Significance
Dengue virus has emerged as one of the most significant arboviruses, affecting millions of people around the world. Here, we show the production of a functional microRNA-like small RNA encoded by Dengue virus, which negatively regulates virus replication by targeting the viral nonstructural protein 1 sequences. The report provides a comprehensive analysis of the biogenesis and function of the viral small RNA. The results also highlight the possibility of utilizing the small RNA in inhibiting at least three serotypes of the virus. The outcomes have a significant impact on our understanding of the biology of the virus (and flaviviruses in general) and small RNA biogenesis, and open up a potential avenue for the control of the virus.
Abstract
MicroRNAs (miRNAs) are small regulatory RNAs that play significant roles in most cellular processes. In the seemingly endless arms race between hosts and pathogens, viruses also encode miRNAs that facilitate successful infection. In search of functional miRNAs or viral small RNAs (vsRNAs) encoded by Dengue virus (DENV), deep sequencing data of virus-infected Aedes aegypti mosquitoes were used. From six vsRNAs, with candidate stem-loop structures in the 5′ and 3′ untranslated regions of the viral genomic RNA, inhibition of DENV–vsRNA-5 led to significant increases in viral replication. Silencing of RNA interference (RNAi)/miRNA pathways’ associated proteins showed that Argonaute 2 is mainly involved in DENV–vsRNA-5 biogenesis. Cloning of the precursor stem loop, immunoprecipitations, ectopic expression and detection in RNAi-deficient C6/36, and the mammalian Vero cell lines further confirmed DENV–vsRNA-5 production. Furthermore, significant impact of synthetic mimic and inhibitor of DENV–vsRNA-5 on DENV RNA levels revealed DENV–vsRNA-5’s role in virus autoregulation by targeting the virus nonstructural protein 1 gene. Notably, DENV–vsRNA-5 homologous mimics from DENV serotypes 1 and 4, but not 3, inhibited DENV-2 replication. The results revealed that DENV is able to encode functional vsRNAs, and one of those, which resembles miRNAs, specifically targets a viral gene, opening an avenue for possible utilization of the small RNA to limit DENV replication.
The mosquito-borne flaviviruses cause widespread deadly diseases, morbidity, and mortality globally (1). The well-known members of this group of single-stranded RNA viruses are Dengue virus (DENV), West Nile virus (WNV), and Yellow fever virus. Every year, nearly 100 million cases of DENV infectious diseases, including half a million of dengue hemorrhagic fever, are reported from more than 100 countries (2). The mosquitoes of the genus Aedes, mainly Aedes aegypti and Aedes albopictus, transmit DENV (3). During the last two to three decades, the incidence and severity of DENV diseases has been on the rise. The major factors that have contributed to this upward trend are human population growth, unavailability of an effective vaccine or antiviral drug, and lack of effective mosquito control measures (4). There are four distinct serotypes of DENV, designated DENV-1, DENV-2, DENV-3, and DENV-4. The DENV genome is an approximately 11-kb positive single-stranded RNA that contains a small 100-nt 5′ untranslated region (UTR), a long ORF encoding 10 genes, and a 384-nt-long 3′UTR region (reviewed in ref. 5). The whole genome is translated into a polypeptide chain that is then cleaved proteolytically into three structural (capsid, membrane, and envelop) and seven nonstructural proteins (reviewed in ref. 4). Both of the 5′ and 3′UTR regions consist of complex secondary stem-loop structures, and their interactions are involved in initiation and regulation of virus translation, replication, and assembly (6). The promoter, called stem-loop A, is located in the 5′UTR that binds and activates viral NS5 and initiates RNA replication at the 3′ end of a circularized virus genome (7). Three different regions have been identified in the 3′UTR: a variable, a core with conserved secondary structure, and a 3′ stem loop (reviewed in ref. 8).
During the last decade, several different classes of small RNAs and the proteins associated with their processing have been discovered from eukaryotes, prokaryotes, and viruses (9). Similar to their hosts, virus derived small RNAs are also classified into short interfering RNA, microRNA (miRNA), PIWI-interacting RNA, viral small RNA (vsRNA), and unusually small RNA (reviewed in ref. 10). Among them, viral miRNAs encoded by DNA viruses have been well studied in comparison with those by RNA viruses. In animal cells, canonical miRNA biogenesis involves nuclear events in which the primary miRNA is cleaved by Drosha and DiGeorge syndrome critical region gene 8 (DGCR8)/Pasha to produce the precursor miRNA (premiRNA), and cytoplasmic events where Dicer-1 cuts the premiRNA to yield a mature miRNA duplex (reviewed in ref. 11). Recently, several other noncanonical pathways have also been reported that depend on Argonaute (AGO) 2 for cleavage of premiRNA to mature miRNA (12, 13). Similar noncanonical pathways might be used to generate functional miRNAs derived from RNA viruses that replicate in the cytoplasm. For example, a cellular miRNA, miR-124, was shown to be expressed from Sindbis virus, a cytoplasmic RNA virus, in which the premiRNA was cloned, and a premiRNA sequence from a DNA virus, Epstein–Barr virus, which was cloned in the 3′UTR of tick-borne encephalitis virus (a flavivirus) that successfully produced the mature miRNA without affecting the viral genome (14, 15). In another example, several miRNAs were found to be produced by a retrovirus, Bovine leukemia virus (BLV) based on DNA polymerase III transcription in vivo as well as in vitro (16). Functional analyses have revealed that viral miRNAs can target both cellular and viral mRNAs to regulate virus replication leading to a successful infection (reviewed in ref. 10).
In this study, we examined the possibility of DENV-2 to encode functional miRNAs/vsRNAs. The effect of inhibition of six vsRNAs, mapped to the virus genome UTR regions, was studied in Aag2 cell line (from A. aegypti) on DENV replication. Based on its significant impact on DENV replication, DENV–vsRNA-5 was further analyzed. In addition, we determined that DENV–vsRNA-5 targets the NS1 gene of DENV-2 and as a consequence regulates viral replication.
Results
Deep Sequencing of DENV-2–Infected Mosquitoes Reveals a Variety of vsRNAs.
Deep sequencing was conducted on total RNA extracted from pools of 10 mosquitoes (A. aegypti) from three biological replicates that were blood fed with and without DENV-2 collected at 1, 3, and 7 d after infection (dpi). The total number of mappable reads from each library was about 4–5 × 106. Small RNA sequences (15–30 nt) were aligned to DENV-2 viral genome (accession no. M29095.1). We found a total of 484 different vsRNA sequences with 41% 15–19 nt, 33% 20–24 nt, and 25% 25–30 nt. After filtering the mappable reads, from 640,994 (1-d-infected sample), 560,828 (3-d-infected), and 671,461 (7-d-infected) unique reads, 0.01%, 0.01%, and 0.09% of the unique reads mapped to viral sequences, which was consistent with a previous study (17). Concentrating on the UTR regions where stem-loop structures are most abundant, one small RNA was found to be derived from the 5′UTR and 22 from the 3′UTR of the virus.
Characterization of DENV-2 Candidate vsRNAs Using a Mosquito Cell Line.
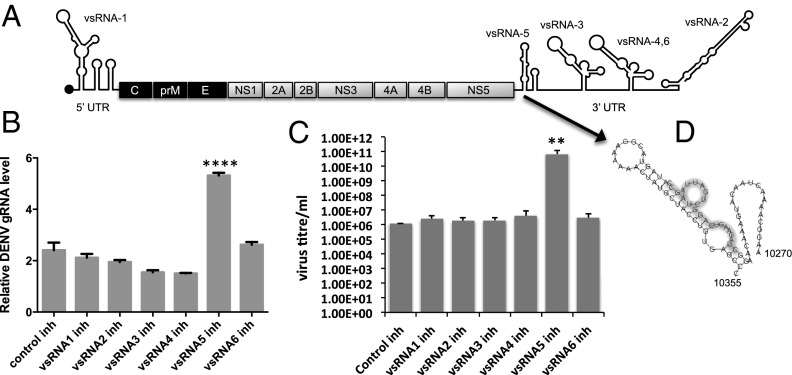
To explore the possibility of any vsRNAs/miRNA(s) derived from DENV-2 RNA in our vsRNA deep sequencing data, RNAfold software was used for prediction of precursor stem-loop structures in the 5′ and 3′UTR regions of the virus genome by using the minimum free energy (mfe) of −21 kcal/mol as a cutoff. Six such precursors containing small RNA sequences found in the deep sequencing data were selected: one from the 5′UTR (DENV–vsRNA-1) and five from the 3′UTR (DENV–vsRNA-2 to -6) (Fig. 1A and Table S1). To find out the impact of the vsRNAs on viral RNA replication, synthetic inhibitors of the six vsRNAs and a control inhibitor with random sequences were separately transfected into Aag2 cells followed by DENV-2 infection. The inhibitors were RNA oligos with reverse complementary sequences to the vsRNA sequences. Quantitative RT-PCR (RT-qPCR) analyses of RNA collected at 7 dpi showed significantly higher viral genomic RNA (gRNA) in only the vsRNA-5 inhibitor transfected cells (Fig. 1B; P < 0.0001; ANOVA), which indicated possible significance of this small RNA in regulating virus RNA replication. This experiment was also replicated in A. albopictus RML-12 cells with similar results at the viral gRNA level (Fig. S1). Virus titer determination from the experiment in Aag2 cells using the 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl-tetrazoliumbromide (MTT) method revealed that in the presence of the vsRNA-5 inhibitor, significantly more viral infectious units were produced (Fig. 1C; P = 0.0043; ANOVA). Therefore, this small RNA was analyzed further. DENV–vsRNA-5 corresponds to the first stem-loop structure at the beginning of the 3′UTR or subgenomic flavivirus RNA (sfRNA) of the virus (Fig. 1D).
Fig. 1.
Locations of DENV-2 vsRNAs in the viral genome and the effect of their inhibitors on virus replication. (A) Diagram showing the genome of DENV with locations of vsRNAs mapped to the genome. (B) RT-qPCR analysis to examine the levels of viral gRNA in Aag2 cells transfected with synthetic inhibitors of the six vsRNAs shown in A followed by DENV-2 infection at 7 dpi. ****P < 0.0001 ANOVA. (C) Virus titer determination in the medium from three replicates of the experiment described in B by using the MTT method; **P = 0.0043 ANOVA. (D) Secondary structure of the stem-loop in DENV-2 3′UTR and the location of DENV–vsRNA-5 shown with a gray shadow.
DENV–vsRNA-5 Expression Analysis.
When total RNA from mock and virus-infected cells at 3, 5, and 7 dpi were analyzed in a small RNA Northern blot hybridization, a 23-nt small RNA was detected at 7 dpi (Fig. 2A), which matched the size of the sequenced small RNA in the deep sequencing. DENV–vsRNA-5 was also detected in C6/36 cell line from A. albopictus with a similar pattern to that of Aag2 cells at 7 dpi (Fig. 2B, 5′ probe). Independent studies have demonstrated that C6/36 cells are RNAi pathway deficient (18–20), assuring that DENV–vsRNA-5 is not an RNAi degradation product. To further confirm that the small RNA is not a degradation product, another probe of 21 nt was used, which was reverse complementary to the precursor stem-loop sequence of DENV–vsRNA-5 covering the terminal loop and a few nucleotides (9 nt) of the other strand of the stem. Using this probe, the small RNA was not detected in the same blot (Fig. 2B, 3′ TL probe). Further, we cloned the precursor stem loop of DENV–vsRNA-5 through RT-PCR from DENV-2–infected Aag2 cells at 7 dpi by using two independent approaches. A sequence of 93 nt was obtained corresponding to nucleotide coordinates 10298–10392 on the DENV-2 genome.
Fig. 2.
DENV–vsRNA-5 expression in Aag2 cells. (A) Northern blot analysis using a specific probe to vsRNA-5 in DENV-2–infected Aag2 cells at 3, 5, and 7 dpi; M, mock-infection. U6 RNA serves as loading control. Arrows indicate the precursor and mature vsRNA-5. (B) Northern blot analysis of vsRNA-5 in C6/36 cells infected with DENV-2 at 3, 5, and 7 dpi using the 5′ probe (Left); the same blot was used for 3′TL probe (Right). (C) Northern blot analysis for detection of vsRNA-5 in Aag2 cells transfected with 3′UTR ssRNA from DENV-2 using vsRNA-5 5′, 3′TL, and vsRNA-2 probes. SS1 and SS2 show two replicates from separate transfections. (D) miRNA RT-qPCR analysis confirming ectopic expression of vsRNA-5 in Aag2 cells transfected with pIZ/pre-5 (72 h after transfection) encoding the precursor of DENV–vsRNA-5. pIZ, empty vector. (E) miRNA RT-qPCR analysis for expression of vsRNA-5 in the mammalian Vero cells infected with DENV-2. Data were normalized against 5S rRNA.
To investigate the processing and production of DENV–vsRNA-5 in the absence of virus infection, a 451-nt single-strand RNA of DENV containing the 3′UTR (3′UTR ssRNA) was synthesized in vitro and transfected into Aag2 cells. Northern blot hybridization with DENV–vsRNA-5 5′ probe showed distinct bands, one corresponding to DENV–vsRNA-5 (Fig. 2C). The same blot was reprobed with 3′ TL and DENV–vsRNA-2 probes, but the small RNA was not detected. Further, the stem-loop precursor of DENV–vsRNA-5 was cloned into the pIZ/V5-His vector and transfected into Aag2 cells. Synthesis of the mature DENV–vsRNA-5 in the cells was confirmed by small RNA-specific RT-qPCR (Fig. 2D). The PCR product was sequenced to confirm the identity of the small RNA. To investigate whether DENV–vsRNA-5 is produced in DENV-infected mammalian cells, Vero cells (from African green monkey) were infected with DENV-2 virus. RT-qPCR confirmed expression of the small RNA in the cells, which increased over time with maximum levels at 7 dpi (Fig. 2E).
DENV–vsRNA-5 Biogenesis Is AGO2 Dependent.
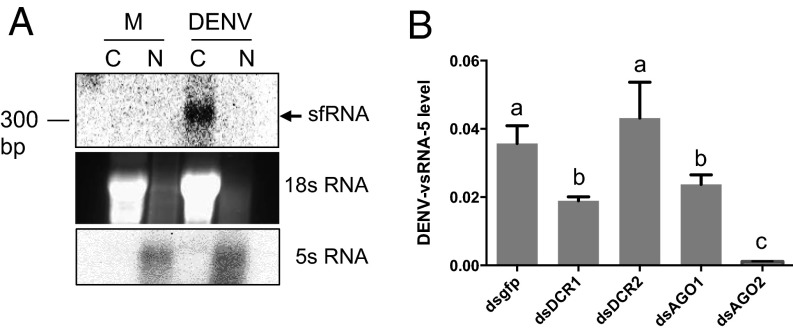
To investigate whether DENV–vsRNA-5 was produced through the canonical biogenesis pathway that involves nuclear event of Drosha processing, we wanted to know whether the primary transcript, the sfRNA, which constitutes most of the 3′UTR (21), enters the nucleus at later stages of infection. Cytoplasmic and nuclear RNA fractions of DENV-2–infected Aag2 cells at 7 dpi were extracted, and Northern blot analysis detected sfRNA only in the cytoplasmic fraction (Fig. 3A). The absence of the primary transcript in the nucleus of infected cells reduced further the chances of it being cleaved by Drosha in the nucleus. To find out which proteins of miRNA/RNAi pathways are involved in the processing of mature DENV–vsRNA-5, Dicer-1, Dicer-2, AGO1, and AGO2 genes were silenced in Aag2 cells followed by DENV infection. Silencing of all of these genes was confirmed by RT-qPCR analyses. Subsequently, the levels of mature DENV–vsRNA-5 using RT-qPCR were analyzed, which showed significantly less DENV–vsRNA-5 levels in AGO2-silenced cells, suggesting its possible role in the biogenesis of the vsRNA (Fig. 3B). In addition, DENV–vsRNA-5 levels were reduced to about half in Dicer-1– and AGO1-silenced cells, but Dicer-2 silencing had no effect on the levels of the small RNA. In insects, Dicer-1 is known to be primarily involved in the miRNA biogenesis, whereas Dicer-2 is implicated mainly in the RNAi response (22). The results further confirmed that DENV–vsRNA-5 is not an RNA degradation product of Dicer-2 cleavage.
Fig. 3.
Biogenesis of DENV–vsRNA-5 is mainly AGO2 dependent. (A) Northern blot detection of DENV sfRNA in the cytoplasmic fraction of Aag2 cells infected with DENV-2 at 7 dpi. C, cytoplasmic; M, mock infection; N, nuclear RNA fractions. To distinguish cytoplasmic and nuclear fractions, 18s rRNA and 5s rRNA are shown, respectively. (B) RT-qPCR analyses to show detection levels of vsRNA-5 in control (dsgfp), Dicer-1, Dicer-2, AGO1, and AGO2 silenced Aag2 cells infected with DENV-2 at 7 dpi. There are statistically significant differences between groups with different letters at P < 0.0001.
Moreover, immunoprecipitation (IP) analysis was carried out by using AGO1 and AGO2 antisera and cell lysate from DENV-infected Aag2 cells. Anti-GFP antibody was used as a control. Using polyadenylation of RNA followed by RT-PCR with an Oligo dT and a specific DENV–vsRNA-5 forward primer, the precursor transcript of 93 nt (10298–10392) and a larger 240-nt fragment (10298–10540; included the precursor transcript) of the DENV-2 genome were enriched in AGO2 IP, whereas the mature vsRNA sequence was found in both AGO1 and AGO2 IP and not in anti-GFP IP. The sequence identity of the PCR products was confirmed by sequencing. The coimmunoprecipitation of precursor transcripts in AGO2 IP suggested AGO2-mediated cleavage of the precursor sequences into the mature vsRNA.
DENV–vsRNA-5 Targets NS1 and Down-Regulates Viral RNA Replication.
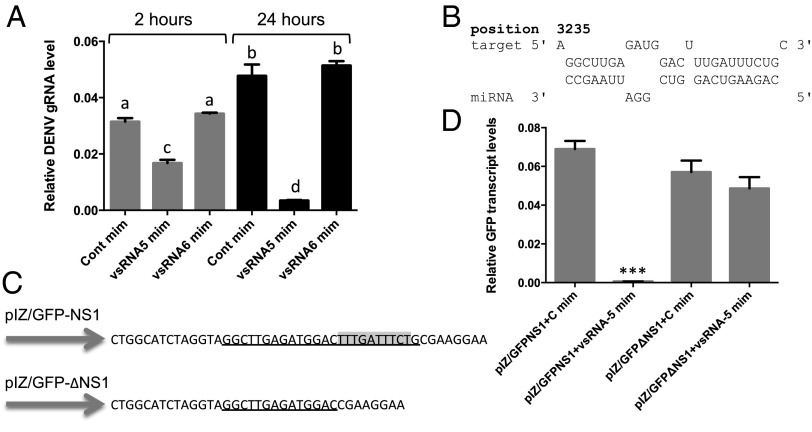
Because transfection of DENV–vsRNA-5 inhibitor led to significant increases in DENV replication (Fig. 1B), we investigated whether any direct or indirect interaction between DENV–vsRNA-5 and DENV-2 gRNA exists. First, synthetic vsRNA mimics were transfected into Aag2 cells followed by DENV infection. The levels of viral gRNA were examined by RT-qPCR at 2 and 24 hpi, which showed significant decreases in cells transfected with DENV–vsRNA-5 mimic compared with control mimic, and further, no effect was observed when another vsRNA mimic (vsRNA-6) was used as a control (Fig. 4A). Next, we analyzed potential target sequences of DENV–vsRNA-5 in DENV RNA genome by using RNAhybrid software that revealed a strong binding site in NS1 at nucleotide coordinates 3235–3261 with the mfe of −25.5 kcal/mol and complete complementarity in the so-called seed region (nucleotides 2–8 from the 5′ end) (Fig. 4B). A second target that met the criteria, but with higher mfe (−23 kcal/mol) and less complementarity was in the NS5 region. Because of higher confidence in NS1 target, we pursued it further.
Fig. 4.
DENV–vsRNA-5 targets the NS1 gene and down-regulates viral gRNA levels. (A) RT-qPCR analysis of DENV gRNA levels in Aag2 cells transfected with the control mimic (Cont mim), vsRNA-5 mimic (vsRNA-5 mim), and vsRNA-6 mimic at 2 and 24 hpi. There are statistically significant differences between groups with different letters at P < 0.01. (B) Predicted target sequences of DENV–vsRNA-5 in the NS1 gene of DENV-2. (C) Cloned sequences of the NS1 target and the mutant target downstream of the GFP gene (pIZ/GFP-NS1). Complementary sequences to vsRNA-5 seed region that are highlighted gray were deleted in the mutant target construct (pIZ/GFP-∆NS1). (D) RT-qPCR analysis of RNA extracted from Aag2 cells cotransfected with the constructs in C and either vsRNA-5 or control mimic (mim) using GFP specific primers. ***P < 0.001.
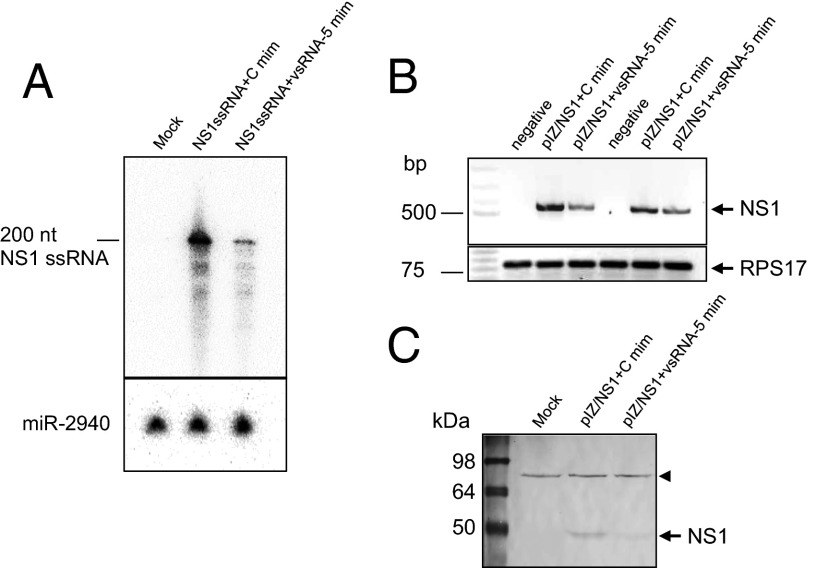
We investigated whether vsRNA-5:NS1 target interaction could be validated by using GFP as a reporter system. For this validation, pIZ/GFP-NS1 (containing the complete target sequence; Fig. 4C) together with DENV–vsRNA-5 mimic or a control mimic were cotransfected into Aag2 cells. In addition, another construct pIZ/GFP-∆NS1 (minus target complementary sequences to the seed region; Fig. 4C) was used as a control. Using RT-qPCR, there was a significant reduction (P < 0.0001) in the GFP transcript levels in the case of pIZ/GFP-NS1 in the presence of DENV–vsRNA-5 mimic compared with the control mimic; however, the GFP levels were unaffected in pIZ/GFP-∆NS1 transfections by both mimics (Fig. 4D). Additionally, the direct impact of DENV–vsRNA-5 mediated down-regulation of NS1 transcript levels was investigated in a couple of experiments. First, a 200-nt ssRNA of NS1 region (NS1 ssRNA) containing DENV–vsRNA-5 target sequences were synthesized in vitro and used in cotransfection with DENV–vsRNA-5 mimic and a control mimic. Northern blot hybridization using an NS1-specific probe showed near-complete degradation of NS1 ssRNA in the presence of DENV–vsRNA-5 mimic (Fig. 5A). Second, the full-length NS1 gene was cloned in the pIZ/V5-His vector (pIZ/NS1) and cotransfected together with DENV–vsRNA-5 mimic and a control mimic into Aag2 cells. RT-PCR revealed substantial down-regulation of NS1 transcripts in the presence of DENV–vsRNA-5 mimic compared with the control mimic (Fig. 5B). Down-regulation of NS1 in the presence of DENV–vsRNA-5 mimic, but not the control mimic, was also confirmed at the protein level by using a monoclonal antibody specific to the protein (Fig. 5C). The results above validated the specific interaction of DENV–vsRNA-5 with NS1 sequences.
Fig. 5.
Validation of DENV–vsRNA-5 and NS1 interaction. (A) Northern blot analysis of RNA from Aag2 cells cotransfected with the synthesized NS1 target ssRNA and control or vsRNA-5 mimic using a specific probe to ssRNA. aae-miR-2940 was used as loading control. (B) RT-PCR analysis of NS1 transcript levels after cotransfection of Aag2 cells with pIZ/NS1-full and control or vsRNA-5 mimics in two experimental replicates. RPS17 amplification is shown as control. (C) Western blot analysis of NS1 protein levels after cotransfection of Aag2 cells with pIZ/NS1-full and control or vsRNA-5 mimics using a monoclonal antibody to NS1. A nonspecific protein (arrowhead) bound to the antibody shows equal loading of the samples.
Sequence alignment was carried out to examine sequence conservation of DENV-2 vsRNA-5 in the other three DENV serotypes. There was greater similarity of the vsRNA sequence to DENV-1 and DENV-4, whereas the first five bases were not highly conserved in DENV-3 (Fig. 6A). These bases fall within the so-called seed region in miRNAs. Subsequently, Aag2 cells were transfected with synthetic mimics based on vsRNA-5 homolog sequences of the other three serotypes followed by DENV-2 infection. The results revealed a similar degree of down-regulation in viral gRNA levels with vsRNA-5 mimics of DENV-1, DENV-2, and DENV-4, whereas no effect was observed with that of the DENV-3 vsRNA-5 mimic (Fig. 6B).
Fig. 6.
(A) Nucleotide sequence alignment to show conservation of DENV–vsRNA-5 among the four DENV serotypes. The accession numbers for the serotypes 1–4 are GU131786.1, M29095.1, JF808129.1, and KF041260.1, respectively. (B) RT-qPCR analysis of RNA from Aag2 cells transfected with vsRNA-5 mimics of DENV-1, -2, -3, and -4 followed by DENV-2 infection at 72 hpi. There are statistically significant differences between groups with different letters at P < 0.0001.
Discussion
In this study, we identified vsRNAs obtained from small RNA deep sequencing of DENV-2–infected A. aegypti mosquitoes. Six vsRNAs mapped to the stem-loop structures in the 5′ and 3′UTR regions of the virus genome. Transfection of synthetic inhibitors of the vsRNAs into mosquito cells followed by DENV infection revealed significant increases in the replication of the virus in the presence of inhibitor of only one of the vsRNAs. This outcome led us to further investigate the miRNA-like vsRNA, named DENV–vsRNA-5, in the context of the host–virus interaction.
To date, almost 500 virus-encoded miRNAs have been deposited on miRBase (23) from various virus families with both DNA and RNA genomes. As a result of extensive research in miRNA field, several new flexible or noncanonical pathways have been revealed for miRNA biogenesis. Viruses may use such pathways to express their miRNAs or miRNA-like small RNAs. A study on processing of miRNAs encoded by BLV, a retrovirus, ruled out Drosha’s (a nuclear protein) involvement in generating premiRNAs, instead they were transcription products of RNA polymerase III (16). Moreover, the secondary stem-loop structures of premiRNAs did not have Drosha cleavage signatures revealed by bioinformatics analysis. In another report, a primary miRNA was cloned into a cytoplasmic virus that produced a functional mature miRNA, which was Dicer dependent and Drosha/DGCR8 independent (15). A followup investigation by the same group recently showed that Drosha could still be involved in processing of virus-derived cytoplasmic primary miRNAs as Drosha was found to be relocalized to the cytoplasm upon virus infection (24). In addition, WNV, a cytoplasmic RNA virus of Flaviviridae family, was shown to encode a miRNA from its terminal stem-loop structure (25).
To explore DENV–vsRNA-5 biogenesis, we were looking for a substitute for Drosha processing given Northern blot analysis indicated that the primary transcript of DENV–vsRNA-5 remains in the cytoplasm. Silencing Dicer-1 (involved in miRNA biogenesis in insects), Dicer-2 (mainly in RNAi response), AGO1 (involved in miRNA biogenesis in insects), and AGO2 (associated with both miRNA and RNAi response), revealed complete inhibition of the synthesis of DENV–vsRNA-5 mature small RNA in AGO2-silenced cells. Silencing of Dicer-2 had no effect on the small RNA levels; however, silencing of Dicer-1 and AGO1 reduced DENV–vsRNA-5 levels to a similar extent, but not as significant as AGO2 silencing, which suggests that Dicer-1 may still function in the processing of the small RNA along with AGO2. Recently, the primary role of AGO2 in miRNA biogenesis has been documented, because in mammals and flies, AGO2 possesses slicer activity and is associated with the biogenesis of miRNA bypassing the Dicer-1 processing step (26–28). We further confirmed that DENV–vsRNA-5 is not a degradation viral product through its detection in RNAi-deficient C6/36 cell line, its ectopic expression in noninfected Aag2 cells by transfection of the 3′UTR ssRNA, and from the plasmid construct (pIZ/pre-5) containing the precursor sequences.
We investigated the functionality of DENV–vsRNA-5 by using its synthetic mimic and inhibitor followed by virus infection in Aag2 cells, which showed significant decreases and increases in viral gRNA production, respectively. The effects on virus gRNA replication prompted us to look for potential targets within the viral RNA genome, which revealed a strong binding site in the NS1 region. The target–vsRNA interaction was confirmed independent of DENV infection through down-regulation of NS1 transcripts in cells transfected with either NS1 ssRNA or pIZ/NS1-full construct together with DENV–vsRNA-5 mimic. In addition, the use of GFP reporter system with complete or mutant target sequences showed the same negative regulatory interaction.
It still remains to be confirmed whether the other three DENV serotypes (1, 3, and 4) express DENV–vsRNA-5, but we found sufficient sequence homology among the serotypes. By using synthetic mimics based on those sequences, similar depletion of DENV-2 viral RNA was found in the case of DENV-1 and DENV-4, but not DENV-3. The lack of inhibitory effect of DENV-3 vsRNA-5 mimic on DENV-2 replication is most likely due to the absence of key nucleotides in the first five bases (at the 5′ end) in vsRNA-5 sequence of DENV-3. In most miRNA–target interactions, the first two to eight bases that make the seed region of a miRNA are important for efficient interaction with the target.
More in-depth functional analyses of several virus-encoded miRNAs are emerging recently that demonstrate their role in the regulation of cellular and viral genes. A number of miRNAs encoded by Epstein–Barr virus (EBV), SV40, JC polyomavirus, BKPyV, and mouse polyomaviruses, carcinomatosis virus type 1 and 2, and Heliothis virescens ascovirus autoregulate viral genes and control replication at certain infection stage (29–33). miRNAs encoded from Kaposi Sarcoma-associated herpesvirus, EBV, and Marek’s disease virus 1 prevent apoptosis by targeting proapoptotic host genes and are also associated with tumorigenesis (34, 35). Moreover, we previously reported that the WNV KUN–miR-1 regulates the host GATA-4 transcription factor that facilitates virus replication (25). miRNAs are believed to be fine tuners of gene expression. Production of DENV–vsRNA-5 in low quantities may benefit the virus by autoregulating virus replication to avoid overreplication of the virus and premature death of the host cells. In particular, this regulation would benefit the virus in the mosquito vector in preventing excessive replication, causing harm to the vector. Autoregulation of virus replication has also been shown in other viruses, such as miR-BART2 from EBV (36), HvAV-miR-1 from Heliothis virescens ascovirus (33), and miR–H2-30 and miR-H6 in Heliothis zea nudivirus 1 (37), all targeting viral genes.
In summary, we have shown that DENV-2 produces at least one functional vsRNA (DENV–vsRNA-5) with characteristics similar to miRNAs with AGO-2, and to some extent Dicer-1 and AGO-1, playing a significant role in its biogenesis. Inhibition of the vsRNA using synthetic inhibitor led to significantly increased DENV replication, suggesting its regulatory role in virus replication. Experiments revealed that DENV–vsRNA-5 interacts with NS1 sequences in the virus genome negatively regulating virus replication. Given the conserved sequences of DENV–vsRNA-5 in at least three serotypes of DENV, broader utilization of the small RNA in inhibition of virus replication may provide a strategy to prevent DENV.
Materials and Methods
Mosquitoes and DENV.
A. aegypti mosquitoes were fed with sheep blood (Applied Biological Products Management) containing DENV-2 (New Guinea strain) at 107 50% tissue-culture lethal dose (TCID50)/mL and were collected at 1, 3, and 7 dpi.
Deep Sequencing of Small RNAs.
Total RNA was extracted from noninfected control and DENV-infected mosquitoes (1, 3, and 7 d after infection) by using Tri-Reagent following the manufacturer’s protocol (Molecular Research Center). RNA concentrations were measured by using a spectrophotometer, and integrity was ensured through analysis of RNA on a 1% (wt/vol) agarose gel and bioanalyzer. Small RNA libraries were generated from both samples by using the Illumina Truseq Small RNA Preparation kit at LC Sciences. The purified cDNA libraries were sequenced on Illumina GAIIx, and raw sequence reads (36 nt) were obtained by using Illumina’s Sequencing Control Studio software version 2.8 followed by real-time sequencing image analysis and base-calling by Illumina's Real-Time Analysis version 1.8.70 (LC Sciences).
RNA Extraction and Northern Blot Hybridization.
A. aegypti (Aag2), A. albopictus C6/36, and RML-12 cells were infected with MOI = 1 of DENV-2. Mock-infected cells were used as control. Total RNA was isolated by using TRI-Reagent (Molecular Research Centre) at different time points after infection, and enriched for small RNAs by using PureLink miRNA isolation kit (Invitrogen). Small RNA samples (20 µg) were separated on 15% (wt/vol) denaturing polyacrylamide gels, electroblotted onto nylon membrane, and UV cross-linked. The probes were generated by labeling of DNA oligonucleotides with [α32P]dCTP by using a terminal nucleotide transferase. All of the probe hybridizations and washings were done at 50 °C. Probe sequences are provided in Table S2. Cytoplasmic and nuclear fractions were isolated by using PARIS kit (Ambion) according to the manufacturer’s instructions, and TRI-Reagent was used for RNA extraction.
Cloning the Precursor of DENV–vsRNA-5.
To determine the right sequences of premiRNA of DENV–vsRNA-5, we used two independent cloning methods. In the first approach, RNA extracted from DENV-2–infected Aag2 cells at 7 dpi was polyadenylated by using the Ncode miRNA first-strand cDNA synthesis kit (Invitrogen) as per the manufacturer’s instructions followed by first-strand cDNA synthesis using a poly-dT primer (Invitrogen). The first-strand cDNA was then used as a template for PCR with DENV–vsRNA-5 specific forward primer and oligo-dT as reverse primer. In the second approach, a 5′ adaptor and a reverse primer designed at the end of the premiRNA sequence were used in combination. PCR products were cloned by using the pGEM-T-Easy vector system (Promega) and sequenced.
Western Blotting.
Protein samples were run on a 12% (wt/vol) SDS/PAGE and transferred onto a nitrocellulose membrane. The membrane was probed with a monoclonal antibody specific to DENV-2 NS1 antibody (1:1,000) and subsequently detected by using an alkaline phosphatase-conjugated anti-mouse antibody.
IPs.
IPs were conducted according to ref. 28 with minor modifications. Briefly, DENV-2–infected Aag2 cells at 7 dpi were collected by centrifugation (500 × g, 10 min, 4 °C) and washed with ice-cold PBS. Cells were lysed in Nonidet P-40 buffer (50 mM Hepes-KOH at pH 7.5, 150 mM KCl, 2 mM EDTA, 0.5% Nonidet P-40, 0.5 mM DTT, 1× Complete EDTA free protease inhibitor; Roche) for 30 min on ice, and the lysates were centrifuged (10 min, 4 °C, 14,000 × g). Supernatants were incubated with G Sepharose beads (GE Healthcare) coated with 3 μg of each antibodies, anti-Ago1, anti-AGO2, and anti-GFP (Abcam), overnight at 4 °C. Beads were washed four times in Nonidet P-40 lysis buffer, and coimmunoprecipitated RNA was extracted from beads by TRI-Reagent.
RNAi-Mediated Silencing of Cellular Genes.
Primer pairs both having T7 promoter sequences (5′-ATACGACTCACTATAGGG-3′) were designed to amplify A. aegypti Dicer-1 (XM_001652162.1), Dicer-2 (AY713296.1), AGO1 (XM_001662504.1), and AGO2 (FJ979880.1) genes. For in vitro dsRNA synthesis, MEGAscript T7 kit (Ambion) was used following the manufacturer’s instructions (Ambion). A total of 5 μg of dsRNA was used in transfection of cells, which were transfected again with the same reagents at 48 h after the first transfection. Cellfectin II (Invitrogen) was used as the transfection reagent. Cells were collected after 96 h for RNA isolation. RT-qPCR was done to confirm transcript levels of silenced genes.
miRNA Target Studies and RT-qPCRs.
RNAHybrid software was used to find potential targets of DENV–vsRNA-5 in DENV RNA genome. A pair of synthetic oligos (59 nt) reverse complementary to each other containing complete target sequence were annealed and cloned into pIZ/V5-His vector (Invitrogen) downstream to GFP resulting in pIZ/GFP-NS1 construct. Another pair of synthetic oligos (49 nt) with mutant target sites was cloned in a similar way resulting in pIZ/GFP-∆NS1. Transcript levels of GFP were analyzed by RT-qPCR while using the mosquito RPS17 gene as reference. For each experiment, three biological replicates with three technical replicates were analyzed in a Rotor-Gene thermal cycler (QIAGEN) under the following conditions: 95 °C for 30 s, and 40 cycles of 95 °C for 10 s and 60 °C for 45 s, followed by the melting curve (68 °C to 95 °C). ANOVA with post hoc Tukey’s test was used to compare differences in means between different treatments.
For full-length (1054 bp) NS1 cloning, specific primers (Table S2) were used for PCR amplification and subsequent cloning into the pIZ/V5-His vector resulting in pIZ/NS1-full. To produce single-stranded RNA (ssRNA, 200 nt) of NS1, a PCR product was first obtained by using T7 promoter-NS1 forward primer and a normal NS1 reverse primer. For in vitro ssRNA synthesis, the MEGAscript T7 kit was used. All of the miRNA mimics, control scrambled sequence mimics (controls), miRNA inhibitors and control scrambled sequence inhibitors (Table S2) were synthesized by Genepharma and used in transfections into insect cells by using the Cellfectin II reagent. RT-qPCR was performed to examine the levels of DENV–vsRNA-5 by using a small RNA-specific oligodT primer (17 nt linker + 12 nt oligo dT and 6 nt complementary to the last six bases of the small RNA) for reverse transcription and a miRNA-specific sense as a forward primer as described (38). The PCR products were sequenced to confirm the identity of small RNAs. For normalizing data, 5S rRNA was used in three biological replicates.
Virus Titration (MTT).
To evaluate the effect of DENV-2 vsRNA inhibitors on virus titer, MTT assay was conducted. In short, BHK21 cells at the density of 1 × 104 were seeded in each well of 96-well plates in 50 µL of DMEM media. Serial tenfold (10−1 to 10−5) dilutions of mature virus collected from supernatants taken from inhibitor transfections experiment were made, and 10 µL of each dilution was used for inoculation (5 wells per dilution), while keeping the same number of wells uninfected. After 5 d of virus infection, 10 µL of MTT stock solution (5 g/L) was added to each well and incubated for 2 h at 37 °C. Plates were centrifuged at 2,000 × g for 10 min, and supernatant was replaced with 50 µL of DMSO. Absorbance at 570 nm was determined in an EPOCH spectrophotometer (BioTek) and plotted against dilution factor D (log-scale) in Sigmaplot 11 software to obtain D0 (dilution factor at which the response is 50%). TLCD50/mL was calculated with the formula [Titer (MTT assay) = 1/D0V], where V is virus inoculum volume (0.01 mL per well).
Supplementary Material
Acknowledgments
We thank Dr. Guangjin Lu from Queensland Institute of Medical Research for providing the DENV-infected mosquitoes and Prof. Paul Young at UQ for the anti-NS1 antibody and DENV-2 inoculum. This work was supported by National Health and Medical Research Council Grant APP1027110 (to S.A.) and an Australian Research Council Discovery Early Career Researcher Award Fellowship DE120101512 (to M.H.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1320123111/-/DCSupplemental.
References
- 1.Mackenzie JS, Gubler DJ, Petersen LR. Emerging flaviviruses: The spread and resurgence of Japanese encephalitis, West Nile and dengue viruses. Nat Med. 2004;10(12) Suppl:S98–S109. doi: 10.1038/nm1144. [DOI] [PubMed] [Google Scholar]
- 2.Gubler DJ. Epidemic dengue/dengue hemorrhagic fever as a public health, social and economic problem in the 21st century. Trends Microbiol. 2002;10(2):100–103. doi: 10.1016/s0966-842x(01)02288-0. [DOI] [PubMed] [Google Scholar]
- 3.Scott TW, Morrison AC. Vector dynamics and transmission of dengue virus: Implications for dengue surveillance and prevention strategies: Vector dynamics and dengue prevention. Curr Top Microbiol Immunol. 2010;338:115–128. doi: 10.1007/978-3-642-02215-9_9. [DOI] [PubMed] [Google Scholar]
- 4.Murray CL, Jones CT, Rice CM. Architects of assembly: Roles of Flaviviridae non-structural proteins in virion morphogenesis. Nat Rev Microbiol. 2008;6(9):699–708. doi: 10.1038/nrmicro1928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Alvarez DE, De Lella Ezcurra AL, Fucito S, Gamarnik AV. Role of RNA structures present at the 3’UTR of dengue virus on translation, RNA synthesis, and viral replication. Virology. 2005;339(2):200–212. doi: 10.1016/j.virol.2005.06.009. [DOI] [PubMed] [Google Scholar]
- 6.Lodeiro MF, Filomatori CV, Gamarnik AV. Structural and functional studies of the promoter element for dengue virus RNA replication. J Virol. 2009;83(2):993–1008. doi: 10.1128/JVI.01647-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Filomatori CV, Iglesias NG, Villordo SM, Alvarez DE, Gamarnik AV. RNA sequences and structures required for the recruitment and activity of the dengue virus polymerase. J Biol Chem. 2011;286(9):6929–6939. doi: 10.1074/jbc.M110.162289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gritsun TS, Gould EA. Origin and evolution of 3’UTR of flaviviruses: Long direct repeats as a basis for the formation of secondary structures and their significance for virus transmission. Adv Virus Res. 2007;69:203–248. doi: 10.1016/S0065-3527(06)69005-2. [DOI] [PubMed] [Google Scholar]
- 9.Ameres SL, Zamore PD. Diversifying microRNA sequence and function. Nat Rev Mol Cell Biol. 2013;14(8):475–488. doi: 10.1038/nrm3611. [DOI] [PubMed] [Google Scholar]
- 10.Kincaid RP, Sullivan CS. Virus-encoded microRNAs: An overview and a look to the future. PLoS Pathog. 2012;8(12):e1003018. doi: 10.1371/journal.ppat.1003018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bartel DP. MicroRNAs: Target recognition and regulatory functions. Cell. 2009;136(2):215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Westholm JO, Ladewig E, Okamura K, Robine N, Lai EC. Common and distinct patterns of terminal modifications to mirtrons and canonical microRNAs. RNA. 2012;18(2):177–192. doi: 10.1261/rna.030627.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yang JS, Lai EC. Alternative miRNA biogenesis pathways and the interpretation of core miRNA pathway mutants. Mol Cell. 2011;43(6):892–903. doi: 10.1016/j.molcel.2011.07.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rouha H, Thurner C, Mandl CW. Functional microRNA generated from a cytoplasmic RNA virus. Nucleic Acids Res. 2010;38(22):8328–8337. doi: 10.1093/nar/gkq681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shapiro JS, Varble A, Pham AM, Tenoever BR. Noncanonical cytoplasmic processing of viral microRNAs. RNA. 2010;16(11):2068–2074. doi: 10.1261/rna.2303610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rosewick N, et al. Deep sequencing reveals abundant noncanonical retroviral microRNAs in B-cell leukemia/lymphoma. Proc Natl Acad Sci USA. 2013;110(6):2306–2311. doi: 10.1073/pnas.1213842110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hess AM, et al. Small RNA profiling of Dengue virus-mosquito interactions implicates the PIWI RNA pathway in anti-viral defense. BMC Microbiol. 2011;11:45. doi: 10.1186/1471-2180-11-45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Brackney DE, et al. C6/36 Aedes albopictus cells have a dysfunctional antiviral RNA interference response. PLoS Negl Trop Dis. 2010;4(10):e856. doi: 10.1371/journal.pntd.0000856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chotkowski HL, et al. West Nile virus infection of Drosophila melanogaster induces a protective RNAi response. Virology. 2008;377(1):197–206. doi: 10.1016/j.virol.2008.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Scott JC, et al. Comparison of dengue virus type 2-specific small RNAs from RNA interference-competent and -incompetent mosquito cells. PLoS Negl Trop Dis. 2010;4(10):e848. doi: 10.1371/journal.pntd.0000848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pijlman GP, et al. A highly structured, nuclease-resistant, noncoding RNA produced by flaviviruses is required for pathogenicity. Cell Host Microbe. 2008;4(6):579–591. doi: 10.1016/j.chom.2008.10.007. [DOI] [PubMed] [Google Scholar]
- 22.Ye X, Liu Q. Expression, purification, and analysis of recombinant Drosophila Dicer-1 and Dicer-2 enzymes. Methods Mol Biol. 2008;442:11–27. doi: 10.1007/978-1-59745-191-8_2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kozomara A, Griffiths-Jones S. miRBase: Integrating microRNA annotation and deep-sequencing data. Nucleic Acids Res. 2011;39(Database issue):D152–D157. doi: 10.1093/nar/gkq1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shapiro JS, Langlois RA, Pham AM, Tenoever BR. Evidence for a cytoplasmic microprocessor of pri-miRNAs. RNA. 2012;18(7):1338–1346. doi: 10.1261/rna.032268.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hussain M, et al. West Nile virus encodes a microRNA-like small RNA in the 3′ untranslated region which up-regulates GATA4 mRNA and facilitates virus replication in mosquito cells. Nucleic Acids Res. 2012;40(5):2210–2223. doi: 10.1093/nar/gkr848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Förstemann K, Horwich MD, Wee L, Tomari Y, Zamore PD. Drosophila microRNAs are sorted into functionally distinct argonaute complexes after production by dicer-1. Cell. 2007;130(2):287–297. doi: 10.1016/j.cell.2007.05.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cifuentes D, et al. A novel miRNA processing pathway independent of Dicer requires Argonaute2 catalytic activity. Science. 2010;328(5986):1694–1698. doi: 10.1126/science.1190809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Maurin T, Cazalla D, Yang S, Jr, Bortolamiol-Becet D, Lai EC. RNase III-independent microRNA biogenesis in mammalian cells. RNA. 2012;18(12):2166–2173. doi: 10.1261/rna.036194.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Barth S, et al. Epstein-Barr virus-encoded microRNA miR-BART2 down-regulates the viral DNA polymerase BALF5. Nucleic Acids Res. 2008;36(2):666–675. doi: 10.1093/nar/gkm1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Broekema NM, Imperiale MJ. miRNA regulation of BK polyomavirus replication during early infection. Proc Natl Acad Sci USA. 2013;110(20):8200–8205. doi: 10.1073/pnas.1301907110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sullivan CS, Ganem D. MicroRNAs and viral infection. Mol Cell. 2005;20(1):3–7. doi: 10.1016/j.molcel.2005.09.012. [DOI] [PubMed] [Google Scholar]
- 32.Sullivan CS, et al. Murine Polyomavirus encodes a microRNA that cleaves early RNA transcripts but is not essential for experimental infection. Virology. 2009;387(1):157–167. doi: 10.1016/j.virol.2009.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hussain M, Taft RJ, Asgari S. An insect virus-encoded microRNA regulates viral replication. J Virol. 2008;82(18):9164–9170. doi: 10.1128/JVI.01109-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Samols MA, et al. Identification of cellular genes targeted by KSHV-encoded microRNAs. PLoS Pathog. 2007;3(5):e65. doi: 10.1371/journal.ppat.0030065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Godshalk SE, Bhaduri-McIntosh S, Slack FJ. Epstein-Barr virus-mediated dysregulation of human microRNA expression. Cell Cycle. 2008;7(22):3595–3600. doi: 10.4161/cc.7.22.7120. [DOI] [PubMed] [Google Scholar]
- 36.Pfeffer S, et al. Identification of virus-encoded microRNAs. Science. 2004;304(5671):734–736. doi: 10.1126/science.1096781. [DOI] [PubMed] [Google Scholar]
- 37.Wu Y-L, et al. A non-coding RNA of insect HzNV-1 virus establishes latent viral infection through microRNA. Sci Rep. 2011;1:60. doi: 10.1038/srep00060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kang K, et al. A novel real-time PCR assay of microRNAs using S-Poly(T), a specific oligo(dT) reverse transcription primer with excellent sensitivity and specificity. PLoS ONE. 2012;7(11):e48536. doi: 10.1371/journal.pone.0048536. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.