Significance
Epigenetic modifications are required for the regulation of hematopoiesis. DNA methyltransferase 3A (DNMT3A), a critical epigenetic modifier responsible for de novo DNA methylation, was reported recently to be a frequently mutated gene in hematopoietic malignancies. However, the role of mutated DNMT3A in hematopoiesis remains largely unknown. Here we show that the Arg882 (R882) mutation of DNMT3A disrupts the normal function of this enzyme and results in chronic myelomonocytic leukemia (CMML) in mice. Meanwhile, the gene expression, DNA methylation, and protein–protein interaction assays suggest that DNMT3A R882 mutation drives CMML by disturbing the transcriptional expression/DNA methylation program and cell-cycle regulation of hematopoietic cells. This study may shed light on the function of DNMT3A mutant in myeloid leukemogenesis.
Keywords: genomic variation, epigenetic abnormality, leukemogenic effect
Abstract
The gene encoding DNA methyltransferase 3A (DNMT3A) is mutated in ∼20% of acute myeloid leukemia cases, with Arg882 (R882) as the hotspot. Here, we addressed the transformation ability of the DNMT3A-Arg882His (R882H) mutant by using a retroviral transduction and bone marrow transplantation (BMT) approach and found that the mutant gene can induce aberrant proliferation of hematopoietic stem/progenitor cells. At 12 mo post-BMT, all mice developed chronic myelomonocytic leukemia with thrombocytosis. RNA microarray analysis revealed abnormal expressions of some hematopoiesis-related genes, and the DNA methylation assay identified corresponding changes in methylation patterns in gene body regions. Moreover, DNMT3A-R882H increased the CDK1 protein level and enhanced cell-cycle activity, thereby contributing to leukemogenesis.
DNA methylation represents one of the major epigenetic modifications and plays a key role in a number of regulatory mechanisms of life processes (1–3). In mammals, the executors of genome methylation are members of the DNA methyltransferase (DNMT) family, including DNMT1, DNMT3A and DNMT3B. It has been well established that DNMT3A forms complex with DNMT3L to catalyze the de novo DNA methylation (4, 5). Both DNMT3A and DNMT3B show high expression levels at the early embryogenesis, and their expressions are down-regulated along with the embryonic development and cell differentiation (6, 7).
It is well known that all blood-cell lineages originate from the multipotent hematopoietic stem cells (HSCs). A number of regulations are involved in directing HSCs activities, and the epigenetic modifications are of great importance (8, 9). It has been shown that loss of Dnmt3a in a Dnmt3a-conditional knockout mouse results in progressive impairment of HSCs differentiation and expansion (10). Notably, DNMT3A recently has been reported to be mutated in up to 20% of cases of acute myeloid leukemia (AML), mostly in cases with monocytic lineage (AML-M5 or -M4) and clinical features including old age, normal karyotype, leukocytosis and thrombocytosis, and poor prognosis (11, 12). Although a variety of DNMT3A mutations have been identified, the majority (∼50%) affect Arg882 (R882) located at the catalytic domain, and the most common substitution is Arg882His (R882H) (11, 13, 14). It also has been suggested that R882 mutation may interfere with oligomerization of DNMT3A and thereby exert an aberrant effect on its enzymatic function (15). Evidence has been obtained supporting DNMT3A mutations as the fundamental genetic event at the initiation stage of AML pathogenesis (16, 17). However, the in vivo transformation power of DNMT3A mutations needs to be addressed, and the relevant molecular and cellular mechanisms of these mutations in AML pathogenesis remain obscure.
In the present work, using retroviral transduction and bone marrow transplantation (BMT) technology, we were able to investigate the in vivo effect of the DNMT3A-R882H mutation on the transforming potentials of hematopoietic stem/progenitor cells (HSPCs). We also examined the possible mechanisms of these abnormal potentials at transcriptome, epigenetics, and protein–protein interaction levels in transformed HSPCs with the DNMT3A-R882H mutation.
Results
DNMT3A-R882H Mutation Enhances Proliferation Potential of Hematopoietic Cells.
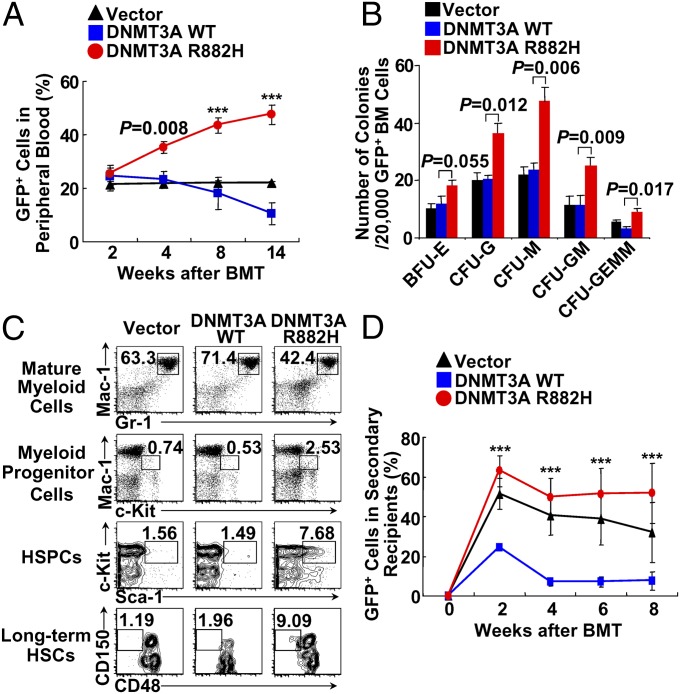
To explore the in vivo transformation effect of DNMT3A mutation, we carried out BMT experiments using murine bone marrow (BM) cells retrovirally transduced with mutated or WT DNMT3A or empty (vehicle) retroviral constructs. GFP was used to trace transduced cells. We found that cells in peripheral blood (PB) with DNMT3A-R882H could compete effectively with the cells without GFP and presented an increasing proliferative potential, whereas proliferative capacities were gradually reduced in cells overexpressing WT DNMT3A (Fig. 1A). However, at 6 mo post-BMT, no obvious changes were observed in hemoglobin level, white blood cell (WBC) count, or platelet count (Fig. S1A). To evaluate the possible effect of R882H on the stemness of HSPCs, GFP+ BM cells from each group were isolated for cfu assay. The DNMT3A protein was overexpressed in sorted GFP+ BM cells from both the WT and mutant DNMT3A groups (Fig. S1B). Of note, cells with DNMT3A-R882H formed statistically more CFU-G (granulocyte), CFU-M (monocyte/macrophage), and CFU-GM colonies than those from the WT and vehicle groups (Fig. 1B). Lin−Sca-1+C-kit+ (LSK) cells and long-term HSCs (Lin−Sca-1+C-kit+CD150+CD48−) were significantly increased in the DNMT3A-R882H group as compared with the WT group. In addition, myeloid progenitor (Mac-1lowC-kit+) cells also showed an apparent increase, and mature myeloid cells were comparatively decreased in DNMT3A-R882H mice (Fig. 1C). To assess further the ability of the DNMT3A mutation to provide HSPCs with an advantage in growth and survival, we transplanted 1 × 106 GFP+ BM cells from the first generation of mice in each group into the secondary recipient mice treated with sublethal irradiation. In the second-generation mice, the percentage of GFP+ cells was maintained at a higher level in the DNMT3A-R882H group but dropped rapidly in the WT group (Fig.1D).
Fig. 1.
Phenotypic analysis in mice at 6 mo post-BMT. (A) Average percent of GFP+ cells in PB after BMT (n = 8–10 recipients per group). (B) Increased myeloid colony formation by GFP+ DNMT3A-R882H BM cells in methylcellulose compared with that by WT or vector groups. (C) Representative flow cytometry plots of BM cells from mice at 6 mo post-BMT. (D) Average ratios of PB GFP+ cells after secondary transplantation (vector: n = 7; DNMT3A WT: n = 3; DNMT3A-R882H: n = 10). ***P < 0.001.
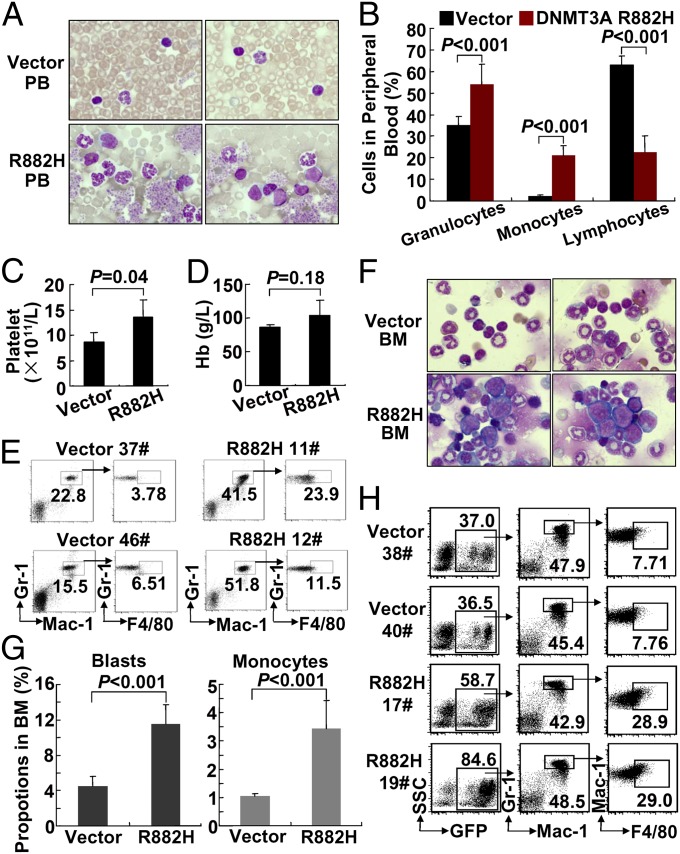
At 1 y after BMT, remarkable differences of GFP+ cells appeared among the three groups: PB cells carrying DNMT3A-R882H increased continuously, GFP+ PB cells with WT DNMT3A almost died out, and those from vehicle group remained at the same level as 6-mo post-BMT (Fig. S1C). Morphological analysis of PB smears with Wright’s staining revealed striking features in the DNMT3A-R882H group, notably significantly increased granulocytes and monocytes, and a few early myeloid elements could be found (Fig. 2A). The number of monocytes was greater than 109/L (Fig. 2B), and the number of platelets increased significantly (Fig. 2C), but no statistically significant changes in hemoglobin were observed (Fig. 2D). In agreement with the morphology data, the sorted GFP+ WBCs showed increased monocytes in all nine DNMT3A-R882H mice investigated (Fig. S1D), and the number of lymphocytes was reduced significantly; thus the ratio between granulocytes/monocytes and lymphocytes was reversed in DNMT3A-R882H mice as compared with the vector group (Fig. 2B). An analysis of the development of myeloid lineages showed that, within GFP+ clusters, the percentage and number of Mac-1+Gr-1+ cells was statistically significantly higher in DNMT3A-R882H mice than in the vehicle group (P < 0.001 and P = 0.003, respectively). Moreover, the proportion and number of GFP+ (Mac-1+Gr-1+F4/80+) monocytes were much higher in DNMT3A-R882H mice than in vehicle group (P = 0.007 and P = 0.018, respectively) (Fig. 2E and Fig. S1E). These results suggested that DNMT3A mutation positively regulates cell growth in monocytic/megakaryocytic lineages.
Fig. 2.
Morphological and immunophenotypical analyses at 1 y post-BMT. (A) Morphological comparison of PB in DNMT3A-R882H and vector groups. (B) Statistical analysis of cell counts in PB. (C and D) Absolute values of platelets (C) and hemoglobin (D). (E) Representative flow cytometry plots of murine PB. (F) Morphological analysis of BM in the DNMT3A-R882H and vector groups. Wright’s staining was used in BM cytospin preparations. (G) Statistical analysis of the numbers of blasts and monocytes in BM. (H) Representative flow cytometry plots of BM from mice at around 1 y post-BMT (vector group: n = 8 mice; DNMT3A-R882H group: n = 9 mice).
When the BM of mice was examined 1 y after BMT, significantly increased numbers of blasts were noted in all nine DNMT3A-R882H mice (11.5% on average in DNMT3A-R882H mice, versus 4.5% on average in the vehicle group; P < 0.001). The blasts in the DNMT3A-R882H mice were characterized by large size, high nuclear/cytoplasmic ratio, and fine chromatin. The nuclei of some blasts were indented, reminiscent of the morphology of monoblasts. Among mature myeloid cells, the percentage of monocytes (3.4%) was significantly increased in the DNMT3A-R882H group as compared with the vehicle group (1.0%) (P < 0.001) (Fig. 2 F and G). As observed in PB at 1 y post-BMT, the percentage of GFP+ cells was much higher in BM cells with the R882H mutation than in the vehicle group. Immunophenotype analysis of these GFP+ BM cells showed that the Mac-1+Gr-1+F4/80+ population was increased significantly in DNMT3A-R882H mice as compared with the vehicle group (P < 0.001) (Fig. 2H), providing strong evidence that the DNMT3A mutation promotes the proliferation of a myeloid compartment, monocytic lineage in particular. We performed bubble PCR to identify the retroviral integrating sites. A single integration site was identified in each sample from three mice at 12 mo post-BMT. These sites were located at different chromosomes (Table S1). Taken as a whole, these features in DNMT3A-R882H mice at 1 y post-BMT represented a disease phenotype mimicking chronic myelomonocytic leukemia (CMML) in humans.
The advantage in cellular proliferation provided by DNMT3A-R882H also was demonstrated in a distinct set of experiments using in vitro NIH3T3 cell lines stably expressing either DNMT3A-R882H or WT DNMT3A. In a colony-formation assay, NIH3T3 cells expressing DNMT3A-R882H formed many more colonies than those expressing WT DNMT3A (Fig. S2).
DNMT3A-R882H Mutation Causes a Transcriptional Alteration.
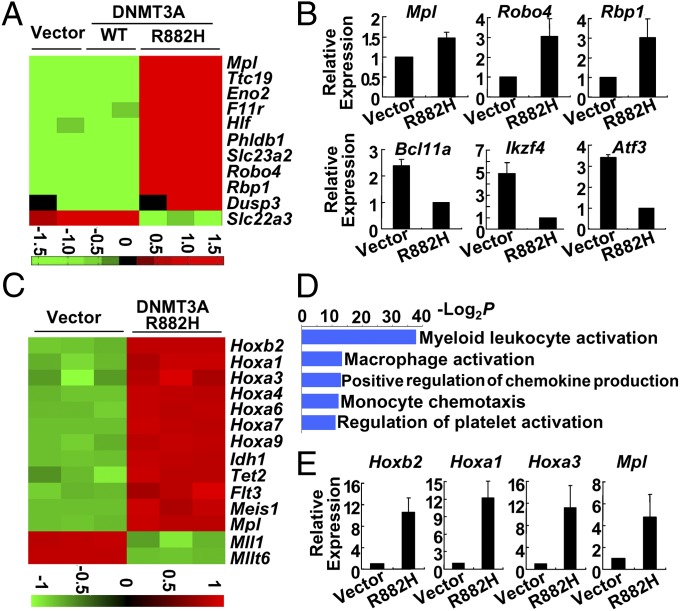
To explore the mechanism underlying DNMT3A-R882H phenotypic defects, we performed gene-expression microarray assay using the GFP+ BM cells harvested at 6 mo and 1 y post-BMT. At 6 mo post-BMT, supervised analysis revealed 1,618 and 1,247 genes with differential expression in the DNMT3A-R882H group as compared with the WT DNMT3A and vehicle groups, respectively (Datasets S1 and S2). The expression levels of 738 genes in DNMT3A-R882H mice were different from the levels in both the WT and vehicle groups (Dataset S3). These results indicate that the DNMT3A mutant exerts a dramatic effect on transcriptional regulation. Analysis based on Gene Ontology (GO) classification and the Hematopoietic Fingerprints Database (18) revealed 14 differentially expressed genes in the “long-term HSC subgroup,” and 11 of these genes were well annotated. Genes such as Mpl and Hlf were reported to be associated with hematopoietic malignancies, as well (19, 20). Notably, 10 of the 11 genes were up-regulated in DNMT3A-R882H cells (Fig. 3A), suggesting that cells transduced with DNMT3A-R882H acquired characters of stemness. Interestingly, a group of genes related to hematopoietic proliferation and differentiation were dysregulated dramatically, with the functions of myeloid lineage being activated and the activities of lymphoid lineage repressed (Table S2). A portion of the differentially expressed genes was validated in murine GFP+ BM samples (Fig. 3B). We also compared the mRNA levels of some differentially expressed genes in the OCI-AML3 cell line, a human AML strain harboring the DNMT3A-Arg882Cys (R882C) mutation, which was infected by lentivirus expressing shRNA against mRNAs encoding both WT and DNMT3A-R882C, with the levels in control cells expressing scramble shRNA. Quantitative RT-PCR results indicated that the expression of genes related to transcriptional regulation, including FOS, MYC, NF-κB, and ATF3, was increased in cells with DNMT3A knockdown (Fig. S3A).
Fig. 3.
Transcriptional changes in murine BM at 6 mo and 1 y post-BMT. (A) Heatmap representation of mouse long-term HSC regulatory genes identified as being differentially expressed in GFP+ vector, WT, and DNMT3A-R882H mouse BM cells at 6 mo post-BMT. Red indicates up-regulated genes; green indicates down-regulated genes. (B) Quantitative RT-PCR results for six genes in the BM samples of distinct groups at 6 mo post-BMT. (C) Heatmap for gene expression in BM samples with DNMT3A-R882H (n = 3) and vector (n = 3) at 1 y post-BMT. Genes associated with myeloid function are clustered. (D) GO analysis of up-regulated genes in mice at 1 y post-BMT found that a series of functional genes positively regulating myeloid lineage development was activated in DNMT3A-R882H mice. (E) Quantitative RT-PCR of representative genes closely associated with myeloid lineage development in murine BM samples at 1 y post-BMT showed the same patterns observed in microarray analysis.
At 1 y post-BMT, 6,544 genes in the DNMT3A-R882H group had an expression pattern that differed from that in the vector group; 3,680 genes were up-regulated, and 2,864 were down-regulated (Dataset S4). We further found that a group of genes known to be activated in leukemia progression was up-regulated dramatically in mutant mice (Fig. 3C). For example, Hox family genes and Idh1, which have been found to be activated in human AML with DNMT3A-R882 mutations, were overexpressed. Mll1 and Mllt6 were down-regulated, supporting a previous observation that DNMT3A mutation and MLL abnormalities are mutually exclusive (11). On the other hand, a series of genes that play key roles in T-cell differentiation was down-regulated (Fig. S3B). The pathways involved in granulocytic/monocytic lineage differentiation were mostly up-regulated (Fig. 3D), whereas gene clusters related to lymphocytes, mainly to T-cell development, were largely repressed (Fig. S3C). Some key genes were validated by quantitative RT-PCR (Fig. 3E and Fig. S3D).
DNMT3A-R882H Mutation Induces Modifications of Genomic Methylation Patterns.
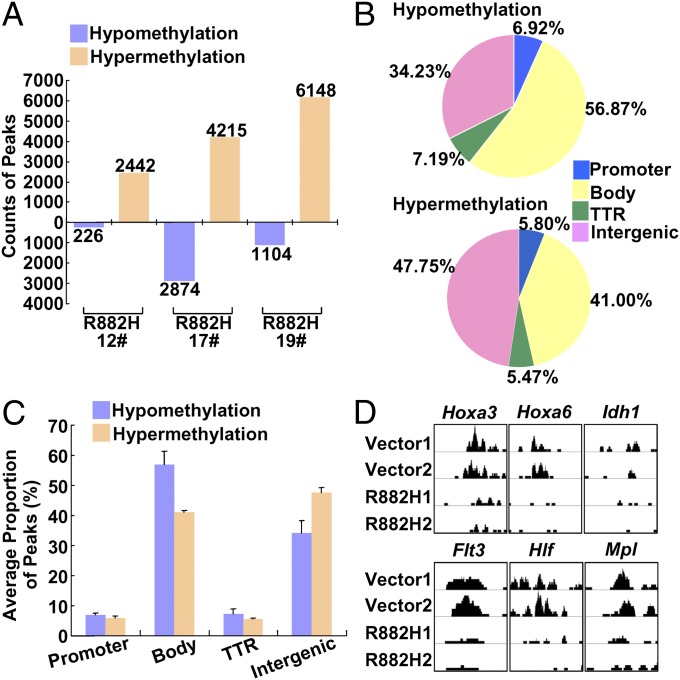
It was predicted previously that the DNMT3A-R882 mutation could disrupt the tetramerization of the enzyme, and that this disruption might explain its reduced activity (15). To evaluate the possible injury to genomic methylation caused by DNMT3A-R882H, methylated DNA immunoprecipitation (MeDIP) sequencing was carried out for murine BM cells transduced with vehicle or mutated DNMT3A constructs. Indeed, unique DNA hypo- and hypermethylation features were identified in murine CMML BM cells. On the whole, there was no significant change between vector and mutant groups (Fig. S4A), but the change in hypo- and hypermethylation patterns took place regionally throughout the genome (Fig. 4A). We then looked at methylation changes in four regions defined by the distance from CpG islands (21), namely the CpG island, Shore, Shelf, and Open Sea regions, the latter three being respectively 2 kb, 2–4 kb, and more than 4 kb from the CpG island. Most hypo- and hypermethylation patterns were detected in the Open Sea region (Fig. S4B). No differences in the distribution of hypo- and hypermethylation status were found in any of these four regions (Fig. S4C). Next, we scrutinized the DNA hypo- and hypermethylation distribution in the context of gene structure. Four relevant regions were defined: promoter, gene body, the transcriptional termination region (TTR), and the intergenic region. The number of methylated cytosines was much higher than the number of demethylated ones in CMML mice as compared with the vehicle group. However, DNA hypo- and hypermethylations were distributed differently in the four regions. As shown in Fig. 4 B and C, hypomethylation was concentrated mainly in gene body areas, and more hypermethylation was detected in the intergenic region, suggesting that DNMT3A-R882H could induce abnormal modifications of methylation selectively. Notably, most hematopoiesis-related genes that displayed up-regulation in the DNMT3A-R882H group, such as Hoxa3, Hoxa6, Idh1, Flt3, and Hlf, were hypomethylated in the gene body (Fig. 4D). In contrast, a cluster of suppressed genes that regulate lymphocyte development, such as Notch1, Notch3, Gata3, and Tcf7, showed more hypermethylated status in gene body regions (Fig. S4D). These results suggest that aberrant activity of DNMT3A-R882H can target mainly the gene body region, where abnormal DNA methylation patterns—either hypomethylation or hypermethylation—can be produced.
Fig. 4.
Genomic methylation patterns at 1 y post-BMT. (A) Hypo- and hypermethylation peak counts obtained from three mice for each group are shown in the histogram. DNMT3A-R882H versus control (vector) peaks were considered as hypermethylation peaks; control (vector) versus DNMT3A-R882H peaks were considered as hypomethylation peaks. (B and C) The average proportions of peaks in each region defined by genomic structure are shown in pie charts (B) and a histogram (C). Peak ratios of hypo- and hypermethylation are shown, respectively. Hypomethylation was concentrated mainly in gene body areas, and hypermethylation was detected more frequently in intergenic regions. (D) Hoxa3, Hoxa6, Idh1, Flt3, and Hlf genes showed a local hypomethylation pattern in the gene body whereas Mpl exhibited a hypomethylation status in promoter region in DNMT3A-R882H mice compared with vector group.
DNMT3A-R882H Mutation Increases CDK1 Protein Level and Enhances Cell-Cycle Activity.
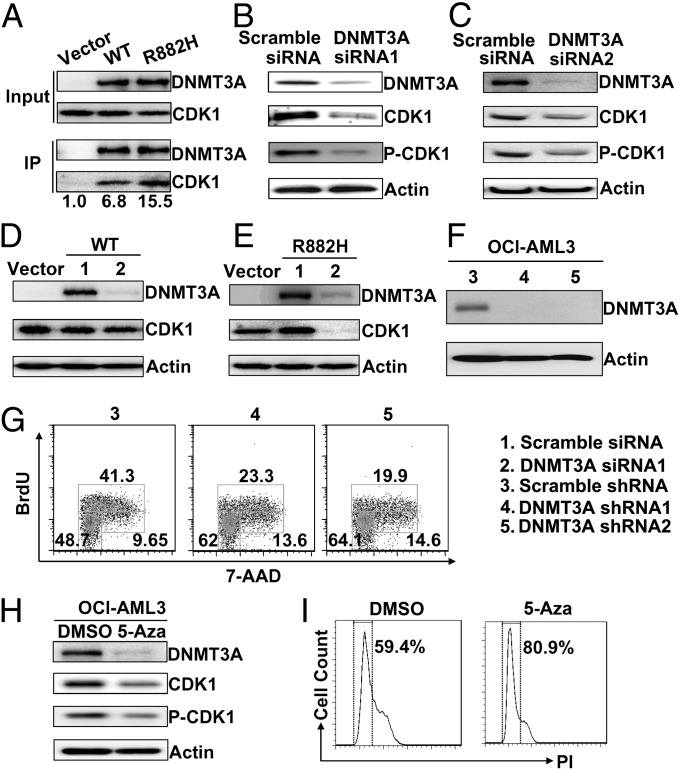
To explore mechanisms for enhanced cell proliferation other than aberrant DNA methylation and gene transcription, we investigated partner proteins interacting with WT DNMT3A and DNMT3A-R882H. We performed immunoprecipitation and mass spectrometry analysis in transfected 293T cells. In total, 1,565 proteins were identified, and 1,562 proteins were quantified by label-free quantification (LFQ) with a false-discovery rate of 0.01 (Dataset S5). Seven candidate proteins including DNMT3A with P < 0.05 were identified (Table S3). Among them, CDK1, one of the key proteins that promote the cell cycle, caught our attention. In the validation test, we found that both WT DNMT3A and DNMT3A-R882H could form a complex with CDK1, but the binding capacity of DNMT3A-R882H to CDK1 was higher than that of WT DNMT3A (Fig. 5A). To test the functional significance of such binding, we knocked down the expression potential of both WT and mutant DNMT3A in the OCI-AML3 cell line by RNAi technology. We found that both total CDK1 and its phosphorylated form were decreased markedly when the DNMT3A mutant was silenced by two different siRNAs (Fig. 5 B and C). Because OCI-AML3 cells can express both WT and mutant DNMT3A, we further investigated whether WT or mutated DNMT3A might exert an effect on CDK1 protein levels. However, specific siRNA targeting the R882 point mutation and the corresponding WT sequence failed to reduce protein-expression levels, and the effort to make an antibody to distinguish WT from mutant DNMT3A protein was not successful. Therefore we tried to address the issue in NIH3T3 cells by transfection assay. Compared with the controls, CDK1 increased in NIH3T3 cells overexpressing DNMT3A-R882H but not in cells overexpressing DNMT3A WT. Interestingly, when we applied RNAi to silence WT or mutant DNMT3A, the CDK1 protein level decreased substantially when DNMT3A-R882H was inhibited but not when WT DNMT3A was inhibited (Fig. 5 D and E). We also examined the possible effect of DNMT3A mutation on cell-cycle status in OCI-AML3 cells. In OCI-AML3 cells with knockdown of DNMT3A mutant, the compartment at the G0/G1 phase was much higher, and that at S phase was much smaller, than in control cells treated with scramble shRNA (Fig. 5 F and G). We found the expression levels of both total and phosphorylated CDK1 proteins were reduced after cells were treated with 5-Aza, an inhibitor of DNA methyltransferases (Fig. 5H). Moreover, 5-Aza–treated OCI-AML3 cells were largely blocked at the G1 phase (Fig. 5I). Together, these data suggest that the aberrant increase of CDK1 in the presence of the DNMT3A mutant can promote cell-cycle activity.
Fig. 5.
Protein–protein and functional interaction between the DNMT3A mutant and CDK1. (A) Interaction of DNMT3A with CDK1. Lysates from 293T cells transfected with plasmids encoding Flag-DNMT3A were immunoprecipitated using anti-Flag and analyzed by Western blot using the indicated antibodies. (B and C) Lysates from OCI-AML3 cells transfected with two siRNAs (siRNA1 and 2) against different DNMT3A regions were Western blotted with the indicated antibodies. (D and E) Analysis of lysates from NIH3T3 cells stably expressing DNMT3A WT or the R882H mutant. These cells were transfected with scramble siRNA or DNMT3A siRNA1, and their lysates were Western blotted with antibodies against the indicated proteins. Vector-transfected cells were used to estimate the endogenous CDK1 level compared with levels in cells transfected with WT or mutant DNMT3A. (F) OCI-AML3 cells were infected with lentiviruses expressing scramble shRNA (control) or two shRNAs (shRNA 1 and 2) targeting different DNMT3A regions. (G) OCI-AML3 cells infected with lentiviruses expressing control or two different DNMT3A shRNAs were labeled with BrdU and 7-AAD for flow cytometry analysis. (H and I) Western blot analysis of indicated proteins (H) and propidium iodide (PI) staining flow cytometry analysis (I) in OCI-AML3 cells treated by DMSO or 5-Aza (0.8 μM) for 48 h.
Discussion
Leukemia represents a group of heterogeneous clonal malignancies of HSPCs. Abnormalities of transcriptional factors and of protein tyrosine kinases have been considered two categories of major molecular events in leukemogenesis. However, over the past few years, a large body of evidence has been generated supporting the involvement of epigenetic abnormalities in both DNA and histone modifications in leukemia and lymphoma (22, 23). High-throughput parallel sequencing has revealed mutations of a third group of genes, such as EZH2, TET2, IDH1/2, and DNMT3A, involved in epigenetic regulations (11, 13, 24–26). Some of these gene mutations have been shown to cause genome-wide aberration of epigenetic modifications (27) and may play a role as disease drivers (17, 28).
Among the leukemia-associated epigenetic regulators, mutations of DNMT3A have drawn particular attention because of their preferential correlation with the important physiological functions of the gene. Several lines of evidence suggest that mutated DNMT3A might be present in the initial clone of leukemia cells (29). In the present work, to address the transforming potential of DNMT3A mutants in a definitive way, we used a murine BMT model with HSPCs transduced by DNMT3A-R882H. Interestingly, by 6 mo post-BMT, murine hematopoietic cells with the DNMT3A-R882H mutation showed an obvious growth advantage, and the unique features of the HSPCs with mutant DNMT3A could be transmitted to other mice via secondary BMT. At 1 y post-BMT, the hematopoietic cells with this hotspot mutant persisted and gradually acquired a growth advantage over the residual normal blood cells. Importantly, a CMML-like disease phenotype appeared at this stage, as defined by the 2008 World Health Organization nomenclature for human setting (>109 monocytes/L in PB, with <20% blasts in both PB and BM and dysplasia in more myeloid lineages, without BCR-ABL1 fusion gene or PDGFRA/PDGFRB rearrangement). Interestingly, this murine CMML phenotype was accompanied by thrombocytosis, which was reminiscent of the clinical features of AML with DNMT3A mutation (12). That the CMML-like phenotype developed in all recipient mice of DNMT3A mutant within a similar latent time and that the retroviral integration sites for this mutant were on different chromosomes in the mice investigated provide strong evidence that this mutation alone is capable of initiating leukemia. DNMT3A-R882H therefore should be considered not only as driving leukemogenesis but also as directing the survival/growth advantage of the myelomonocytic lineage, with specific clinical manifestations. However, this phenotype corresponds to a chronic hematopoietic disorder, and further genetic events, in addition to DNMT3A mutations, may be required for full-blown AML to occur.
Now that the transformational ability of DNMT3A mutation has been established, how might the DNMT3A mutant impact genome-wide transcriptional expression and epigenetic regulation? We systematically investigated gene-expression profiles of hematopoietic cells at 6 or 12 mo post-BMT and found that the expression of a series of genes associated with long-term hematopoiesis, such as Mpl and Hlf, was up-regulated significantly at 6 mo post-BMT. Of note, Mpl codes for thrombopoietin receptor and regulates growth not only of the megakaryocytic lineage-generating platelets but also other myeloid progenitors (30, 31). Interestingly, at 1 y post-BMT, in addition to continuous overexpression of Mpl, pathways involved in granulocytic/monocytic lineage proliferation, including Hoxb2, Hoxa9, and Meis1, were activated, and those involved in platelet activation, including Pdgfb, and Itgb3, were up-regulated. In contrast, gene clusters related to lymphocytes, and mainly to T-cell development, were largely repressed. Thus, a dynamic change in the gene-expression program seems to emerge over the disease process. At a relatively early stage, DNMT3A-R882H may exert an effect on gene expression at whole-genome level and initiate abnormal proliferation of HSPCs. With the clonal expansion of HSPCs harboring the DNMT3A mutant, the relative balance of transcriptional controls between proliferation and differentiation of hematopoietic cells may be broken, leading to disease progression.
Notably, the pattern of DNA methylation also was found to be aberrant in DNMT3A-R882H–transformed cells. Previous work suggested that R882 is important for the oligomerization of DNMT3A because it is located at the surface of protein–protein interaction of the enzyme, and the oligomer form of the enzyme is essential for its function (4, 15). It is reasonable to hypothesize that DNMT3A-R882H could affect DNA methylation, probably via a dominant-negative effect (32). In fact, an altered genomic DNA methylation pattern characterized by regionally modified patterns of hypo- and hypermethylation existed in mouse HSPCs. Notably, there was no major change in the methylation pattern at CpG islands, and the abnormal hypo- and hypermethylation status existed mostly in regions where CpG islands were sparse. This observation was consistent with a recent study on genomics and epigenomics analysis in a large series of AML (33). One interesting finding in our work is that hypomethylation is present in the gene body regions of a number of genes that are key in hematopoietic regulation and whose expression is up-regulated significantly in transformed HSPCs. It was hypothesized previously that DNMT3A might exert a selective effect on DNA methylation (7), and our data support this notion. Moreover, the methylation pattern in gene body regions seems to correlate well with a gene-expression profile in which myeloid genes are up-regulated and lymphoid genes are down-regulated. Therefore we speculate that DNMT3A may be responsible for the balance between myeloid and lymphoid commitment in HSPCs.
It has been proposed that the DNMT3A mutant also may function by forming a protein complex with other components of the cell transcription machinery to change the expression of certain genes (34). Indeed, CDK1 is among proteins interacting with DNMT3A-R882H. It is well known that, as one of the key regulators of the cell cycle, CDK1 binds to cycline A and cycline B and regulates the mitotic division of the cell (35, 36). Interestingly, our experiments showed that DNMT3A-R882H forms a complex with CDK1 more readily than does WT DNMT3A. Moreover, the stability of both total CDK1 and its active form is increased in OCI-AML3 cells that have the potential of expressing both WT and mutant DNMT3A. In the NIH3T3 cells, overexpression of DNMT3A-R882H, but not WT DNMT3A, induces a higher level of CDK1, whereas knockdown of the DNMT3A mutant, but not the WT protein, leads to a significant reduction of CDK1. A possible explanation for this observation might be that the long-time existence of DNMT3A-R882H changes the protein expression/stability program in the cells to some extent, so that the level of CDK1 becomes dependent on to the level of the DNMT3A mutant. Of note, knocking down the potential of expressing both WT and the DNMT3A mutant in OCI-AML3 strains reduces the cell-cycle activity. Finally, our study provides evidence that 5-Aza, as DNMTs inhibitor, might be used in the clinical treatment of AML with DNMT3A mutations.
Materials and Methods
Plasmid Construction.
The full-length coding sequence of human DNMT3A-WT cDNA was cloned into the BglII and EcoRI multiple cloning sites of retroviral vector MSCV-IRES-GFP (MigR1) or CMV4-Flag. The constructs containing the DNMT3A R882H mutant were generated by site mutagenesis (Quick Change).
Retroviral Preparations and Protein Expression Analysis.
Retrovirus was produced by transfection of the retroviral construct into the packaging cell line Plat-E by calcium phosphate precipitation. Retroviral supernatants were harvested at 48 h after transfection, and the viral titer was determined by flow cytometry using infected NIH3T3 cells. The cell lysates were processed for Western blot analysis. The following primary antibodies were used: anti-DNMT3A (Cell Signaling), anti-CDK1 (Cell Signaling), phospho-CDK1 (Cell Signaling), and β-actin (Santa Cruz Biotechnology).
Murine Bone Marrow Transduction and Transplantation.
BM cells were isolated from the 6- to 8-wk-old BALB/c donor mice which were pretreated with 5-fluorouracil (250 mg/kg) 5 d before operation. The primary murine BM cells then were infected with retroviruses and transplanted into lethally irradiated recipient mice (details are given in SI Materials and Methods). Recipient mice in secondary transplantation assay were treated by sublethally irradiation. All animal experiments were approved by the Department of Animal Experimentation at Shanghai Jiao Tong University School of Medicine.
Mouse Bone Marrow Colony-Forming Cell Assays.
Assays were performed according to the manufacturer’s instructions (MethoCult GF M3434; STEMCELL Technologies). Briefly, 2 × 104 GFP+ BM cells were mixed with 1.1 mL MethoCult medium and plated in a 35-mm culture dish. Triplicate cultures for each sample were performed. Colonies were counted at day 8 after plating.
Morphological and Immunophenotypic Analysis of Hematopoietic Cells.
Murine PB cells and BM cells were prepared for analysis. Smears and cytospin preparations were subject to Wright's staining for routine cell morphology. Single-cell suspensions were incubated with antibodies and harvested for flow cytometry, as described in SI Materials and Methods.
Cell-Cycle Detection.
Cells were stained with propidium iodide or BrdU/7-AAD, and the assays were performed using flow cytometry according to the manufacturers’ instructions (BD Sciences and BD Pharmingen).
Lentivirus-Mediated shRNA Infection and siRNA Transfection Assay.
Lentiviruses expressing scramble shRNA or DNMT3A shRNA were generated, and cells were transfected with siRNA oligomer using Lipofectamine 2000 Transfection Reagent (Invitrogen). Details are given in SI Materials and Methods.
Immunoprecipitation and Immunoblotting.
293T cells (1 × 107) overexpressing DNMT3A-flag were washed twice with cold PBS and suspended in 1 mL lysis buffer (Thermo). Lysates were harvested and incubated overnight with anti-FLAG M2 Affinity Gel (Sigma-Aldrich). Gels with immunoprecipitated products were washed for immunoblotting.
Microarray Expression Profiling.
Total RNAs from murine GFP+ BM samples were prepared and, after quality control, subjected to the Agilent whole mouse genome 4 × 44K Array GeneChip microarray test according to the manufacturer’s instructions (Agilent). The scanning procedure was performed using an Agilent Microarray Scanner (Agilent G2565BA) at the KangCheng Bio-Tech Corporation in Shanghai, China. Agilent Feature Extraction Software was used for data analysis.
Methylation Analysis by MeDIP Sequencing.
A total of 3 μg DNA prepared from sorted murine GFP+ BM for each mouse was used for MeDIP sequencing. Detailed procedures are available in SI Materials and Methods.
Mass Spectrometry Analysis.
Lysate of 293T cells transfected with pFLAG-CMV4 or DNMT3A WT-pFLAG-CMV4 constructs were immunoprecipitated by anti-Flag M2 Affinity Gel (Sigma-Aldrich) and analyzed by mass spectrometry. Details are available in SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank colleagues from Shanghai Institute of Hematology, particularly Dr. R.-B. Ren for constructive discussions, T. Zhen and J.-Y. Shi for assistance in biological experiments, and J.-Q. Mi and B. Chen for hematological morphological analysis, and. C. Li from Shanghai Institutes for Biological Science for technical support. This work was supported by the Chinese National Key Basic Research Project 973 Grants 2010CB529200 and 2013CB966800; Ministry of Health Grant 201202003; the Mega-projects of Scientific Research for the 12th Five-Year Plan Grant 2013ZX09303302; the State Key Laboratories Project of Excellence Grant 81123005; the National Natural Science Foundation of China for Excellent Young Scholars Grant 81222004; the Shanghai Municipal Commission for the Rising-Star Program (follow-up support) Grant 12QH1401500; the Special Foundation for Doctor Discipline of Ministry of Education of China Grant 20120073110074; the Innovation Foundation for Doctoral Students of Shanghai Jiao Tong University School of Medicine Grant BXJ201208; and the Samuel Waxman Cancer Research Foundation Co-Principal Investigator Program.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1400150111/-/DCSupplemental.
References
- 1.Feng S, Jacobsen SE, Reik W. Epigenetic reprogramming in plant and animal development. Science. 2010;330(6004):622–627. doi: 10.1126/science.1190614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Reik W, Dean W, Walter J. Epigenetic reprogramming in mammalian development. Science. 2001;293(5532):1089–1093. doi: 10.1126/science.1063443. [DOI] [PubMed] [Google Scholar]
- 3.Jones PA, Baylin SB. The epigenomics of cancer. Cell. 2007;128(4):683–692. doi: 10.1016/j.cell.2007.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jia D, Jurkowska RZ, Zhang X, Jeltsch A, Cheng X. Structure of Dnmt3a bound to Dnmt3L suggests a model for de novo DNA methylation. Nature. 2007;449(7159):248–251. doi: 10.1038/nature06146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gowher H, Liebert K, Hermann A, Xu G, Jeltsch A. Mechanism of stimulation of catalytic activity of Dnmt3A and Dnmt3B DNA-(cytosine-C5)-methyltransferases by Dnmt3L. J Biol Chem. 2005;280(14):13341–13348. doi: 10.1074/jbc.M413412200. [DOI] [PubMed] [Google Scholar]
- 6.Xie S, et al. Cloning, expression and chromosome locations of the human DNMT3 gene family. Gene. 1999;236(1):87–95. doi: 10.1016/s0378-1119(99)00252-8. [DOI] [PubMed] [Google Scholar]
- 7.Okano M, Bell DW, Haber DA, Li E. DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell. 1999;99(3):247–257. doi: 10.1016/s0092-8674(00)81656-6. [DOI] [PubMed] [Google Scholar]
- 8.Bröske AM, et al. DNA methylation protects hematopoietic stem cell multipotency from myeloerythroid restriction. Nat Genet. 2009;41(11):1207–1215. doi: 10.1038/ng.463. [DOI] [PubMed] [Google Scholar]
- 9.Trowbridge JJ, Snow JW, Kim J, Orkin SH. DNA methyltransferase 1 is essential for and uniquely regulates hematopoietic stem and progenitor cells. Cell Stem Cell. 2009;5(4):442–449. doi: 10.1016/j.stem.2009.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Challen GA, et al. Dnmt3a is essential for hematopoietic stem cell differentiation. Nat Genet. 2012;44(1):23–31. doi: 10.1038/ng.1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yan XJ, et al. Exome sequencing identifies somatic mutations of DNA methyltransferase gene DNMT3A in acute monocytic leukemia. Nat Genet. 2011;43(4):309–315. doi: 10.1038/ng.788. [DOI] [PubMed] [Google Scholar]
- 12.Thol F, et al. Incidence and prognostic influence of DNMT3A mutations in acute myeloid leukemia. J Clin Oncol. 2011;29(21):2889–2896. doi: 10.1200/JCO.2011.35.4894. [DOI] [PubMed] [Google Scholar]
- 13.Ley TJ, et al. DNMT3A mutations in acute myeloid leukemia. N Engl J Med. 2010;363(25):2424–2433. doi: 10.1056/NEJMoa1005143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ribeiro AF, et al. Mutant DNMT3A: A marker of poor prognosis in acute myeloid leukemia. Blood. 2012;119(24):5824–5831. doi: 10.1182/blood-2011-07-367961. [DOI] [PubMed] [Google Scholar]
- 15.Holz-Schietinger C, Matje DM, Reich NO. Mutations in DNA methyltransferase (DNMT3A) observed in acute myeloid leukemia patients disrupt processive methylation. J Biol Chem. 2012;287(37):30941–30951. doi: 10.1074/jbc.M112.366625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Welch JS, et al. The origin and evolution of mutations in acute myeloid leukemia. Cell. 2012;150(2):264–278. doi: 10.1016/j.cell.2012.06.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ding L, et al. Clonal evolution in relapsed acute myeloid leukaemia revealed by whole-genome sequencing. Nature. 2012;481(7382):506–510. doi: 10.1038/nature10738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chambers SM, et al. Hematopoietic fingerprints: An expression database of stem cells and their progeny. Cell Stem Cell. 2007;1(5):578–591. doi: 10.1016/j.stem.2007.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chou FS, Mulloy JC. The thrombopoietin/MPL pathway in hematopoiesis and leukemogenesis. J Cell Biochem. 2011;112(6):1491–1498. doi: 10.1002/jcb.23089. [DOI] [PubMed] [Google Scholar]
- 20.Smith KS, Rhee JW, Naumovski L, Cleary ML. Disrupted differentiation and oncogenic transformation of lymphoid progenitors in E2A-HLF transgenic mice. Mol Cell Biol. 1999;19(6):4443–4451. doi: 10.1128/mcb.19.6.4443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sandoval J, et al. Validation of a DNA methylation microarray for 450,000 CpG sites in the human genome. Epigenetics. 2011;6(6):692–702. doi: 10.4161/epi.6.6.16196. [DOI] [PubMed] [Google Scholar]
- 22.Figueroa ME, et al. DNA methylation signatures identify biologically distinct subtypes in acute myeloid leukemia. Cancer Cell. 2010;17(1):13–27. doi: 10.1016/j.ccr.2009.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shaknovich R, et al. DNA methylation signatures define molecular subtypes of diffuse large B-cell lymphoma. Blood. 2010;116(20):e81–e89. doi: 10.1182/blood-2010-05-285320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mardis ER, et al. Recurring mutations found by sequencing an acute myeloid leukemia genome. N Engl J Med. 2009;361(11):1058–1066. doi: 10.1056/NEJMoa0903840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Delhommeau F, et al. Mutation in TET2 in myeloid cancers. N Engl J Med. 2009;360(22):2289–2301. doi: 10.1056/NEJMoa0810069. [DOI] [PubMed] [Google Scholar]
- 26.Morin RD, et al. Somatic mutations altering EZH2 (Tyr641) in follicular and diffuse large B-cell lymphomas of germinal-center origin. Nat Genet. 2010;42(2):181–185. doi: 10.1038/ng.518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sasaki M, et al. IDH1(R132H) mutation increases murine haematopoietic progenitors and alters epigenetics. Nature. 2012;488(7413):656–659. doi: 10.1038/nature11323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wakita S, et al. Mutations of the epigenetics-modifying gene (DNMT3a, TET2, IDH1/2) at diagnosis may induce FLT3-ITD at relapse in de novo acute myeloid leukemia. Leukemia. 2013;27(5):1044–1052. doi: 10.1038/leu.2012.317. [DOI] [PubMed] [Google Scholar]
- 29.Walter MJ, et al. Recurrent DNMT3A mutations in patients with myelodysplastic syndromes. Leukemia. 2011;25(7):1153–1158. doi: 10.1038/leu.2011.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gurney AL, Carver-Moore K, de Sauvage FJ, Moore MW. Thrombocytopenia in c-mpl-deficient mice. Science. 1994;265(5177):1445–1447. doi: 10.1126/science.8073287. [DOI] [PubMed] [Google Scholar]
- 31.Akashi K, Traver D, Miyamoto T, Weissman IL. A clonogenic common myeloid progenitor that gives rise to all myeloid lineages. Nature. 2000;404(6774):193–197. doi: 10.1038/35004599. [DOI] [PubMed] [Google Scholar]
- 32.Kim SJ, et al. A DNMT3A mutation common in AML exhibits dominant-negative effects in murine ES cells. Blood. 2013;122(25):4086–4089. doi: 10.1182/blood-2013-02-483487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Network TCGAR. Cancer Genome Atlas Research Network Genomic and epigenomic landscapes of adult de novo acute myeloid leukemia. N Engl J Med. 2013;368(22):2059–2074. doi: 10.1056/NEJMoa1301689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Trowbridge JJ, Orkin SH. Dnmt3a silences hematopoietic stem cell self-renewal. Nat Genet. 2012;44(1):13–14. doi: 10.1038/ng.1043. [DOI] [PubMed] [Google Scholar]
- 35.Katsuno Y, et al. Cyclin A-Cdk1 regulates the origin firing program in mammalian cells. Proc Natl Acad Sci USA. 2009;106(9):3184–3189. doi: 10.1073/pnas.0809350106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hara M, et al. Greatwall kinase and cyclin B-Cdk1 are both critical constituents of M-phase-promoting factor. Nat Commun. 2012;3:1059. doi: 10.1038/ncomms2062. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.