Significance
Cell division is driven by the cell cycle machinery, which responds in an unknown fashion to the metabolic and nutrient state of the cell. We uncovered a previously unknown link between the cell cycle machinery and the tricarboxylic acid (TCA) cycle (also known as the Krebs cycle), which forms intermediates required for ATP production and other anabolic pathways. We show that in Caenorhabditis elegans embryos, down-regulation of the TCA cycle inhibits entry into the first mitotic division by preventing the removal of inhibitory phosphorylation on cyclin-dependent kinase 1. Our data suggest that in the one-cell stage embryo, the cell cycle machinery is sensitive to the metabolic state of the cell, a phenomenon that may also exist in mammalian embryos.
Abstract
The cell cycle is a highly regulated process that enables the accurate transmission of chromosomes to daughter cells. Here we uncover a previously unknown link between the tricarboxylic acid (TCA) cycle and cell cycle progression in the Caenorhabditis elegans early embryo. We found that down-regulation of TCA cycle components, including citrate synthase, malate dehydrogenase, and aconitase, resulted in a one-cell stage arrest before entry into mitosis: pronuclear meeting occurred normally, but nuclear envelope breakdown, centrosome separation, and chromosome condensation did not take place. Mitotic entry is controlled by the cyclin B–cyclin-dependent kinase 1 (Cdk1) complex, and the inhibitory phosphorylation of Cdk1 must be removed in order for the complex to be active. We found that following down-regulation of the TCA cycle, cyclin B levels were normal but CDK-1 remained inhibitory-phosphorylated in one-cell stage-arrested embryos, indicative of a G2-like arrest. Moreover, this was not due to an indirect effect caused by checkpoint activation by DNA damage or replication defects. These observations suggest that CDK-1 activation in the C. elegans one-cell embryo is sensitive to the metabolic state of the cell, and that down-regulation of the TCA cycle prevents the removal of CDK-1 inhibitory phosphorylation. The TCA cycle was previously shown to be necessary for the development of the early embryo in mammals, but the molecular processes affected were not known. Our study demonstrates a link between the TCA cycle and a specific cell cycle transition in the one-cell stage embryo.
The developmental program of any organism must be precisely executed. In Caenorhabditis elegans embryos, immediately after fertilization, two pronuclei form at opposite poles of the embryo: one containing the maternal chromosomes and the other containing the paternal ones (1, 2). These pronuclei then move toward each other, and at the same time centrosomes separate and begin to assemble a spindle. After pronuclear meeting, the cell enters its first mitosis, resulting in nuclear envelope breakdown, chromatin condensation, and the subsequent alignment of chromosomes on the metaphase plate, followed by chromosome segregation (2). Entry into mitosis depends on the mitotic cyclin B–cyclin-dependent kinase 1 (Cdk1) complex. The activity of this complex is regulated by both cyclin B levels and regulatory phosphorylation of Cdk1. In particular, Cdk1 activity is inhibited by Wee1 phosphorylation, which is removed at the onset of mitosis by the Cdc25 phosphatase (3, 4). Cdk1 activation is also subjected to various checkpoints that inhibit mitotic progression in the presence of intracellular damage (5). However, in organisms that undergo rapid embryonic divisions, including C. elegans, checkpoints are inoperative during the first few cell cycles (6).
Although it is clear that cell cycle progression requires energy, the link, if any, between metabolic pathways and progression through mitosis is poorly understood. Genes and proteins involved in various aspects of metabolism (e.g., nucleotide biosynthesis and lipid metabolism) are regulated by the cell cycle machinery, and cells will not commit to a new cell cycle if nutrients are scarce (7). However, to what extent the metabolic state of the cell is sensed by the cell cycle machinery once cells have passed into S phase is not clear (8).
We have previously conducted a visual screen in C. elegans embryos for genes that when down-regulated by RNAi lead to an abnormal nuclear morphology (9). Most genes whose inactivation affected early embryonic development did so without arresting cell cycle progression. It was therefore striking when we came across a set of genes, coding for enzymes of the tricarboxylic acid (TCA) cycle, that when down-regulated, led to a one-cell stage arrest with paired nuclei. The TCA cycle, also known as the Krebs cycle, uses the oxidation of acetate (in the form of acetyl CoA) derived from carbohydrates, proteins, or lipids, to generate intermediates (i.e., NADH and FADH2) that are used by the electron transport chain for ATP production. Intermediates of the TCA cycle are also important for various anabolic pathways, such as fatty acid synthesis, and the synthesis of nucleotides. In this study, we examine the relationship between TCA cycle down-regulation and cell cycle progression in the one-cell C. elegans embryo. Our data suggest that down-regulation of the TCA cycle leads to a G2-like arrest at the one-cell stage embryo by preventing the activation of cyclin B–Cdk1.
Results
Down-Regulation of the C. elegans Citrate Synthase Ortholog Leads to a One-Cell Stage Embryonic Arrest Before Nuclear Envelope Breakdown.
Recently we conducted an RNAi screen in C. elegans for genes that affect nuclear morphology by targeting genes that were reported to cause embryonic lethality when mutated or down-regulated by RNAi (9). In the course of these studies we uncovered an unusual phenotype caused by the down-regulation of ORF T20G5.2 that codes for CTS-1, an ortholog of the eukaryotic citrate synthase (Fig. S1). When cts-1 was down-regulated by RNAi, embryos accumulated at the one-cell stage with paired nuclei (Fig. 1A).
Fig. 1.
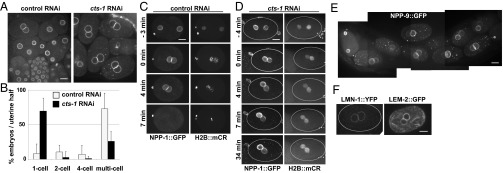
Down-regulation of cts-1 results in an accumulation of one-cell stage embryos with paired nuclei. (A) Embryos from animals after 40 h of RNAi treatment with either control RNAi (Left) or cts-1 RNAi (Right). The animals (strain OCF3) expressed NPP-1 fused to GFP (NPP-1::GFP) and histone H2B fused to mCherry (H2B::mCR). (B) The percentages of one-, two-, four-, or multicell stage embryos per uterine half of OCF3 animals treated with control or cts-1 RNAi (n = 16 and n = 19 animals, respectively) are shown. For cts-1 RNAi-treated animals, only those that accumulated one-cell stage embryos (i.e., two or more per gonad arm) were included. Shown are the results from a typical experiment, where 50% of animals accumulated one-cell stage-arrested embryos following cts-1 RNAi (see Fig. S2 for additional experiments). Error bars indicate SD. The difference in the percentage of embryos in the one-cell stage between control and cts-1(RNAi) animals was statistically significant (P < 0.05, Student t test). (C) Time-lapse images of an embryo from a control RNAi-treated OCF3 animal. Time 0 was defined as the time point when the maternal and paternal pronuclei met. (D) Time-lapse images of an embryo from a cts-1 RNAi-treated OCF3 animal. Outlines indicate the edges of the embryos. (E) The uterus of a cts-1 RNAi-treated animal expressing NPP-9::GFP. (F) Embryos from cts-1 RNAi-treated animals expressing the indicated fusion proteins, imaged 30 min following pronuclear meeting. (Scale bar: 10 µm.)
During normal development, embryos at the one-cell stage with paired nuclei are relatively rare: they are present transiently following maternal and paternal pronuclear meeting and before nuclear envelope breakdown (1). Adult C. elegans animals have one or no such embryos on each side of the uterus (that is, the uterine half that is between a spermatheca and the vulva); most embryos in the uterus contain four or more cells, as was also observed in our control RNAi-treated animals (Fig. 1 A and B and Fig. S2). In contrast, a 40 h treatment with RNAi against cts-1 resulted in a dramatic increase in one-cell stage embryos with paired nuclei (Fig. 1 A and B and Fig. S2). The percentage of animals exhibiting this phenotype (i.e., at least two or more one-cell stage embryos with paired nuclei per uterine half; henceforth, “animals with arrested embryos”) varied from experiment to experiment, ranging from 50% to 90%. The reason for this partial penetrance is not known. The RNAi treatment resulted in a ∼60% reduction in CTS-1 levels (Fig. S3). Although CTS-1 levels in embryos from cts-1 RNAi-treated animals that did not accumulate one-cell embryos was slightly higher than that of arrested embryos (Fig. S3), the difference was very small. This difference may be sufficient to bypass the one-cell stage arrest, but it is also possible that embryos that can progress past the one-cell stage despite low CTS-1 levels have somehow adapted to low CTS-1 levels. In animals with arrested embryos, the average fraction of embryos at the one-cell stage with paired nuclei was over 50% of the total number of embryos per each uterine half (Fig. 1B and Fig. S2). This was not accompanied by an accumulation of two- and four-cell stage embryos (Fig. 1B and Fig. S2), arguing that the down-regulation of cts-1 did not cause a general slowing down of embryonic development. Embryos from all cts-1 RNAi-treated animals that did not arrest at the one-cell stage continued to develop but arrested before hatching, with the exception of a small fraction of embryos that hatched but arrested as larvae. Thus, down-regulation of cts-1 resulted in one of two fates: an arrest at the one-cell stage with paired nuclei, or a developmental arrest at a multicellular embryo or early larva stage.
Cell cycle events that preceded fertilization were also examined. The germ line of cts-1 RNAi animals containing one-cell stage-arrested embryos were indistinguishable from the germ line of control animals (Fig. S4A). Meiosis appeared to have taken place normally, as all six chromosome pairs connected by chiasmata were visible at diakinesis in oocytes from animals treated with either control RNAi or cts-1 RNAi (Fig. S4B). Moreover, 100% of the one-cell stage-arrested embryos (n = 60) contained two polar bodies, indicating that both meiosis I and II divisions took place. Finally, polarity cues, as judged by the distribution of the asymmetrically localized P granules (10) and the polo-like kinase PLK-1 (11), appeared normal in one-cell stage-arrested embryos (Fig. S5). The one difference we noticed between control and cts-1 RNAi-treated animals at the point when the one-cell stage-arrested embryos were assayed (i.e., 40 h of RNAi treatment) was a reduced rate of egg laying in the cts-1 RNAi-treated worms: 39% fewer eggs laid per animal per 40 h in the cts-1 RNAi-treated worms (n = 38) compared with control worms (n = 16). The reduced rate of egg laying could be indicative of starvation or muscle/neuronal dysfunction (12), both plausible consequences of TCA cycle down-regulation. Nonetheless, the cell division events leading to the formation of one-cell stage embryos appeared to be overall normal.
To examine mitotic cell cycle events after fertilization, we followed chromosomes using histone H2B fused to mCherry (H2B::mCR) and nuclear envelope dynamics using nuclear pore protein NPP-1::GFP. In embryos from animals treated with control RNAi, chromosome condensation happened as the maternal and paternal pronuclei met, followed immediately by nuclear envelope breakdown and metaphase plate formation (less than 7 min after pronuclear meeting; Fig. 1C). In contrast, in embryos from cts-1(RNAi) animals, maternal and paternal pronuclear meeting occurred normally, but nuclear envelope breakdown did not happen, even after a prolonged time (greater than 30 min, n = 8; for example, Fig. 1D). In these animals, chromatin condensation did not take place and a metaphase plate never formed (compare Fig. 1 C with D).
We also examined nuclear envelope dynamics using additional markers, including LMN-1 (the single C. elegans B-type lamin homolog) (13), NPP-9 (another nuclear pore complex component) (14), and LEM-2 (a LEM-domain protein that resides in the nuclear envelope and is a component of the nuclear lamina) (15). Following cts-1 RNAi, all three proteins remained at the nuclear envelopes of the paired nuclei in one-cell stage-arrested embryos for at least 30 min following pronuclear meeting (Fig. 1 E and F). Thus, down-regulation of cts-1 causes a general defect in nuclear envelope breakdown.
Down-Regulation of Additional TCA Cycle Components Leads to an Embryonic One-Cell Stage Premitotic Arrest.
To determine whether the one-cell stage embryonic arrest observed following cts-1 RNAi was due to CTS-1’s function in the TCA cycle, we examined whether down-regulation of other TCA cycle components resulted in a similar phenotype. Acetyl CoA is made from pyruvate by the pyruvate dehydrogenase (PDH) complex. A reduction in PDH complex activity would likely limit the amount of acetyl CoA available for conversion to citrate by citrate synthase, thus reducing overall TCA cycle activity (16). The PDH complex consists of three enzymes: PDH, dihydrolipoyl transacetylase, and dihydrolipoyl dehydrogenase (17). The sequence of C. elegans ORF F23B12.5, coding for DLAT-1, is highly similar to vertebrate dihydrolipoyl dehydrogenase, which is also part of the oxoglutarate dyhedrogenase complex (also in the TCA cycle) and the alpha-keto acid dehydrogenase complex. Down-regulation of dlat-1 by RNAi resulted in a paired-nuclei phenotype in one-cell stage-arrested embryos, similar to the phenotype observed following cts-1 down-regulation (Fig. 2 A and B). Moreover, the fraction of animals with arrested embryos and the percentage of one-cell stage-arrested embryos in these animals was similar to that of cts-1(RNAi) animals (Fig. 2B). A double RNAi treatment against dlat-1 and cts-1 resulted in a similar phenotype as cts-1 RNAi or dlat-1 RNAi alone (Fig. 2B and Fig. S6), suggesting that DLAT-1 and CTS-1 affect mitotic progression in the one-cell stage embryo through the same metabolic pathway. Consistent with this, we found that down-regulation of additional TCA cycle components, malate dehydrogenase, aconitase, and succinate dehydrogenase, also resulted in an accumulation of one-cell stage embryos with paired nuclei (Fig. S7). Thus, down-regulating the TCA cycle blocks the first embryonic cell cycle before mitotic entry.
Fig. 2.
Down-regulation of TCA cycle components results in a paired-nuclei phenotype in a one-cell stage-arrested embryo. (A) Embryos from an OCF3 animal treated with RNAi against dlat-1 (Left) or dlat + cts-1 (Right). Control and cts-1 RNAi alone are the same as shown in Fig. 1A. (Scale bar: 10 μm.) (B) OCF3 animals were treated with control RNAi or RNAi against cts-1, dlat-1, or both, and the percentages of embryos in the indicated stages in each uterine half were scored. Shown is a typical RNAi experiment (additional experiments are in Fig. S6). For each treatment, uterine halves from at least 10 animals were scored. The percentages of animals with arrested embryos in this experiment were 50%, 61.9%, and 40% for RNAi against cts-1, dlat-1, and cts-1 + dlat-1, respectively. Error bars indicate SD. cts-1, dlat-1, and cts-1 + dlat-1 RNAi treatments led to the accumulation of one-cell stage embryos that was statistically significantly higher than in the control (P < 0.05, Student t test). The difference between control and the three RNAi treatments in the percentage of multistage embryos was also statistically significant (P < 0.05, Student t test).
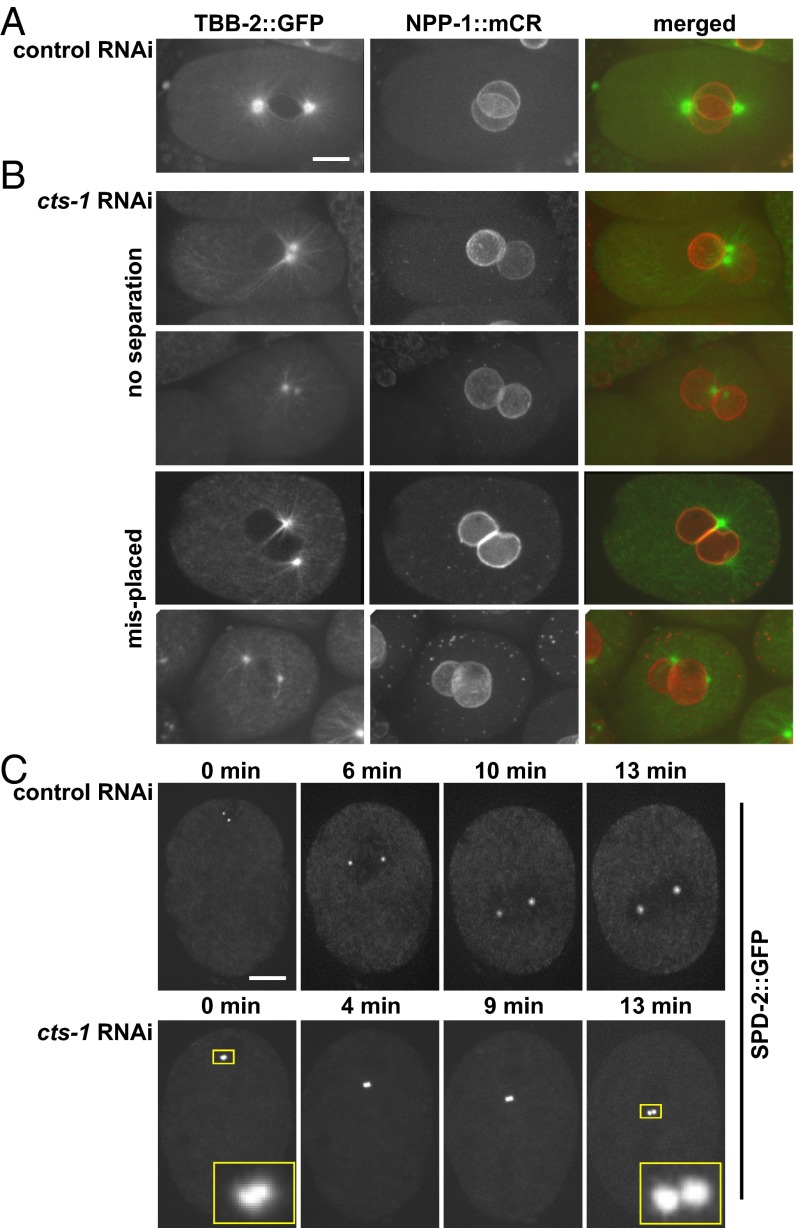
Down-Regulation of the TCA Cycle Affects Centrosome Separation in the C. elegans One-Cell Stage Embryos.
Centrosomes are necessary for the formation of a bipolar spindle in the C. elegans embryo (2, 18). After fertilization, the paternally contributed centrioles duplicate, generating two centrosomes that separate and migrate to the center of the embryo, where they nucleate microtubules that ultimately form a bipolar spindle (2). To follow spindle dynamics in vivo we used transgenic animals expressing β-tubulin fused to GFP (TBB-2::GFP) and NPP-1 fused to mCherry (NPP-1::mCR). In control RNAi-treated animals, before nuclear envelope breakdown, the two centrosomes align on either side of the plane where the two pronuclei meet (Fig. 3A). In contrast, in embryos from cts-1 RNAi-treated animals, the position of the centrosomes was abnormal: over half of the embryos had either one or two closely associated centrosomes (53.8%, n = 78), whereas in the remaining embryos the centrosomes were separated but abnormally positioned (Fig. 3B). Live imaging of transgenic animals expressing a centrosomal component, SPD-2, fused to GFP (SPD-2::GFP) (19) confirmed that cts-1 down-regulation prevented the timely separation of centrosomes in the one-cell stage embryo (n = 4; Fig. 3C, Insets). A similar trend was observed following dlat-1 down-regulation by RNAi: of the 57 embryos arrested in the one-cell stage with paired nuclei, 50.9% had centrosomes that failed to separate, 7% had centrosomes that separated but were abnormally positioned, and in 42.1% the centrosomes were separated and positioned correctly. Thus, down-regulation of the TCA cycle not only blocked nuclear envelope breakdown and chromosome condensation, but also inhibited the timely separation of centrosomes, another event associated with mitotic entry.
Fig. 3.
Down-regulation of cts-1 results in failure in centrosome separation. Embryos from a control (A) or cts-1 (B) RNAi-treated animals (strain OCF28) expressing β-tubulin fused to GFP and NPP-1 fused to mCherry. (C, Upper) An embryo from a control RNAi-treated animal expressing SPD-2::GFP (strain OC534) showing two centrosomes shortly following duplication (0 min), and as they move to the center of the embryo. Images were taken at the indicated time points. (C, Lower) An embryo from the same strain treated with cts-1(RNAi), showing duplicated centrosomes (see Insets, which are an enlargement of the boxed areas in the first and last time points). (Scale bar: 10 μm.)
Down-Regulation of the TCA Cycle Prevents CDK-1 Activation in the One-Cell Stage Embryo.
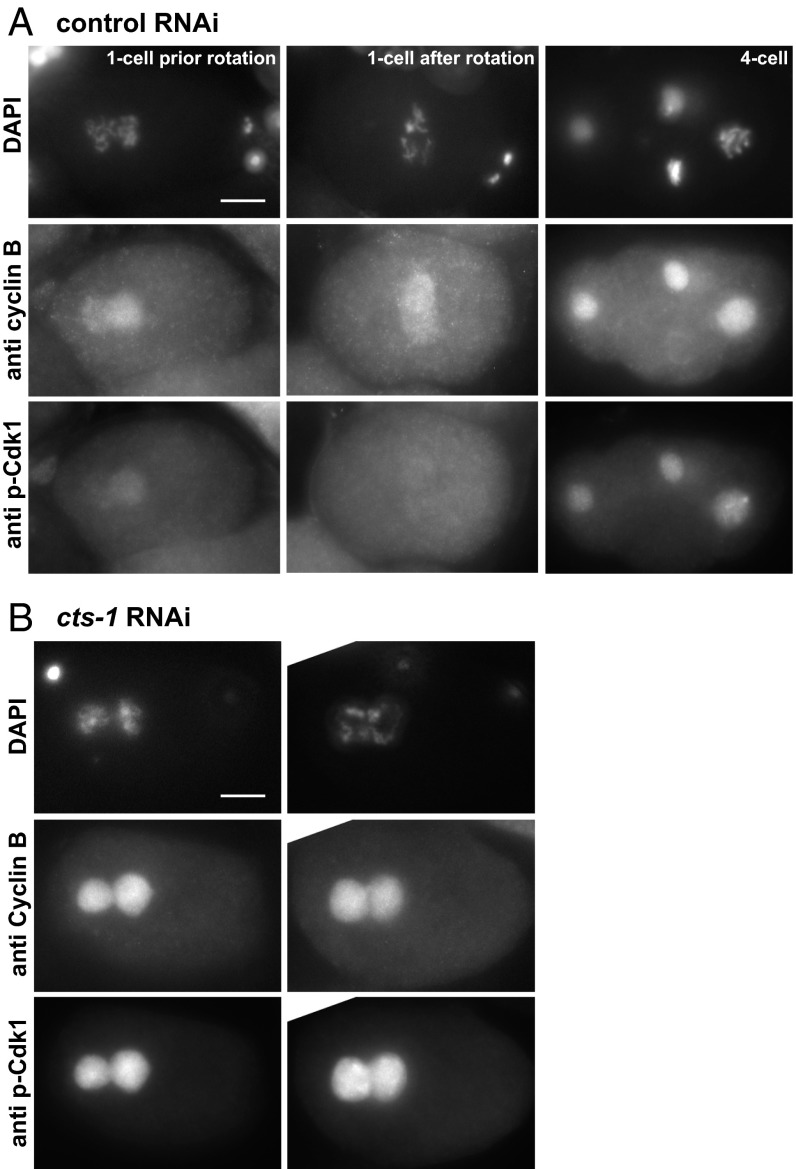
Because centrosome separation, chromosome condensation and nuclear envelope breakdown are mechanistically independent (20, 21), it was likely that down-regulation of the TCA cycle prevented mitotic entry by blocking a common upstream regulatory event. In metazoans, entry into mitosis is largely dependent on increased activity of Cdk1, which involves the accumulation of nuclear cyclin B and the removal of inhibitory phosphorylation from threonine (Thr14) and tyrosine (Tyr15) residues of Cdk1 (residue numbers according to the human Cdk1) by the Cdc25 phosphatase (3, 4). Cyclin B builds up in the nucleus in G2 and begins to be degraded upon entry into mitosis (22). Thus, in G2 cells, Cdk1 activity is held in check primarily by inhibitory phosphorylation on Thr14/Tyr15 (4). To examine the status of Cdk1 in the one-cell stage arrest induced by down-regulation of the TCA cycle, we examined the levels of C. elegans cyclin B (CYB-1) and the phosphorylation state of CDK-1 in embryos following control or cts-1 RNAi using antibodies against CYB-1 (23) and antibodies against human inhibitory phosphorylated Cdk1(Thr14/Tyr15) that also recognize the C. elegans phospho-Cdk1(Thr32/Tyr33) (Fig. S8).
In control RNAi-treated animals, cyclin B was visible at the one-cell stage before nuclear envelope breakdown (Fig. 4A), and it exhibited the expected pattern in later cell cycle stages (see, for example, its absence from a metaphase cell in the four-cell stage embryo). One-cell stage-arrested embryos from cts-1(RNAi) animals also had significant levels of CYB-1 (in 59 embryos of 65 analyzed; Fig. 4B), comparable to interphase nuclei in embryos from control RNAi-treated animals. Thus, the inability to enter mitosis following cts-1 down-regulation was not due to a defect in CYB-1 accumulation. However, although in one-cell stage embryos from the control RNAi-treated animals the levels of inhibitory phosphorylated CDK-1 diminished as the two pronuclei rotated (Fig. 4A, Center) and were absent once mitosis had begun (Fig. S9), the levels of inhibitory phosphorylated CDK-1 in one-cell stage-arrested embryos from cts-1(RNAi) animals were unusually high (Fig. 4B). This suggested that the one-cell stage arrest is due to persistent inhibitory phosphorylation of CDK-1. The high level of inhibitory phosphorylated CDK-1, along with the simultaneous presence of nuclear localized CYB-1, suggests that the one-cell stage-arrested embryos from cts-1 RNAi-treated animals are in a G2-like state.
Fig. 4.
Down-regulation of cts-1 results in a G2-arrest in the one-cell stage embryo. (A) One-cell stage embryos before (Left) or after (Center) pronuclear rotation, and a four-cell stage embryo, all from control RNAi-treated animals, were coimmunostained with antibodies against cyclin B (Middle) and phosphospecific antibodies against Cdk1 inhibitory phosphorylation at Thr14 and Tyr15 (Bottom). Note that neither cyclin B nor phospho-Cdk1 is detected in the metaphase cell in the four-cell stage embryo, as shown by DAPI staining (Top). (B) Two typical examples of embryos arrested in the one-cell stage from cts-1 RNAi-treated animal. Embryos were coimmunostained as in A. (Scale bar: 10 μm.)
The One-Cell Stage Embryonic Arrest Following cts-1 Down-Regulation Is Not Due to the Activation of the DNA Damage Checkpoint or the Spindle Assembly Checkpoint.
The G2-like arrest could have been the result of a cell cycle defect that led to checkpoint activation and consequently inhibition of CDK-1 activation. The G2/M transition is subjected to regulation by DNA damage response pathways, and specifically by the Chk1 protein kinase (24, 25). In the presence of DNA damage, activated Chk1, which is phosphorylated on Ser345 (residue number according to the human protein), inhibits the Cdc25 protein phosphatase (26). This, in turn, prevents the removal of inhibitory phosphorylation of Cdk1 that is critical for Cdk1 activation, as discussed in the Introduction. In C. elegans, a similar CHK-1 activation pathway, through the phosphorylation of the corresponding Ser344, has been reported (27, 28). Although the DNA damage and replication checkpoints are thought to be inactive in the one-cell stage embryo (28, 29), it was formally possible that down-regulation of the TCA cycle led to unscheduled CHK-1 activation at this stage. To test this, we examined one-cell stage-arrested embryos from cts-1 RNAi-treated animals for the presence of CHK-1 phosphorylated at Ser344 (p-CHK-1S344) by immunofluorescence using antibodies against human phospho-Chk1 at Ser345 (27). As a positive control for the presence of p-CHK-1S344, we down-regulated the large subunit of ribonucleotide reductase (rnr-1) by RNAi, thereby reducing de novo synthesis of deoxyribonucleotides, leading to stalling of DNA replication fork progression and delaying mitotic entry via CHK-1 activation (30, 31). As expected, down-regulation of rnr-1 resulted in activation of CHK-1 (Fig. S10 A and B). In contrast, p-CHK-1S344 was largely absent in embryos from control RNAi-treated animals and never detected in the one-cell–arrested embryos from cts-1(RNAi) animals (Fig. S10 A and B). Thus, the persistence of Cdk1 inhibitory phosphorylation following cts-1 down-regulation is not due to the activation of the DNA damage or replication checkpoint through CHK-1 activation.
We also examined whether the one-cell stage arrest following TCA cycle down-regulation depended on the spindle assembly checkpoint, which prevents progression through mitosis in the absence of chromosome-microtubule attachments (32). If that were the case, then the inactivation of this checkpoint would bypass the one-cell arrest. To test this possibility we inactivated the C. elegans MAD3 homolog by using a san-1 mutant (33, 34) and treated the worms with RNAi against cts-1. The absence of a functional spindle assembly checkpoint did not bypass the one-cell stage arrest following cts-1 RNAi (Fig. S10C), consistent with a cell cycle arrest before mitotic entry.
Discussion
Metabolism affects cell cycle progression, but the extent to which this happens during embryogenesis, and the pathways that link metabolism and the cell cycle circuitry, are largely unknown. Here we show that in C. elegans, down-regulation of TCA cycle components results in a one-cell stage embryonic arrest with paired nuclei. The arrested embryos fail to break down the nuclear envelope, chromosomes remain decondensed, and centrosomes are unseparated or abnormally separated. Taken together, these phenotypes are consistent with a defect in mitotic entry. Indeed, the arrested embryos had high levels of both cyclin B and inhibitory-phosphorylated CDK-1, indicative of a G2-like stage arrest. Thus, our data suggest that the cell cycle machinery of the one-cell stage embryo is sensitive to the metabolic state of the cell, and that when the TCA cycle is down-regulated, the one-cell stage embryo fails to enter the first mitotic division following fertilization.
One of the earliest defects caused by TCA cycle down-regulation was a defect in centrosome separation. Previously, Hachet et al. proposed that centrosomes affect timely entry into mitosis in the C. elegans early embryo (35). However, it is unlikely that centrosomes are the main target affected by down-regulation of the TCA cycle because defects in centrosome maturation, caused by down-regulation of SPD-2 or SPD-5 and which blocks centrosome separation and spindle assembly, do not prevent nuclear envelope breakdown or entry into mitosis (19, 35, 36).
At present we do not know how the TCA cycle affects the cell cycle machinery; it may cause a metabolic imbalance in the embryo that blocks Cdk1 activation, but it may also cause an imbalance earlier, in the germ line, which is maintained postfertilization. Even if the TCA cycle perturbation occurs in the germ line, our data suggest that its effect on the cell cycle machinery occurs in the embryo rather than earlier. This is based on the observations that the germ line and oocytes of cts-1 RNAi animals appear morphologically normal and that all of the arrested embryos had two polar bodies, indicating that the completion of meiosis was not perturbed. Had Cdk1 been inactivated in the germ line, meiosis would not have been completed (37). Thus, the most straightforward explanation for the one-cell embryonic arrest is that down-regulation of TCA cycle activity led, either directly or indirectly, to a defect in mitotic Cdk1 activation in the one-cell stage embryo, and this, in turn, blocked further progression through the cell cycle.
An interesting observation is that embryos that did not arrest in the one-cell stage following TCA cycle down-regulation continued to develop to multicellular embryos. This indicates that although the first mitotic division is sensitive to perturbations in the TCA cycle, subsequent divisions are not. One possibility is that the first mitotic division is inherently different from later divisions, either in its regulation of Cdk-1 activation or in its dependence on TCA cycle activity. For example, in mammalian embryos the TCA cycle is active during early embryonic divisions whereas glycolysis is activated only postimplantation (see below). Down-regulation of enzymes in other metabolic pathways, including glycolysis [the C. elegans homologs of hexokinase (coded by F14B4.2), glyceraldehyde 3-phosphoate dehydrogenase and 6-phosphofructokinase], the electron transport chain (including the CYC-1 and UCR-1 subunits of complex III and the CCO-1 and CCO-2 subunits of complex IV) and the PHI-37, ATP-4, and ATP-2 subunits of ATP synthase, all led to embryonic lethality but did not lead to an increase above control of one-cell stage embryos. This suggests that either the TCA cycle is predominant during this phase of development, or that it has a unique intermediate that affects cell cycle progression. It was previously shown that depolarizing mitochondria in human colorectal carcinoma HTC116 tissue culture cells (38) or reducing ATP levels in Drosophila cells (39) blocks the G1/S transition. We, however, found that down-regulating the TCA cycle arrests cells in G2-like state. It is possible that the first cell division cycle in the C. elegans embryo has unique features that allow it to progress past G1/S and on to G2 in the presence of low ATP levels. Alternatively, the arrest in the one-cell stage embryo following TCA cycle down-regulation could be due to changes in the levels of a metabolite other than ATP; either the accumulation of a toxic intermediate or the absence of a critical one. Alterations in the TCA cycle may also affect intracellular pH or the redox state of the cytoplasm, both of which can have a profound effect on many different processes.
The Cdc25 phosphatase, which removes the Wee1-dependent inhibitory phosphates from Cdk1, is an obvious candidate for being affected by the TCA cycle (40, 41). In C. elegans, CDC-25.1, which is homologous to the mammalian Cdc25A, is maternally contributed and is proposed to promote mitotic progression in the early embryo (42, 43). If our model is correct, then down-regulation of CDC-25 should phenocopy cts-1 RNAi whereas inactivation of WEE-1 should bypass the one-cell stage arrest. Unfortunately, in C. elegans, the activities of both CDC-25 and WEE-1 are necessary for gametogenesis (44, 45), precluding us from testing these predictions at this time.
To what extent can our findings in C. elegans embryos extend to mammalian systems? Studies conducted mostly in the 1960s and 1970s have shown that two-cell stage mammalian embryos use pyruvate and lactate through the TCA cycle, whereas glycolysis is activated at a later embryonic, postimplantation, stage (46–48). Moreover, inhibitors of the TCA cycle block mammalian embryonic development at the one-cell stage, whereas inhibition of glycolysis affects the morula to blastocyst transition (49). When these studies were conducted, very little was known about the molecular details of the cell cycle machinery, and thus the molecular link between the TCA cycle and the ability to execute the early embryonic divisions was not known. The present study gains insight into the link between metabolism and cell cycle progression in the early embryo; at least in C. elegans, down-regulation of the TCA cycle blocks Cdk1 activation by preventing the removal of its inhibitory phosphorylation.
Materials and Methods
Strains.
All C. elegans strains were maintained at 20 °C using standard methods except where noted otherwise (50). A list of strains and their genotypes is in Table S1.
RNAi Experiments.
RNAi constructs were isolated from the RNAi feeding library (Open Biosystems) and performed using standard feeding methods. The identity of each RNAi clone was verified by sequencing. L4-stage larvae were transferred to RNAi plates, and embryos were examined after 40 h of RNAi treatment at 20 °C. For each treatment, at least 10 animals were scored, and each experiment was repeated multiple times. Because the two gonad arms act independently of each other, we scored the phenotypes of embryos per uterine half, that is, between each spermatheca and the vulva. To determine embryonic lethality, 8–10 animals were transferred to new RNAi plates following 40 h treatment and removed 3–6 h thereafter. Hatching was scored 24 h later.
Immunofluorescence.
Animals from RNAi plates were dissected on poly-l-lysine–coated slides and embryos were opened by the freeze-cracking method as described previously (9). Samples were fixed in −20 °C cold methanol for 2 min and blocked in 5% (vol/vol) BSA in PBS for 30 min at 4 °C before overnight incubation at 4 °C with primary antibody diluted in 1% BSA. The following antibodies were used: anti-cyclin B at 1:50 dilution (Development Studies Hybridoma Bank, University of Iowa, Iowa City, IA), anti–phospho-Cdk1 Thr14/Tyr15 1:200 dilution (Santa Cruz Biotechnology; sc-28435-R). Samples were washed three times in PBS containing 0.1% Tween20 followed by 45 min incubation in secondary antibodies, Alexa Fluor 488 and 568 (Invitrogen), each at a 1:2,500 dilution. Samples were then washed three times in PBS containing 0.1% Tween20 and mounted in Vectashield with DAPI (Vector Laboratories).
Microscopy.
Confocal images of live embryos were taken on a Nikon Eclipse TE2000U (spinning-disk confocal) microscope using IPLab 4.0.8 software (BioVision Technologies). The microscope is equipped with a 60 × 1.4 N.A. Apo objective, four LMM5 laser merge module with diode lasers (excitation at 405, 491, 561, and 655 nm) from Applied Research, a CSU10 spinning-disk unit by Yokogawa, and a C9100-13 EM-CCD camera by Hamamatsu. Immunostained embryo images were taken on a Nikon E800 microscope using IPLab 3.9.5 software (BioVision Technologies). The microscope is equipped with a 60 × 1.4 N.A. Apo objective and a C4742-95 CCD camera by Hamamatsu. Images were processed with IPLab 3.9.5 and 4.0.8, ImageJ 1.44o (http://imagej.nih.gov/ij), Adobe Photoshop CS Version 8.0, and Adobe Illustrator CS5 Version 15.1.0.
Supplementary Material
Acknowledgments
We thank Andy Golden, Amy Fabritius, and Edward Kipreos for comments on the manuscript. We also thank Andy Golden and Kevin O’Connell for advice, strains, and antibodies; and Geraldine Seydoux for the PGL-1::GFP strain. Some strains were provided by the Caenorhabditis Genetics Center, which is funded by the National Institutes of Health Office of Research Infrastructure Programs (P40 OD010440). M.M.R., S.R., D.J.-S. and O.C.-F. were funded by an intramural grant from National Institute of Diabetes and Digestive and Kidney Diseases.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1311635111/-/DCSupplemental.
References
- 1. Oegema K, Hyman AA (January 19, 2006) Cell division. WormBook, 10.1895/wormbook.1.72.1. Available at www.wormbook.org. [DOI] [PMC free article] [PubMed]
- 2.Begasse ML, Hyman AA. The first cell cycle of the Caenorhabditis elegans embryo: Spatial and temporal control of an asymmetric cell division. Results Probl Cell Differ. 2011;53:109–133. doi: 10.1007/978-3-642-19065-0_6. [DOI] [PubMed] [Google Scholar]
- 3.Takizawa CG, Morgan DO. Control of mitosis by changes in the subcellular location of cyclin-B1-Cdk1 and Cdc25C. Curr Opin Cell Biol. 2000;12(6):658–665. doi: 10.1016/s0955-0674(00)00149-6. [DOI] [PubMed] [Google Scholar]
- 4.O’Farrell PH. Triggering the all-or-nothing switch into mitosis. Trends Cell Biol. 2001;11(12):512–519. doi: 10.1016/s0962-8924(01)02142-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bartek J, Lukas C, Lukas J. Checking on DNA damage in S phase. Nat Rev Mol Cell Biol. 2004;5(10):792–804. doi: 10.1038/nrm1493. [DOI] [PubMed] [Google Scholar]
- 6.Kipreos ET. C. elegans cell cycles: Invariance and stem cell divisions. Nat Rev Mol Cell Biol. 2005;6(10):766–776. doi: 10.1038/nrm1738. [DOI] [PubMed] [Google Scholar]
- 7.Buchakjian MR, Kornbluth S. The engine driving the ship: Metabolic steering of cell proliferation and death. Nat Rev Mol Cell Biol. 2010;11(10):715–727. doi: 10.1038/nrm2972. [DOI] [PubMed] [Google Scholar]
- 8.Cai L, Tu BP. Driving the cell cycle through metabolism. Annu Rev Cell Dev Biol. 2012;28:59–87. doi: 10.1146/annurev-cellbio-092910-154010. [DOI] [PubMed] [Google Scholar]
- 9.Joseph-Strauss D, et al. Sm protein down-regulation leads to defects in nuclear pore complex disassembly and distribution in C. elegans embryos. Dev Biol. 2012;365(2):445–457. doi: 10.1016/j.ydbio.2012.02.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kawasaki I, et al. The PGL family proteins associate with germ granules and function redundantly in Caenorhabditis elegans germline development. Genetics. 2004;167(2):645–661. doi: 10.1534/genetics.103.023093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nishi Y, Rogers E, Robertson SM, Lin R. Polo kinases regulate C. elegans embryonic polarity via binding to DYRK2-primed MEX-5 and MEX-6. Development. 2008;135(4):687–697. doi: 10.1242/dev.013425. [DOI] [PubMed] [Google Scholar]
- 12. Schafer WR (December 14, 2005) Egg-laying. WormBook, 10.1895/wormbook.1.38.1. Available at www.wormbook.org. [DOI] [PMC free article] [PubMed]
- 13.Liu J, et al. Essential roles for Caenorhabditis elegans lamin gene in nuclear organization, cell cycle progression, and spatial organization of nuclear pore complexes. Mol Biol Cell. 2000;11(11):3937–3947. doi: 10.1091/mbc.11.11.3937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Voronina E, Seydoux G. The C. elegans homolog of nucleoporin Nup98 is required for the integrity and function of germline P granules. Development. 2010;137(9):1441–1450. doi: 10.1242/dev.047654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu J, et al. MAN1 and emerin have overlapping function(s) essential for chromosome segregation and cell division in Caenorhabditis elegans. Proc Natl Acad Sci USA. 2003;100(8):4598–4603. doi: 10.1073/pnas.0730821100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mackay N, Robinson BH. Measurement of the ratio of lactate to pyruvate in skin fibroblast cultures. Methods Cell Biol. 2007;80:173–178. doi: 10.1016/S0091-679X(06)80008-7. [DOI] [PubMed] [Google Scholar]
- 17.Hucho F. The pyruvate dehydrogenase multienzyme complex. Angew Chem Int Ed Engl. 1975;14(9):591–601. doi: 10.1002/anie.197505911. [DOI] [PubMed] [Google Scholar]
- 18.Nigg EA, Stearns T. The centrosome cycle: Centriole biogenesis, duplication and inherent asymmetries. Nat Cell Biol. 2011;13(10):1154–1160. doi: 10.1038/ncb2345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kemp CA, Kopish KR, Zipperlen P, Ahringer J, O’Connell KF. Centrosome maturation and duplication in C. elegans require the coiled-coil protein SPD-2. Dev Cell. 2004;6(4):511–523. doi: 10.1016/s1534-5807(04)00066-8. [DOI] [PubMed] [Google Scholar]
- 20.Rattner JB, Berns MW. Centriole behavior in early mitosis of rat kangaroo cells (PTK2) Chromosoma. 1976;54(4):387–395. doi: 10.1007/BF00292817. [DOI] [PubMed] [Google Scholar]
- 21.Silkworth WT, Nardi IK, Paul R, Mogilner A, Cimini D. Timing of centrosome separation is important for accurate chromosome segregation. Mol Biol Cell. 2012;23(3):401–411. doi: 10.1091/mbc.E11-02-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sullivan M, Morgan DO. Finishing mitosis, one step at a time. Nat Rev Mol Cell Biol. 2007;8(11):894–903. doi: 10.1038/nrm2276. [DOI] [PubMed] [Google Scholar]
- 23.Shakes DC, et al. Spermatogenesis-specific features of the meiotic program in Caenorhabditis elegans. PLoS Genet. 2009;5(8):e1000611. doi: 10.1371/journal.pgen.1000611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ciccia A, Elledge SJ. The DNA damage response: Making it safe to play with knives. Mol Cell. 2010;40(2):179–204. doi: 10.1016/j.molcel.2010.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liu Q, et al. Chk1 is an essential kinase that is regulated by Atr and required for the G(2)/M DNA damage checkpoint. Genes Dev. 2000;14(12):1448–1459. [PMC free article] [PubMed] [Google Scholar]
- 26.Reinhardt HC, Yaffe MB. Kinases that control the cell cycle in response to DNA damage: Chk1, Chk2, and MK2. Curr Opin Cell Biol. 2009;21(2):245–255. doi: 10.1016/j.ceb.2009.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lee SJ, Gartner A, Hyun M, Ahn B, Koo HS. The Caenorhabditis elegans Werner syndrome protein functions upstream of ATR and ATM in response to DNA replication inhibition and double-strand DNA breaks. PLoS Genet. 2010;6(1):e1000801. doi: 10.1371/journal.pgen.1000801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Holway AH, Kim SH, La Volpe A, Michael WM. Checkpoint silencing during the DNA damage response in Caenorhabditis elegans embryos. J Cell Biol. 2006;172(7):999–1008. doi: 10.1083/jcb.200512136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Brauchle M, Baumer K, Gönczy P. Differential activation of the DNA replication checkpoint contributes to asynchrony of cell division in C. elegans embryos. Curr Biol. 2003;13(10):819–827. doi: 10.1016/s0960-9822(03)00295-1. [DOI] [PubMed] [Google Scholar]
- 30.Elford HL. Effect of hydroxyurea on ribonucleotide reductase. Biochem Biophys Res Commun. 1968;33(1):129–135. doi: 10.1016/0006-291x(68)90266-0. [DOI] [PubMed] [Google Scholar]
- 31.Zeng Y, et al. Replication checkpoint requires phosphorylation of the phosphatase Cdc25 by Cds1 or Chk1. Nature. 1998;395(6701):507–510. doi: 10.1038/26766. [DOI] [PubMed] [Google Scholar]
- 32.Lara-Gonzalez P, Westhorpe FG, Taylor SS. The spindle assembly checkpoint. Curr Biol. 2012;22(22):R966–R980. doi: 10.1016/j.cub.2012.10.006. [DOI] [PubMed] [Google Scholar]
- 33.Nystul TG, Goldmark JP, Padilla PA, Roth MB. Suspended animation in C. elegans requires the spindle checkpoint. Science. 2003;302(5647):1038–1041. doi: 10.1126/science.1089705. [DOI] [PubMed] [Google Scholar]
- 34.Stein KK, Davis ES, Hays T, Golden A. Components of the spindle assembly checkpoint regulate the anaphase-promoting complex during meiosis in Caenorhabditis elegans. Genetics. 2007;175(1):107–123. doi: 10.1534/genetics.106.059105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hachet V, Canard C, Gönczy P. Centrosomes promote timely mitotic entry in C. elegans embryos. Dev Cell. 2007;12(4):531–541. doi: 10.1016/j.devcel.2007.02.015. [DOI] [PubMed] [Google Scholar]
- 36.Hamill DR, Severson AF, Carter JC, Bowerman B. Centrosome maturation and mitotic spindle assembly in C. elegans require SPD-5, a protein with multiple coiled-coil domains. Dev Cell. 2002;3(5):673–684. doi: 10.1016/s1534-5807(02)00327-1. [DOI] [PubMed] [Google Scholar]
- 37.Boxem M, Srinivasan DG, van den Heuvel S. The Caenorhabditis elegans gene ncc-1 encodes a cdc2-related kinase required for M phase in meiotic and mitotic cell divisions, but not for S phase. Development. 1999;126(10):2227–2239. doi: 10.1242/dev.126.10.2227. [DOI] [PubMed] [Google Scholar]
- 38.Mitra K, Wunder C, Roysam B, Lin G, Lippincott-Schwartz J. A hyperfused mitochondrial state achieved at G1-S regulates cyclin E buildup and entry into S phase. Proc Natl Acad Sci USA. 2009;106(29):11960–11965. doi: 10.1073/pnas.0904875106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mandal S, Guptan P, Owusu-Ansah E, Banerjee U. Mitochondrial regulation of cell cycle progression during development as revealed by the tenured mutation in Drosophila. Dev Cell. 2005;9(6):843–854. doi: 10.1016/j.devcel.2005.11.006. [DOI] [PubMed] [Google Scholar]
- 40.Sebastian B, Kakizuka A, Hunter T. Cdc25M2 activation of cyclin-dependent kinases by dephosphorylation of threonine-14 and tyrosine-15. Proc Natl Acad Sci USA. 1993;90(8):3521–3524. doi: 10.1073/pnas.90.8.3521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Johnson ES, Kornbluth S. Phosphatases driving mitosis: Pushing the gas and lifting the brakes. Prog Mol Biol Transl Sci. 2012;106:327–341. doi: 10.1016/B978-0-12-396456-4.00008-0. [DOI] [PubMed] [Google Scholar]
- 42.Ashcroft N, Golden A. CDC-25.1 regulates germline proliferation in Caenorhabditis elegans. Genesis. 2002;33(1):1–7. doi: 10.1002/gene.10083. [DOI] [PubMed] [Google Scholar]
- 43.Hebeisen M, Roy R. CDC-25.1 stability is regulated by distinct domains to restrict cell division during embryogenesis in C. elegans. Development. 2008;135(7):1259–1269. doi: 10.1242/dev.014969. [DOI] [PubMed] [Google Scholar]
- 44.Burrows AE, et al. The C. elegans Myt1 ortholog is required for the proper timing of oocyte maturation. Development. 2006;133(4):697–709. doi: 10.1242/dev.02241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kim J, Kawasaki I, Shim YH. cdc-25.2, a C. elegans ortholog of cdc25, is required to promote oocyte maturation. J Cell Sci. 2010;123(Pt 6):993–1000. doi: 10.1242/jcs.060442. [DOI] [PubMed] [Google Scholar]
- 46.Clough JR. Energy metabolism during mammalian embryogenesis. Biochem Soc Trans. 1985;13(1):77–79. doi: 10.1042/bst0130077. [DOI] [PubMed] [Google Scholar]
- 47.McKiernan SH, Bavister BD, Tasca RJ. Energy substrate requirements for in-vitro development of hamster 1- and 2-cell embryos to the blastocyst stage. Hum Reprod. 1991;6(1):64–75. doi: 10.1093/oxfordjournals.humrep.a137260. [DOI] [PubMed] [Google Scholar]
- 48.Biggers JD, Whittingham DG, Donahue RP. The pattern of energy metabolism in the mouse oöcyte and zygote. Proc Natl Acad Sci USA. 1967;58(2):560–567. doi: 10.1073/pnas.58.2.560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kane MT, Buckley NJ. The effects of inhibitors of energy metabolism on the growth of one-cell rabbit ova to blastocysts in vitro. J Reprod Fertil. 1977;49(2):261–266. doi: 10.1530/jrf.0.0490261. [DOI] [PubMed] [Google Scholar]
- 50.Brenner S. The genetics of Caenorhabditis elegans. Genetics. 1974;77(1):71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.