Significance
Understanding loci nominated by genome-wide association studies (GWASs) is challenging. Here, we show, using the specific example of Parkinson disease, that identification of protein–protein interactions can help determine the most likely candidate for several GWAS loci. This result illustrates a significant general principle that will likely apply across multiple diseases.
Keywords: BAG5, GAK, trans-Golgi, autophagy
Abstract
Mutations in leucine-rich repeat kinase 2 (LRRK2) cause inherited Parkinson disease (PD), and common variants around LRRK2 are a risk factor for sporadic PD. Using protein–protein interaction arrays, we identified BCL2-associated athanogene 5, Rab7L1 (RAB7, member RAS oncogene family-like 1), and Cyclin-G–associated kinase as binding partners of LRRK2. The latter two genes are candidate genes for risk for sporadic PD identified by genome-wide association studies. These proteins form a complex that promotes clearance of Golgi-derived vesicles through the autophagy–lysosome system both in vitro and in vivo. We propose that three different genes for PD have a common biological function. More generally, data integration from multiple unbiased screens can provide insight into human disease mechanisms.
Genetics contribute to the pathogenesis of Parkinson disease (PD) in two ways. Mutations in several genes can cause inherited PD (1), and risk factor variants contribute to the risk of sporadic PD (2). Some genes contribute to both mechanisms. These pleomorphic risk loci (3) include genes that encode α-synuclein and leucine-rich repeat kinase 2 (LRRK2) (4). However, risk factors for sporadic PD identified by genome-wide association studies (GWASs) (5–9) actually nominate large genomic loci with multiple candidate genes (10). These loci may include variants that change amino acids or affect disease risk through gene expression (11). Also, whether all of the genes for PD converge on a small number of biological pathways is unknown (1). It is, therefore, important to develop unbiased approaches that would resolve whether genes for PD have similar biological functions and understand the mechanism(s) of disease risk. Here, we examine one genetic cause of PD (LRRK2) and show that identifying protein interaction partners can clarify disease mechanisms.
Results
LRRK2 contains several protein–protein interaction motifs (shown diagrammatically in Fig. 1A). To identify protein interactions, we probed protein–protein arrays with biotinylated Glutathione-S transferase (GST)-LRRK2970–2527, which was kinase-active and therefore, likely folded (SI Appendix, Fig. S1). Biotinylated GST was used as a negative control to exclude false positives that might nonspecifically bind to many proteins. We also included arrays where LRRK2 was coincubated with GDP or GMPPcP. These experiments recovered several candidate interactors, including three members of the Bcl2-associated athanogene (BAG) domain cochaperones (12) (SI Appendix, Table S1 and Datasets S1 and S2). Of these candidates, we chose to focus on BAG5, because it is expressed in neurons and enhances dopaminergic neuronal death (13).
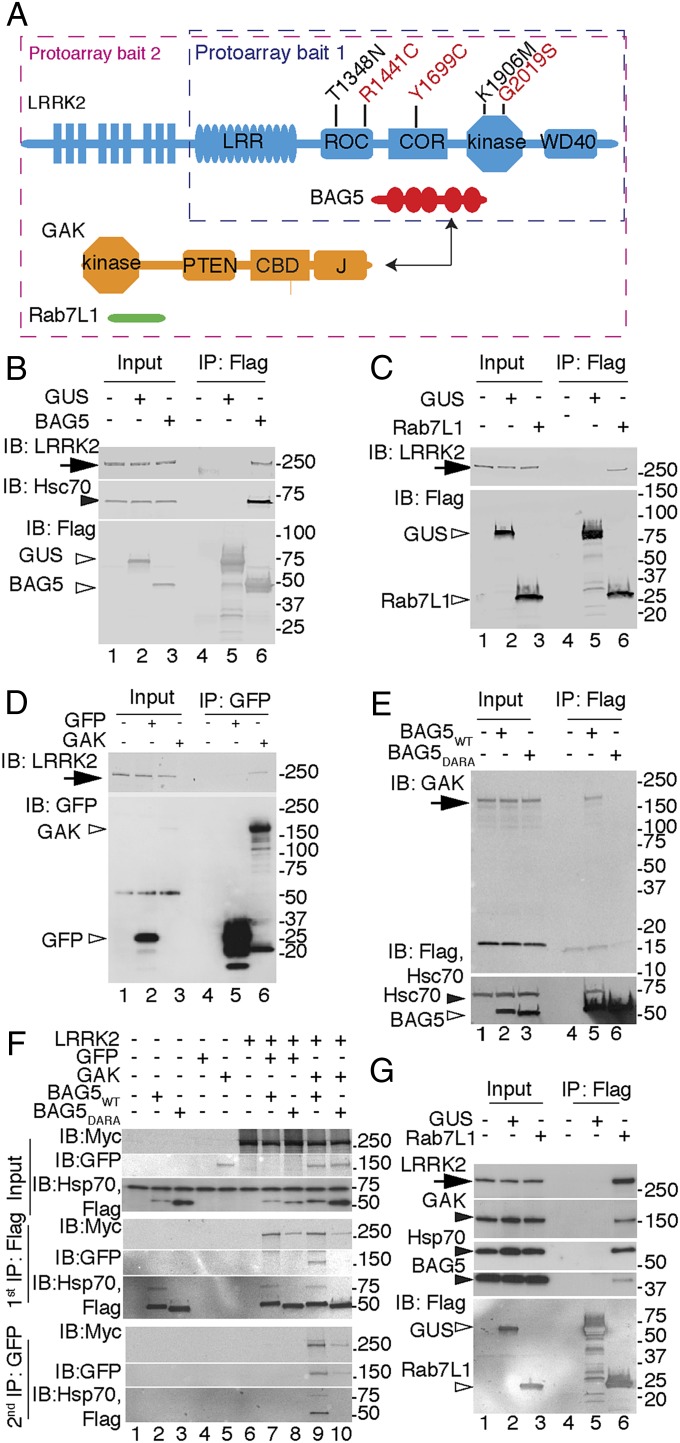
Fig. 1.
A LRRK2 protein complex. (A) Ideograms of LRRK2 and candidate protein interactors. LRRK2 shows the location of the mutations used in this study, including pathogenic mutations (red) and T1348N or K1906M hypothesis-testing mutants (black). Domains include leucine-rich repeat (LRR), Ras of complex protein (ROC), and COR as well as kinase and WD40 domains. The protoarray baits used in each experiment are boxed. BAG5 has five BAG domains. GAK has PTEN-like domain (PTEN), Clathrin binding domain (CBD), and J domain. Rab7L1 has a single Ras-like domain. (B–E) Single-protein interactions. Coimmunoprecipitations using antibodies to (B and C) Flag or (D) GFP from cell lysates expressing (B) BAG5, (C) Rab7L1, or (D) GAK with (B) mock-transfected cells, (C) Flag-β-glucoronidase (GUS), or (D) GFP as negative controls probed for endogenous LRRK2 (arrows), Hsc70 (closed arrowhead), and tagged proteins (open arrowheads). IB, immunoblotting; IP, immunoprecipitation. (E) IP of BAG5 probed for endogenous GAK (arrowheads). (F and G) Cocomplex formation between LRRK2, GAK, BAG5, and Rab7L1. (F) Lysates were immunoprecipitated first with Flag for BAG5 and then with GFP for GAK, and they were probed for LRRK2 and interacting partners. (G) Flag-tagged Rab7L1 complexes probed for endogenous LRRK2, GAK, Hsc70, and Bag5. For all panels, markers on the right of the blots are in kilodaltons, and results are representative of at least three independent experiments.
Some known interactors of LRRK2, such as 14-3-3 proteins (14, 15), could not have been recovered with the truncated LRRK2 probe that lacks the S910/S935 14-3-3 binding sites. Therefore, we repeated the experiments with full-length Flag-tagged LRRK2 (16) with Flag-tagged green fluorescent protein (GFP) as a negative control. We confirmed interaction with the BAG cochaperones (SI Appendix, Table S2 and Dataset S3) and recovered 14-3-3 proteins as well as another well-characterized LRRK2 interactor, STUB1/CHIP (17). This experiment also yielded several additional candidates, including two candidates that are in risk loci for sporadic PD: Rab7L1 (5, 6) and GAK (7, 8). Because of our interest in the genetic basis of PD, we chose to consider Rab7L1 (RAB7, member RAS oncogene family-like 1) and Cyclin-G–associated kinase (GAK) in subsequent experiments.
We first validated individual interactions between LRRK2 and each candidate. Flag-tagged BAG5 coimmunoprecipitated endogenous LRRK2 (Fig. 1B), and reciprocally, endogenous BAG5 coimmunoprecipitated with LRRK2 in stable cell lines (SI Appendix, Fig. S2A). Endogenous BAG5 coimmunoprecipitated with endogenous LRRK2 in the mouse brain (SI Appendix, Fig. S2B). Similarly, we found that endogenous LRRK2 coimmunoprecipitated Flag-tagged Rab7L1 (Fig. 1C) or GFP-tagged GAK (Fig. 1D). Collectively, these results show that LRRK2 can interact with three candidate proteins (BAG5, Rab7L1, and GAK), including endogenous protein in cells and the brain.
It is likely that different proteins have different sequence requirements for binding LRRK2. For example, when 14-3-3 proteins were dissociated from LRRK2 by treating with the LRRK2 kinase inhibitor LRRK2-IN1 (18), endogenous BAG5 remained bound (SI Appendix, Fig. S2C). To address the sequence requirements for BAG5-LRRK2 interactions further, we used a DARA mutant BAG5, where key aspartate and arginine residues in the BAG domains were replaced with alanine, diminishing heat shock cognate protein 70 (Hsc70) binding (13). DARA-BAG5 had lower binding to LRRK2 than WT BAG5 (SI Appendix, Fig. S2D) [mean intensity of BAG5 relative to LRRK2 (± SEM) of 0.49 ± 0.023 for WT and 0.21 ± 0.037 for DARA; P = 0.0097, two-tailed paired t test, n = 3 independent experiments], showing that recruitment of Hsc70/heat shock protein 70 (Hsp70) stabilizes the BAG5-LRRK2 complex.
Considering LRRK2, the C-terminal Ras of complex protein (COR) domain was sufficient for interaction with BAG5 (SI Appendix, Fig. S2E). LRRK1, with 46% similarity in the COR domain to LRRK2, also interacted with BAG5 (SI Appendix, Fig. S2F). These results show that the BAG5-Hsc70/Hsp70 complex is recruited to the COR domain of LRRK proteins. Adding recombinant BAG5 to LRRK2 did not affect kinase activity (SI Appendix, Fig. S2G).
To confirm the interactions using an additional independent technique, GST-BAG5 was used to pull down myc-LRRK2 from cells (SI Appendix, Fig. S3A). This technique also confirmed that the COR domain of LRRK2 was sufficient for BAG5 interaction (SI Appendix, Fig. S3B). Therefore, the LRRK2-BAG5 interactions were supported by protoarrays, coimmunoprecipitation, and GST pull downs.
Some LRRK2 interactors are also substrates for LRRK2 (19, 20). When testing whether BAG5 was a LRRK2 substrate, we recovered a copurifying autophosphorylated protein of ∼150 kDa (SI Appendix, Fig. S4 A and B). Given the results in the second protoarray experiment with full-length LRRK2, we considered GAK (143 kDa) as a candidate for this unknown kinase. Supporting this hypothesis, we found that GAK coimmunoprecipitated with WT BAG5 but not DARA mutant, suggesting that this interaction is also Hsp70-mediated (Fig. 1E). Furthermore, endogenous BAG5 was coimmunoprecipitated with GAK in the mouse brain (SI Appendix, Fig. S4C). Deletion of the PTEN-like domain of GAK disrupted and expression of the PTEN-like domain was sufficient for interaction with both BAG5 and LRRK2 (SI Appendix, Fig. S4D).
These results do not discriminate whether BAG5 and LRRK2 bind independently to GAK or whether the three proteins are present in a cocomplex. To address these possibilities, we performed sequential coimmunoprecipitation experiments. GAK was recovered from BAG5 purification like in the single coimmunoprectipitation; then, LRRK2 was coimmunprecipitated from the GAK-BAG5 complexes (Fig. 1F). Furthermore, the cocomplex was more easily recovered with WT than DARA BAG5, again suggesting that Hsp70/Hsc70 stabilizes the overall assembly.
To address whether the interactions require all components or can be formed in the absence of any one member, we knocked down each component in turn and performed coimmunoprecipitation experiments for the remaining partners. These results (SI Appendix, Fig. S4 E–G) showed that interactions remained even when BAG5, GAK, or LRRK2 was depleted from cells.
We next considered whether Rab7L1 might be part of the same protein complex. Rab7L1 was coimmunoprecipitated with LRRK2, and this binding was not significantly altered by mutations in LRRK2 (SI Appendix, Fig. S5 A and B). However, deletion of the N-terminal HEAT [Huntingtin, elongation factor 3 (EF3), protein phosphatase 2A (PP2A), TOR1] region of LRRK2 weakened interaction dramatically (SI Appendix, Fig. S5 C and D).
Rab7L1 is a single-domain protein, and therefore, we did not generate truncation constructs that would likely be misfolded. Instead, we asked whether LRRK2 interactions depended on the nucleotide state of Rab7L1. To this end, we generated T21N and Q67L mutants in Rab7L1 that were predicted to lack GTP binding and show impaired GTP hydrolysis based on structural modeling (SI Appendix, Fig. S6 A–C). As expected, T21N was deficient in GTP binding (SI Appendix, Fig. S6D) [F(4,15), P = 4.5 × 10−13, one-way ANOVA, n = 4 independent experiments]. Unexpectedly, Q67L had an ∼10-fold higher GTP/GDP dissociation rate than WT Rab7L1 (SI Appendix, Fig. S6 E and F), and both mutants showed altered mobility on size exclusion chromatography (SI Appendix, Fig. S5 G and H). Therefore, both Rab7L1 mutants act as loss-of-function variants, likely because of failure to retain GTP. Both T21N and Q67L Rab7L1 showed enhanced interaction with LRRK2 (SI Appendix, Fig. S6 I and J) [one-way ANOVA; F(4,10), P = 4.71 × 10−9, n = 3 independent experiments], suggesting that Rab7L1 binds LRRK2 in both the GTP- and GDP-bound states. However, GTP binding seemed to be important for some functions of Rab7L1, because both Rab7L1 mutants showed diminished (Q67L) or abolished (T21N) Golgi localization (SI Appendix, Fig. S6K). Endogenous Rab7L1 could simultaneously coimmunoprecipitate endogenous LRRK2, GAK, Hsp70, and BAG5 (Fig. 1G). Collectively, these results suggest that all five proteins are in a single complex.
We next asked whether the formation of a single-protein complex implied that these proteins have similar effects in cells. We used neurite shortening, which is exaggerated by pathogenic LRRK2 mutations (21–24). WT BAG5 caused neurite shortening, but this effect was diminished with the DARA mutant BAG5 (SI Appendix, Fig. S7 A–C) [F(2,158) = 109, P < 2 × 10−16 by one-way ANOVA; n > 140 cells across three independent experiments]. Expression of full-length GAK, but not the deletion constructs incapable of binding LRRK2 (SI Appendix, Fig. S7D) [F(3,215) = 5.97, P = 6.3 × 10−4 by one-way ANOVA; n > 49 cells per construct across two independent experiments), and expression of WT Rab7L1, but not the GTP binding-deficient Q67L variant, also decreased neurite length (SI Appendix, Fig. S7E) [F(2,435) = 55, P < 2 × 10−16 by one-way ANOVA; n > 125 cells across three independent experiments]. The effect of Rab7L1 was diminished in LRRK2 KO neurons (SI Appendix, Fig. S7F) [two-way ANOVA, F(1,281) = 21, P = 0.00076 for genotype; F(1,281) = 2.7, P = 0.06 for Rab7L1 variant; F(1,281) = 21, P = 2.04 × 10−5 for interaction; n > 70 cells counted per condition), suggesting that the effects of Rab7L1 require expression of LRRK2.
We next asked where each protein might be in the cell. LRRK2 was present in microsomes as reported (25), and endogenous BAG5 and GAK were also present in the same subcellular fractions (SI Appendix, Fig. S8A). To provide additional resolution, we expressed each interactor as tagged proteins in neurons, allowing us to use the same antibody for visualization. LRRK2 was cytosolic and vesicular (18, 26), and BAG5 was cytosolic and nuclear, whereas GAK and Rab7L1 were largely vesicular (SI Appendix, Fig. S8B). When coexpressed, LRRK2 and GAK showed vesicular colocalization. WT Rab7L1 promoted vesicular LRRK2 localization, but the Q67L Rab7L1 variant that was deficient in GTP binding prevented this effect (SI Appendix, Fig. S8C). These results show that a cocomplex of these proteins may form in vesicular compartments of the cell and that the effects require GTP binding by Rab7L1.
To further understand the possible function of the complex, we considered what type of vesicular compartments to which it might localize. Both Rab7L1 (27, 28) and GAK (29, 30) are reported to localize to the trans-Golgi network (TGN). WT Rab7L1 expressed alone (SI Appendix, Fig. S6K) or with LRRK2 (SI Appendix, Fig. S9 A and B) partially colocalized with markers of the TGN but not retromer (31) (SI Appendix, Fig. S9C). When we coexpressed both Rab7L1 and LRRK2, TGN staining was diminished and therefore, appeared clustered in the perinuclear area. Quantification showed that this phenotype was stronger for TGN (TGN46, trans-Golgi network integral membrane protein 2) than cis-Golgi (GM130, Golgin subfamily A member 2) markers (SI Appendix, Fig. S6 A and B) [F(3,8) = 56.9, P = 9.7 × 10−6 for TGN46; F(3,8) = 9.5, P = 0.0051 for GM130; one-way ANOVA). These results suggest that Rab7L1 directs LRRK2 to TGN-derived vesicles.
In LRRK2-transfected neurons, the TGN was again clustered, or dramatically, only residual staining was visible (Fig. 2A). Counting cells showed that there was a significant change in the proportions of cells with clustered or cleared Golgi after expression of LRRK2 or each interaction partner—BAG5, Rab7L1, or GAK (Fig. 2B, P values) (two-sided Fisher exact test corrected for the number of tests performed). Validating these results biochemically, HEK cells expressing LRRK2 had lower amounts of the TGN marker TGN46 (SI Appendix, Fig. S10 A and B) (P = 0.00038, t test with Welch correction, n = 5 samples) but not the cis-Golgi marker GM130 (SI Appendix, Fig. S10C) (P = 0.07). Considering the timing of these events, the recruitment of LRRK2 to TGN46-positive vesicles by Rab7L1 was complete by 24 h after transfection, but clearance of Golgi increased up to 72 h (SI Appendix, Fig. S11). Therefore, physical recruitment of LRRK2 to the TGN precedes removal of TGN-derived vesicles.
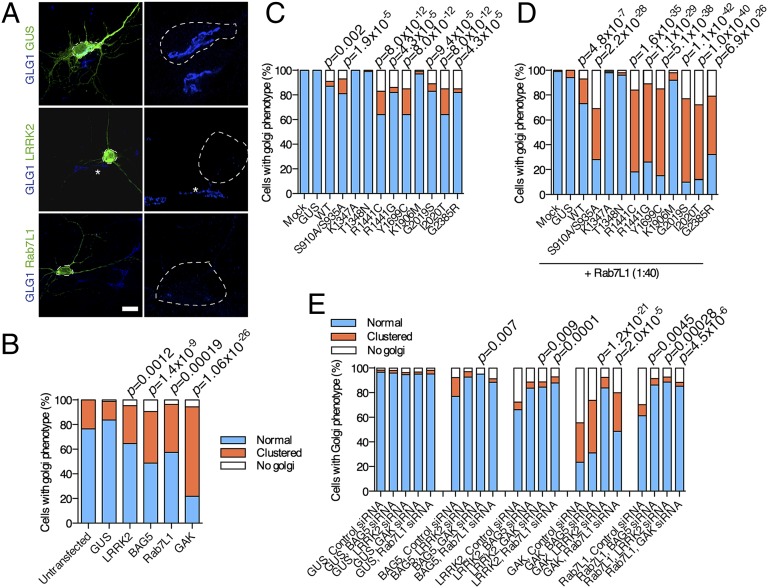
Fig. 2.
(A) LRRK2 complex and LRRK2 mutations promote clearance of trans-Golgi–derived vesicles. Clustering and clearance of trans-Golgi by LRRK2 and interacting partners. Primary neurons were transfected with the indicated constructs and stained for transfection markers (green) and trans-Golgi (GLG1; blue). Right shows higher magnification of the circled neurons, whereas adjacent cells (asterisks) show normal Golgi morphology, including clustered Golgi in Rab7L1 (Lower Right) transfections and a LRRK2-transfected neuron with minimal residual Golgi staining (Center Right). (B) Blinded counts of cells with normal Golgi (blue), clustered Golgi (orange), or minimal or no apparent Golgi staining (white) as a proportion of all counted cells (n > 50 cells in three independent cultures per construct, P values are by Fisher exact test for proportions against GUS-transfected cultures corrected for the number of tests applied). (C and D) Pathogenic mutations in LRRK2 increase clearance of the trans-Golgi network. HEK cells were transfected with indicated LRRK2 variants or GUS as a negative control and stained with antibodies to TGN46. Counting of different Golgi phenotypes after transfections with different LRRK2 variants (C) without or (D) with cotransfection of Rab7L1 and statistical analysis were performed as in B. (E) Minimal functional complex includes LRRK2, BAG5 GAK, and Rab7L1. Cells were transfected with GUS (negative control for transfection), BAG5, LRRK2, GAK, or Rab7L1 and simultaneously treated with siRNA against the other components. Counting of different Golgi phenotypes after transfections with different combinations was performed as in C and D.
The clearance of the TGN potentially provides a cellular measure of the function of LRRK2 and binding partners. We next asked whether point mutations in LRRK2 affect Golgi clearance, thus allowing us to infer what functional roles of LRRK2 are important. All pathogenic mutations in LRRK2, including those mutations that increase kinase activity (G2019S) or decrease GTP turnover (R1441C and Y1699C), enhanced Golgi clearance (Fig. 2C, P values). However, hypothesis-testing mutations that diminish GTP binding (T1348N) or are kinase-inactive (K1906M) did not support Golgi clearance. The effects of LRRK2 mutations on Golgi clearance were enhanced by Rab7L1 (Fig. 2D, P values). These results suggest that the known enzymatic functions of LRRK2 (GTP binding and kinase activity) are important in the clearance of Golgi by a LRRK2 complex.
To define the minimum functional unit of the complex, we transfected cells with BAG5, LRRK2, GAK, or Rab7L1 and knocked down each of the other components in turn. We found that knockdown of any single component was able to rescue the effects of increased expression of other components on Golgi turnover (Fig. 2E, P values). Given that knockdown of each component did not disrupt protein interactions (SI Appendix, Fig. S4 E–G), these results show that, even if a complex forms, full function requires all components.
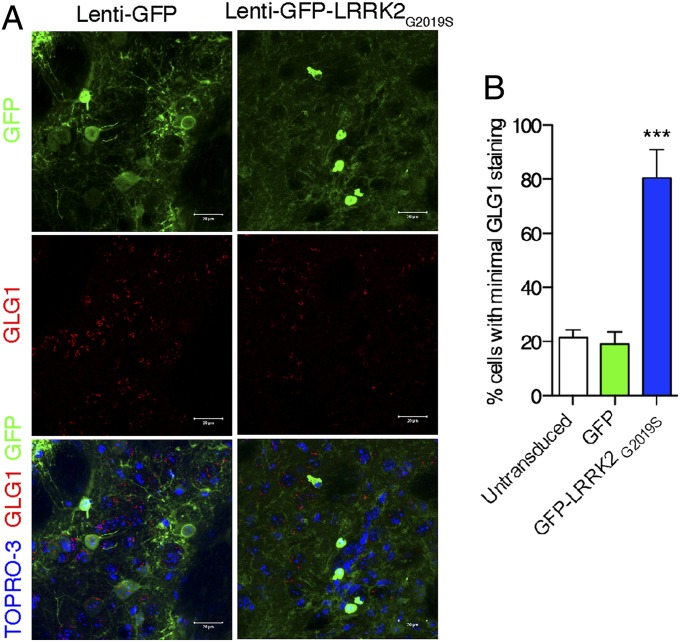
To confirm these results in vivo, we used lentiviral vectors to express enhanced green fluorescent protein (eGFP)-tagged G2019S LRRK2 in the striatum of adult mice. We found more cells with minimal staining for GLG1, a TGN marker suitable for immunostaining in rodent tissue, after expression of mutant LRRK2 compared with controls (Fig. 3) [F(2,12) = 131.4, P = 6.93 × 10−9; one-way ANOVA, n = 3 animals per group]. These results suggest that increased expression of LRRK2 has similar functional effects in the mouse brain as in cellular systems.
Fig. 3.
Acute expression of mutant LRRK2 in adult mouse brain results in diminished staining of TGN markers in vivo. (A) Mouse striatum sections from animals 2 wk after injection with lentiviral vectors expressing either (Left) eGFP or (Right) G2019S mutant eGFP-LRRK2 stained for GFP (green), GLG1 (red), and TOPRO3 (blue in Bottom). (B) Counting of cells with minimal GLG1 staining in animals transduced with the vectors, with untransduced cells in the same sections also counted as controls. Error bars show SEM. ***P < 0.001 (Tukey posthoc test from one-way ANOVA compared with eGFP control).
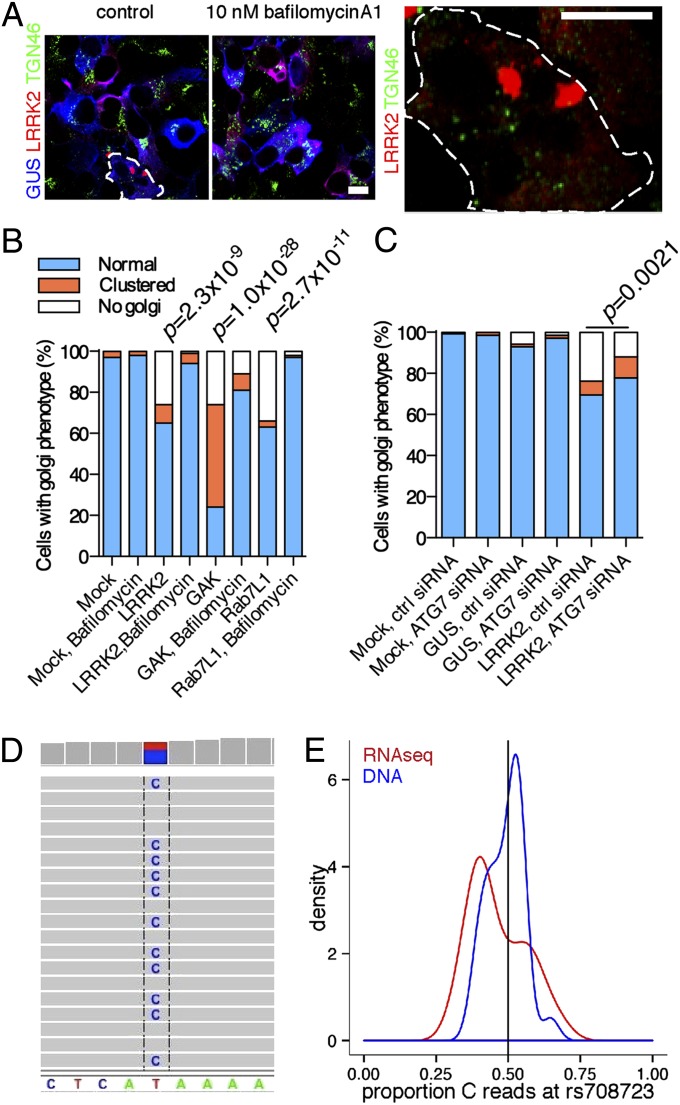
We next sought to understand the mechanisms involved in TGN turnover. The above results are reminiscent of the effects of expression and activity of another Parkinson-related protein, Parkin, that promotes removal of mitochondria by autophagy (32). LRRK2 has previously been reported to control autophagy in several systems (24, 33, 34). To test whether the autophagy–lysosome system contributed to LRRK2-dependent phenotypes, we treated cells with bafilomycin-A1, which prevents lysosomal acidification. Bafilomycin-A1 blocked the effect of LRRK2 (Fig. 4 A and B, P values) and also prevented the turnover of TGN46-positive vesicles when LRRK2 was cotransfected with GAK (SI Appendix, Fig. S12A).
Fig. 4.
Mechanism of turnover of Golgi-derived vesicles involves the autophagy–lysosomal system and is consistent with altered gene expression in the human brain. (A–C) Selective Golgi clearance involves the autophagy–lysosome system. (A) Cells coexpressing LRRK2 (red) and GUS (blue) stained for TGN46 (green) after treatment for 24 h with DMSO or 10 nM bafilomycin A1. A cell with only residual TGN46 is shown in Right. (B and C) Blinded counts of the proportions of Golgi phenotypes in cells treated with (B) bafilomycin A1 or (C) knockdown of Atg7. (D and E) Gene expression in the human brain. (D) Direct assay of rs708723 in Rab7L1 from the brain of an individual who was heterozygous at this marker using RNA-Seq. (E) Density plots for the proportion of reads with the C allele for RNA-Seq (red) and exome data for DNA (blue) in brains from individuals heterozygous for rs708723. There is departure from the expected proportion of 0.5 (black vertical line) for RNA-Seq data.
Asking whether the effects were specifically related to autophagy, we found that the effects of LRRK2 were diminished by siRNA against the autophagy protein ATG7 (Fig. 4C). Morphologically, cells expressing LRRK2 and treated with bafilomycin-A1 contained larger LRRK2-positive structures adjacent to lysosomes (SI Appendix, Fig. S12B). When cotransfected, both Rab7L1 and LRRK2 were found in swollen vesicles near lysosomes (SI Appendix, Fig. S12C). Some areas of TGN46 immunoreactivity were enclosed by swollen LRRK2/Rab7L1-positive vesicles (SI Appendix, Fig. S12D). Collectively, these results support the hypothesis that turnover of TGN vesicles depends on the autophagy–lysosome system.
The above results make the assumption that increasing the expression of LRRK2 or its binding partners models the direction of effect of genetic risk in PD. To evaluate whether expression of these genes might explain the risk of PD in the human brain, we assayed a series of postmortem samples (n ∼ 60) using exome sequencing and RNA-Seq. We examined the loci containing Rab7L1 (SI Appendix, Fig. S13A), GAK (SI Appendix, Fig. S13B), or LRRK2 (SI Appendix, Fig. S13C) for sequence variants. Although we reliably detected expression of LRRK2, GAK, and Rab7L1 in the human cerebral cortex, many pathogenic variants were too rare to be analyzed or in regions that did not code for RNA. However, rs708723 in the 3′ UTR of Rab7L1 could be identified directly from RNA-Seq data (Fig. 4D). There was lower expression of the C allele in RNA-Seq data in heterozygous carriers, and the proportions of C alleles in the aggregate counts were significantly different between exome and RNA-Seq data (Fig. 4E) (χ2 test, χ2 = 5.9, P = 0.015). The C allele of rs708723 was associated with a lower risk of PD (odds ratio = 0.914, P = 0.000554 under an additive model). Therefore, higher expression of normal sequence variants is a plausible mechanism for risk of PD, at least for Rab7L1.
Discussion
Here, we used an unbiased survey of protein interactions of LRRK2 to identify binding partners and show that these proteins form a cocomplex that promotes TGN removal by autophagy-dependent mechanisms. TGN clearance also occurs in the brain in vivo, and expression levels of some of the protein components are a reasonable mechanism for risk of sporadic PD. Our results overall lead us to propose that several PD genes act in a common pathway (SI Appendix, Fig. S14).
Because LRRK2 has several proposed interaction domains, it may act as a scaffold for multiple interaction partners. Although we recovered known direct binding partners of LRRK2 (15, 35) and inferred indirect interaction with Hsp70 (17), not all proposed literature candidates were identified. This result may indicate a false-negative rate with arrays, which do not include all splice variants of all proteins in the human genome or all posttranslational modifications. The approach used here should be combined with other methods, which has been suggested more generally for understanding protein function (36).
With these caveats, our approach led us to show that LRRK2 promotes the relocalization to and clearance of trans-Golgi–derived vesicles. This effect is presumably related to the known function of LRRK2 in autophagy (18, 24, 33, 37, 38) but can be tied mechanistically to disruption of TGN. Interestingly, autophagic vesicles may derive from the trans-Golgi network (37); hence, altered turnover of Golgi-derived vesicles would impact autophagy function over time, consistent with effects of LRRK2 deficiency in animal models (34).
Critically, this activity is enhanced by all pathogenic mutations in LRRK2. However, LRRK2 mutants do not alter binding to its partner proteins. This observation suggests that the enzymatic activity of LRRK2, including GTP binding and kinase activity, may be critical for activity of the complex. Supporting this idea, hypothesis-testing mutations lacking GTP binding or kinase activity do not promote clearance of the TGN-derived structures. It is possible that, after being bound, protein interaction partners also influence either LRRK2 kinase activity or GTP affinity. Defining the stoichiometry of the complex and effects on LRRK2 is an important future goal.
The other proteins each seem to be required for LRRK2 to affect Golgi clearance by autophagy. We have shown that removal of BAG5, GAK, or Rab7L1 does not prevent LRRK2 binding with other partners, implying that formation of a partial complex is not sufficient for Golgi clearance. Within the complex, we speculate that each protein may play a specific role. It is of interest that Rab7L1 is localized to the trans-Golgi network, likely in its GTP-bound form, where it might recruit LRRK2 and other components to cooperatively cause TGN to be engulfed by the autophagosomes. The presence of BAG5 and Hsp70/Hsc70 suggests that this process requires remodeling of proteins or protein complexes. GAK is also known to cooperate to remodel clathrin complexes (39).
Our results have the broadest implications for mechanisms of understanding disease risk for loci identified by GWASs. Higher expression of Rab7L1 is associated with higher risk of PD in humans, expression quantitative trait locus identified by microarrays, thus providing a reasonable mechanism for the action of the WT allele. We cannot exclude that other genes at each GWAS locus may contribute to disease risk, because several of these loci are complex genetically (9). However, identification of GAK and Rab7L1 as binding partners of LRRK2 from an unbiased screen raises the probability that these genes explain some of the GWAS signal. This concept has important implications for other diseases that have been studied by GWASs, because it supports the idea that integrating datasets from multiple unbiased approaches leads to unexpected insights into human diseases.
Materials and Methods
Detailed materials and methods are included in SI Appendix. Briefly, identification of protein interactors was achieved by screening two batches of Protoarrays (Invitrogen) with either truncated GST-LRRK2970–2527 (Invitrogen) or full-length protein prepared as described (16). Protein interactions were validated using coimmunoprecipitation in HEK293FT cells or mouse brain and also GST pull downs in cells or brain. Subcellular fractionation was performed as described (25) using LRRK2-transfected HEK293FT cells. Protein localization and effects on turnover of TGN were performed using primary postnatal cortical neurons with calcium–phosphate transfection (40) or in HEK293FT cells. All counting experiments were performed by an operator blinded to the transfection construct used; details of blinding and statistical approaches are included in SI Appendix.
Supplementary Material
Acknowledgments
We thank the Genomic Technologies Section at the National Institute of Allergy and Infectious Diseases for help with protein array scans. This research was supported, in part, by the Intramural Research Program of the National Institutes of Health, National Institute on Aging; the Michael J. Fox Foundation for Parkinson’s Disease (V.B., J.-M.T., and E.G.); KU Leuven Grant OT/08/052A (to V.B.); Fonds Wetenschappelijk Onderzoek (FWO)-Vlaanderen Project G.0666.09 (to V.B.); Agentschap voor Innovatie door Wetenschap en Technologie-Vlaanderen Grants SBO/80020 (to V.B.) and SBO/100042 (to V.B.); an FWO-Vlaanderen postdoctoral fellowship (to J.-M.T.); an FWO-Vlaanderen PhD fellowship (to E.L.); the Fund Druwé-Eerdekens managed by the King Baudouin Foundation (J.-M.T.); the Rientro dei Cervelli Program (lncentivazione alla mobilità di studiosi stranieri e italiani residenti all’estero) from the Italian Ministry of Education, University and Research (E.G.), Fondazione Cariplo Grant 2011 0540 (to E.G.); and Fondazione Telethon Grant GGP12237 (to E.G.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
1Consortia members are listed in Supporting Information.
See Commentary on page 2402.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1318306111/-/DCSupplemental.
References
- 1.Cookson MR, Bandmann O. Parkinson’s disease: Insights from pathways. Hum Mol Genet. 2010;19(R1):R21–R27. doi: 10.1093/hmg/ddq167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lill CM, Bertram L. Towards unveiling the genetics of neurodegenerative diseases. Semin Neurol. 2011;31(5):531–541. doi: 10.1055/s-0031-1299791. [DOI] [PubMed] [Google Scholar]
- 3.Singleton A, Hardy J. A generalizable hypothesis for the genetic architecture of disease: Pleomorphic risk loci. Hum Mol Genet. 2011;20(R2):R158–R162. doi: 10.1093/hmg/ddr358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cookson MR. The role of leucine-rich repeat kinase 2 (LRRK2) in Parkinson’s disease. Nat Rev Neurosci. 2010;11(12):791–797. doi: 10.1038/nrn2935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Satake W, et al. Genome-wide association study identifies common variants at four loci as genetic risk factors for Parkinson’s disease. Nat Genet. 2009;41(12):1303–1307. doi: 10.1038/ng.485. [DOI] [PubMed] [Google Scholar]
- 6.Simón-Sánchez J, et al. Genome-wide association study reveals genetic risk underlying Parkinson’s disease. Nat Genet. 2009;41(12):1308–1312. doi: 10.1038/ng.487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.International Parkinson’s Disease Genomics Consortium (IPDGC) Wellcome Trust Case Control Consortium 2 (WTCCC2) A two-stage meta-analysis identifies several new loci for Parkinson’s disease. PLoS Genet. 2011;7(6):e1002142. doi: 10.1371/journal.pgen.1002142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Do CB, et al. Web-based genome-wide association study identifies two novel loci and a substantial genetic component for Parkinson’s disease. PLoS Genet. 2011;7(6):e1002141. doi: 10.1371/journal.pgen.1002141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nalls MA, et al. Imputation of sequence variants for identification of genetic risks for Parkinson’s disease: A meta-analysis of genome-wide association studies. Lancet. 2011;377(9766):641–649. doi: 10.1016/S0140-6736(10)62345-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ku CS, Loy EY, Pawitan Y, Chia KS. The pursuit of genome-wide association studies: Where are we now? J Hum Genet. 2010;55(4):195–206. doi: 10.1038/jhg.2010.19. [DOI] [PubMed] [Google Scholar]
- 11.Latourelle JC, Dumitriu A, Hadzi TC, Beach TG, Myers RH. Evaluation of Parkinson disease risk variants as expression-QTLs. PLoS One. 2012;7(10):e46199. doi: 10.1371/journal.pone.0046199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kabbage M, Dickman MB. The BAG proteins: A ubiquitous family of chaperone regulators. Cell Mol Life Sci. 2008;65(9):1390–1402. doi: 10.1007/s00018-008-7535-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kalia SK, et al. BAG5 inhibits parkin and enhances dopaminergic neuron degeneration. Neuron. 2004;44(6):931–945. doi: 10.1016/j.neuron.2004.11.026. [DOI] [PubMed] [Google Scholar]
- 14.Dzamko N, et al. Inhibition of LRRK2 kinase activity leads to dephosphorylation of Ser(910)/Ser(935), disruption of 14-3-3 binding and altered cytoplasmic localization. Biochem J. 2010;430(3):405–413. doi: 10.1042/BJ20100784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li X, et al. Phosphorylation-dependent 14-3-3 binding to LRRK2 is impaired by common mutations of familial Parkinson’s disease. PLoS One. 2011;6(3):e17153. doi: 10.1371/journal.pone.0017153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Civiero L, et al. Biochemical characterization of highly purified leucine-rich repeat kinases 1 and 2 demonstrates formation of homodimers. PLoS One. 2012;7(8):e43472. doi: 10.1371/journal.pone.0043472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ko HS, et al. CHIP regulates leucine-rich repeat kinase-2 ubiquitination, degradation, and toxicity. Proc Natl Acad Sci USA. 2009;106(8):2897–2902. doi: 10.1073/pnas.0810123106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Alegre-Abarrategui J, et al. LRRK2 regulates autophagic activity and localizes to specific membrane microdomains in a novel human genomic reporter cellular model. Hum Mol Genet. 2009;18(21):4022–4034. doi: 10.1093/hmg/ddp346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Stafa K, et al. GTPase activity and neuronal toxicity of Parkinson’s disease-associated LRRK2 is regulated by ArfGAP1. PLoS Genet. 2012;8(2):e1002526. doi: 10.1371/journal.pgen.1002526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Xiong Y, Yuan C, Chen R, Dawson TM, Dawson VL. ArfGAP1 is a GTPase activating protein for LRRK2: Reciprocal regulation of ArfGAP1 by LRRK2. J Neurosci. 2012;32(11):3877–3886. doi: 10.1523/JNEUROSCI.4566-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.MacLeod D, et al. The familial Parkinsonism gene LRRK2 regulates neurite process morphology. Neuron. 2006;52(4):587–593. doi: 10.1016/j.neuron.2006.10.008. [DOI] [PubMed] [Google Scholar]
- 22.Winner B, et al. Adult neurogenesis and neurite outgrowth are impaired in LRRK2 G2019S mice. Neurobiol Dis. 2011;41(3):706–716. doi: 10.1016/j.nbd.2010.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dächsel JC, et al. A comparative study of Lrrk2 function in primary neuronal cultures. Parkinsonism Relat Disord. 2010;16(10):650–655. doi: 10.1016/j.parkreldis.2010.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Plowey ED, Cherra SJ, 3rd, Liu Y-J, Chu CT. Role of autophagy in G2019S-LRRK2-associated neurite shortening in differentiated SH-SY5Y cells. J Neurochem. 2008;105(3):1048–1056. doi: 10.1111/j.1471-4159.2008.05217.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rudenko IN, et al. The G2385R variant of leucine-rich repeat kinase 2 associated with Parkinson’s disease is a partial loss-of-function mutation. Biochem J. 2012;446(1):99–111. doi: 10.1042/BJ20120637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Biskup S, et al. Localization of LRRK2 to membranous and vesicular structures in mammalian brain. Ann Neurol. 2006;60(5):557–569. doi: 10.1002/ana.21019. [DOI] [PubMed] [Google Scholar]
- 27.Helip-Wooley A, Thoene JG. Sucrose-induced vacuolation results in increased expression of cholesterol biosynthesis and lysosomal genes. Exp Cell Res. 2004;292(1):89–100. doi: 10.1016/j.yexcr.2003.09.003. [DOI] [PubMed] [Google Scholar]
- 28.MacLeod DA, et al. RAB7L1 interacts with LRRK2 to modify intraneuronal protein sorting and Parkinson’s disease risk. Neuron. 2013;77(3):425–439. doi: 10.1016/j.neuron.2012.11.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lee D-W, Wu X, Eisenberg E, Greene LE. Recruitment dynamics of GAK and auxilin to clathrin-coated pits during endocytosis. J Cell Sci. 2006;119(Pt 17):3502–3512. doi: 10.1242/jcs.03092. [DOI] [PubMed] [Google Scholar]
- 30.Kametaka S, et al. Canonical interaction of cyclin G associated kinase with adaptor protein 1 regulates lysosomal enzyme sorting. Mol Biol Cell. 2007;18(8):2991–3001. doi: 10.1091/mbc.E06-12-1162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cullen PJ, Korswagen HC. Sorting nexins provide diversity for retromer-dependent trafficking events. Nat Cell Biol. 2012;14(1):29–37. doi: 10.1038/ncb2374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Youle RJ, Narendra DP. Mechanisms of mitophagy. Nat Rev Mol Cell Biol. 2011;12(1):9–14. doi: 10.1038/nrm3028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gómez-Suaga P, et al. Leucine-rich repeat kinase 2 regulates autophagy through a calcium-dependent pathway involving NAADP. Hum Mol Genet. 2012;21(3):511–525. doi: 10.1093/hmg/ddr481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tong Y, et al. Loss of leucine-rich repeat kinase 2 causes age-dependent bi-phasic alterations of the autophagy pathway. Mol Neurodegener. 2012;7:2. doi: 10.1186/1750-1326-7-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nichols RJ, et al. 14-3-3 binding to LRRK2 is disrupted by multiple Parkinson’s disease-associated mutations and regulates cytoplasmic localization. Biochem J. 2010;430(3):393–404. doi: 10.1042/BJ20100483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ryan CJ, et al. High-resolution network biology: Connecting sequence with function. Nat Rev Genet. 2013;14(12):865–879. doi: 10.1038/nrg3574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Manzoni C, et al. Pathogenic Parkinson’s disease mutations across the functional domains of LRRK2 alter the autophagic/lysosomal response to starvation. Biochem Biophys Res Commun. 2013;441(4):862–866. doi: 10.1016/j.bbrc.2013.10.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ramonet D, et al. Dopaminergic neuronal loss, reduced neurite complexity and autophagic abnormalities in transgenic mice expressing G2019S mutant LRRK2. PLoS One. 2011;6(4):e18568. doi: 10.1371/journal.pone.0018568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lee DW, Zhao X, Zhang F, Eisenberg E, Greene LE. Depletion of GAK/auxilin 2 inhibits receptor-mediated endocytosis and recruitment of both clathrin and clathrin adaptors. J Cell Sci. 2005;118(Pt 18):4311–4321. doi: 10.1242/jcs.02548. [DOI] [PubMed] [Google Scholar]
- 40.Jiang M, Chen G. High Ca2+-phosphate transfection efficiency in low-density neuronal cultures. Nat Protoc. 2006;1(2):695–700. doi: 10.1038/nprot.2006.86. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.