (primers used are listed in Table S4) and cloned into the expression vector pET28a (Merck Biosciences). The tdm gene from Pelagibacteraceae strain HIMB59 was chemically synthesized (GenScript Corporation) and cloned into pET28a. The resulting plasmids were transformed into the expression host E. coli BLR(DE3) pLysS (Merck Biosciences). Detailed protocols for protein expression and DMA quantification are described in SI Materials and Methods.
Identification of Tmm and GmaS Homologs in the GOS Metagenome.
The Tdm and Tmm sequences of R. pomeroyi were used as query sequences for a BLASTP search of the GOS peptides at CAMERA [https://portal.camera.calit2.net/gridsphere/gridsphere?cid=]; GOS: all ORF peptides (P) database [e−60], and this resulted in 2,274 and 1,177 unique sequences, respectively. For Tdm, sequences were further grouped into 122 unique groups (identity >80% within each group) using the CD-HIT program (51). Representative sequences from each group were aligned using MEGA 5.1 (52). To estimate the frequency of Tdm-containing cells, the data were processed as described previously (13, 51). To compare the distribution of Tdm and Tmm against each other in the GOS dataset, both proteins were normalized to RecA by the following: Tdm = RecA (376)/Avg. Tdm length (778); Tmm = RecA (376)/Avg Tmm length (445). The number of reads at each site were normalized per 100,000 reads.
Supplementary Material
Acknowledgments
We thank Dr. J. Todd and Dr. J Christie-Oleza for providing the plasmids pBIO1878 and pBBR1MCS-km, respectively. This work was supported by the Natural and Environment Research Council through a research studentship (to I.L.) and a fellowship award (NE/H016236/1).
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1317834111/-/DCSupplemental.
References
- 1.Yancey PH, Clark ME, Hand SC, Bowlus RD, Somero GN. Living with water stress: Evolution of osmolyte systems. Science. 1982;217(4566):1214–1222. doi: 10.1126/science.7112124. [DOI] [PubMed] [Google Scholar]
- 2.Seibel BA, Walsh PJ. Trimethylamine oxide accumulation in marine animals: Relationship to acylglycerol storage. J Exp Biol. 2002;205(Pt 3):297–306. doi: 10.1242/jeb.205.3.297. [DOI] [PubMed] [Google Scholar]
- 3.Ballantyne JS. Jaws: The inside story. the metabolism of elasmobranch fishes. Comp Biochem Physiol B Biochem Mol Biol. 1997;118(4):703–742. [Google Scholar]
- 4.Treberg JR, et al. The accumulation of methylamine counteracting solutes in elasmobranchs with differing levels of urea: A comparison of marine and freshwater species. J Exp Biol. 2006;209(Pt 5):860–870. doi: 10.1242/jeb.02055. [DOI] [PubMed] [Google Scholar]
- 5.Quinn PK, Charlson RJ, Bates TS. Simultaneous observations of ammonia in the atmosphere and ocean. Nature. 1988;335(6188):336–338. [Google Scholar]
- 6.King GM. Metabolism of trimethylamine, choline, and glycine betaine by sulfate-reducing and methanogenic bacteria in marine sediments. Appl Environ Microbiol. 1984;48(4):719–725. doi: 10.1128/aem.48.4.719-725.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Carpenter LJ, Archer SD, Beale R. Ocean-atmosphere trace gas exchange. Chem Soc Rev. 2012;41(19):6473–6506. doi: 10.1039/c2cs35121h. [DOI] [PubMed] [Google Scholar]
- 8.Gibb SW, Hatton AD. The occurrence and distribution of trimethylamine-N-oxide in Antarctic coastal waters. Mar Chem. 2004;91(1–4):65–75. [Google Scholar]
- 9.Arata H, Shimizu M, Takamiya K. Purification and properties of trimethylamine N-oxide reductase from aerobic photosynthetic bacterium Roseobacter denitrificans. J Biochem. 1992;112(4):470–475. doi: 10.1093/oxfordjournals.jbchem.a123923. [DOI] [PubMed] [Google Scholar]
- 10.Gon S, Giudici-Orticoni M-T, Méjean V, Iobbi-Nivol C. Electron transfer and binding of the c-type cytochrome TorC to the trimethylamine N-oxide reductase in Escherichia coli. J Biol Chem. 2001;276(15):11545–11551. doi: 10.1074/jbc.M008875200. [DOI] [PubMed] [Google Scholar]
- 11.Sun J, et al. One carbon metabolism in SAR11 pelagic marine bacteria. PLoS ONE. 2011;6(8):e23973. doi: 10.1371/journal.pone.0023973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Halsey KH, Carter AE, Giovannoni SJ. Synergistic metabolism of a broad range of C1 compounds in the marine methylotrophic bacterium HTCC2181. Environ Microbiol. 2012;14(3):630–640. doi: 10.1111/j.1462-2920.2011.02605.x. [DOI] [PubMed] [Google Scholar]
- 13.Chen Y, Patel NA, Crombie A, Scrivens JH, Murrell JC. Bacterial flavin-containing monooxygenase is trimethylamine monooxygenase. Proc Natl Acad Sci USA. 2011;108(43):17791–17796. doi: 10.1073/pnas.1112928108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Young J, Holland IB. ABC transporters: Bacterial exporters-revisited five years on. Biochim Biophys Acta Biomembranes. 1999;1461(2):177–200. doi: 10.1016/s0005-2736(99)00158-3. [DOI] [PubMed] [Google Scholar]
- 15.Sowell SM, et al. Transport functions dominate the SAR11 metaproteome at low-nutrient extremes in the Sargasso Sea. ISME J. 2009;3(1):93–105. doi: 10.1038/ismej.2008.83. [DOI] [PubMed] [Google Scholar]
- 16.Sowell SM, et al. Environmental proteomics of microbial plankton in a highly productive coastal upwelling system. ISME J. 2011;5(5):856–865. doi: 10.1038/ismej.2010.168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ottesen EA, et al. Metatranscriptomic analysis of autonomously collected and preserved marine bacterioplankton. ISME J. 2011;5(12):1881–1895. doi: 10.1038/ismej.2011.70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Davidson AL, Chen J. ATP-binding cassette transporters in bacteria. Annu Rev Biochem. 2004;73(1):241–268. doi: 10.1146/annurev.biochem.73.011303.073626. [DOI] [PubMed] [Google Scholar]
- 19.Chen C, Malek AA, Wargo MJ, Hogan DA, Beattie GA. The ATP-binding cassette transporter Cbc (choline/betaine/carnitine) recruits multiple substrate-binding proteins with strong specificity for distinct quaternary ammonium compounds. Mol Microbiol. 2010;75(1):29–45. doi: 10.1111/j.1365-2958.2009.06962.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Albers SV, et al. Glucose transport in the extremely thermoacidophilic Sulfolobus solfataricus involves a high-affinity membrane-integrated binding protein. J Bacteriol. 1999;181(14):4285–4291. doi: 10.1128/jb.181.14.4285-4291.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Thomas GH. Homes for the orphans: Utilization of multiple substrate-binding proteins by ABC transporters. Mol Microbiol. 2010;75(1):6–9. doi: 10.1111/j.1365-2958.2009.06961.x. [DOI] [PubMed] [Google Scholar]
- 22.Berntsson RPA, Smits SHJ, Schmitt L, Slotboom D-J, Poolman B. A structural classification of substrate-binding proteins. FEBS Lett. 2010;584(12):2606–2617. doi: 10.1016/j.febslet.2010.04.043. [DOI] [PubMed] [Google Scholar]
- 23.May G, Faatz E, Villarejo M, Bremer E. Binding protein dependent transport of glycine betaine and its osmotic regulation in Escherichia coli K12. Mol Gen Genet. 1986;205(2):225–233. doi: 10.1007/BF00430432. [DOI] [PubMed] [Google Scholar]
- 24.Raymond JA, Plopper GE. A bacterial TMAO transporter. Comp Biochem Physiol B Biochem Mol Biol. 2002;133(1):29–34. doi: 10.1016/s1096-4959(02)00097-0. [DOI] [PubMed] [Google Scholar]
- 25.Buchan A, González JM, Moran MA. Overview of the marine roseobacter lineage. Appl Environ Microbiol. 2005;71(10):5665–5677. doi: 10.1128/AEM.71.10.5665-5677.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen Y. Comparative genomics of methylated amine utilization by marine Roseobacter clade bacteria and development of functional gene markers (tmm, gmaS) Environ Microbiol. 2012;14(9):2308–2322. doi: 10.1111/j.1462-2920.2012.02765.x. [DOI] [PubMed] [Google Scholar]
- 27.Tripp HJ, et al. SAR11 marine bacteria require exogenous reduced sulphur for growth. Nature. 2008;452(7188):741–744. doi: 10.1038/nature06776. [DOI] [PubMed] [Google Scholar]
- 28.Giovannoni SJ, et al. Genome streamlining in a cosmopolitan oceanic bacterium. Science. 2005;309(5738):1242–1245. doi: 10.1126/science.1114057. [DOI] [PubMed] [Google Scholar]
- 29.Tripp HJ, et al. Unique glycine-activated riboswitch linked to glycine-serine auxotrophy in SAR11. Environ Microbiol. 2009;11(1):230–238. doi: 10.1111/j.1462-2920.2008.01758.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Grote J, et al. Streamlining and core genome conservation among highly divergent members of the SAR11 Clade. MBio. 2012;3(5):e00252-12. doi: 10.1128/mBio.00252-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Newton RJ, et al. Genome characteristics of a generalist marine bacterial lineage. ISME J. 2010;4(6):784–798. doi: 10.1038/ismej.2009.150. [DOI] [PubMed] [Google Scholar]
- 32.Moran MA, et al. Genome sequence of Silicibacter pomeroyi reveals adaptations to the marine environment. Nature. 2004;432(7019):910–913. doi: 10.1038/nature03170. [DOI] [PubMed] [Google Scholar]
- 33.Wagner-Döbler I, et al. The complete genome sequence of the algal symbiont Dinoroseobacter shibae: A hitchhiker’s guide to life in the sea. ISME J. 2010;4(1):61–77. doi: 10.1038/ismej.2009.94. [DOI] [PubMed] [Google Scholar]
- 34.Alonso C, Pernthaler J. Roseobacter and SAR11 dominate microbial glucose uptake in coastal North Sea waters. Environ Microbiol. 2006;8(11):2022–2030. doi: 10.1111/j.1462-2920.2006.01082.x. [DOI] [PubMed] [Google Scholar]
- 35.González JM, et al. Silicibacter pomeroyi sp. nov. and Roseovarius nubinhibens sp. nov., dimethylsulfoniopropionate-demethylating bacteria from marine environments. Int J Syst Evol Microbiol. 2003;53(Pt 5):1261–1269. doi: 10.1099/ijs.0.02491-0. [DOI] [PubMed] [Google Scholar]
- 36.Todd JD, Kirkwood M, Newton-Payne S, Johnston AWB. DddW, a third DMSP lyase in a model Roseobacter marine bacterium, Ruegeria pomeroyi DSS-3. ISME J. 2012;6(1):223–226. doi: 10.1038/ismej.2011.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sebastian M, Ammerman JW. Role of the phosphatase PhoX in the phosphorus metabolism of the marine bacterium Ruegeria pomeroyi DSS-3. Environ Microbiol Rep. 2011;3(5):535–542. doi: 10.1111/j.1758-2229.2011.00253.x. [DOI] [PubMed] [Google Scholar]
- 38.Cunliffe M. Physiological and metabolic effects of carbon monoxide oxidation in the model marine bacterioplankton Ruegeria pomeroyi DSS-3. Appl Environ Microbiol. 2013;79(2):738–740. doi: 10.1128/AEM.02466-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Christie-Oleza JA, Fernandez B, Nogales B, Bosch R, Armengaud J. Proteomic insights into the lifestyle of an environmentally relevant marine bacterium. ISME J. 2012;6(1):124–135. doi: 10.1038/ismej.2011.86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kovach ME, et al. Four new derivatives of the broad-host-range cloning vector pBBR1MCS, carrying different antibiotic-resistance cassettes. Gene. 1995;166(1):175–176. doi: 10.1016/0378-1119(95)00584-1. [DOI] [PubMed] [Google Scholar]
- 41.Rusch DB, et al. The Sorcerer II Global Ocean Sampling expedition: Northwest Atlantic through eastern tropical Pacific. PLoS Biol. 2007;5(3):e77. doi: 10.1371/journal.pbio.0050077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gon S, Patte J-C, Dos Santos J-P, Méjean V. Reconstitution of the trimethylamine oxide reductase regulatory elements of Shewanella oneidensis in Escherichia coli. J Bacteriol. 2002;184(5):1262–1269. doi: 10.1128/JB.184.5.1262-1269.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Swan BK, et al. Prevalent genome streamlining and latitudinal divergence of planktonic bacteria in the surface ocean. Proc Natl Acad Sci USA. 2013;110(28):11463–11468. doi: 10.1073/pnas.1304246110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ottesen EA, et al. Pattern and synchrony of gene expression among sympatric marine microbial populations. Proc Natl Acad Sci USA. 2013;110(6):E488–E497. doi: 10.1073/pnas.1222099110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gifford SM, Sharma S, Booth M, Moran MA. Expression patterns reveal niche diversification in a marine microbial assemblage. ISME J. 2013;7(2):281–298. doi: 10.1038/ismej.2012.96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Williams TJ, et al. A metaproteomic assessment of winter and summer bacterioplankton from Antarctic Peninsula coastal surface waters. ISME J. 2012;6(10):1883–1900. doi: 10.1038/ismej.2012.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Swan BK, et al. Potential for chemolithoautotrophy among ubiquitous bacteria lineages in the dark ocean. Science. 2011;333(6047):1296–1300. doi: 10.1126/science.1203690. [DOI] [PubMed] [Google Scholar]
- 48.Sheik CS, Jain S, Dick GJ. Metabolic flexibility of enigmatic SAR324 revealed through metagenomics and metatranscriptomics. Environ Microbiol. 2014;16(1):304–317. doi: 10.1111/1462-2920.12165. [DOI] [PubMed] [Google Scholar]
- 49.Kirkwood M, Le Brun NE, Todd JD, Johnston AWB. The dddP gene of Roseovarius nubinhibens encodes a novel lyase that cleaves dimethylsulfoniopropionate into acrylate plus dimethyl sulfide. Microbiology. 2010;156(Pt 6):1900–1906. doi: 10.1099/mic.0.038927-0. [DOI] [PubMed] [Google Scholar]
- 50.Crombie A, Murrell JC. Development of a system for genetic manipulation of the facultative methanotroph Methylocella silvestris BL2. Methods Enzymol. 2011;495:119–133. doi: 10.1016/B978-0-12-386905-0.00008-5. [DOI] [PubMed] [Google Scholar]
- 51.Li W, Godzik A. Cd-hit: A fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics. 2006;22:1658–1659. doi: 10.1093/bioinformatics/btl158. [DOI] [PubMed] [Google Scholar]
- 52.Tamura K, et al. MEGA5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 2011;28(10):2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Saitou N, Nei M. The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4(4):406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.