Significance
Plant organs, such as leaves, petals, or fruits, are shaped by the behavior of their constituent cells: cell growth, oriented extension of cell walls, and cell division. However, we know little about how these processes are coordinated by regulatory genes that shape plant organs, such as JAGGED (JAG) in Arabidopsis. By identifying the genes bound by JAG throughout the genome and the consequent changes in gene expression, we reveal that JAG functions as a direct mediator between genes that control the identity of organs and tissues and the cellular activities required for organ growth. In particular, we show that JAG sculpts floral organs by directly repressing genes that control entry into DNA replication.
Keywords: plant development, flower development, shoot organogenesis
Abstract
Plant morphogenesis requires coordinated cytoplasmic growth, oriented cell wall extension, and cell cycle progression, but it is debated which of these processes are primary drivers for tissue growth and directly targeted by developmental genes. Here, we used ChIP high-throughput sequencing combined with transcriptome analysis to identify global target genes of the Arabidopsis transcription factor JAGGED (JAG), which promotes growth of the distal region of floral organs. Consistent with the roles of JAG during organ initiation and subsequent distal organ growth, we found that JAG directly repressed genes involved in meristem development, such as CLAVATA1 and HANABA TARANU, and genes involved in the development of the basal region of shoot organs, such as BLADE ON PETIOLE 2 and the GROWTH REGULATORY FACTOR pathway. At the same time, JAG regulated genes involved in tissue polarity, cell wall modification, and cell cycle progression. In particular, JAG directly repressed KIP RELATED PROTEIN 4 (KRP4) and KRP2, which control the transition to the DNA synthesis phase (S-phase) of the cell cycle. The krp2 and krp4 mutations suppressed jag defects in organ growth and in the morphology of petal epidermal cells, showing that the interaction between JAG and KRP genes is functionally relevant. Our work reveals that JAG is a direct mediator between genetic pathways involved in organ patterning and cellular functions required for tissue growth, and it shows that a regulatory gene shapes plant organs by releasing a constraint on S-phase entry.
Morphogenesis is fundamentally different in plants and animals: Plants have to contend with mechanical restrictions imposed by cell walls, do not use cell migration, and generally do not rely on programmed cell death to shape tissues. Instead, tissue growth requires cytoplasmic growth, oriented cell wall extension, and cell division. These processes are functionally interconnected: Manipulation of each affects the others and can modify plant growth and organ shape. For example, overexpression of the cell cycle inhibitor KIP RELATED PROTEIN 2 (KRP2) results in smaller organs with larger cells (1); enzymes that facilitate cell wall extensibility promote the initiation of organ primordia, including the required cell divisions (2, 3), and overall organ growth can be modified by manipulating the target of rapamycin signaling pathway, which promotes general anabolism (4–6). However, it remains unclear which of these processes, singly or in combination, are the primary targets of developmental regulatory genes to produce the localized patterns of growth that result in the shape and size of plant organs.
Plant organs, such as leaves and floral organs, are initiated on the flanks of the apical meristems, which contain the stem cell populations that sustain the continuous production of new organs. One of the key regulators of shoot organ growth in Arabidopsis is the single C2H2 zinc finger transcription factor JAGGED (JAG), which is activated in the emerging organ primordia and in the distal region of immature organs (7, 8). JAG has been proposed to stimulate organ growth by promoting cell proliferation (7, 8), but quantitative 3D imaging of floral organ primordia showed that the changes in cell behavior induced by JAG are more complex, including increased proliferation, cell enlargement, changes in cell size homeostasis, and a shift to oriented anisotropic growth (9). Computer modeling of the changes in organ growth in response to JAG also supported a role in polarized tissue growth (10). The molecular mechanisms that mediate the growth functions of JAG, however, remain unknown.
Here, we combined ChIP high-throughput sequencing (ChIP-Seq) and transcriptome analysis to reveal the links between JAG and organ identity, patterning, and cellular effectors of growth. Our results show that direct control of genes that regulate the Growth 1/synthesis (S) transition of the cell cycle is one of the key mechanisms by which JAG controls organ size and shape.
Results
To reveal the genome-wide JAG binding sites, we used anti-GFP antibodies to pull down JAG-bound DNA from jag-2 inflorescences complemented with a genomic JAG-GFP fusion (JAG:JAG-GFP) (9). ChIP-Seq was performed and analyzed in triplicate, with WT inflorescences used as epitope-negative controls. As expected for the function of JAG as a transcription factor, binding sites were enriched within the 1.5-kb regions upstream and downstream of coding sequences (Table 1). A total of 1,634 genes contained binding peaks with a false discovery rate (FDR) of less than 1% in all three replicates, and were therefore selected as high-confidence JAG targets (Dataset S1).
Table 1.
Number of peaks with ChIP-Seq scores higher than the threshold for FDR < 1% for each ChIP-Seq replicate in different gene regions relative to the TAIR10 annotated genes
| Score greater than | −3 to −1.5 kb | −1.5 to 0 kb | Within gene | 0 to +1.5 kb | Total | |
| Replicate 1 | 1.85 | 1,107 | 1,635 | 632 | 1,384 | 4,235 |
| Replicate 2 | 1.88 | 863 | 1,395 | 500 | 1,054 | 3,449 |
| Replicate 3 | 1.86 | 976 | 1,442 | 521 | 1,097 | 3,652 |
| Combined | 1,634 |
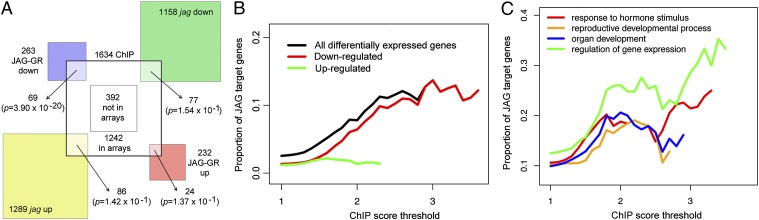
To resolve possible ambiguities in assigning gene models to ChIP-Seq peaks and to select functionally relevant direct JAG targets, we next overlapped the ChIP-Seq data with changes in gene expression detected with Affymetrix ATH1 oligonucleotide arrays (Fig. 1). In addition to comparing WT and jag-1 mutant buds, we looked for expression changes shortly after widespread JAG activation to facilitate detection of early JAG targets and of genes regulated by JAG in only a limited number of cells. For this, we used plants in which the constitutively expressed 35S promoter drove expression of a fusion between JAG and the rat glucocorticoid receptor (GR), which complemented the jag-2 mutant upon treatment with dexamethasone (9). In accordance with the suggestion that JAG functions as a transcriptional repressor (8), as discussed below, the overlap between genes repressed by JAG-GR and up-regulated in the mutant was higher than expected by chance, whereas genes activated by JAG-GR were not significantly enriched for lower expression in the mutant (Fig. S1 and Dataset S2). Surprisingly, there was also a significant enrichment for genes that responded in the same way to JAG-GR activation and to loss of endogenous JAG function (Fig. S1), although very few of these were directly bound by JAG (Dataset S3), suggesting that this overlap corresponded to indirect, downstream effects on gene expression. Especially in the case of genes repressed by JAG-GR but also down-regulated in the mutant, these indirect effects could result from inhibited growth of the relevant tissues in the jag mutant.
Fig. 1.
Combined JAG ChIP-Seq and expression array analyses reveal that JAG functions mostly as a repressor and targets gene regulation, hormone functions, and cellular functions required for growth. (A) Overlap between the sets of differentially expressed genes and the combined set of 1,634 ChIP-Seq targets shown in Table 1. To avoid detecting correlations due to the choice of tissue (inflorescence tips for both arrays and ChIP-Seq), when calculating P values for the overlaps (Fisher’s exact test), only genes whose expression was above the minimum expression value detectable on the set of arrays and present in the ChIP-Seq list were considered (16,024 genes in total). (B) Enrichment for repressed or activated genes with ChIP-Seq scores above the threshold shown on the horizontal axis, within the set of JAG-GR–responsive genes. The graph does not display the proportion of JAG targets when the total number of genes above a ChIP-Seq score was 3 or lower. (C) Enrichment for GO terms associated with differentially expressed, direct JAG target genes with ChIP-Seq scores above the threshold shown on the horizontal axis. The single ChIP-Seq score attributed to each gene was from the replicate with the lowest value. The graphs do not display the proportion of genes with a given GO term when the total number of genes above the ChIP-Seq score is 3 or lower.
Comparison of the ChiP-Seq and expression data showed that as expected for genes whose expression changed a short time after activating JAG function, the set of JAG-GR–responsive genes (Dataset S2) was strongly enriched for ChIP-Seq targets (Fig. 1A). This enrichment was significant for genes repressed by JAG-GR (P = 3.90 × 10−20, Fisher’s exact test) but not for activated genes (P = 1.37 × 10−1, Fisher’s exact test). Furthermore, the proportion of genes repressed by JAG-GR, but not the proportion of those activated, rose with increasing ChIP-Seq peak scores (Fig. 1B). Together, these data suggest that JAG functions preferentially as a transcriptional repressor, in accordance with the presence of the ethylene-responsive element binding factor-associated amphiphilic repression motif near its N terminus (8). However, we have also confirmed direct, positively regulated targets (Dataset S4). In contrast to the JAG-GR–responsive genes, and as expected for the presence of a large number of indirect effects, the sets of genes that were differentially expressed between jag-1 and the WT (Dataset S2) did not show significant enrichment for JAG-bound genes (Fig. 1A). Nevertheless, ChIP-Seq targets that were differentially expressed in the mutant vs. WT comparison, but did not respond to ectopic JAG-GR, may correspond to genes that are regulated by JAG in combination with cell type-specific factors; thus, the overlap with these differentially expressed genes was also included in the list of target genes.
The resulting set of 235 direct, transcriptionally responsive target genes (Dataset S3) showed a strong enrichment for gene ontology (GO) terms related to transcriptional control and hormone responses (Fig. 1C and Dataset S5), and included numerous known regulators of organ identity and growth. In addition to known direct target genes (9, 10), such as PETAL LOSS (PTL) (11) and BELL1 (BEL1) (12), JAG directly repressed floral patterning genes, such as CLAVATA1 (CLV1) (13) and HANABA TARANU (HAN) (14); genes involved in floral identity, such as LEAFY (LFY) (15) and its direct targets LATE MERISTEM IDENTITY 1 (LMI1) (16) and LMI2 (17); and BLADE ON PETIOLE 2 (BOP2), which regulates organ development along the proximodistal and adaxial-abaxial axes (18, 19). Consistent with the role of JAG in development of the distal region of floral organs, JAG interacted directly with AtMyb16, which has been implicated in the differentiation of conical cells that are characteristic of the petal lobes (20). Another regulatory network with multiple nodes targeted by JAG is the GROWTH REGULATING FACTORS (GRF) organ growth pathway, which preferentially promotes growth of the basal region of shoot organs (21, 22): JAG repressed GRF8 and its cofactor ANGUSTIFOLIA 3 (AN3) (23, 24) and activated TEOSINTE-BRANCHED 1, CYCLOIDEA, and PROLIFERATING CELL FACTORS 1 and 2 (TCP) 4, which, in turn, antagonizes GRF expression (25).
Apart from the gene regulatory networks mentioned above, JAG also targeted cellular functions required for tissue growth. Direct links to cell cycle control included repression of the KRP genes KRP4 and KRP2, which is analyzed in detail below, and of CYCLIN D3;3 (26). Target genes implicated in cell wall extension included ARABIDOPSIS H+-ATPase 2 (AHA2), which encodes a plasma membrane proton ATPase that participates in the acidification of the apoplast during the initial stages of wall extension (27, 28), and PROTEIN KINASE SOS2-LIKE 5 (PKS5), which controls AHA2 activity (29). Other JAG targets were homologous to genes implicated in cell wall synthesis or modification, including XYLOGLUCAN ENDOTRANSGLUCOSYLASE/HYDROLASE 28 (XTH28) (30), TRICHOME BIREFRINGENCE-LIKE 37 (TBL37) (31), and CELLULOSE SYNTHASE LIKE A11 (CSLA11) (32). Relevant to the role of JAG in promoting polarized tissue growth (9, 10), JAG activated UNICORN (UCN), which encodes a protein kinase implicated in oriented tissue growth (33), and inhibited PINOID (PID), which regulates the direction of polar auxin transport and, consequently, tissue polarity (34, 35).
In summary, the combined ChIP-Seq and expression data showed that JAG is intimately connected with gene networks that control organ identity and organ patterning; at the same time, it directly controls genes required to execute tissue growth. To confirm differential expression in response to JAG independently, representative genes for different functional categories were also tested by quantitative RT-PCR (qRT-PCR) in an independent expression experiment (Dataset S4).
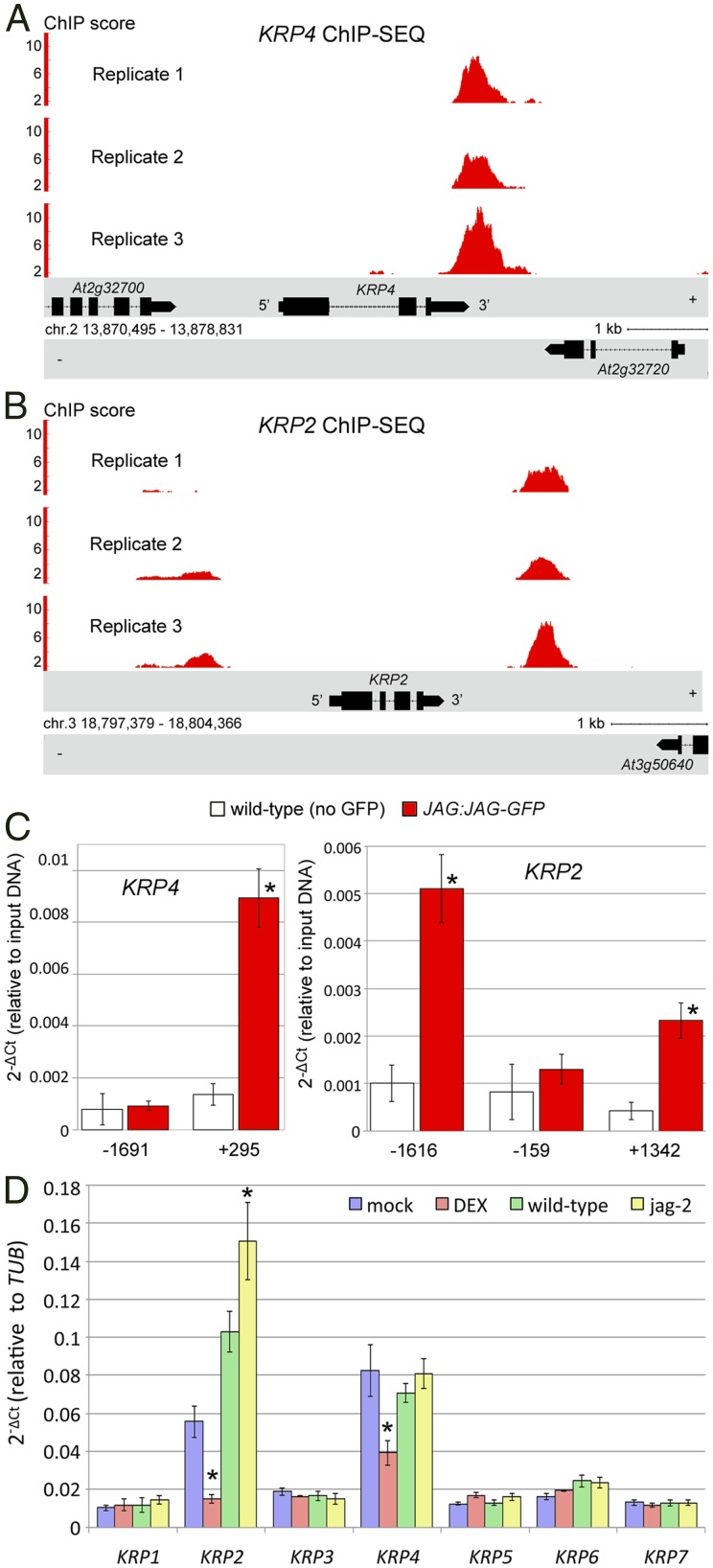
Among the genes directly repressed by JAG, the two genes with the highest ChIP-Seq scores were KRP4 and KRP2 (Dataset S3). KRP2 and KRP4 encode cyclin-dependent kinase (CDK) inhibitors, which play a key role in controlling the transition to the S-phase of the cell cycle (36). Repression of KRP genes is consistent with the role of JAG in promoting cell proliferation (7, 8) and with the premature entry into the S-phase induced by ectopic JAG (9); thus, we focused next on the interaction between JAG and KRP genes.
Both KRP2 and KRP4 showed strong ChIP-Seq peaks in all replicates, particularly in their 3′ regions (Fig. 2 A and B), and significant binding to the regions corresponding to the ChIP-Seq peaks was confirmed independently by ChIP-quantitative PCR (qPCR) using both the JAG-GFP fusion and anti-GFP antibodies (Fig. 2C) and JAG-GR with anti-GR antibodies (Fig. S2). To test differential expression independent of the array experiments, we used qRT-PCR to measure how each KRP family member responded to changes in JAG function (Fig. 2D). Of the seven family members, only KRP4 and KRP2 were repressed upon JAG-GR activation. In the jag mutant compared with the WT, both the arrays and qRT-PCR showed increased expression of KRP2 but not KRP4. This suggested that KRP4 could be widely expressed and repressed by endogenous JAG only in specific tissues or that there could be feedback regulation of KRP4 in the jag mutant. We could not test these possibilities by in situ hybridization or reporter genes, which were not sensitive enough to detect KRP4 expression reliably in floral buds. However, the phenotypic effects of krp4 mutations, which are described below, imply that KRP4 is expressed during floral organogenesis.
Fig. 2.
KRP4 and KRP2 are directly targeted by JAG in floral buds. ChIP-Seq peaks detected in each replicate within 3 kb upstream or downstream of the coding sequences for KRP4 (A) and KRP2 (B) are shown. (C) Binding of JAG-GFP to the regions of KRP4 and KRP2 confirmed by ChIP-qPCR. The numbers on the horizontal axis below the bars correspond to the left border of the amplified region (average amplicon size = 100 bp) relative to the coding sequence, and bars indicate means and SDs; asterisks indicate significant difference to the negative control (P value < 0.01, Student t test). (D) Expression of all seven KRP genes in JAG-GR inflorescence apices 4 h after treatment with dexamethasone (red) or mock treatment (blue) or in WT apices (green) compared with jag-2 (yellow). Bars indicate means and SDs, and asterisks indicate a significant difference from the negative control (P < 0.01, Student t test).
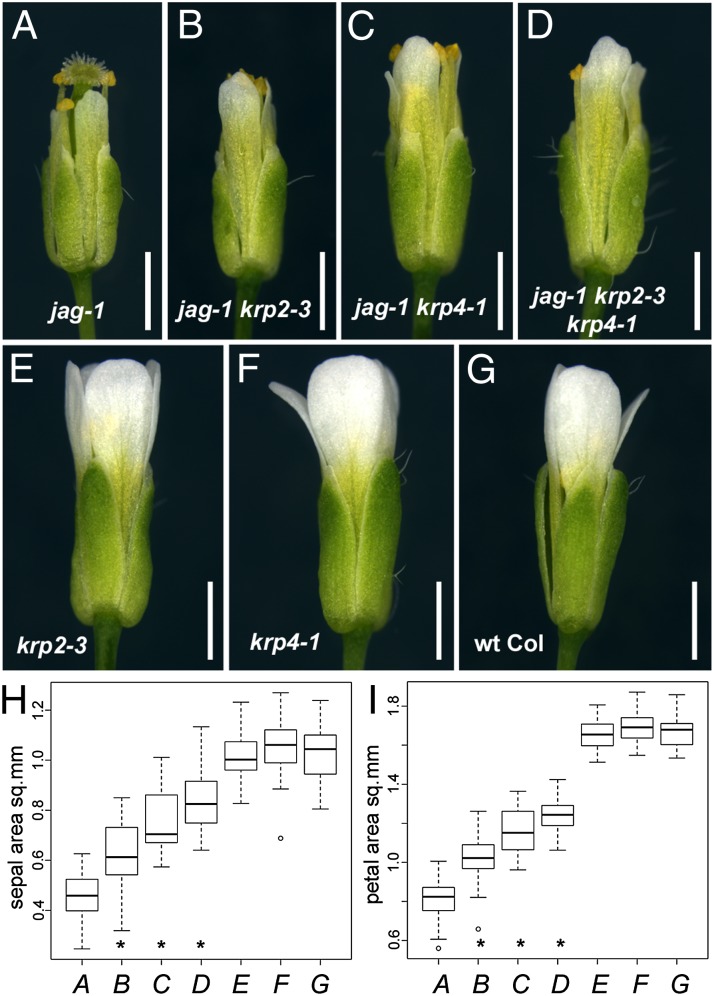
To test whether repression of KRP4 or KRP2 was required to promote floral organ growth, we compared mature organ sizes in plants with different combinations of the jag-1, krp2-3, and krp4-1 mutations with the WT Columbia-0 (Col-0) control. As reported previously (37), the krp2 and krp4 single mutants did not show any obvious defects or significant differences in organ size compared with the Col-0 control (Fig. 3 E–G). In the jag-1 mutant, sepals and petals were shorter and narrower than in the WT (Fig. 3 A and G). Both phenotypes were partially suppressed in the jag-1 krp2-3 and jag-1 krp4-1 double mutants and in the triple mutant jag-1 krp2-3 krp4-1 (Fig. 3 B–D). Measurement of the areas of mature sepals and petals confirmed that the recovery of growth in the double and triple mutants was statistically significant (P < 0.05, Student t test) (Fig. 3 H and I).
Fig. 3.
Suppression of jag organ growth defects by krp2 and krp4 mutations. Representative mature flowers of jag-1 (A), jag-1 krp2-3 (B), jag-1 krp4-1 (C), jag-1 krp2-3 krp4-1 (D), krp2-3 (E), krp4-1 (F), and WT Col-0 (G) are shown; note the defective sepal and petal growth in jag-1 and the partial recovery of growth in the jag-1 krp2-3 and jag-1 krp4-1 double mutants. (Scale bars: 1 mm.) Distribution of sepal area (H) and petal area (I) for the same genotypes shown in A–G (indicated in italics on the horizontal axis); box plots show median (thick line) second to third quartiles (box), minimum and maximum ranges (dashed line), and outliers (single points). Asterisks indicate significantly different means for jag-1 krp2-3, jag-1 krp4-1, and jag-1 krp2-3 krp4-1 (B–D) compared with jag-1 (A) (P < 0.05, one-way ANOVA; n = 40). The means of the single krp2-3 (E) and krp4-1 (F) mutants were not significantly different from the Col-0 control (G) (one-way ANOVA; n = 40). sq. mm, square millimeters.
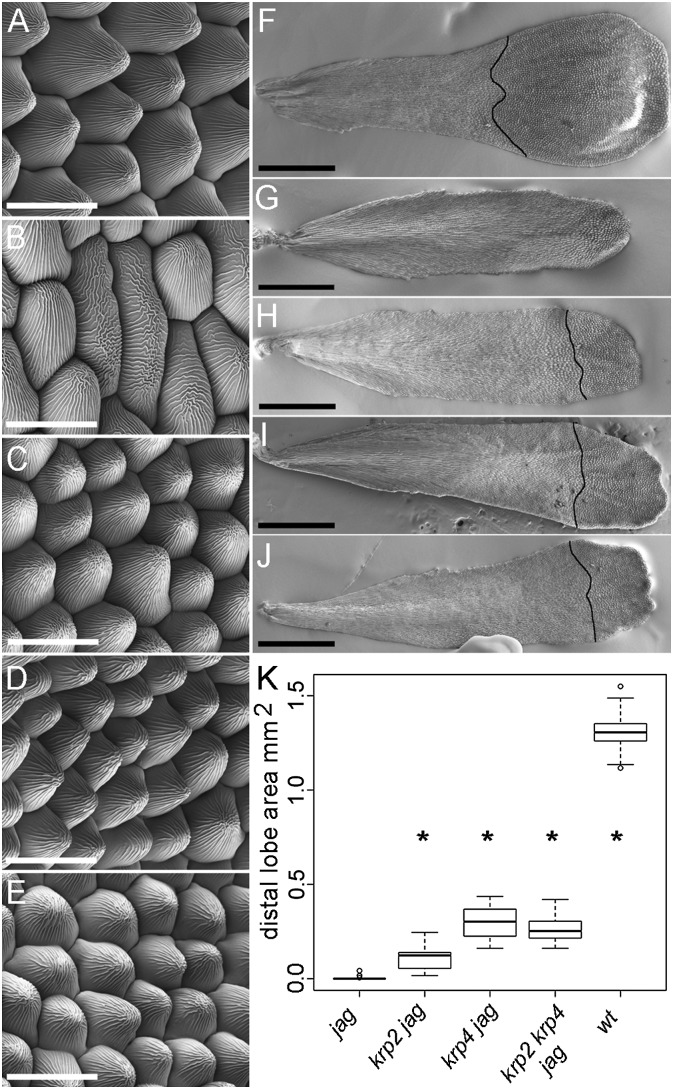
In addition to rescuing organ growth partially in jag-1, the krp2-3 and krp4-1 mutations restored cell morphology in the petal epidermis. The characteristic conical epidermal cells seen in the distal region of WT petals are replaced in jag-1 by elongated epidermal cells resembling those found near the petal base in the WT (7, 8) (Fig. 4 A and B). In contrast, the krp2-3 jag-1, krp4-1 jag-1 double mutants and the krp2-3 krp4-1 jag-1 triple mutant developed WT conical cells in the petal lobe, although the area occupied by these cells was smaller than in the WT (Fig. 4 C–K). The restoration of conical cells in the petals of the jag-1 krp2-3 and jag-1 krp4-1 double mutants is consistent with a recovery of growth specifically in the distal region of petals, which is preferentially inhibited in jag mutants.
Fig. 4.
The krp2 and krp4 mutations recover epidermal cell morphology in jag petals. Scanning electron micrographs of the epidermis of the distal region of mature petals from WT Col (A), jag-1 (B), jag-1 krp2-3 (C), jag-1 krp4-1 (D), and jag-1 krp2-3 krp4-1 (E); note the characteristic conical cells in A and C–E. Scanning electron micrographs of mature petals from WT Col (F), jag-1 (G), jag-1 krp2-3 (H), jag-1 krp4-1 (I), and jag-1 krp2-3 krp4-1 (J). The black lines across the distal region of the petals show the boundary of the distal petal lobe, where conical epidermal cells are seen. (Scale bars: A–E, 20 μm, F–J, 500 μm.) (K) Area of the distal petal lobe for the genotypes indicated. Box plots show median (thick line) second to third quartiles (box), minimum and maximum ranges (dashed line), and outliers (single points). Asterisks indicate that the mean is significantly different from jag-1 (P < 0.05, Student’s t test). wt, wild type.
Discussion
Our data reveal direct links between JAG and other genetic pathways that control organ patterning and growth. Examples include repression of meristem development genes, such as CLV1 and HAN, corroborating the previous finding that one of the roles of JAG is to antagonize meristem genes in cells that are changing from meristem to organ primordium identity (9). Another example is LFY, which activates floral homeotic genes that, in turn, activate JAG (38–40); our results suggest that JAG, in turn, antagonizes LFY activity, in line with the observation that ectopic JAG expression in the jag-5D mutant converts flowers into leafy shoots (7). Connected to LFY function, BOP2 is expressed in the basal region of lateral organs, regulates organ development along the proximodistal and adaxial-abaxial axes, and antagonizes JAG (18, 41). We found that JAG directly repressed BOP2, suggesting that mutual antagonism between JAG and BOP2 is involved in establishing proximodistal organ polarity.
The direct links between JAG and cellular effectors of growth also give insight into what cellular processes are directly targeted by regulatory genes to shape plant organs. Cytoplasmic growth, cell wall extension, and cell division are interdependent processes required for plant organ growth, but it is not clear how they are coordinated (42); at present, no plant equivalent is known of the well-studied Myc and Hippo pathways that coordinate growth-related cellular activities in animals (43). It has been debated which cellular processes are primary drivers for plant organ growth and directly targeted by developmental genes and which are likely to be subordinate; tissue mechanics is a current focus in plant morphogenesis (44), and it has been disputed whether control of cell cycle progression can drive plant organ growth (45). The interaction between JAG and KRP4/KRP2 shows that direct developmental control of regulators of S-phase entry is important for growth and morphogenesis, consistent with the observation that meristem growth is rapidly inhibited by suppressing DNA synthesis but continues when mitosis is inhibited (46). The results are also consistent with the idea that the growth of plant tissues is actively restrained below the levels that would be physiologically possible. This idea was initially proposed as an adaptive response to environmental stress, during which growth is restrained by transcriptional repressors of the DELLA family (25). In this respect, it is interesting that DELLA proteins have also been shown to regulate KRP2 expression, although the functional relevance of this interaction has not been demonstrated (47). Our results suggest that in addition to a potential role in modulating growth in response to environmental conditions, localized release of a growth restraint imposed by the KRP CDK inhibitors can be used to generate the differential tissue growth required for morphogenesis.
The suppression of the jag growth defects by the krp mutations was not complete; therefore, additional targets of JAG are required for full organ growth. Plausible candidates include genes implicated in cell wall modification and cell wall extension. In addition, JAG controls not only growth rates but also polarized tissue growth (9, 10). The molecular targets of JAG provide a starting point to address the molecular mechanisms behind these other key aspects of plant tissue growth.
Materials and Methods
Plant Material.
Arabidopsis thaliana Landsberg-erecta (L-er) and Col were used as WTs; jag-1 (7), jag-2 (8), and krp2-3 (48) have been described. The krp4-1 (SALK 102417, Col background with a T-DNA insertion in the second exon) was obtained from the Nottingham Arabidopsis Stock Centre (NASC); loss of KRP4 expression in krp4-1 (37) was confirmed by RT-PCR (Fig. S3). 35S:JAG-GR and pJAG:JAG-GFP have been described (9). Plants were grown under long-day conditions (16 h light and 8 h dark, 80% humidity) in John Innes Centre Arabidopsis Soil Mix (Levington F2 compost with Intercept and grit at a 6:1 ratio), at 18 °C for ChIP and array experiments and at 22 °C for floral organ measurements.
ChIP.
pJAG:JAG-GFP jag-2 and WT L-er control plants were used. Inflorescence apices [1,300–1,500 mg (fresh weight) per sample] were fixed in 35 mL of fixation buffer [0.4 M sucrose, 10 mM Tris (pH 8), 1 mM EDTA (pH 8.5), 1% formaldehyde, 100 μM PMSF] under vacuum for 20 min on ice. Cross-linking was stopped with 100 μM glycine for 10 min on ice. After two washes with sterile water, the tissue was blotted dry and frozen in liquid nitrogen. Nuclei were purified as described (49) and resuspended in 1 mL of sonication buffer containing 500 mM Hepes, 150 mM NaCl, 5 mM MgCl2, 10% (vol/vol) TRITON X-100, and one-half of a tablet of protease inhibitor mixture complete Mini, EDTA-free (Roche). Sonication was performed in a Bioruptor water bath sonicator at 4 °C (2 × 5 min high-power level with 30 s on/30 s off cycles), resulting in an average fragment size of 500 bp. After centrifugation, the supernatant was mixed with 500 μL of immunoprecipitation buffer containing 0.5 M Hepes, 150 mM NaCl, 5 mM MgCl2, 10% (vol/vol) TRITON X-100, 1 mg/mL BSA, and 25 μL of anti-GFP μMACS Microbeads (Miltenyi Biotec); incubated on ice for 30 min; and loaded on a µ Column (Miltenyi Biotec) that had been equilibrated with 200 μL of immunoprecipitation buffer and placed into a magnetic µMACS separator (Miltenyi Biotec). After washing twice with 400 μL and twice with 200 μL of immunoprecipitation buffer and then twice with 200 μL of TE buffer [100 mM Tris (pH 8), 10 mM EDTA (pH 8)], DNA was eluted once with 20 μL and twice with 50 μL of preheated (96 °C) elution buffer containing 50 mM Tris (pH 8), 10 mM EDTA, 50 mM DTT, and 1% SDS. One hundred microliters of TE buffer and 9 μL of 25 mg/mL Proteinase K (Sigma) were added to the eluted samples and to the input control samples. Cross-linking was reverted in eluted and input samples at 37 °C overnight, followed by addition of 9 μL of 25 mg/mL Proteinase K and 8 h of incubation at 65 °C, phenol-chloroform extraction, and precipitation with ethanol overnight at −20 °C. After washing in 70% (vol/vol) EtOH, the air-dried DNA was resuspended in 100 μL of PCR-grade water (Roche), purified using a PCR purification Kit (catalog no. 18104; Qiagen), and stored at −80 °C.
ChIP-Seq and Data Analysis.
Six Illumina TruSeq ChIP-Seq libraries (three pJAG:JAG-GFP replicates and three WT controls) were produced as described (39) and sequenced (50-bp single-end reads) using an Illumina HiSEq 2500 (Rapid-Run mode) as described by the manufacturer (Illumina). Sequence reads that passed the Consensus Assessment of Sequence and Variation (CASAVA) sequencing quality filter were mapped to the unmasked Arabidopsis genome [The Arabidopsis Information Resource (TAIR) 10; ftp://ftp.arabidopsis.org/] using the Short Oligonucleotide Analysis Package (SOAPaligner, version 2) (50), allowing a maximum of two mismatches and no gaps. Reads mapping in multiple genomic locations, to the chloroplast, or to the mitochondrial genome were discarded. The primary ChIP-Seq data were deposited at the Gene Expression Omnibus database (accession no. GSE51537).
ChIP-Seq peaks were detected using ChIP-Seq Analysis in R (CSAR) (51) with default parameter values except for “backg,” which was set to 20. Each JAG-GFP library was analyzed independently in comparison to a single negative control with all three WT libraries combined. Mapped reads were extended directionally to 300 bp, and the distribution of the number of extended mapped reads overlapping each nucleotide in the JAG-GFP library and in the negative control was normalized to have the same mean and variance. Enrichment relative to control was calculated as the ratio of normalized extended reads between JAG-GFP and the control sample. Regions having less than 20 reads mapped in the control were set to 20 (parameter backg = 20 in CSAR) to avoid false-positive results due to the low coverage of the control in some regions. FDR thresholds were estimated by permutation of reads between samples and controls using CSAR for each biological replicate independently. Candidate JAG target genes were defined as genes containing a significant (FDR < 0.01) binding event in all three replicates in the region between 3 kb upstream of the beginning and 1.5 kb downstream of the annotated gene. Because multiple copies of the JAG promoter are present in our pJAG:JAG-GFP plant in contrast to the unique copy in our WT plants, JAG was excluded from the ChIP-Seq target list.
GO Analysis.
GO term enrichment analysis was performed with the module BiNGO (52) from Cytoscape (53). Plots of GO term enrichment vs. ChIP-Seq score threshold were produced in R (www.r-project.org/). The GO database (GO.db, version 2.9.0) and the mapping of Arabidopsis genes to GO terms (org.At.tair.db, version 2.9.0) were downloaded from Bioconductor (www.bioconductor.org/). The ChIP-Seq score attributed to each gene was the minimum value for the ChIP-Seq score of each of the three biological replicates.
Global Expression Analysis.
For JAG-GR activation, inflorescences were treated for 5 h with 10 μM dexamethasone or mock-treated, and RNA was extracted as described (9) from 12 inflorescence apices per sample in three biological replicates per treatment. Probe synthesis and hybridization to Affymetrix gene chip ATH1 were performed at the NASC; the raw data and metadata are available at http://affymetrix.arabidopsis.info/ (experiment ID NASCARRAYS-605).
To select differentially expressed genes, raw expression values obtained from each hybridized chip were imported in an R session (www.r-project.org/). The probe set to gene annotation ath1121501cdf was downloaded from Bioconductor (www.bioconductor.org/). Data were normalized using the package GCRMA (54), and differential expression was tested using a t test statistic. The FDR, based on the Benjamini and Hochberg method (55), was calculated using the Bioconductor package multtest. Probe sets targeting more than one TAIR10 gene and genes associated with multiple probe sets were discarded from the analysis. Because of the confounding effect of 35S:JAG-GR transcripts, JAG was also excluded. A gene was considered differentially expressed when FDR < 0.01 and the absolute value of the log2 ratio was larger than 0.5.
qPCR.
For ChIP-PCR of individual genes, 1 μL of the immunoprecipitated DNA and 1 μL of the purified input sample were used per 10-μL PCR reaction to perform qPCR in technical triplicates with the LightCycler 480 System (Roche) and SYBR Green I (Roche), as well as the primers listed in Table S1. Enrichment from cycle threshold (ΔCt) values (Ct immunoprecipitated DNA − Ct input DNA) was evaluated using the 2-ΔΔCt method as described (56). qRT-PCR was performed as published (9) using the LightCycler System as described above. Data were normalized to TUBULIN alpha 4 chain (TUB4) expression amplified with primers TUB4-RT_1-F and TUB4-RT_1-R (Table S1) as described by Livak and Schmittgen (56). For both ChIP-qPCR and qRT-PCR, unpaired two-sample Student t tests were used to test for statistical significance of differences between treatments.
Imaging.
To measure mature organs, flowers at full anthesis were detached from the inflorescences and dehydrated in a 15%, 30%, 50%, and 70% (vol/vol) ethanol series. Sepals and petals were dissected in 70% (vol/vol) ethanol, imaged with a Leica DM6000 microscope, and measured using Fiji (57). Petal epidermal cells were imaged by cryoscanning EM using a Zeiss Supra 55 VP field emission gun scanning electron microscope (Zeiss SMT). Images of single flowers at anthesis were taken with a Leica 205A stereomicroscope.
For statistical analysis of petal and sepal measurements, the RCommander package (www.rcommander.com) was used for box plots and to test for normal distribution using the “Shapiro–Wilk” test for normality. Subsequently, one-way ANOVA with “Multiple Comparisons of means” using “Tukey Contrasts” was used.
Supplementary Material
Acknowledgments
We thank Kerstin Kaufmann (University of Potsdam, Germany) and Anna Hows (Miltenyi Biotec, United Kingdom) for advice on ChIP-Seq; Florian Pantin and Pauline Haleux for advice on statistics; Matthew Hartley for advice on Python scripts; Kim Findlay for help with SEM; Ben White (The Genome Analysis Centre, Norwich, United Kingdom) for help with Illumina sequencing; the NASC for mutant seeds; and Vinod Kumar, Lars Østergaard, and Antonio Mislata for critical reading. K.S. received a Marie-Curie fellowship (Grant 237909). This work was supported by Biotechnology and Biological Sciences Research Council Grants BB/F005571/1, BB/J007056/1, and BB/J004588/1 and by the John Innes Foundation.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Database deposition: The ChIP-Seq data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (accession no. GSE51537). Expression array data in this paper have been deposited in the Nottingham Arabidopsis Stock Centre NASCArrays database, http://affymetrix.arabidopsis.info/ (experiment ID NASCARRAYS-605).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1320457111/-/DCSupplemental.
References
- 1.De Veylder L, et al. Functional analysis of cyclin-dependent kinase inhibitors of Arabidopsis. Plant Cell. 2001;13(7):1653–1668. doi: 10.1105/TPC.010087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Peaucelle A, et al. Pectin-induced changes in cell wall mechanics underlie organ initiation in Arabidopsis. Curr Biol. 2011;21(20):1720–1726. doi: 10.1016/j.cub.2011.08.057. [DOI] [PubMed] [Google Scholar]
- 3.Fleming AJ, McQueen Mason S, Mandel T, Kuhlemeier C. Induction of leaf primordia by the cell wall protein expansin. Science. 1997;276(5317):1415–1418. [Google Scholar]
- 4.Deprost D, et al. The Arabidopsis TOR kinase links plant growth, yield, stress resistance and mRNA translation. EMBO Rep. 2007;8(9):864–870. doi: 10.1038/sj.embor.7401043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Xiong Y, et al. Glucose-TOR signalling reprograms the transcriptome and activates meristems. Nature. 2013;496(7444):181–186. doi: 10.1038/nature12030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Caldana C, et al. Systemic analysis of inducible target of rapamycin mutants reveal a general metabolic switch controlling growth in Arabidopsis thaliana. Plant J. 2013;73(6):897–909. doi: 10.1111/tpj.12080. [DOI] [PubMed] [Google Scholar]
- 7.Dinneny JR, Yadegari R, Fischer RL, Yanofsky MF, Weigel D. The role of JAGGED in shaping lateral organs. Development. 2004;131(5):1101–1110. doi: 10.1242/dev.00949. [DOI] [PubMed] [Google Scholar]
- 8.Ohno CK, Reddy GV, Heisler MGB, Meyerowitz EM. The Arabidopsis JAGGED gene encodes a zinc finger protein that promotes leaf tissue development. Development. 2004;131(5):1111–1122. doi: 10.1242/dev.00991. [DOI] [PubMed] [Google Scholar]
- 9.Schiessl K, Kausika S, Southam P, Bush M, Sablowski R. JAGGED controls growth anisotropy and coordination between cell size and cell cycle during plant organogenesis. Curr Biol. 2012;22(19):1739–1746. doi: 10.1016/j.cub.2012.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sauret-Güeto S, Schiessl K, Bangham A, Sablowski R, Coen E. JAGGED controls Arabidopsis petal growth and shape by interacting with a divergent polarity field. PLoS Biol. 2013;11(4):e1001550. doi: 10.1371/journal.pbio.1001550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brewer PB, et al. PETAL LOSS, a trihelix transcription factor gene, regulates perianth architecture in the Arabidopsis flower. Development. 2004;131(16):4035–4045. doi: 10.1242/dev.01279. [DOI] [PubMed] [Google Scholar]
- 12.Reiser L, et al. The BELL1 gene encodes a homeodomain protein involved in pattern formation in the Arabidopsis ovule primordium. Cell. 1995;83(5):735–742. doi: 10.1016/0092-8674(95)90186-8. [DOI] [PubMed] [Google Scholar]
- 13.Clark SE, Williams RW, Meyerowitz EM. The CLAVATA1 gene encodes a putative receptor kinase that controls shoot and floral meristem size in Arabidopsis. Cell. 1997;89(4):575–585. doi: 10.1016/s0092-8674(00)80239-1. [DOI] [PubMed] [Google Scholar]
- 14.Zhao YX, et al. HANABA TARANU is a GATA transcription factor that regulates shoot apical meristem and flower development in Arabidopsis. Plant Cell. 2004;16(10):2586–2600. doi: 10.1105/tpc.104.024869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Weigel D, Alvarez J, Smyth DR, Yanofsky MF, Meyerowitz EM. LEAFY controls floral meristem identity in Arabidopsis. Cell. 1992;69(5):843–859. doi: 10.1016/0092-8674(92)90295-n. [DOI] [PubMed] [Google Scholar]
- 16.Saddic LA, et al. The LEAFY target LMI1 is a meristem identity regulator and acts together with LEAFY to regulate expression of CAULIFLOWER. Development. 2006;133(9):1673–1682. doi: 10.1242/dev.02331. [DOI] [PubMed] [Google Scholar]
- 17.Pastore JJ, et al. LATE MERISTEM IDENTITY2 acts together with LEAFY to activate APETALA1. Development. 2011;138(15):3189–3198. doi: 10.1242/dev.063073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jun JH, Ha CM, Fletcher JC. BLADE-ON-PETIOLE1 coordinates organ determinacy and axial polarity in arabidopsis by directly activating ASYMMETRIC LEAVES2. Plant Cell. 2010;22(1):62–76. doi: 10.1105/tpc.109.070763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ha CM, Jun JH, Nam HG, Fletcher JC. BLADE-ON-PETIOLE 1 and 2 control Arabidopsis lateral organ fate through regulation of LOB domain and adaxial-abaxial polarity genes. Plant Cell. 2007;19(6):1809–1825. doi: 10.1105/tpc.107.051938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Baumann K, et al. Control of cell and petal morphogenesis by R2R3 MYB transcription factors. Development. 2007;134(9):1691–1701. doi: 10.1242/dev.02836. [DOI] [PubMed] [Google Scholar]
- 21.Kim JH, Choi D, Kende H. The AtGRF family of putative transcription factors is involved in leaf and cotyledon growth in Arabidopsis. Plant J. 2003;36(1):94–104. doi: 10.1046/j.1365-313x.2003.01862.x. [DOI] [PubMed] [Google Scholar]
- 22.Rodriguez RE, et al. Control of cell proliferation in Arabidopsis thaliana by microRNA miR396. Development. 2010;137(1):103–112. doi: 10.1242/dev.043067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kim JH, Kende H. A transcriptional coactivator, AtGIF1, is involved in regulating leaf growth and morphology in Arabidopsis. Proc Natl Acad Sci USA. 2004;101(36):13374–13379. doi: 10.1073/pnas.0405450101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Horiguchi G, Kim G-T, Tsukaya H. The transcription factor AtGRF5 and the transcription coactivator AN3 regulate cell proliferation in leaf primordia of Arabidopsis thaliana. Plant J. 2005;43(1):68–78. doi: 10.1111/j.1365-313X.2005.02429.x. [DOI] [PubMed] [Google Scholar]
- 25.Achard P, et al. Integration of plant responses to environmentally activated phytohormonal signals. Science. 2006;311(5757):91–94. doi: 10.1126/science.1118642. [DOI] [PubMed] [Google Scholar]
- 26.Dewitte W, et al. Arabidopsis CYCD3 D-type cyclins link cell proliferation and endocycles and are rate-limiting for cytokinin responses. Proc Natl Acad Sci USA. 2007;104(36):14537–14542. doi: 10.1073/pnas.0704166104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Haruta M, Sussman MR. The effect of a genetically reduced plasma membrane protonmotive force on vegetative growth of Arabidopsis. Plant Physiol. 2012;158(3):1158–1171. doi: 10.1104/pp.111.189167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wolf S, Hématy K, Höfte H. Growth control and cell wall signaling in plants. Annu Rev Plant Biol. 2012;63(1):381–407. doi: 10.1146/annurev-arplant-042811-105449. [DOI] [PubMed] [Google Scholar]
- 29.Fuglsang AT, et al. Arabidopsis protein kinase PKS5 inhibits the plasma membrane H+ -ATPase by preventing interaction with 14-3-3 protein. Plant Cell. 2007;19(5):1617–1634. doi: 10.1105/tpc.105.035626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rose JKC, Braam J, Fry SC, Nishitani K. The XTH family of enzymes involved in xyloglucan endotransglucosylation and endohydrolysis: Current perspectives and a new unifying nomenclature. Plant Cell Physiol. 2002;43(12):1421–1435. doi: 10.1093/pcp/pcf171. [DOI] [PubMed] [Google Scholar]
- 31.Gille S, et al. O-acetylation of Arabidopsis hemicellulose xyloglucan requires AXY4 or AXY4L, proteins with a TBL and DUF231 domain. Plant Cell. 2011;23(11):4041–4053. doi: 10.1105/tpc.111.091728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liepman AH, Wilkerson CG, Keegstra K. Expression of cellulose synthase-like (Csl) genes in insect cells reveals that CslA family members encode mannan synthases. Proc Natl Acad Sci USA. 2005;102(6):2221–2226. doi: 10.1073/pnas.0409179102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Enugutti B, et al. Regulation of planar growth by the Arabidopsis AGC protein kinase UNICORN. Proc Natl Acad Sci USA. 2012;109(37):15060–15065. doi: 10.1073/pnas.1205089109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Friml J, et al. A PINOID-dependent binary switch in apical-basal PIN polar targeting directs auxin efflux. Science. 2004;306(5697):862–865. doi: 10.1126/science.1100618. [DOI] [PubMed] [Google Scholar]
- 35.Huang F, et al. Phosphorylation of conserved PIN motifs directs Arabidopsis PIN1 polarity and auxin transport. Plant Cell. 2010;22(4):1129–1142. doi: 10.1105/tpc.109.072678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhao XA, et al. A general G1/S-phase cell-cycle control module in the flowering plant Arabidopsis thaliana. PLoS Genet. 2012;8(8):e1002847. doi: 10.1371/journal.pgen.1002847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cheng Y, et al. Downregulation of multiple CDK inhibitor ICK/KRP genes upregulates the E2F pathway and increases cell proliferation, and organ and seed sizes in Arabidopsis. Plant J. 2013;75(4):642–655. doi: 10.1111/tpj.12228. [DOI] [PubMed] [Google Scholar]
- 38.Kaufmann K, et al. Orchestration of floral initiation by APETALA1. Science. 2010;328(5974):85–89. doi: 10.1126/science.1185244. [DOI] [PubMed] [Google Scholar]
- 39.Kaufmann K, et al. Target genes of the MADS transcription factor SEPALLATA3: Integration of developmental and hormonal pathways in the Arabidopsis flower. PLoS Biol. 2009;7(4):e1000090. doi: 10.1371/journal.pbio.1000090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wagner D, Sablowski RW, Meyerowitz EM. Transcriptional activation of APETALA1 by LEAFY. Science. 1999;285(5427):582–584. doi: 10.1126/science.285.5427.582. [DOI] [PubMed] [Google Scholar]
- 41.Norberg M, Holmlund M, Nilsson O. The BLADE ON PETIOLE genes act redundantly to control the growth and development of lateral organs. Development. 2005;132(9):2203–2213. doi: 10.1242/dev.01815. [DOI] [PubMed] [Google Scholar]
- 42.Sablowski R, Carnier Dornelas M. Interplay between cell growth and cell cycle in plants. J Exp Bot. 2013 doi: 10.1093/jxb/ert354. [DOI] [PubMed] [Google Scholar]
- 43.Pan D. The hippo signaling pathway in development and cancer. Dev Cell. 2010;19(4):491–505. doi: 10.1016/j.devcel.2010.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Routier-Kierzkowska A-L, Smith RS. Measuring the mechanics of morphogenesis. Curr Opin Plant Biol. 2013;16(1):25–32. doi: 10.1016/j.pbi.2012.11.002. [DOI] [PubMed] [Google Scholar]
- 45.John PCL, Qi R. Cell division and endoreduplication: doubtful engines of vegetative growth. Trends Plant Sci. 2008;13(3):121–127. doi: 10.1016/j.tplants.2008.01.004. [DOI] [PubMed] [Google Scholar]
- 46.Grandjean O, et al. In vivo analysis of cell division, cell growth, and differentiation at the shoot apical meristem in Arabidopsis. Plant Cell. 2004;16(1):74–87. doi: 10.1105/tpc.017962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Achard P, et al. Gibberellin signaling controls cell proliferation rate in Arabidopsis. Curr Biol. 2009;19(14):1188–1193. doi: 10.1016/j.cub.2009.05.059. [DOI] [PubMed] [Google Scholar]
- 48.Sanz L, et al. The Arabidopsis D-type cyclin CYCD2;1 and the inhibitor ICK2/KRP2 modulate auxin-induced lateral root formation. Plant Cell. 2011;23(2):641–660. doi: 10.1105/tpc.110.080002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gómez-Mena C, de Folter S, Costa MMR, Angenent GC, Sablowski R. Transcriptional program controlled by the floral homeotic gene AGAMOUS during early organogenesis. Development. 2005;132(3):429–438. doi: 10.1242/dev.01600. [DOI] [PubMed] [Google Scholar]
- 50.Li R, Li Y, Kristiansen K, Wang J. SOAP: Short oligonucleotide alignment program. Bioinformatics. 2008;24(5):713–714. doi: 10.1093/bioinformatics/btn025. [DOI] [PubMed] [Google Scholar]
- 51.Muiño JM, Kaufmann K, van Ham RC, Angenent GC, Krajewski P. ChIP-seq Analysis in R (CSAR): An R package for the statistical detection of protein-bound genomic regions. Plant Methods. 2011;7(11):11. doi: 10.1186/1746-4811-7-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Maere S, Heymans K, Kuiper M. BiNGO: A Cytoscape plugin to assess overrepresentation of gene ontology categories in biological networks. Bioinformatics. 2005;21(16):3448–3449. doi: 10.1093/bioinformatics/bti551. [DOI] [PubMed] [Google Scholar]
- 53.Saito R, et al. A travel guide to Cytoscape plugins. Nat Methods. 2012;9(11):1069–1076. doi: 10.1038/nmeth.2212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gharaibeh RZ, Fodor AA, Gibas CJ. Background correction using dinucleotide affinities improves the performance of GCRMA. BMC Bioinformatics. 2008;9(1):452. doi: 10.1186/1471-2105-9-452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Series B Stat Methodol. 1995;57(1):289–300. [Google Scholar]
- 56.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25(4):402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 57.Schindelin J, et al. Fiji: An open-source platform for biological-image analysis. Nat Methods. 2012;9(7):676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.