Significance
The mechanisms underlying odorant receptor (OR) monoallelic and monogenic expression are unclear. We show for the first time that the nuclei of olfactory neurons have a characteristic organization of facultative heterochromatin: it is highly concentrated around a large constitutive heterochromatin block located centrally in the nucleus. In addition, we show that the homologous alleles of a given OR gene are located in different chromatin compartments in the olfactory nucleus: while one of the alleles is associated with deeply repressed constitutive heterochromatin, the other one is located in or close to the more plastic facultative heterochromatin. This spatial organization is likely to be important for both the monoallelic and monogenic expression of these genes.
Keywords: H3K27me3, chromatin, OR gene choice
Abstract
Odorants are detected by odorant receptors, which are located on olfactory sensory neurons of the nose. Each olfactory sensory neuron expresses one single odorant receptor gene allele from a large family of odorant receptor genes. To gain insight into the mechanisms underlying this monogenic and monoallelic expression, we examined the 3D nuclear organization of olfactory sensory neurons and determined the positions of homologous odorant receptor gene alleles in relation to different nuclear compartments. Our results show that olfactory neurons exhibit a singular nuclear architecture that is characterized by a large centrally localized constitutive heterochromatin block and by the presence of prominent facultative heterochromatin domains that are localized around this constitutive heterochromatin block. We also found that the two homologous alleles of a given odorant receptor gene are frequently segregated to separate compartments in the nucleus, with one of the alleles localized to the constitutive heterochromatin block and the other one localized to the more plastic facultative heterochromatin, or next to it. Our findings suggest that this nuclear compartmentalization may play a critical role in the expression of odorant receptor genes.
Odorants are detected by a large family of odorant receptors (ORs) located on olfactory sensory neurons in the nasal olfactory epithelium. In the mouse, each olfactory sensory neuron expresses one out of ∼1,000 different OR genes, which are dispersed throughout the genome (1–3). In addition, OR genes are monoallelically expressed, that is, only one allele of the gene (maternal or paternal) is transcribed per neuron (1, 4, 5).
The mechanisms underlying both monoallelic and monogenic expression of OR genes are unclear. It has been clearly demonstrated that OR gene promoters are necessary for OR gene expression (6, 7); however, because these promoters show similar cis-regulatory elements (8–12), it is not clear how monogenic OR gene expression is achieved. Previous studies have identified cis-acting enhancers located upstream of OR gene clusters (denominated H and P elements) which can select one single gene for expression (13–15). These elements would ensure the expression of a single OR gene from their respective clusters, but would not preclude the expression of genes located in other OR gene clusters (16, 17). How then would monogenic expression of OR genes be established? It has been demonstrated that the expression of a functional OR gene (but not of a nonfunctional OR gene) prevents expression of other OR genes (13, 18). Therefore, it has been proposed that once the first odorant receptor gene is stochastically expressed, a feedback mechanism is established, ensuring the mutually exclusive expression of OR genes (19, 20).
The vast majority of OR genes are exclusively expressed by olfactory sensory neurons. These highly specialized neurons must therefore have unique regulatory mechanisms that allow for the characteristic expression of the OR genes. Different cell types show different nuclear architectures, with different chromosome arrangements and, importantly, distinct patterns of chromatin condensation (21–25). There are two types of chromatin: heterochromatin and euchromatin. Heterochromatin is highly condensed and transcriptionally inactive, whereas euchromatin is less condensed and transcriptionally active. In addition, heterochromatin can be subdivided into constitutive heterochromatin and facultative heterochromatin. Both types of heterochromatin are repressive; however, whereas constitutive heterochromatin is stably heterochromatized and transcriptionally silent, facultative heterochromatin can decondense and become transcriptionally active (26). Constitutive heterochromatin is rich in repetitive DNA and includes centromeric, pericentromeric, and telomeric domains (27). Facultative heterochromatin genomic areas can vary largely in size, comprising an entire chromosome (such as the inactive X chromosome), large genomic domains (such as the Hox gene clusters), imprinted autosomal genes, or a few nucleosomes within a particular genomic region (26, 27).
Previous chromatin immunoprecipitation (ChIP) experiments showed that OR genes are marked with the repressive constitutive heterochromatin marks H3K9me3 and H4K20me3 in the nuclei of olfactory neurons (28). In addition, experiments using a complex DNA FISH probe that recognizes the complete OR gene repertoire have shown that the OR genes are aggregated in large constitutive heterochromatic foci in the nucleus of olfactory sensory neurons (29). This aggregation would organize the OR genes for monogenic activation, so that only a single OR gene allele would escape from chromatin repression and become active (29).
Here we have investigated the positions of the two homologous alleles of OR genes in the nucleus of olfactory sensory neurons. To do this, we performed 3D immuno-DNA FISH to determine the nuclear positions of the OR gene alleles in relation to the different types of chromatin compartments. In agreement with previous work, we found that olfactory neurons have a characteristic nuclear architecture that differs from other cell types. Odorant receptor genes that are located on different chromosomes are frequently associated with a large common constitutive heterochromatin block (HB), which is located in a central region of the nucleus. In contrast, the olfactory marker protein (OMP) gene alleles, which are biallelically expressed in all mature olfactory neurons, are not associated with this heterochromatic block. We also found that nuclei from olfactory sensory neurons show a characteristic organization of facultative heterochromatin, which is absent from the nuclei from supporting cells or from the progenitor cells located in the basal cell layer of the olfactory epithelium. Interestingly, facultative heterochromatin is predominantly localized around the large constitutive heterochromatin blocks. Moreover, we found that in a given nucleus, the two homologous alleles of a given OR gene are frequently segregated to separate nuclear compartments, whereas one of the alleles is associated with the constitutive heterochromatin block, the other one localizes with the more plastic facultative heterochromatin.
Results
Nuclear Organization of Olfactory Neurons.
We first analyzed the distribution of euchromatin (H3K4me3) and heterochromatin (H3K9me3) marks in interphase nuclei of olfactory neurons. Accordingly, as detected by immunofluorescence in nuclei of olfactory neurons, H3K9me3 colocalizes with the DAPI-stained heterochromatin blocks, whereas H3K4me3 is localized to the euchromatic regions, excluding the regions that are densely stained by DAPI and opposite to the H3K9me3 staining (Fig. S1A). We also analyzed the distribution of active RNA polymerase II (RNAPII) in the nuclei of olfactory neurons. Discrete RNAPII foci can be visualized, and as expected, they are localized to the euchromatic regions, but absent from the constitutive heterochromatic regions (Fig. S1B).
Distinct constitutive heterochromatin blocks can be visualized in the nuclei of olfactory neurons (Fig. S2A). These nuclei show a smaller number of blocks compared with nuclei of the supporting cells of the olfactory epithelium or liver cells (Fig. S2B). Strikingly, most of the nuclei of olfactory neurons have one large constitutive heterochromatin block, which occupies on average 9% of the nuclear volume and is located more centrally (Fig. S2C). In contrast, in the nuclei of supporting cells and liver cells, the heterochromatin blocks are proportionally smaller (Fig. S2C). These results indicate that olfactory neurons have a distinct pattern of heterochromatin distribution compared with other cell types.
Spatial Organization of Odorant Receptor Gene Alleles in the Nucleus of Olfactory Neurons.
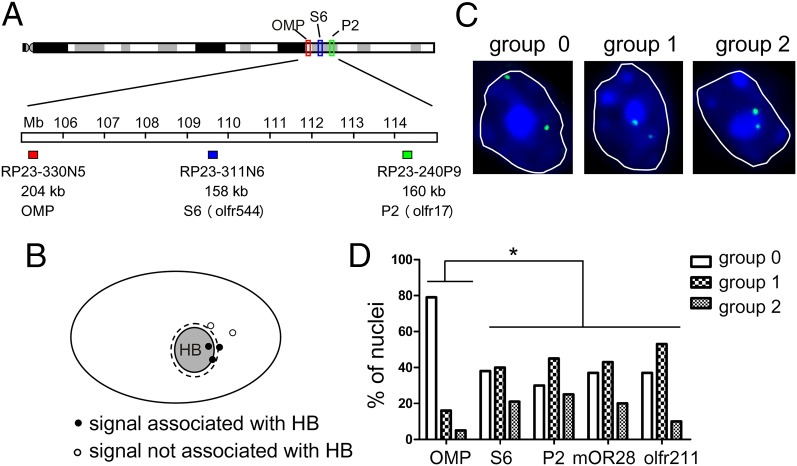
We next analyzed the nuclear positioning of OR gene alleles. Three-dimensional DNA FISH experiments were carried out to determine the positions of the loci containing the OR genes P2 or S6 or the OMP gene in the nuclei of olfactory neurons. These three loci are located close together (∼4 Mb apart one from another) on mouse chromosome 7 (Fig. 1A), but show different patterns of gene expression: whereas the OR genes are monoallelically expressed in ∼0.1% of the mature olfactory neurons (30, 31), the OMP gene is biallelically expressed in all mature olfactory neurons (32). Three BAC clones containing these genes (Fig. 1A and Fig. S3) were used as probes that were hybridized to sections cut through the olfactory epithelium.
Fig. 1.
Relative positioning of OR and OMP gene loci to heterochromatin blocks. (A) Genomic positions of the OMP, S6, and P2 gene loci in chromosome 7. The chromosome regions covered by BACs used in the 3D DNA FISH experiments are indicated. (B) Schematic representation of a nucleus with a constitutive heterochromatin block shown in gray. DNA FISH signals located >0.35 μm away from the periphery of the heterochromatin block (represented by the dashed line) are not associated with heterochromatin block. (C) Scoring system used to analyze the nuclei. Representative DNA FISH images are shown of nuclei where no allele (group 0), one allele (group 1), or two alleles (group 2) are associated with heterochromatin blocks. DNA FISH signals are shown in green, heterochromatin is visualized by DAPI (blue). The white outline shows the boundary of a single nucleus. Images are deconvolved single z-sections. In these images, both alleles are seen in the same optical plane. (D) Three-dimensional DNA FISH was used to determine the positions of the OMP, P2, S6, mOR28, and olfr211 loci relative to the heterochromatin blocks in olfactory neurons. A total of 60–70 nuclei (for OMP, P2, and S6) or 30 nuclei (for mOR28 and olfr211) were scored and classified into one of the three groups. Group distribution of OR gene loci was significantly different from OMP loci (*P < 0.0001, χ2 test).
Initial inspection of the 3D DNA FISH signals in the olfactory neurons indicated that the OR gene loci are localized near the constitutive heterochromatin blocks in a large number of nuclei (Movie S1). We therefore analyzed the spatial proximity of each one of the two OR gene alleles to the heterochromatin blocks (Fig. 1B). Nuclei were subdivided into three groups: nuclei that have no allele (group 0), nuclei that have one allele (group 1), and nuclei that have the two alleles (group 2) associated with heterochromatin blocks (Fig. 1C). Nuclei of olfactory neurons were analyzed for the S6, P2, and OMP FISH signals and classified into one of the three groups (Fig. 1D). We observed that in 69% of the nuclei analyzed for the P2 loci, at least one of the P2 alleles is associated with a heterochromatin block. Similarly, in 61% of the nuclei analyzed for the S6 loci, at least one of the S6 alleles is associated with a heterochromatin block. In contrast, in only 21% of the nuclei analyzed for OMP, at least one of the alleles is associated with a heterochromatin block (Fig. 1D). We also analyzed the nuclear position of two additional OR gene loci that are located on different chromosomes: the one containing mOR28 (on chromosome 14) and the one containing olfr211 (on chromosome 6) (Fig. S3). As shown in Fig. 1D, we found that these OR gene loci are also predominantly associated with the large heterochromatin block, and their group distribution resembles the ones observed for the S6 and P2 OR gene loci. However, we can not exclude the possibility that other OR genes, which were not analyzed in this study, show different nuclear positions.
We next analyzed the positions of the P2 OR gene alleles in the nuclei of liver cells, where they are not expressed. In this case, in 66% of the nuclei, none of the two alleles was found to be associated with a heterochromatin block. Curiously, whereas the P2 alleles are associated with heterochromatin blocks in nuclei of olfactory neurons, they are predominantly associated with the nuclear periphery in nuclei of liver cells, where they are not expressed (Fig. S4). The nuclear periphery has been generally considered to be repressive to gene expression, although this effect is not necessarily absolute (23, 33). These results show that these OR alleles are differently positioned in the nuclei of olfactory neurons and liver cells and further support the involvement of nuclear positioning in the specific pattern of the P2 OR gene expression in olfactory neurons.
Interestingly, a large percentage of the nuclei of olfactory neurons (∼45%) have one OR gene allele associated with a heterochromatin block, whereas the corresponding homolog allele is not (group 1) (Fig. 1D and Table S1). The same is not observed for the OMP gene, which is biallelically expressed: only 16% of the nuclei were classified as group 1 (Fig. 1D and Table S1). These results suggest that in each one of these nuclei, the two OR gene alleles are segregated to separate locations, one that is repressive (constitutive heterochromatin block) and the other one that is transcriptionally permissive (within the euchromatic environment). They also suggest that this differential segregation of the two OR gene alleles in one same nucleus could be related to the characteristic monoallelic expression of these genes.
Positioning of OR Genes Relative to Constitutive and Facultative Heterochromatin.
To better characterize the localization of the OR gene alleles in the different nuclear compartments, we first analyzed whether the OR gene loci colocalize with H3K9me3, a marker for constitutive heterochromatin. We performed immuno-DNA FISH experiments to costain H3K9me3 and the P2 OR gene loci. We found that 45% of the total DNA FISH signals colocalize with H3K9me3 (Fig. 2A). These results confirmed that not all of the P2 alleles colocalize with H3K9me3. We next asked whether the alleles that are not associated with H3K9me3 colocalize with the euchromatin mark H3K4me3. We found, however, that only 17.5% of the total DNA FISH signals colocalize with H3K4me3 (Fig. 2B).
Fig. 2.
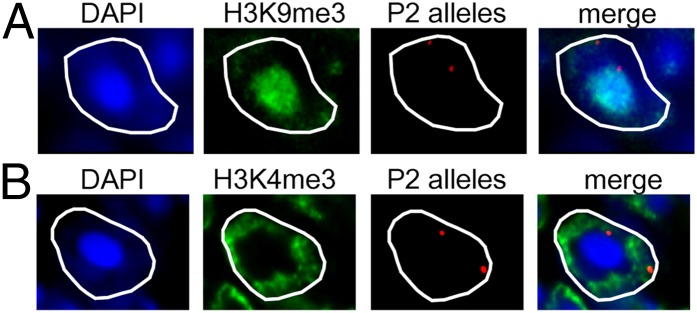
Localization of P2 OR gene loci in constitutive heterochromatin and euchromatin. (A) Representative immuno-DNA FISH images of an olfactory nucleus showing the staining for H3K9me3 (in green) and the P2 OR gene loci (in red). Only one of the P2 OR gene alleles is associated with H3K9me3. (B) Same as in A but staining for H3K4me3 is shown in green. One of the P2 OR gene alleles is associated with the constitutive heterochromatin block, and the other one is associated with H3K4me3.
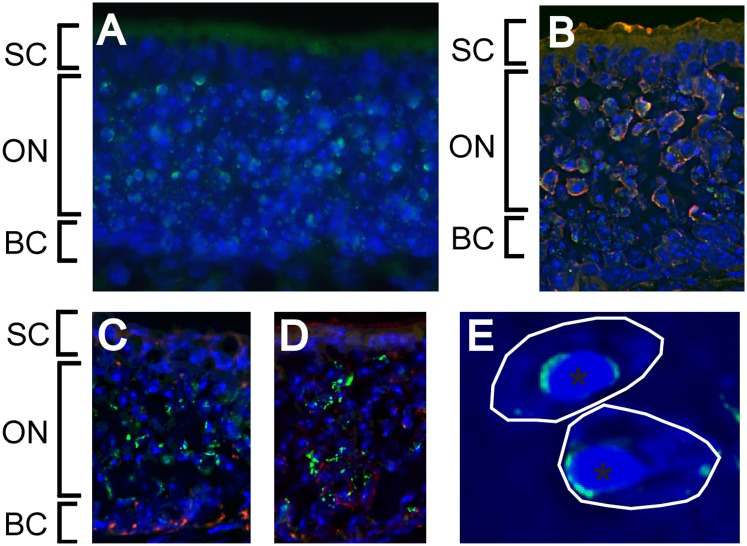
We therefore analyzed the localization of the OR gene loci that are not colocalized with H3K9me3 or with H3K4me3 relative to the more plastic facultative heterochromatin. Immunofluorescence staining of the olfactory epithelium for H3K27me3, a mark for facultative heterochromatin that is absent from constitutive heterochromatin (34), showed that this type of heterochromatin is clearly present in the nuclei of mature and immature olfactory neurons but not visible in the nuclei of supporting and basal cells in the olfactory epithelium (Fig. 3 A–D). In the olfactory nuclei, the H3K27me3 staining is not distributed all over the nucleus, but is highly concentrated in one or a few domains in the euchromatic environment. In the majority of the nuclei these domains are localized around or close to the large constitutive heterochromatin block, forming a cap-like structure (Fig. 3E).
Fig. 3.
Facultative chromatin compartments in olfactory nuclei. (A) Immunostaining of the olfactory epithelium for H3K27me3 (in green), DAPI (blue). (B–D) Immunostaining of the olfactory epithelium for H3K27me3 (in green), and the markers for: mature olfactory neurons (OMP, B), horizontal basaI cells (CK14, C), and immature olfactory neurons (GAP43, D) (in red). (E) Magnified region from A, showing the distribution of H3K27me3 around the large HBs (asterisks) within the nuclei of olfactory neurons. The layers contaning the supporting cells (SCs), mature olfactory neurons (ONs), and basal cells (BCs) are indicated.
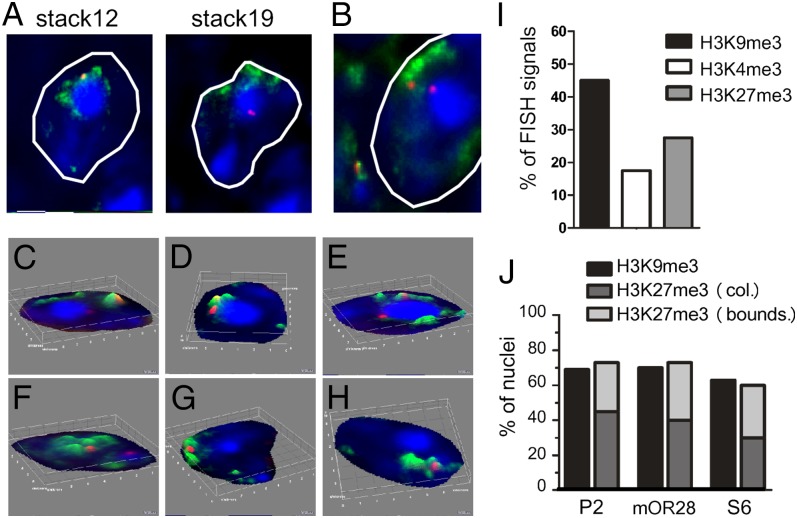
Interestingly, we observed that a significant number of the P2 OR gene loci colocalize with H3K27me3 (Fig. 4 A–H). Altogether, we found that 27.5% of the total DNA FISH signals colocalize with H3K27me3 (Fig. 4I). It is important to note that we also observed that even though in some of the nuclei the P2 FISH signal is not colocalized with H3K27me3, it is located in the boundaries of H3K27me3 domains and not randomly distributed within the euchromatic environment in the nucleus (see for example nuclei shown in Fig. 4 G and H).
Fig. 4.
Localization of OR gene loci in facultative heterochromatin. (A and B) Representative immuno-DNA FISH images of olfactory nuclei showing the staining for H3K27me3 (in green) and the P2 OR gene loci (in red). In the nucleus shown in A, the alleles can be visualized in different stacks; one allele is inside the heterochromatin block (stack 19), and the other one colocalizes with H3K27me3 (stack 12). In the nucleus shown in B, both of the alleles are visualized in the same stack. (C–H) Three-dimensional surface plots from different olfactory nuclei showing H3K27me3 (in green) and the P2 OR gene loci (in red). In the nuclei shown in C–F, both the alleles can be visualized; in the nuclei shown in G and H, only one of the alleles can be visualized. (I) The graph shows the percentages of the total number of P2 OR DNA FISH signals that colocalize with H3K9me3 (n = 40), with H3K4me3 (n = 40), or with H3K27me3 (n = 120). (J) The graph shows the percentages of the total number of nuclei that have at least one OR gene allele associated with constitutive heterochromatin (H3K9me3), colocalized with facultative heterochromatin (H3K27me3, col.), or located in the boundaries of the facultative heterochromatin domains (H3K27me3, bounds.). P2, n = 75; mOR28, n = 30; S6, n = 30.
Analysis of the distribution of the P2 DNA FISH signals in nuclei showed that in 45% of the nuclei, at least one of the P2 alleles is colocalized to H3K27me3 domains, and in an additional 28% of the nuclei, at least one of the P2 alleles is localized in the boundaries of a H3K27me3 domain (Fig. 4J). These results show that, whereas 69% of the nuclei have at least one allele associated with H3K9me3, 73% of the nuclei have at least one allele colocalized or near H3K27me3 domains (Fig. 4J). We next analyzed whether the alleles of different OR genes, mOR28 and S6, also colocalize with H3K27me3. Similar to the P2 OR gene, we found that in 73% (for mOR28) and 60% (for S6) of the nuclei, at least one of the alleles is colocalized or near H3K27me3 domains (Fig. 4J). Altogether these results show that in a large number of nuclei, the two homologous OR gene alleles are segregated to different compartments: one is associated with constitutive heterochromatin and the other one is localized to facultative heterochromatin.
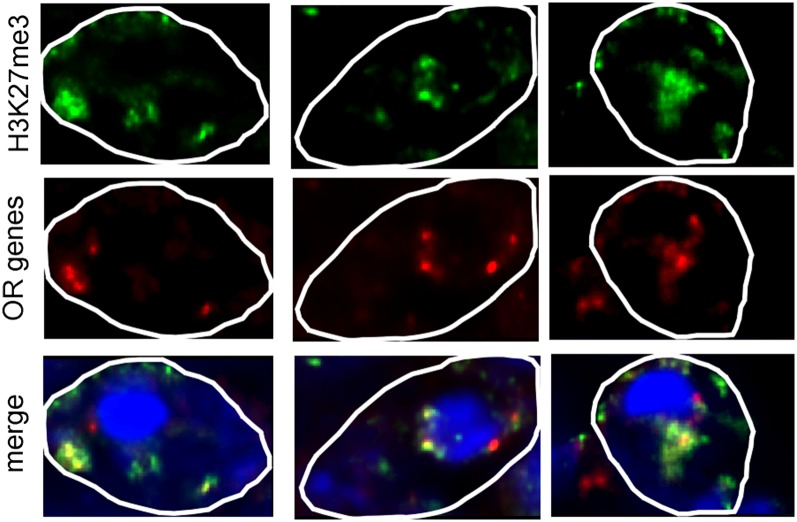
We next asked how different OR genes are organized relative to facultative and constitutive heterochromatin in one same given olfactory nucleus. To do this, we used a pool of the four different OR gene-containing BACs (Fig. S3) in the immuno-DNA FISH experiments. We detected only four to six spots of hybridization per nucleus (instead of eight), indicating that some of the DNA FISH signals are located so close together in the nuclear space that they cannot be distinguished one from another. Strikingly, in many nuclei, we found clusters of colocalized FISH signals that are located within the H3K27me3 domains (Fig. 5). Analysis of 20 nuclei indicated that on average, half of the spots colocalized with DAPI and half of the spots colocalized with H3K27me3.
Fig. 5.
Relative positioning of different OR genes in the nucleus of olfactory neurons. Representative immuno-DNA FISH images of three olfactory nuclei showing the staining for H3K27me3 (in green) and the OR gene loci (a pool of BACs containing the P2, S6, mOR28, and olfr211 OR genes was used for the DNA FISH, shown in red).
Nuclear Positioning of Active OR Genes.
It has been demonstrated that some genes are repositioned in the nucleus upon activation. For example, it was shown that gene-dense loci can form giant loops that extend outside of their corresponding chromosome territories (35–37). We next asked whether OR genes are also repositioned in the nucleus upon activation. To do this, we used P2-internal ribosome entry site (IRES)-tauGFP mice (38) to identify the olfactory neurons that express the P2 OR gene and determined the positions of the P2 alleles in the nuclei of these neurons. In these experiments, the DNA FISH procedure was performed exactly as described above on sections cut through the olfactory epithelium, except that it was now followed by immunostaining using anti-GFP to detect the P2 expressing neurons. A representative GFP+ neuron is shown in Fig. S5A and Movie S2. First, we measured the 3D distances (in micrometers) of the P2, S6, mOR28, and OMP gene loci to the heterochromatin blocks in GFP− neurons. We found that, whereas the OR genes are located at equivalent distances from the center of the heterochromatin blocks, the median distance between the OMP alleles and the center of the heterochromatin blocks is significantly larger than that of the OR gene loci (Fig. S5B). The 3D distances between the P2 alleles and the center of the heterochromatin block were then measured in GFP+ neurons. As shown in Fig. S5B, the median distance between the alleles and the center of the heterochromatin block was only slightly different in the GFP+ and GFP− neurons.
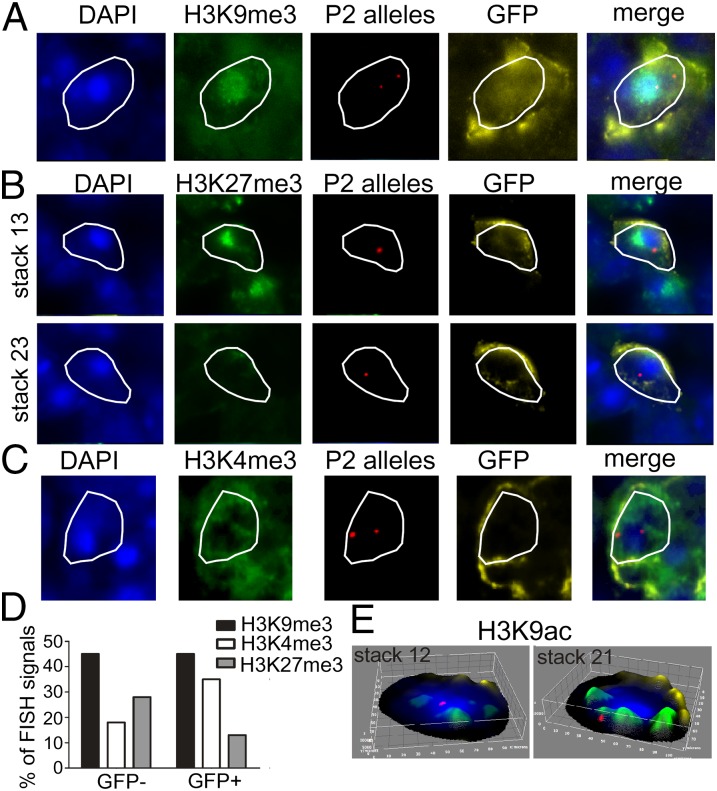
Because both constitutive and facultative heterochromatins are repressive, we would expect that active OR gene alleles would not be associated with them. We next asked what are the locations of the P2 OR gene alleles in relation to the chromatin compartments in neurons that express the P2 OR gene, and therefore must contain one active P2 OR gene allele (Fig. 6 A–D and Fig. S5C). The percentages of P2 DNA FISH signals that colocalize with H3K9me3, H3K4me3, or with H3K27me3 in GFP− or GFP+ neurons were analyzed. We found that, whereas equivalent percentages (∼45%) of the P2 DNA FISH signals colocalized with H3K9me3 in both GFP+ and GFP− neurons, the number of signals colocalizing with H3K27me3 is significantly smaller in GFP+ neurons than in GFP− neurons (12.5% and 27.5%, respectively), and the number of signals colocalizing with H3K4me3 is larger in GFP+ neurons than in GFP− neurons (35% and 17.5%, respectively) (Fig. 6D). Next, we asked whether the P2 alleles in GFP+ neurons colocalize with H3K9ac, a histone mark for actively transcribed genes. As shown in Fig. 6E, we found that in the GFP+ neuron, whereas one of the P2 alleles is localized to constitutive heterochromatin, the homologous one is localized near the H3K9ac compartments.
Fig. 6.
Positioning of the active P2 OR genes. (A–C) Representative immuno-DNA FISH images of GFP positive olfactory neurons (yellow) showing the P2 OR gene loci (red), heterochromatin marks as indicated (green), and DAPI (blue). In the neuron in A, one P2 allele is colocalized with H3K9me3 and the other one is not. In the neuron in B, one allele is colocalized with the constitutive heterochromatin block (in stack 13) and the other one is not colocalized with the constitutive heterochromatin block nor with H3K27me3 (in stack 23). In the neuron in C, one P2 allele is colocalized with H3K4me3 and the other one with the constitutive heterochromatin block. (D) The graph shows the percentages of the total number of P2 OR DNA FISH signals that colocalize with H3K9me3 (n = 40 and 20), H3K4me3 (n = 40 and 20), or with H3K27me3 (n = 120 and 40) in GFP− or GFP+ neurons. (E) Surface plots of an olfactory nucleus showing H3K9ac in green, GFP in yellow, and the P2 OR gene loci in red. The allele visualized in stack 12 is associated with the constitutive heterochromatin block, and the allele visualized in stack 21 is localized near H3K9ac.
Altogether, these results suggest that in a given olfactory neuron, the OR gene allele that is associated with constitutive heterochromatin is permanently repressed and the remaining one is the one available for transcription. They also show that the P2 OR gene alleles do not undergo large-scale repositioning upon activation.
Discussion
In this study, we analyzed the spatial distribution of homologous OR gene alleles in the nuclei of olfactory neurons. Usually, in eukaryotic cells, constitutive heterochromatin is found at the nuclear periphery, whereas euchromatin is mostly located in the nuclear center (39). However, we found that in the nuclei of olfactory neurons, the organization of constitutive heterochromatin is different: even though heterochromatin clusters can be visualized at the nuclear periphery, the major bulk of heterochromatin is concentrated in large blocks of constitutive heterochromatin, which are located more centrally in the nucleus. Similar results were recently obtained by Clowney et al. (29).
We show that OR genes that are located on different chromosomes can be found associated with the same central heterochromatin block. These results are in agreement with previous experiments in which a complex DNA FISH probe that recognizes the complete OR gene repertoire was used (29).
Chromatin immunoprecipitation experiments have shown that OR genes are marked with the constitutive heterochromatin marks H3K9me3 and H4K20me3 (28, 29). These marks would be removed later from a single OR gene allele to assure monogenic and monoallelic OR gene expression. Here we analyzed in detail how the two homologous alleles of a given OR gene are positioned in the nucleus relative to the different chromatin marks. Our immuno-DNA FISH experiments show that, whereas ∼45–50% of the OR gene alleles colocalize with H3K9me3, the remaining ones do not, indicating that not all of the OR gene alleles are deeply repressed within constitutive heterochromatin.
It has been previously shown that the two alleles of the Ig Igh and Igk genes and of the T-cell receptor Tcrb gene, which are monoallelically expressed, are segregated to different compartments in the nucleus (40, 41). Our observation that in the majority of nuclei the two homologous OR gene alleles are segregated to different nuclear compartments suggests that this segregation may be critical for the characteristic OR gene monoallelic expression. Accordingly, it is known that OR genes are replicated asynchronously, like all other monoallelically expressed genes (1, 42). Asynchronous replication is established through epigenetic marks early in development, in such a way that one of the homologous alleles is inactivated. Therefore, our results indicate that there is a separate mechanism for OR gene allelic exclusion: one of the alleles would be “permanently” inactive, whereas the remaining one would be available for trancription.
Olfactory neurons that express a nonfunctional OR gene can switch expression to a second OR gene (43). Switching is important because it assures that all neurons will express a functional OR. It has also been shown that neurons are able to switch expression to the homologous OR gene allele (43), indicating that in these neurons the two alleles have the potential to be transcribed. It is known that switching can occur only early during neuron development and during a very narrow window of time (43). It is possible that during this critical period the repressive chromatin compartments are not yet stably established, allowing for switching until a functional OR gene is expressed and OR gene choice is stabilized. In agreement with this is the recent observation that biallelic expression of an OR gene is possible but very infrequent and occurs predominantly in younger neurons (44).
We found that nuclei of olfactory neurons possess prominent facultative heterochromatin domains that are localized around the large constitutive heterochromatin block. Interestingly, we found that in a given nucleus, whereas one of the two homologous OR gene alleles is associated with constitutive heterochromatin, the other one is frequently associated with facultative heterochromatin, and that alleles from different OR genes can colocalize within these domains. Differently from the constitutive heterochromatin, facultative heterochromatin is involved in transcriptional regulation and can both have a role in stable gene inactivation (such as inactivation of the X chromosome) or can be reversed to promote gene expression of genes during development. It remains to be determined when and how these facultative heterochromatin domains are established in the olfactory nuclei, for example, are they established before or after OR gene expression? The fact that they are present also in immature olfactory neurons, and that OR gene choice probably occurs during terminal differentiation to neurons from neuronal precursors or in immature olfactory neurons, suggests that they are formed before or concomitant with OR gene expression. One intriguing possibility is that they could be formed as a result of the feedback signal that is elicited by an expressed OR gene to prevent the expression of additional OR genes, and thereby contribute to the stability of OR gene choice (44). Alternatively, we cannot yet exclude the possibility that repression of the OR gene alleles by facultative heterochromatin is established before an OR gene is chosen for expression, and removed during neuronal development, as it has been shown for several neural lineage genes (45).
It also remains to be determined how facultative heterochromatin would be targeted to OR genes or to OR gene loci. H3K27 methylation is mediated by the Polycomb repressive complex 2 (PRC2) (46), and it has been shown that the PRC2 subunit EED, which is necessary for H3K27 methylation, is required for asynchronous replication of OR gene loci in mouse ES cells (47). These results indicate that facultative heterochromatin is involved in establishing differences between homologous OR gene alleles in these cells. In addition, binding sites for transcription factors that are known to recruit PRC2, such as YY1 (which is also a PRC2 subunit) and Th-POK, were found in OR genes (10, 48). Altogether, these results indicate that facultative heterochromatin must play an important role in OR gene expression. It has been recently demonstrated that the transient expression of the histone demethylase LSD1, which is able to catalyze the demethylation of both H3K9 and H3K4, is involved in stable OR gene choice (49). It will be interesting to determine whether enzymes involved in H3K27 methylation or demethylation can also affect OR gene expression or the stability of OR gene choice.
The colocalization of OR genes to the same compartments must contribute to the coordination of the mutually exclusive expression of these genes. In this way, unrepressed OR alleles (alleles which are not associated with either type of heterochromatin) may be exposed to a particular subnuclear environment where limiting factors that regulate OR gene expression, such as the H or P elements, are available. These enhancers, would now select an OR gene allele from this limited pool of available OR genes. Alternatively, an OR gene allele would be stochastically and limitedly selected from the pool of the 1,000 unrepressed, available OR gene alleles, with the help of one of the enhancers (44). The expression of this allele would lead to a feedback regulation that would cause the formation of facultative heterochromatin in the additional OR genes. Because they are colocalized in the nucleus, these OR genes could be more efficiently targeted and repressed by locally acting regulatory RNAs transcribed from one neighbor OR gene, which has initiated transcription first. Interestingly, it is known that formation of facultative heterochromatin may require the action of noncoding RNAs (46).
In conclusion, our results show that the homologous OR genes are segregated to different chromatin compartments in the nucleus. This spatial segregation is likely to be important for the monoallelic expression of these genes. Our results also show that a significant fraction of the OR alleles are repressed in facultative heterochromatin. The understanding of the mechanisms involved in the function of facultative heterochromatin repression in olfactory neurons should contribute to the understanding of OR gene expression.
Materials and Methods
BAC Clones and Nick Translation.
RPCI-23 mouse BAC clones (http://bacpac.chori.org/femmouse23.htm) were ordered from Invitrogen (no. RPCI23.C). The BAC clones were labeled by nick translation with biotin-16-dUTP or digoxigenin 11-dUTP using the Nick Translation kit (Roche).
Animal Procedures.
All animal procedures in this study were approved by the University of São Paulo Chemistry Institute's Animal Care and Use Committee, under the protocol number 01/2013.
DNA FISH.
DNA FISH in interphase nuclei was performed basically as described (50). See SI Materials and Methods for details.
Immuno-DNA FISH.
Immuno detection of GFP, H3K9me3, H3K4me3, H3K9ac, and H3K27me3 was combined with DNA FISH. DNA FISH was performed as described above, and then slides were incubated with the primary antibodies: policlonal chicken anti-GFP (1:200; Life Technologies, no. A10262), policlonal rabbit anti-H3K9me3 (1:1,000; Millipore, no. 07–442), anti-H3K27me3 (1:500; Millipore, no. 07–449), anti- H3K4me3 (1:500; Abcam, no. Ab8580), and anti-H3K9ac (1:50; Abcam, no. Ab4441). Slides were washed and incubated with the secondary antibodies Alexa Fluor 514 or Alexa Fluor 488 (Molecular Probes) and mounted with Vectashield mounting media (Vectorlabs).
Three-Dimensional Microscopy and Image Analysis.
Slides were examined by using an inverted Nikon Eclipse Ti microscope equipped with a XYZ motorized stage and a 100× PlanFluor objective lens (oil, NA 1.3). Z-stack images were collected every 0.2 μM with a Roper CoolSnap HQ camera (1 pixel = 0.08 μm). Deconvolved images were generated using Huygens Essential 3.2. Distances between signals were measured on 3D-reconstructed image stacks using FIJI ImageJ software (51) (http://fiji.sc/wiki/index.php/Downloads). The number and volume of heterochromatic blocks shown in Fig. S2 were calculated using the 3D object counter tool from FIJI. Distances between the OR gene alleles and the gravity center of the heterochromatin blocks were measured by using Sync Measure 3D.
Supplementary Material
Acknowledgments
We thank Frederico Gueiros-Filho and Sergio Schenkmann for help with acquisition of microscope images; Sergio Verjovski-Almeida for providing reagents; Daniel Strongin and Tobias Ragoczy for helpful comments and suggestions; Stevens Rehen for help with the initial DNA FISH experiments; and Isaías Glezer, Luisa Kfouri Ribeiro, and Ana Carolina Bottura de Barros for help with the acquisition of images and image analysis. This work was supported by grants from Fundação de Amparo à Pesquisa do Estado de São Paulo, Conselho Nacional de Desenvolvimento Científico e Tecnológico, and Coordenação de Aperfeiçoamento de Pessoal de Nível Superior.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1317036111/-/DCSupplemental.
References
- 1.Chess A, Simon I, Cedar H, Axel R. Allelic inactivation regulates olfactory receptor gene expression. Cell. 1994;78(5):823–834. doi: 10.1016/s0092-8674(94)90562-2. [DOI] [PubMed] [Google Scholar]
- 2.Malnic B, Hirono J, Sato T, Buck LB. Combinatorial receptor codes for odors. Cell. 1999;96(5):713–723. doi: 10.1016/s0092-8674(00)80581-4. [DOI] [PubMed] [Google Scholar]
- 3.Godfrey PA, Malnic B, Buck LB. The mouse olfactory receptor gene family. Proc Natl Acad Sci USA. 2004;101(7):2156–2161. doi: 10.1073/pnas.0308051100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Serizawa S, et al. Mutually exclusive expression of odorant receptor transgenes. Nat Neurosci. 2000;3(7):687–693. doi: 10.1038/76641. [DOI] [PubMed] [Google Scholar]
- 5.Ishii T, et al. Monoallelic expression of the odourant receptor gene and axonal projection of olfactory sensory neurones. Genes Cells. 2001;6(1):71–78. doi: 10.1046/j.1365-2443.2001.00398.x. [DOI] [PubMed] [Google Scholar]
- 6.Vassalli A, Rothman A, Feinstein P, Zapotocky M, Mombaerts P. Minigenes impart odorant receptor-specific axon guidance in the olfactory bulb. Neuron. 2002;35(4):681–696. doi: 10.1016/s0896-6273(02)00793-6. [DOI] [PubMed] [Google Scholar]
- 7.McIntyre JC, Bose SC, Stromberg AJ, McClintock TS. Emx2 stimulates odorant receptor gene expression. Chem Senses. 2008;33(9):825–837. doi: 10.1093/chemse/bjn061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Clowney EJ, et al. High-throughput mapping of the promoters of the mouse olfactory receptor genes reveals a new type of mammalian promoter and provides insight into olfactory receptor gene regulation. Genome Res. 2011;21(8):1249–1259. doi: 10.1101/gr.120162.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Michaloski JS, Galante PA, Malnic B. Identification of potential regulatory motifs in odorant receptor genes by analysis of promoter sequences. Genome Res. 2006;16(9):1091–1098. doi: 10.1101/gr.5185406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Michaloski JS, et al. Common promoter elements in odorant and vomeronasal receptor genes. PLoS ONE. 2011;6(12):e29065. doi: 10.1371/journal.pone.0029065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Young JM, Luche RM, Trask BJ. Rigorous and thorough bioinformatic analyses of olfactory receptor promoters confirm enrichment of O/E and homeodomain binding sites but reveal no new common motifs. BMC Genomics. 2011;12:561. doi: 10.1186/1471-2164-12-561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Plessy C, et al. Promoter architecture of mouse olfactory receptor genes. Genome Res. 2012;22(3):486–497. doi: 10.1101/gr.126201.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Serizawa S, et al. Negative feedback regulation ensures the one receptor-one olfactory neuron rule in mouse. Science. 2003;302(5653):2088–2094. doi: 10.1126/science.1089122. [DOI] [PubMed] [Google Scholar]
- 14.Bozza T, et al. Mapping of class I and class II odorant receptors to glomerular domains by two distinct types of olfactory sensory neurons in the mouse. Neuron. 2009;61(2):220–233. doi: 10.1016/j.neuron.2008.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Khan M, Vaes E, Mombaerts P. Regulation of the probability of mouse odorant receptor gene choice. Cell. 2011;147(4):907–921. doi: 10.1016/j.cell.2011.09.049. [DOI] [PubMed] [Google Scholar]
- 16.Fuss SH, Omura M, Mombaerts P. Local and cis effects of the H element on expression of odorant receptor genes in mouse. Cell. 2007;130(2):373–384. doi: 10.1016/j.cell.2007.06.023. [DOI] [PubMed] [Google Scholar]
- 17.Nishizumi H, Kumasaka K, Inoue N, Nakashima A, Sakano H. Deletion of the core-H region in mice abolishes the expression of three proximal odorant receptor genes in cis. Proc Natl Acad Sci USA. 2007;104(50):20067–20072. doi: 10.1073/pnas.0706544105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lewcock JW, Reed RR. A feedback mechanism regulates monoallelic odorant receptor expression. Proc Natl Acad Sci USA. 2004;101(4):1069–1074. doi: 10.1073/pnas.0307986100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lane RP, et al. Genomic analysis of orthologous mouse and human olfactory receptor loci. Proc Natl Acad Sci USA. 2001;98(13):7390–7395. doi: 10.1073/pnas.131215398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shykind BM. Regulation of odorant receptors: One allele at a time. Hum Mol Genet. 2005;14(Spec No 1):R33–R39. doi: 10.1093/hmg/ddi105. [DOI] [PubMed] [Google Scholar]
- 21.Misteli T. Beyond the sequence: Cellular organization of genome function. Cell. 2007;128(4):787–800. doi: 10.1016/j.cell.2007.01.028. [DOI] [PubMed] [Google Scholar]
- 22.Lanctôt C, Cheutin T, Cremer M, Cavalli G, Cremer T. Dynamic genome architecture in the nuclear space: Regulation of gene expression in three dimensions. Nat Rev Genet. 2007;8(2):104–115. doi: 10.1038/nrg2041. [DOI] [PubMed] [Google Scholar]
- 23.Rajapakse I, Groudine M. On emerging nuclear order. J Cell Biol. 2011;192(5):711–721. doi: 10.1083/jcb.201010129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Francastel C, Schübeler D, Martin DI, Groudine M. Nuclear compartmentalization and gene activity. Nat Rev Mol Cell Biol. 2000;1(2):137–143. doi: 10.1038/35040083. [DOI] [PubMed] [Google Scholar]
- 25.Takizawa T, Meshorer E. Chromatin and nuclear architecture in the nervous system. Trends Neurosci. 2008;31(7):343–352. doi: 10.1016/j.tins.2008.03.005. [DOI] [PubMed] [Google Scholar]
- 26.Trojer P, Reinberg D. Facultative heterochromatin: Is there a distinctive molecular signature? Mol Cell. 2007;28(1):1–13. doi: 10.1016/j.molcel.2007.09.011. [DOI] [PubMed] [Google Scholar]
- 27.Campos EI, Reinberg D. Histones: Annotating chromatin. Annu Rev Genet. 2009;43:559–599. doi: 10.1146/annurev.genet.032608.103928. [DOI] [PubMed] [Google Scholar]
- 28.Magklara A, et al. An epigenetic signature for monoallelic olfactory receptor expression. Cell. 2011;145(4):555–570. doi: 10.1016/j.cell.2011.03.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Clowney EJ, et al. Nuclear aggregation of olfactory receptor genes governs their monogenic expression. Cell. 2012;151(4):724–737. doi: 10.1016/j.cell.2012.09.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ressler KJ, Sullivan SL, Buck LB. A zonal organization of odorant receptor gene expression in the olfactory epithelium. Cell. 1993;73(3):597–609. doi: 10.1016/0092-8674(93)90145-g. [DOI] [PubMed] [Google Scholar]
- 31.Vassar R, Ngai J, Axel R. Spatial segregation of odorant receptor expression in the mammalian olfactory epithelium. Cell. 1993;74(2):309–318. doi: 10.1016/0092-8674(93)90422-m. [DOI] [PubMed] [Google Scholar]
- 32.Lomvardas S, et al. Interchromosomal interactions and olfactory receptor choice. Cell. 2006;126(2):403–413. doi: 10.1016/j.cell.2006.06.035. [DOI] [PubMed] [Google Scholar]
- 33.Zuleger N, Robson MI, Schirmer EC. The nuclear envelope as a chromatin organizer. Nucleus. 2011;2(5):339–349. doi: 10.4161/nucl.2.5.17846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Peters AH, et al. Partitioning and plasticity of repressive histone methylation states in mammalian chromatin. Mol Cell. 2003;12(6):1577–1589. doi: 10.1016/s1097-2765(03)00477-5. [DOI] [PubMed] [Google Scholar]
- 35.Volpi EV, et al. Large-scale chromatin organization of the major histocompatibility complex and other regions of human chromosome 6 and its response to interferon in interphase nuclei. J Cell Sci. 2000;113(Pt 9):1565–1576. doi: 10.1242/jcs.113.9.1565. [DOI] [PubMed] [Google Scholar]
- 36.Williams RR, Broad S, Sheer D, Ragoussis J. Subchromosomal positioning of the epidermal differentiation complex (EDC) in keratinocyte and lymphoblast interphase nuclei. Exp Cell Res. 2002;272(2):163–175. doi: 10.1006/excr.2001.5400. [DOI] [PubMed] [Google Scholar]
- 37.Mahy NL, Perry PE, Bickmore WA. Gene density and transcription influence the localization of chromatin outside of chromosome territories detectable by FISH. J Cell Biol. 2002;159(5):753–763. doi: 10.1083/jcb.200207115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mombaerts P, et al. Visualizing an olfactory sensory map. Cell. 1996;87(4):675–686. doi: 10.1016/s0092-8674(00)81387-2. [DOI] [PubMed] [Google Scholar]
- 39.Ragoczy T, Groudine M. The nucleus inside out—through a rod darkly. Cell. 2009;137(2):205–207. doi: 10.1016/j.cell.2009.04.006. [DOI] [PubMed] [Google Scholar]
- 40.Skok JA, et al. Nonequivalent nuclear location of immunoglobulin alleles in B lymphocytes. Nat Immunol. 2001;2(9):848–854. doi: 10.1038/ni0901-848. [DOI] [PubMed] [Google Scholar]
- 41.Goldmit M, et al. Epigenetic ontogeny of the Igk locus during B cell development. Nat Immunol. 2005;6(2):198–203. doi: 10.1038/ni1154. [DOI] [PubMed] [Google Scholar]
- 42.Singh N, et al. Coordination of the random asynchronous replication of autosomal loci. Nat Genet. 2003;33(3):339–341. doi: 10.1038/ng1102. [DOI] [PubMed] [Google Scholar]
- 43.Shykind BM, et al. Gene switching and the stability of odorant receptor gene choice. Cell. 2004;117(6):801–815. doi: 10.1016/j.cell.2004.05.015. [DOI] [PubMed] [Google Scholar]
- 44.Fleischmann A, Abdus-Saboor I, Sayed A, Shykind B. Functional interrogation of an odorant receptor locus reveals multiple axes of transcriptional regulation. PLoS Biol. 2013;11(5):e1001568. doi: 10.1371/journal.pbio.1001568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hirabayashi Y, Gotoh Y. Epigenetic control of neural precursor cell fate during development. Nat Rev Neurosci. 2010;11(6):377–388. doi: 10.1038/nrn2810. [DOI] [PubMed] [Google Scholar]
- 46.Simon JA, Kingston RE. Occupying chromatin: Polycomb mechanisms for getting to genomic targets, stopping transcriptional traffic, and staying put. Mol Cell. 2013;49(5):808–824. doi: 10.1016/j.molcel.2013.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Alexander MK, et al. Differences between homologous alleles of olfactory receptor genes require the Polycomb Group protein Eed. J Cell Biol. 2007;179(2):269–276. doi: 10.1083/jcb.200706053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Faulk CD, Kim J. YY1’s DNA-binding motifs in mammalian olfactory receptor genes. BMC Genomics. 2009;10:576. doi: 10.1186/1471-2164-10-576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lyons DB, et al. An epigenetic trap stabilizes singular olfactory receptor expression. Cell. 2013;154(2):325–336. doi: 10.1016/j.cell.2013.06.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Foster HA, Abeydeera LR, Griffin DK, Bridger JM. Non-random chromosome positioning in mammalian sperm nuclei, with migration of the sex chromosomes during late spermatogenesis. J Cell Sci. 2005;118(Pt 9):1811–1820. doi: 10.1242/jcs.02301. [DOI] [PubMed] [Google Scholar]
- 51.Schindelin J, et al. Fiji: An open-source platform for biological-image analysis. Nat Methods. 2012;9(7):676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.