Significance
Oncogenic mutations in the BRAF kinase occur in 6–8% of nonsmall cell lung cancers (NSCLCs), but the biological and clinical relevance of these mutations is unclear. We uncovered mechanisms of resistance to BRAF inhibition in NSCLC using an integrated functional chemical genetics approach in human BRAF-mutant NSCLC cells and clinical specimens. Our results provide biological insights into the regulation of BRAF oncogene dependence and identify strategies to optimize outcomes in BRAF-mutant NSCLC patients.
Keywords: targeted therapy, combination therapy
Abstract
Oncogenic mutations in the BRAF kinase occur in 6–8% of nonsmall cell lung cancers (NSCLCs), accounting for more than 90,000 deaths annually worldwide. The biological and clinical relevance of these BRAF mutations in NSCLC is incompletely understood. Here we demonstrate that human NSCLC cells with BRAFV600E, but not other BRAF mutations, initially are sensitive to BRAF-inhibitor treatment. However, these BRAFV600E NSCLC cells rapidly acquire resistance to BRAF inhibition through at least one of two discrete molecular mechanisms: (i) loss of full-length BRAFV600E coupled with expression of an aberrant form of BRAFV600E that retains RAF pathway dependence or (ii) constitutive autocrine EGF receptor (EGFR) signaling driven by c-Jun–mediated EGFR ligand expression. BRAFV600E cells with EGFR-driven resistance are characterized by hyperphosphorylated protein kinase AKT, a biomarker we validated in BRAF inhibitor-resistant NSCLC clinical specimens. These data reveal the multifaceted molecular mechanisms by which NSCLCs establish and regulate BRAF oncogene dependence, provide insights into BRAF–EGFR signaling crosstalk, and uncover mechanism-based strategies to optimize clinical responses to BRAF oncogene inhibition.
The discovery of genetic alterations that drive tumor growth in a wide variety of tumor types and the development of targeted therapies acting against these oncogenic drivers have revolutionized the management of many cancer patients (1). Paradigmatic examples of the successful use of oncogene-targeted therapy include the identification and treatment of patients who have EGF receptor (EGFR)-mutant and ALK fusion-positive lung cancer with the tyrosine kinase inhibitors erlotinib and crizotinib, respectively, and of patients who have BRAFV600E-melanoma with the selective BRAF kinase inhibitor vemurafenib. The clinical success of driver oncogene-targeted therapy arises because of tumor cell oncogene dependence that is established during tumorigenesis, but the mechanistic basis of this dependence remains incompletely understood. Filling this knowledge gap is critical, because the clinical success of driver oncogene-targeted therapies is limited by the almost inevitable escape from oncogene dependence and drug resistance that occur in patients with solid tumors, including lung cancer, the leading cause of cancer mortality worldwide (2, 3). The identification of the signaling events driving resistance provides insights into the molecular mechanisms underlying oncogene dependence and a rationale for mechanism-based polytherapy strategies to subvert resistance in patients (2, 4, 5).
The BRAF gene is mutated in ∼7% of human cancers, including melanoma, colorectal, papillary thyroid, and NSCLC (6, 7). The BRAFV600E variant is the most frequent mutant allele and has been used to match patients genetically to BRAF-inhibitor therapy. The clinical success and approval of the BRAF inhibitors vemurafenib and dabrafenib in BRAFV600E melanoma have provided a rationale for testing BRAF inhibition in nonmelanoma patients whose tumors harbor BRAF mutations (8–10). The success of such efforts has been limited, with either BRAF-inhibitor treatment or downstream MAPK blockade failing to produce the desired clinical activity in patients with colorectal and thyroid cancers harboring BRAFV600E; in both cases the failure is caused by intrinsic resistance (11–13). These observations indicate that tumor cell oncogene dependence is context specific and underscore the need to define the molecular events that regulate oncogene dependence in each tumor type to optimize clinical responses.
Somatic mutations in BRAF (both V600E and non-V600E variants) are found in 6–8% of NSCLCs, accounting for more than 90,000 deaths annually worldwide. BRAF-mutant NSCLCs frequently harbor the V600E allele (∼55%); additional highly recurrent activating BRAF variants reported in NSCLC include G469A (∼35%) and D594G (∼10%) (14–17). The sensitivity of NSCLC cells across the spectrum of BRAF mutant alleles to BRAF-inhibitor treatment has not been characterized. Despite this uncertainty regarding allelotype specificity, the clinical efficacy of BRAF-inhibitor treatment in BRAFV600E-melanoma has prompted a clinical trial testing the efficacy of BRAF-inhibitor treatment in patients with BRAFV600E NSCLC. Given the emerging biological and clinical importance of mutant BRAF and the success (and limitations) of other oncogene-targeted therapies, including EGFR and ALK kinase inhibitors, in NSCLC patients, we sought to define the molecular basis of BRAF oncogene dependence in NSCLC. We investigated and uncovered critical events driving response and resistance to BRAF-inhibitor treatment in models of human BRAF-mutant NSCLC. Our findings provide insight into the regulation of BRAF oncogene dependence and reveal rational strategies for immediate clinical use to enhance patients’ responses to BRAF inhibitors.
Results
Mutant BRAF Oncogene Dependence Is Variable and Transient in NSCLCs.
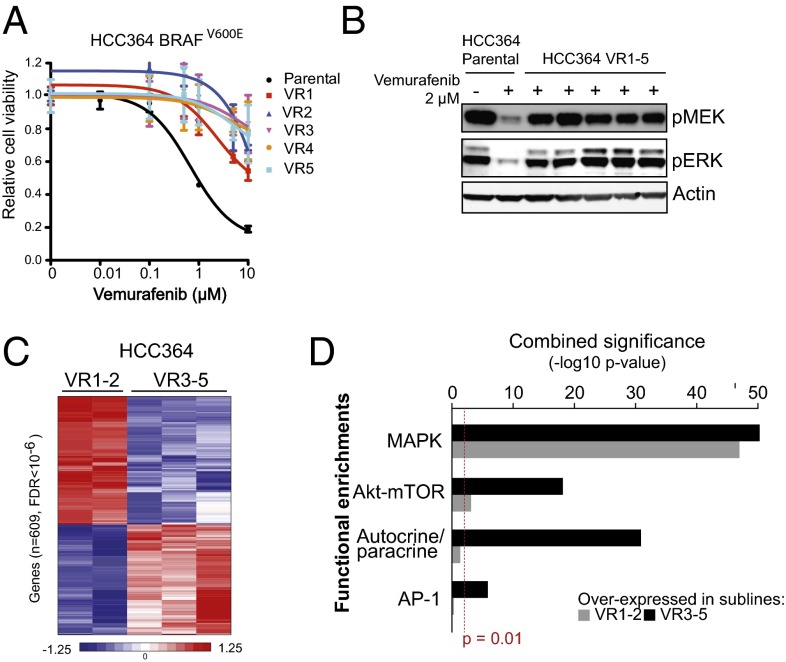
We first characterized the degree to which human BRAF-mutant NSCLC models are dependent on the oncogene for growth. We tested the effects of vemurafenib treatment in NSCLC models derived from seven BRAF-mutant patients that accurately reflect the spectrum of recurrent activating BRAF mutations, including V600E and non-V600E variants, observed in human NSCLCs (Fig. S1A). In agreement with prior work (18), we found that HCC364 cells, unique as the only available human BRAFV600E NSCLC tumor cell line, were the most sensitive of the cell lines tested to vemurafenib (IC50 0.7 μM), the BRAF inhibitor dabrafenib, and the mitogen activated protein kinase kinase (MEK) inhibitor AZD6244 (selumetinib) (Fig. S1A). Consistent with the cell-viability data, Western blot analysis showed that vemurafenib treatment suppressed MEK and ERK phosphorylation (pMEK, pERK) significantly in HCC364 BRAFV600E cells but not in the other non-V600E NSCLC cell lines tested (Fig. S1B). Thus, pharmacologic inhibition of BRAF or MEK was more effective against the NSCLC cells with BRAFV600E than against the cells with the non-V600E BRAF variants.
Having established that BRAFV600E NSCLC cells are sensitive to BRAF inhibition, we reasoned that chronic BRAF-inhibitor treatment would result in the development of models of acquired resistance that could be used to define the molecular determinants of BRAF oncogene dependence. Indeed, the use of an individual, genetically accurate patient-derived cell line has proven successful in several tumor models used recently by our group and others to discover clinically important mechanisms of resistance to targeted therapy in human tumors (4, 19–21). Continuous treatment of initially sensitive HCC364 BRAFV600E cells with vemurafenib resulted in the outgrowth of five sublines with acquired resistance (VR1–VR5, IC50 >10 μM each) (Fig. 1A and Table S1). Each of these sublines exhibited broad RAF kinase-inhibitor resistance, because they also were insensitive to dabrafenib therapy (Fig. S1C and Table S1). Accordingly, MEK–ERK signaling was not diminished by BRAF-inhibitor treatment in each resistant subline, in contrast to the drug-sensitive parental HCC364 cells (Fig. 1B). Thus, the selective pressure exerted by BRAF oncogene inhibition resulted in the outgrowth of resistant NSCLC cells that exhibited persistent MEK–ERK activation in the continued presence of a BRAF inhibitor.
Fig. 1.
BRAFV600E NSCLC models respond to BRAF-inhibitor treatment transiently and acquire drug resistance. (A) Dose–response curve showing the effect of vemurafenib on the viability of HCC364 parental cells and five vemurafenib-resistant lines. Data shown (±SEM) are normalized to the vehicle treatment (n = 3). (B) Western blot analysis of components of MAPK signaling in lysates from the HCC364 parental and each isogenic vemurafenib-resistant subline (VR1–VR5). Data represent three independent experiments. (C) Supervised hierarchical clustering using transcriptome datasets obtained by RNA-seq from HCC364 VR1–VR5 sublines. (D) Plot of functional gene set enrichment analysis indicating pathways significantly activated in the HCC364, VR1-VR2, and VR3–VR5 sublines.
We next conducted coordinated exome and transcriptome sequence analysis in the HCC364 parental cell line and in the VR1–VR5 sublines to define the mechanism(s) of acquired resistance in each case. Exome sequencing indicated that neither mutations nor overt copy number amplifications or deletions were acquired in RAS GTPase–RAF–MEK–ERK or PI3K–AKT signaling components or in upstream receptor tyrosine kinases (RTKs) that could explain drug resistance in any of the models. Nevertheless, unsupervised clustering of mRNA expression levels inferred from RNA sequencing in these models revealed striking segregation of the BRAFV600E drug-resistant sublines into two distinct classes: (i) the VR1-VR2 sublines and (ii) the VR3–VR5 sublines (Fig. S2A). Therefore we conducted a supervised transcriptional analysis and identified 609 genes whose significant differential expression (false discovery rate <10−6) robustly distinguished these two groups of drug-resistant sublines (Fig. 1C). Reasoning that this expression signature may reveal MAPK pathway reactivation or compensatory hyperactivity of other pathways that could contribute to resistance, we performed functional enrichment analysis of this gene set. This analysis revealed multiple established, experimentally derived genetic signatures in each of the two subgroups. Although genes that form, in part, the output of RAS/RAF signaling were represented in both clusters of resistant sublines, the VR1-VR2 subgroup was characterized by significantly up-regulated expression of well-characterized members of MAPK transcriptional output, indicating Ras/Raf activation. These genes included transcription factors such as ETV1, and the feedback regulator DUSP6, among others (Fig. 1D). Conversely, the VR3–VR5 subgroup of resistant lines was characterized by up-regulated expression of genes that are part of AKT-activation signatures as well as those that mediate autocrine/paracrine receptor signaling pathways and both pathway members and transcriptional targets of the AP-1 transcription factor complex (Fig. 1D). Many of these functional associations were internally validated by enrichments with multiple independent published signatures derived from diverse perturbations of the associated pathways. Together, the data suggest that distinct molecular events drive resistance to BRAF inhibition in the VR1-VR2 and the VR3–VR5 sublines, establishing two classes of drug-resistant BRAFV600E NSCLC models with complementary but distinct biological output.
A Switch from Full-Length to Aberrant BRAFV600E Causes BRAF-Inhibitor Resistance in NSCLC.
We set out to determine the molecular basis for the functional and expression-based segregation of the two subgroups of resistant tumor cells. Although the expression analyses indicated that the two subgroups (VR1-2 and VR3–VR5) were highly distinct in their transcriptional output, the VR1 and VR2 sublines were far more similar to each other than were the VR3, VR4, and VR5 sublines (Fig. S2A). The similar transcriptional signatures of VR1 and VR2 suggested a shared molecular abnormality that was absent from each of the VR3, VR4, and VR5 sublines. Moreover, because our exome analysis indicated no evidence of acquired somatic mutations or copy number alterations in the protein-coding regions of the genomes of the VR1 and VR2 sublines that could explain resistance, and because Ras/Raf pathway activation was evident by both expression profiling and Western blot analysis, we hypothesized that these sublines could harbor an aberrant form of BRAF, as described previously in a subset of vemurafenib-resistant melanomas (21). Therefore, we mined the RNA-sequencing data to determine whether the VR1 and VR2 sublines specifically harbored aberrant forms of BRAF. This analysis revealed that both the VR1 and the VR2 sublines expressed an alternative form of BRAF that was absent from the VR3–VR5 sublines and from parental HCC364 cells (Fig. 2A). This structurally abnormal BRAF transcript expressed an aberrant form that lacks exons 4–8, identical to the BRAF splice-form found in a subset of BRAF inhibitor-resistant melanoma (Fig. 2B and Fig. S2B) (21). We confirmed that this pattern of aberrant BRAF expression was exclusive to the VR1 and VR2 sublines by PCR and gel electrophoresis of cDNA derived from the cellular models (Fig. S2C) and verified with Sanger sequencing that the cDNA predicted to give rise to aberrant BRAF exclusively contained the V600E mutation (Fig. S2D). Deep sequencing of genomic DNA isolated from the HCC364 parental cells and VR1-VR2 models did not reveal mutations in the introns adjacent to the spliced reads, suggesting that splice-site alterations are not responsible for the alternative splicing of the aberrant BRAFV600E that we uncovered. Western blot analysis indicated that the aberrant BRAF migrated as a 61-kD protein in VR1 and VR2 cells (denoted “p61VE”) and was not detected in parental or VR3–VR5 cells (Fig. 2C). Strikingly, we noted that, unlike in melanoma (21), p61VE expression occurred simultaneously with the loss of expression of full-length BRAFV600E in the VR1-VR2 sublines (Fig. 2C). The expression of p61VE and the accompanying loss of the full-length BRAFV600E protein were not reversed upon removing vemurafenib from the culture medium of VR1 cells, indicating that vemurafenib treatment led to an irreversible switch to selective expression of p61VE in this system (Fig. S2E). These results indicate that a stable switch to selective and exclusive expression of p61VE occurred upon the development of BRAF-inhibitor resistance in the BRAFV600E VR1-VR2 NSCLC models.
Fig. 2.
A switch to p61VE is necessary and sufficient to drive BRAF-inhibitor resistance in the NSCLC models. (A) Expression of full-length (FL) or aberrant (p61) BRAF shown as fragments per kilobase of exon per million fragments mapped (FPKM) in each HCC364 cell line. (B) RNA sequencing reads that reflect splicing among exons is shown in VR1–VR5 lines demonstrating aberrant splicing of exons 3 and 9 indicated the absence of exons 4–8 in the VR1 and VR2 sublines only. Line widths reflect relative expression levels and in VR1-VR2 mirror the results in A. (C) Western blot analysis of each indicated protein in lysates from the HCC364 parental cell line and each isogenic vemurafenib-resistant subline (VR1–VR5). Data represent three independent experiments. (D) Western blot analysis of each indicated protein in lysates from HCC364 parental cells and from the isogenic vemurafenib-resistant subline VR1 transfected with p61 and full-length BRAF-specific siRNA. Data represent three independent experiments. (E) Effect of each indicated siRNA on the viability of HCC364 VR1 cells. Data shown (±SEM) are normalized to the vehicle treatment (n = 3). (F) Dose–response curve showing the effect of vemurafenib on the viability of HCC364 parental cells overexpressing either empty vector or p61VE plus either nontargeting siRNA (control) or BRAF siRNA targeting only full-length BRAF. Data are shown as ± SEM (n = 3). (G) Western blot analysis (Upper) and quantification (Lower) of each indicated protein in lysates from HCC364 parental cells overexpressing empty vector or p61VE plus either siRNA or BRAF siRNA targeting only full-length BRAF. Data represent three independent experiments. (H) (Upper) Relative cell number of HCC364 VR1 cells after transient transfection of empty vector, wild-type BRAF, or BRAF-V600E (BRAF-VE) constructs and treatment with 2 µM of vemurafenib. (Lower) Western blots for the indicated proteins in lysates from the VR1 cells analyzed. All cell-viability data shown are normalized to the control siRNA or vehicle treatment (n = 3, *P < 0.05).
To determine whether this irreversible switch to p61VE expression is necessary and sufficient for vemurafenib resistance, we knocked down the expression of each form of BRAF in the parental and VR1 cell lines using siRNAs to silence selectively either p61VE or full-length BRAF (two independent duplexes per target), with validation by Western blot analysis (Fig. 2D). As expected, knockdown of full-length BRAF in parental HCC364 cells suppressed MEK–ERK signaling and enhanced the levels of the proapoptotic protein BIM, phenocopying the effects of vemurafenib treatment (Fig. 2D, Left). We observed no effect of the p61VE-directed siRNAs in the parental cells that lack p61VE expression (Fig. 2D, Left). Conversely, knockdown of p61VE in the p61VE+ VR1 cells decreased MEK–ERK signaling and led to BIM induction, effects not observed upon vemurafenib treatment (Fig. 2D, Right). Moreover, no effect of the full-length BRAF-directed siRNAs was observed in the VR1 cells (Fig. 2D, Right). Consistent with these effects on signaling, silencing p61VE significantly diminished the viability of the VR1 subline, whereas treatment with siRNAs directed against full-length BRAF had no effect (Fig. 2E). Indeed, a dose–response relationship existed between the degree of knockdown achieved by each independent p61VE-directed siRNA and its effects on both signaling and viability, suggesting that the effects observed resulted from p61VE silencing in this system (Fig. 2 D and E). Taken together, the data show that p61VE is necessary for BRAF-inhibitor resistance in the VR1 model.
Having noted the aforementioned switch from full-length BRAFV600E to p61VE expression and having established that p61VE is required for BRAF-inhibitor resistance, we investigated whether simultaneous loss of full-length BRAFV600E together with p61VE is essential to confer vemurafenib resistance in treatment-naive, vemurafenib-sensitive BRAFV600E NSCLC cells. Therefore, we introduced p61VE into parental HCC364 cells, which natively express full-length BRAFV600E, either in the presence or absence of the siRNA silencing full-length BRAF. We found that introduction of p61VE induced vemurafenib resistance only in cells in which full-length BRAFV600E was silenced (Fig. 2 F and G). Consistent with these effects on cell viability, vemurafenib treatment significantly diminished MEK–ERK signaling only in the presence, but not in the absence, of full-length BRAF in the HCC364 cells we engineered to express p61VE (Fig. 2G). Because in these cells p61VE partially rescued pMEK and pERK levels during vemurafenib treatment but did not increase pMEK or pERK levels in the absence of vemurafenib (Fig. 2G), the data indicate that p61VE is specifically required for escape from BRAF inhibition. Based on these data, we predicted that reexpression of full-length BRAFV600E would diminish vemurafenib resistance in VR1 cells. Indeed, we found that expression of full-length BRAFV600E enhanced vemurafenib sensitivity in VR1 cells that also express p61VE (Fig. 2H), further indicating a critical role for the loss of full-length BRAFV600E in p61VE-driven resistance. To explore the mechanism by which p61VE may promote this resistance in NSCLC, we reasoned that, as in melanoma, p61VE-driven resistance to vemurafenib could occur through constitutive dimerization of the aberrant BRAF isoform (21). Indeed, we found that expression of the dimerization-impaired p61VE-R509H mutant form rendered greater sensitivity to the effects of vemurafenib treatment in cells that either lack or possess endogenous BRAFV600E (NIH 3T3 and parental HCC364 cells, respectively) than in cells engineered to express p61VE (Fig. S3). Taken together, our data demonstrate that loss of dependence on the native BRAF oncogene and the consequent resistance to BRAF-inhibitor treatment we observed occurred through a switch from the expression of full-length BRAFV600E to p61VE in the NSCLC models. This functional switch distinguishes p61VE-mediated regulation of BRAF oncogene dependence and BRAF-inhibitor resistance in NSCLC from melanoma, indicating that the regulation of BRAF oncogene dependence is context-specific across different human tumor types.
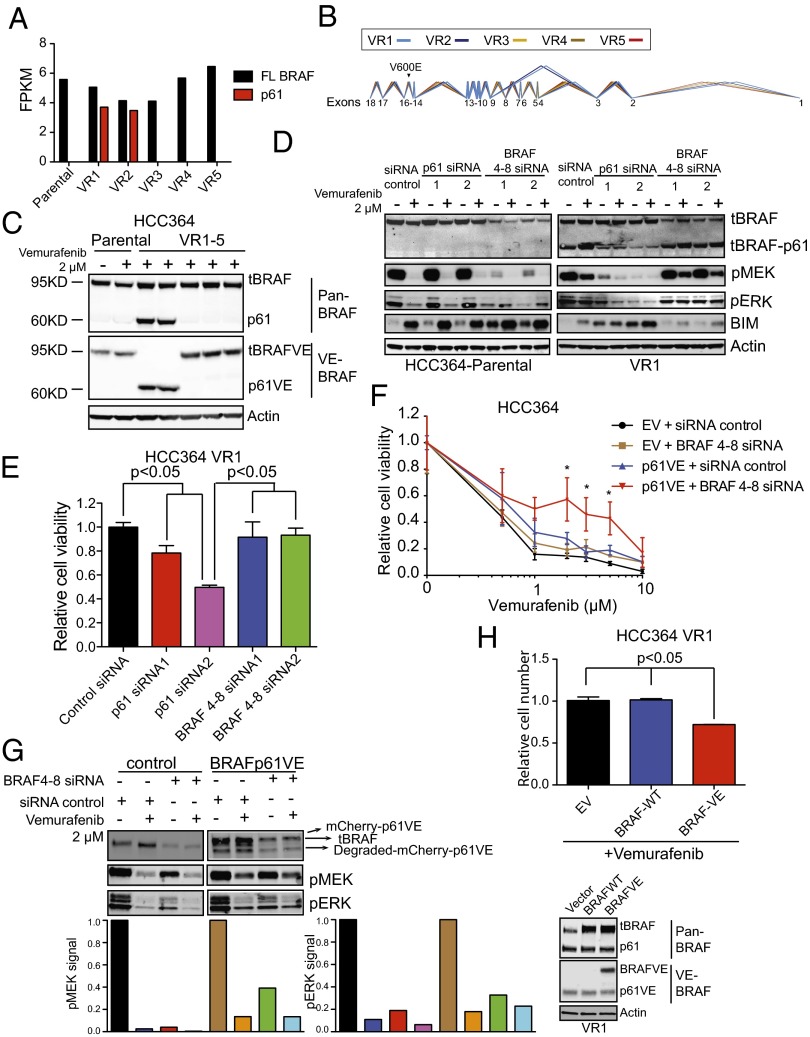
MAPK Pathway Inhibition Suppresses p61VE and Tumor Cell Escape from BRAFV600E Oncogene Inhibition in NSCLC.
Based on our findings of Ras/Raf pathway enrichment by gene-expression profiling and of MEK–ERK activation by Western blot analysis in the VR1 and VR2 sublines, we reasoned that p61VE functions to maintain MAPK pathway dependence. Therefore we hypothesized that inhibition of MEK–ERK signaling downstream of p61VE would overcome and potentially suppress the development of vemurafenib resistance in BRAFV600E NSCLCs. We treated p61VE-expressing VR1 cells with the MEK inhibitor AZD6244. AZD6244 treatment alone significantly decreased the viability of VR1 cells, suppressed MEK–ERK signaling, and increased BIM levels (Fig. 3 A and B). Moreover, treatment of the parental HCC364 cells with AZD6244 alone or in combination with vemurafenib significantly suppressed the development of acquired resistance (Fig. 3C). Notably, in the HCC364 cells that developed acquired resistance to AZD6244 alone or in combination with vemurafenib, we found no evidence of p61VE expression (Fig. S2 F and G), suggesting that the emergence of p61VE can be prevented by MEK inhibition in these cells. Similar therapeutic experiments using the HCC364 parental cells and the resistant cell lines we derived as tumor xenografts in immunocompromised mice were unsuccessful, because these cell lines were incapable of forming tumors of sufficient size for treatment studies. Nevertheless, taken together, our data indicate that a switch to a dimerized p61VE from full-length BRAFV600E drives resistance to BRAF-inhibitor treatment in a subset of tumors through maintenance of MAPK pathway dependence, which can be subverted by MEK inhibition in BRAFV600E NSCLCs.
Fig. 3.
Overriding p61VE-mediated signaling to suppress acquired resistance. (A and B) Dose–response curve showing the effect of each indicated inhibitor on cell viability (A) and activation of the indicated signaling components (B) in VR1 cells. Data in A are normalized to vehicle treatment, ±SEM (n = 3). (C) Effect of treatment with each indicated inhibitor on the development of acquired drug resistance in HCC364 cells as measured by clonal outgrowth assays. Data shown (±SEM) are normalized to vemurafenib treatment (n = 3, P < 0.01).
Engagement of EGFR Signaling Diminishes Dependence on BRAFV600E in NSCLC.
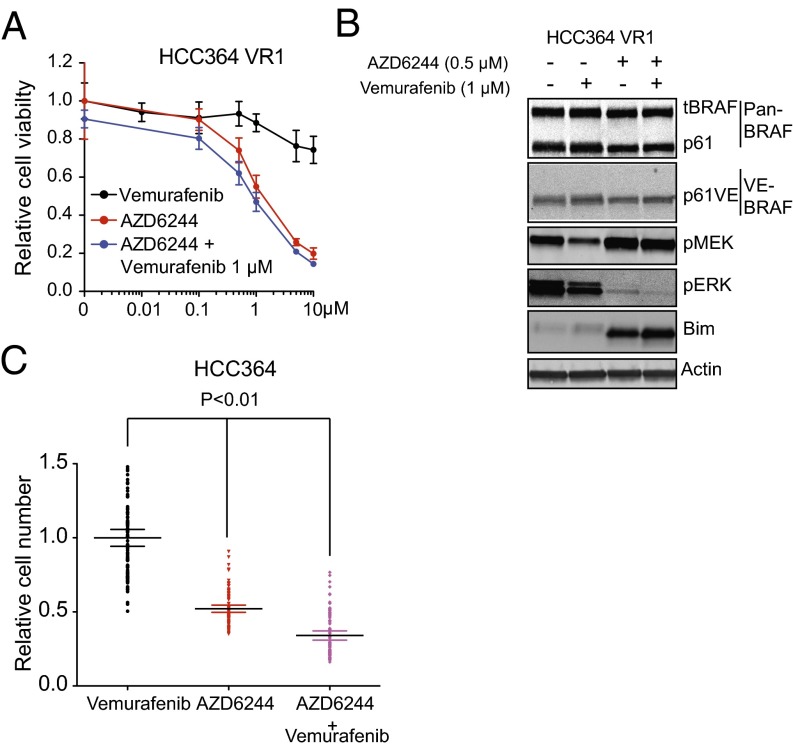
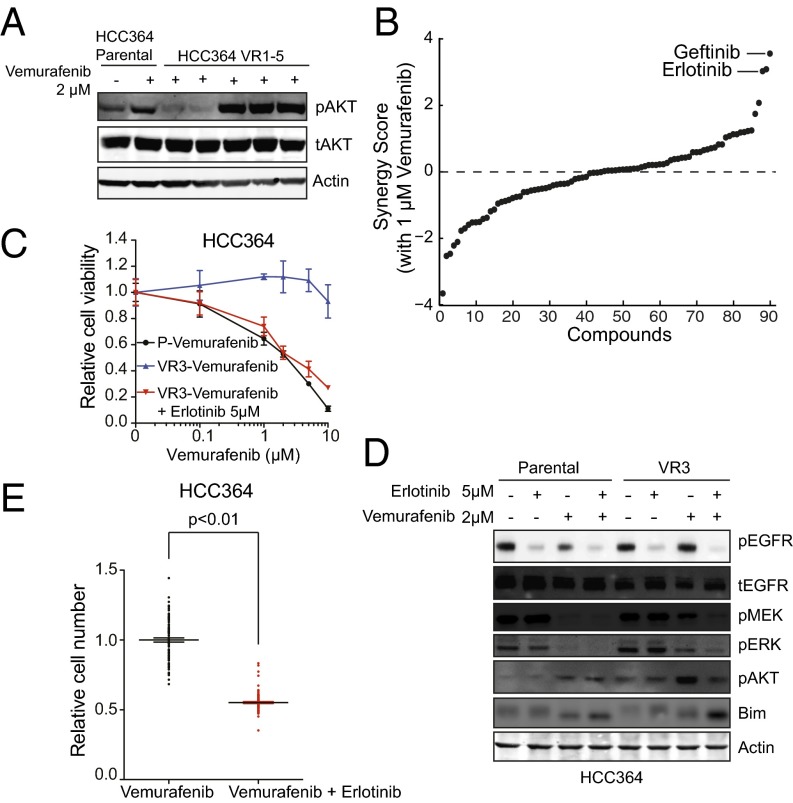
The differential expression and functional enrichment analyses of the transcriptome-sequencing data in vemurafenib-resistant models indicated that the molecular events regulating BRAF oncogene dependence and BRAF-inhibitor resistance are multifaceted. Having established p61VE as the underlying driver of resistance in the transcriptionally distinct VR1-VR2 cluster of resistant sublines, we sought to determine the lesion responsible for BRAF-inhibitor resistance in the VR3–VR5 cluster. The VR3–VR5 sublines all lacked p61VE and expressed full-length BRAFV600E (Fig. 2 A and B). Nevertheless, a defining feature of the gene signature that segregated the VR1-VR2 and VR3–VR5 subgroups of resistant tumor models was the enrichment of multiple established, experimentally derived genetic signatures associated with AKT activation, in addition to signatures of activated Ras/Raf that are consistent with the MEK–ERK activation we observed in the VR3–VR5 sublines (Fig. 1 B and D). Therefore we investigated AKT pathway activation by exploring whether the levels of phosphorylated AKT (pAKT) were increased in VR3–VR5 as compared with VR1-VR2 sublines. Indeed, we observed increased levels of pAKT in the VR3–VR5 sublines as compared with the VR1-VR2 sublines and parental cells (Fig. 4A). Our exome and transcriptome analysis indicated that this activation of the AKT signaling pathway in VR3–VR5 cells could not be explained by acquired mutations, copy number alterations, or transcriptional changes in genes that regulate AKT phosphorylation (including PTEN). Because we observed functional enrichment of autocrine/paracrine receptor signaling genes in the VR3–VR5 sublines and because AKT (and MEK–ERK) activation can occur downstream of RTK signaling (22), we conducted phospho-RTK array profiling to determine whether RTK hyperphosphorylation was present in the VR3–VR5 sublines as compared with parental cells. We did not observe increased RTK phosphorylation in these resistant cell lines as compared with sensitive (parental) cell lines, although some RTKs, including EGFR, were phosphorylated to a similar degree in both the parental and VR3–VR5 sublines (Fig. S4). Reasoning that the RTK phosphorylation that we detected in the VR3–VR5 sublines, although not increased as compared with the parental cells, could contribute to resistance, we devised a functional pharmacological screening approach. To identify agents that acted synergistically with vemurafenib to reduce the viability of VR3 but not of parental HCC364 cells, we designed a chemical screen in which we tested 94 small-molecule–targeted inhibitors that act against RTKs and signaling components connected functionally to AKT and MAPK signaling and that are either clinically approved or in clinical testing. The screening revealed that treatment with selective EGFR kinase inhibitors overcame vemurafenib resistance in the VR3 cells (Fig. 4B and Table S2). We validated this finding both pharmacologically and genetically. First, independent pharmacological assays confirmed that treatment with the top screen hits erlotinib and gefitinib as well as the clinically approved irreversible EGFR kinase inhibitor afatinib overcame vemurafenib resistance in the VR3 and VR4 sublines (Fig. 4C and Fig. S5). Second, silencing EGFR expression with both si- and shRNAs in the VR3 and VR4 sublines verified that EGFR knockdown restored vemurafenib sensitivity in this system (Fig. S6).
Fig. 4.
Engagement of EGFR signaling is necessary for BRAF-inhibitor resistance in the NSCLC models. (A) Western blot analysis of AKT phosphorylation in lysates from the HCC364 parental cell line and each isogenic vemurafenib-resistant subline (VR1–VR5). Data represent three independent experiments. (B) Synergy score plot for the chemical screening revealing small-molecule inhibitors that act synergistically with vemurafenib to diminish the viability specifically in the VR3 subline. Inhibitors targeting EGFR are labeled. (C) Dose–response curve showing the effects of vemurafenib on the viability of HCC364 parental cells (P) and VR3 cells treated with vemurafenib or with vemurafenib and erlotinib. Data shown (±SEM) are normalized to DMSO treatment (n = 3). (D) Western blot analysis of each indicated protein in lysates from HCC364 parental cells and VR3 cells treated with vemurafenib, erlotinib, or the combination as indicated. Data represent three independent experiments. (E) Effect of treatment with each indicated inhibitor on the development of acquired drug resistance in HCC364 cells as measured by clonal outgrowth assays. Data shown (±SEM) are normalized to the vemurafenib treatment (n = 3, P < 0.01).
Based on these findings, we hypothesized that EGFR activation promoted AKT and MAPK signaling that drove BRAF-inhibitor resistance. To explore the biochemical effects of combined of EGFR and BRAF inhibition, we used Western blot analysis to measure the activation status of AKT and MAPK signaling components as well as EGFR in the HCC364 parental and VR3 cells during treatment with vemurafenib alone or in combination with erlotinib. Vemurafenib treatment, either alone or in combination with erlotinib, produced the expected decrease in the levels of pMEK and pERK, as well as increased BIM in parental cells (Fig. 4D). pAKT also was induced in parental cells, likely resulting from feedback activation of AKT signaling observed upon MAPK pathway inhibition (23, 24). In contrast, similar changes in pMEK, pERK, and BIM levels were observed in VR3 and VR4 cells only after combined treatment with vemurafenib and erlotinib (Fig. 4D and Fig. S5E). Furthermore, we found that erlotinib treatment, either alone or in combination with vemurafenib, decreased the levels of pAKT in VR3 and VR4 cells, indicating that EGFR activation contributed to increased AKT phosphorylation in these resistant cells (Fig. 4D and Fig. S5E). As expected, erlotinib treatment decreased the levels of pEGFR in the parental cells and in the VR3 and VR4 sublines, both in the presence and absence of vemurafenib (Fig. 4D and Fig. S5E). Although we noted that the levels of pEGFR were not increased in untreated VR3 cells as compared with parental cells (a result that was consistent with the RTK array profiling data), we unexpectedly found that vemurafenib treatment led to a substantial decrease in the levels of pEGFR in the parental HCC364 cells but not in the VR3 and VR4 sublines (Fig. 4D and Fig. S5E). This decrease appears likely to be driven by BRAF suppression, because the effect was phenocopied by siRNA knockdown of BRAFV600E in HCC364 cells (Fig. S7A). Together, these data demonstrate that EGFR activation causes BRAF-inhibitor resistance, at least in part, through MEK–ERK and AKT signaling and uncover a functional role for MAPK signaling in EGFR activation in BRAF-mutant tumor cells.
Based on these findings, we reasoned that combined inhibition of BRAF and EGFR would suppress the development of acquired resistance. Indeed, we found that vemurafenib and erlotinib combination therapy suppressed the development of acquired resistance in HCC364 cells (Fig. 4E). Together, these data show that resistance to BRAF inhibition occurs through relief of BRAFV600E dependence by the engagement of EGFR signaling in the NSCLC models distinguished by increased AKT activation and that this resistance is overcome by combined inhibition of EGFR and BRAF.
Ligand-Mediated EGFR Signaling Downstream of MAPK Pathway Activation Regulates BRAFV600E Oncogene Dependence in NSCLC.
We sought to characterize the mechanistic basis for the EGFR dependence observed in the VR3–VR5 BRAF inhibitor-resistant sublines. This EGFR dependence was unexpected, given the absence of either activating mutations in or focal amplification of EGFR or increased baseline levels of pEGFR in these resistant sublines as compared with parental HCC364 cells. Prior work has shown that c-RAF overexpression can promote the expression of some EGFR ligands in some nonmalignant breast, ovarian, and murine cell lines (25). Given these previous observations and our data linking BRAF–MEK–ERK signaling with EGFR activation in NSCLC (Fig. 4D), we investigated whether MAPK signaling promotes EGFR activation by up-regulating the expression of EGFR ligands in the NSCLC tumor models.
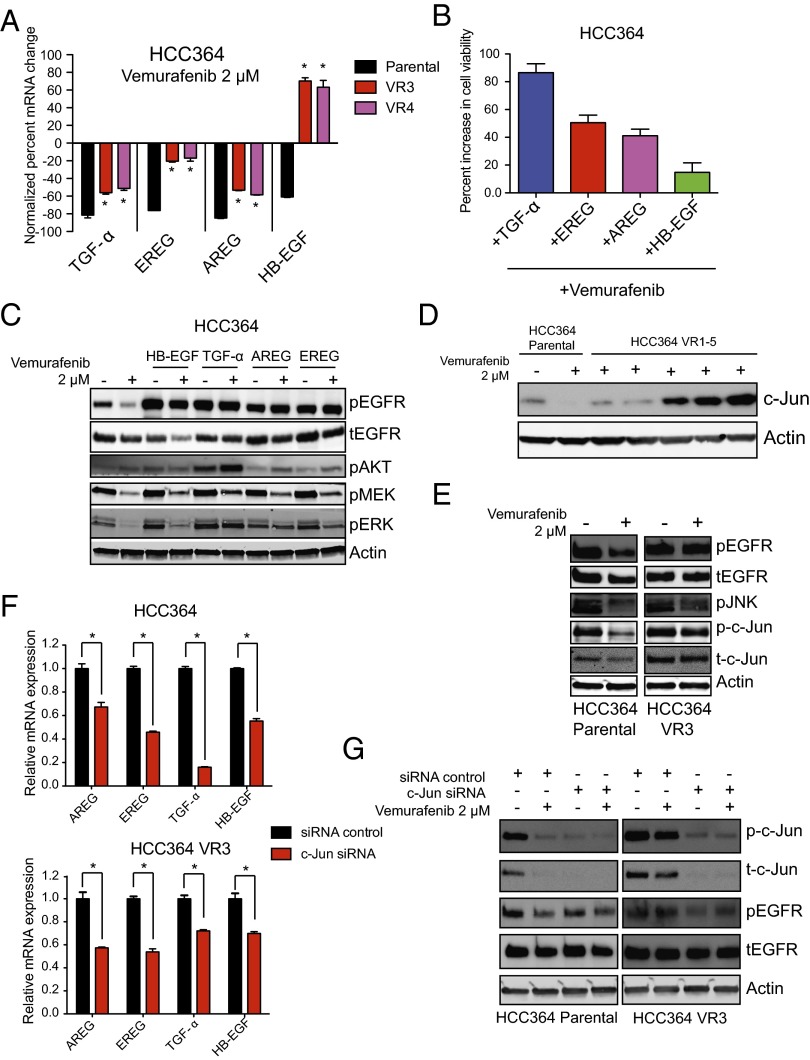
First, we examined whether the expression of EGFR ligands was diminished upon vemurafenib treatment in HCC364 cells. We found that the mRNA levels of several EGFR ligands highly expressed in HCC364 cells, including TGF-α, EREG, AREG, and HB-EGF, were all suppressed significantly upon short-term treatment with vemurafenib in parental HCC364 cells but to a lesser extent or not at all in the VR3 and VR4 sublines (Fig. 5A and Fig. S7B). Indeed, in contrast to the parental cells, the levels of HB-EGF in the VR3 and VR4 cells were increased upon vemurafenib treatment (Fig. 5A). We confirmed these effects of pharmacologic BRAF inhibition on the expression of the EGFR ligands using a genetic approach in which we silenced BRAF expression with two independent siRNAs (Fig. S7C). Based on these findings, we reasoned that ligand-mediated hyperactivation of EGFR might promote de novo resistance to BRAF oncogene inhibition. Indeed, we found that treatment of parental HCC364 cells with each of the EGFR ligands that were modulated by BRAF inhibition promoted resistance to vemurafenib (Fig. 5B) and increased the levels of phosphorylated EGFR, MEK, ERK, and, critically, AKT, particularly in the presence of vemurafenib (Fig. 5C).
Fig. 5.
Ligand-mediated EGFR signaling acting downstream of MAPK pathway activation regulates BRAFV600E oncogene dependence in NSCLC. (A) mRNA expression levels of each indicated EGFR ligand after 24 h of treatment with vemurafenib in HCC364 parental, VR3, and VR4 cells, expressed as normalized to HCC364 parental cells treated with control vehicle (n = 3, *P < 0.05). (B) Effect of growth factor stimulation on cell viability in HCC364 parental cells upon treatment with 2 µM vemurafenib. Data (±SEM) are expressed as the relative increase in viability observed during treatment with each ligand compared with vehicle control in cells treated with 2 µM vemurafenib (n = 3). (C) Western blot analysis of each indicated protein in lysates from the HCC364 parental cell line treated with the EGFR ligands TGF-α, AREG, EREG, and HB-EGF in the presence or absence of vemurafenib. Data represent three independent experiments. (D) Western blot analysis of c-Jun in lysates from the HCC364 parental cell line and each isogenic vemurafenib-resistant subline (VR1–VR5). Data represent three independent experiments. (E) Western blot analysis of each indicated protein in lysates from the HCC364 parental line and the VR3 cell line treated with vemurafenib. Data represent three independent experiments. (F) mRNA expression levels (±SEM) of the indicated genes as measured by quantitative RT-PCR in the HCC364 parental cells and in VR3 cells upon treatment with either nontargeting siRNA (control) or c-Jun siRNA (n = 3). (G) Western blot analysis of each indicated protein in lysates from the HCC364 parental cell line and from the VR3 cell line treated with vemurafenib alone or upon silencing of c-Jun by siRNA. Data shown (±SEM) are normalized to the control siRNA treatment (n = 3, *P < 0.05).
We next investigated the mechanism by which MAPK pathway signaling regulates EGFR ligand expression in the BRAFV600E NSCLC models. In addition to the aforementioned AKT pathway enrichment among differentially expressed genes up-regulated in the VR3–VR5 cluster of sublines (Fig. 1C), we identified a significant enrichment of genes associated with AP-1 signaling that included both AP-1 pathway members and transcription factor targets (Fig. 1D). Given these data and prior work demonstrating that MAPK signaling can regulate c-Jun expression in melanoma (26), we hypothesized that AP-1 signaling via c-Jun promotes the expression of EGFR ligands downstream of MAPK signaling in NSCLC, as is the case in NIH 3T3 cells (27). Indeed, we noted that the levels of c-Jun were increased in the VR3–VR5 sublines as compared with parental HCC364 cells and that the levels of phosphorylated c-Jun and total c-Jun were not diminished by BRAF inhibition as significantly in the VR3 subline as in the parental cells (Fig. 5 D and E). Furthermore, we found that the levels of phosphorylated JNK (pJNK) also were increased in the VR3 cells and were not decreased upon BRAF inhibition in these cells to the degree observed in the parental cells (Fig. 5E). These findings suggest that c-Jun can function downstream of MAPK signaling to regulate EGFR activation and therefore BRAF oncogene dependence in NSCLC.
Based on these observations, we tested whether c-Jun could regulate EGFR ligand expression and thereby EGFR phosphorylation levels in the BRAFV600E NSCLC models. Suppression of c-Jun by siRNA decreased the expression of the EGFR ligands in the parental and VR3 cells (Fig. 5F). Furthermore, silencing c-Jun expression diminished the levels of pEGFR in parental cells, an effect that was even more pronounced in vemurafenib-resistant VR3 cells in which vemurafenib had no effect on pEGFR, phosphorylated Jun, or c-Jun levels (Fig. 5G). Our findings suggest that MAPK signaling promotes EGFR ligand expression in part through the regulation of the AP-1 component c-Jun. Therefore, acquired resistance to BRAF inhibition can occur through enhanced expression of EGFR ligands that potentiate autocrine (or paracrine) activation of EGFR in the presence of BRAF-inhibitor treatment.
pAKT Is a Potential Biomarker of Relief from BRAFV600E Oncogene Dependence and the Acquisition of BRAF-Inhibitor Resistance in Human NSCLC.
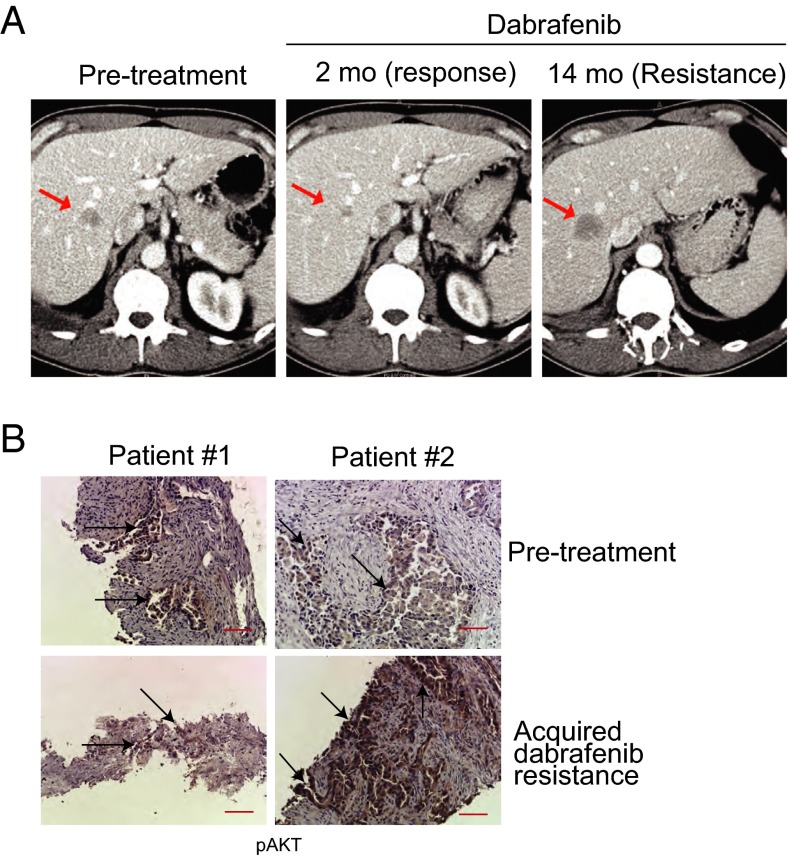
Our collective findings indicate that escape from BRAFV600E oncogene dependence and BRAF inhibition is multifaceted, caused either by compensatory reactivation of the MAPK pathway via p61VE or, alternatively, by ligand-mediated EGFR activation in the NSCLC models. To validate these findings clinically, we sought to examine specimens from BRAFV600E- NSCLC patients obtained before BRAF-inhibitor therapy and after the onset of acquired resistance. We obtained such matched, paired clinical specimens from the only two patients who consented to both pretreatment and resistance biopsies. (These patients, designated patient #1 and patient #2, were enrolled in a recently initiated multicenter, prospective, early-phase clinical trial to test dabrafenib therapy in patients with NSCLC.) These patients experienced an initial response and then acquired resistance, as defined by established clinical and radiographic criteria including conventional CT scanning, as shown in Fig. 6A for one of the two cases we profiled (patient #2). Patient #1 was analyzed using a commercially available targeted sequencing assay, and the resistant specimen was found to harbor a KRASG12D mutation (28). This observation indicates that MAPK pathway reactivation was the mechanism of resistance to dabrafenib in this patient and is consistent with our findings indicating a critical role for reactivation of MAPK pathway signaling in acquired resistance through genetic activation of individual MAPK pathway components. Although the clinical material available was scant, and therefore RNA analysis to detect p61VE was not possible, our preclinical data demonstrated that, although the levels of pEGFR were not different in the treatment-naive and BRAF inhibitor-resistant tumor cells, pAKT levels may be able to distinguish the resistant tumors with EGFR activation from those with p61VE (and exclusive MAPK pathway dependence). Therefore, using a validated immunohistochemistry assay in these two cases, we examined whether pAKT levels could be a potential biomarker of acquired BRAF-inhibitor resistance in NSCLC. Increased pAKT was not observed in the tumor cells of the dabrafenib-resistant specimen that harbored KRASG12D as compared with the treatment-naive matched sample (Fig. 6B, patient #1). However, we found significantly increased expression of pAKT and of the downstream substrate pS6 in tumor cells in the other resistant specimen as compared with the matched, pretreatment sample (Fig. 6B, patient #2 and Fig. S8). In this case with increased pAKT, we confirmed the BRAFV600E mutation in both the pretreatment and resistant specimen and did not find canonical somatic mutations in MAPK or AKT signaling components (including RAS, RAF, MEK, and ERK or PI3K, AKT, and PTEN) by exome sequencing. These clinical findings, although anecdotal, are consistent with our preclinical data in which increased pAKT was found in the setting of resistance not associated with a secondary genetic alteration in a MAPK pathway component such as KRAS (G12D) or RAF (p61VE). Taken together, our observations identify pAKT as a potential resistance biomarker in a subset of NSCLC patients suffering acquired BRAF-inhibitor resistance.
Fig. 6.
pAKT is a potential biomarker of acquired BRAF-inhibitor resistance in human NSCLC. (A) CT scans obtained before treatment and upon dabrafenib resistance in a patient with BRAFV600E NSCLC. Arrows indicate a metastatic liver lesion in this patient that was confirmed as BRAFV600E NSCLC and that initially responded and then progressed on continuous dabrafenib therapy. (B) Immunohistochemistry staining for pAKT in pretreatment and acquired resistance biopsies from two BRAFV600E NSCLC patients treated with the BRAF inhibitor dabrafenib. Arrows indicate pAKT+ tumor cells. (Scale bars: 50 μM.)
Discussion
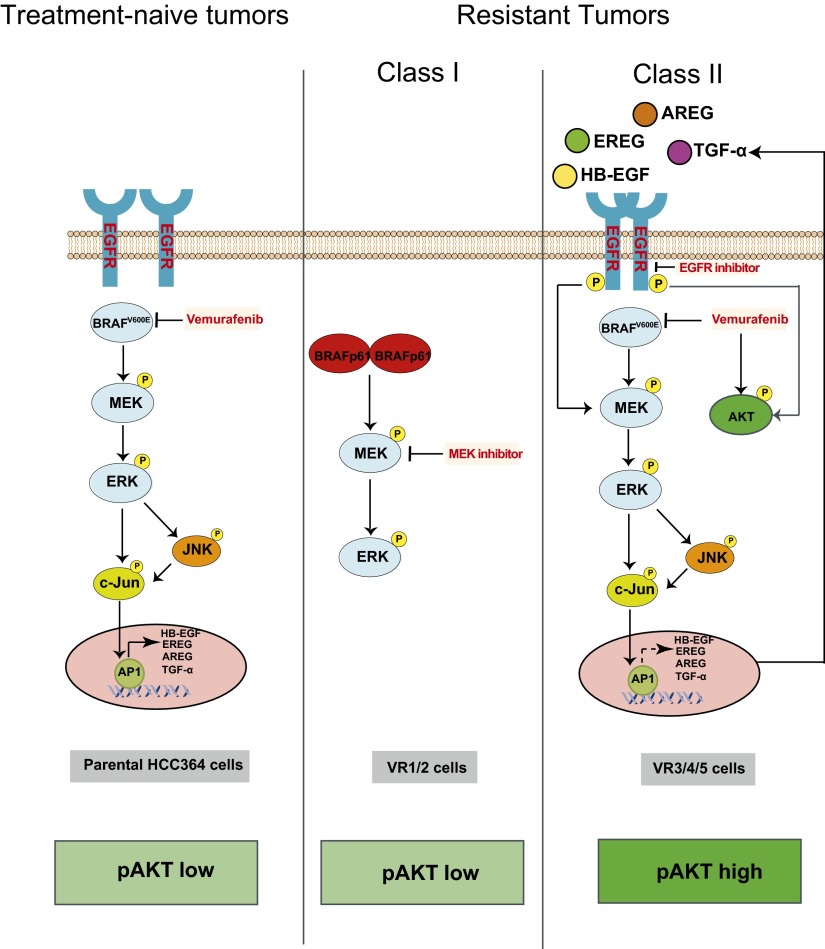
Our investigation of the molecular basis of BRAF oncogene dependence in lung cancer indicates that, although NSCLC patients with non-V600E mutant forms of activated BRAF are unlikely to respond to BRAF-inhibitor treatment alone, those with BRAFV600E mutant NSCLCs are likely to respond initially but then develop resistance. This resistance could occur through loss of dependence on the native BRAF oncogene by one of several distinct and nonoverlapping mechanisms. Based on these findings, we propose a model for the molecular mechanisms that regulate BRAFV600E oncogene dependence in NSCLC (Fig. 7). The presence of BRAFV600E in NSCLC cells drives activation of MEK–ERK signaling, which is required for tumor cell survival. BRAF signaling also simultaneously promotes the expression of EGFR ligands, partially through c-Jun, leading to phosphorylation of EGFR in a MAPK pathway-dependent manner. Therefore, BRAFV600E renders tumor cells oncogene dependent, in part through the control of multiple signaling pathways that regulate survival and growth, including MAPK and EGFR signaling. Hence, oncogene blockade by selective BRAF-inhibitor therapy simultaneously suppresses MAPK and EGFR signaling and induces death in treatment-naive BRAFV600E NSCLC cells, forming the basis for tumor-cell dependence on oncogenic BRAF.
Fig. 7.
Model for the regulation of BRAF oncogene dependence in NSCLC. (Left) Model for BRAFV600E dependence in NSCLC in which BRAF–MEK–ERK signaling controls cell growth and survival and activation of EGFR by regulating EGFR ligand expression. BRAF oncogene inhibition suppresses MEK–ERK and c-Jun signaling, thereby simultaneously downregulating MAPK signaling and EGFR ligand expression and EGFR activation. (Center) Class I resistant cells in which a switch from full-length BRAFV600E to aberrant BRAFV600E (p61VE) relieves dependence on the native oncogene and promotes BRAF-inhibitor resistance. (Right) Class II resistant cells in which engagement of EGFR signaling via c-Jun–mediated up-regulation of EGFR ligand expression and activation drives BRAF-inhibitor resistance. BRAF oncogene dependence is diminished in these resistant cells by compensatory EGFR activation that likely occurs through the loss of exclusive BRAFV600E control over c-Jun signaling and, consequently, EGFR ligand expression and EGFR activation. Hyperphosphorylation of AKT occurs in these cells as a consequence of both BRAF inhibition and EGFR activation and therefore is a biomarker of EGFR-driven acquired resistance.
Our findings indicate that EGFR phosphorylation is not sufficient to promote primary resistance to BRAF inhibition in BRAFV600E NSCLC. This result is in contrast to BRAFV600E colon and thyroid cancers, which demonstrate primary resistance to BRAF (or MEK) inhibitor therapy as a result of baseline EGFR or HER3 activation, respectively (11–13). We propose instead that patients with BRAFV600E NSCLCs are likely to respond initially to BRAF-inhibitor treatment independent of baseline pEGFR status, because BRAF inhibition simultaneously suppresses MAPK and EGFR signaling in treatment-naive tumor cells. Indeed, this prediction is confirmed by case reports of an initial response to treatment with selective BRAF inhibitors observed in individual patients with BRAFV600E NSCLC, findings that also validate the HCC364 cell line we studied as an accurate model of BRAFV600E NSCLC (28, 29).
Beyond its potential clinical relevance, the crosstalk between BRAF and EGFR we uncovered that regulates BRAF oncogene dependence in NSCLC offers insight into the mechanisms through which tumor cells rewire and control canonical signaling networks. The link between BRAF and EGFR activation through c-Jun revealed here in NSCLC is consistent with prior studies demonstrating that c-RAF overexpression can increase the expression of some EGFR ligands, in part through AP-1/Ets transcription factors, and consequently promote activation of EGFR signaling in NIH 3T3 and nonmalignant breast and ovarian epithelial cells (25, 27, 30). Our data also are consistent with previous work showing that MAPK signaling can promote JNK/Jun signaling in melanoma (26). However, our findings reveal a previously unknown essential function for a feed-forward circuit that connects BRAF signaling and subsequent EGFR ligand expression and EGFR activation to the regulation of oncogene dependence and therapeutic response in human tumor cells. This signaling circuit operates mechanistically through context-specific regulatory control of c-Jun, revealing a previously undescribed functional role for this pleiotropic transcription factor.
Our data indicate that BRAFV600E NSCLCs are likely to respond to BRAF- inhibitor treatment only transiently because of drug resistance. BRAF-inhibitor resistance occurred through two distinct, mutually exclusive molecular mechanisms that enabled escape from selective BRAF inhibition and BRAF oncogene dependence, each operating as an effective bypass switch circumventing BRAF inhibition. In Class I, simultaneous loss of full-length BRAFV600E and expression of an aberrant BRAF (p61VE) maintained MAPK pathway dependence and drove resistance. In Class II, the engagement of EGFR signaling caused by sustained EGFR ligand expression through c-Jun led to AKT and MAPK signaling and resistance (Fig. 7).
In tumor cells with class I BRAF-inhibitor resistance (VR1-VR2 sublines), escape from dependence on the native BRAFV600E oncogene and its inhibition occurred through expression of an aberrant form of BRAF (p61VE). The aberrant form of BRAF we uncovered in NSCLC was shown previously to cause BRAF-inhibitor resistance in some melanomas (21). Thus, our data demonstrate that aberrant forms of BRAF are selected for by BRAF oncogene inhibition, independent of tissue of origin and cellular context. In contrast to melanoma, we found that in NSCLC BRAF-inhibitor resistance required not only p61VE but also simultaneous loss of native, full-length BRAFV600E. This switch from expression of the native oncogene (full-length BRAFV600E) to selective expression of an aberrant, alternative form of the oncogene (p61VE) for resistance to oncogene inhibition through pathway reactivation is unexpected and indicates selection against the expression of native BRAFV600E during BRAF-inhibitor therapy in NSCLC. From a translational standpoint, our data indicate that combined BRAF and MEK inhibition with targeted drugs in clinical use can overcome acquired resistance in the BRAFV600E NSCLC models and provide a rationale for testing BRAF/MEK-directed polytherapy in patients with BRAFV600E NSCLC.
In tumor cells with class II BRAF-inhibitor resistance that did not harbor aberrant, truncated BRAF (the VR3–VR5 sublines), escape from exclusive dependence on the native BRAFV600E oncogene and from oncogene inhibition occurred through EGFR activation that resulted from sustained EGFR ligand expression driven, in part, by c-Jun. This EGFR activation promoted not only MAPK pathway reactivation but also AKT signaling. Indeed, we confirmed that AKT hyperphosphorylation selectively marked class II resistant tumor cells with EGFR-mediated resistance, distinguishing them from class I resistant tumor cells with p61VE. Our analysis of both the human BRAFV600E NSCLC tumor models and clinical specimens indicated that pAKT levels could be used as a potential clinical biomarker to differentiate BRAF inhibitor-resistant BRAFV600E NSCLCs with EGFR dependence from those that harbor p61VE. The further validation and potential clinical use of pAKT (and pS6) levels as a biomarker of resistance is particularly important, given that pEGFR status alone is likely insufficient to distinguish responder and nonresponder populations of BRAFV600E NSCLCs because of the crosstalk between MAPK and EGFR signaling we uncovered. Thus, the use of pAKT levels as a clinical biomarker may facilitate the selection of patients based on alternative mechanisms directed against either EGFR (for tumors with high pAKT) or MEK (for tumors with low pAKT) in the setting of acquired BRAF-inhibitor resistance.
We noted that the BRAFV600E NSCLC cells with EGFR-mediated resistance and concurrent MAPK and AKT signaling also exhibited evidence of activation of AP-1. Our data uncover a potentially important role for the AP-1 component c-Jun in the control of EGFR ligand expression and EGFR activation and, consequently, BRAF-inhibitor response and resistance in NSCLC. Our data indicate that c-Jun mediates crosstalk between MAPK and EGFR signaling in NSCLC cells. The rewired signaling architecture that emerges under the selective pressure of BRAF oncogene inhibition establishes a positive feedback loop that decreases dependence on oncogenic BRAF through constitutive EGFR ligand expression and autocrine (or paracrine) EGFR signaling that ultimately drives BRAF-inhibitor resistance. Based on our data, it is possible that factors other than EGFR could contribute to BRAF-inhibitor resistance in class II resistant tumor cells. Nevertheless, our data indicate that polytherapy with vemurafenib and an EGFR tyrosine kinase inhibitor could delay and suppress acquired resistance in patients with BRAFV600E NSCLC.
Collectively, our findings uncover critical molecular determinants of BRAF oncogene dependence in NSCLC and, more broadly, provide mechanistic insight into the regulation of oncogene dependence. Our findings could be used in the selection and treatment of responder and nonresponder populations of patients with BRAF-mutant NSCLC early in the clinical deployment of BRAF inhibitors to improve outcomes by subverting the onset of lethal, drug-resistant disease in these patients.
Methods
Generation of Drug-Resistant Cell Lines.
To generate drug-resistant lines, HCC364 cells were exposed to increasing concentration of vemurafenib from 500 nM to 10 μM. Individual populations of cells were confirmed to be drug resistant by standard cell-viability assays. The genetic identity of each subclone with that of the parental cells was verified by short-tandem repeat (STR) analysis conducted according to established protocols at The Johns Hopkins University.
RNA Sequencing and Analysis.
RNA from each of the indicated cell lines was extracted by the RNeasy kit (Qiagen). In total, 2 μg of total RNA was used for deep sequencing library preparation using Illumina Truseq sample preparation kits (Illumina) according to the manufacturer’s protocols. Sequencing libraries with different indices were pooled and sequenced in paired-end format to a length of 100 bp using the HiSeq2000 platform at the Center for Advanced Technology at the University of California, San Francisco. Reads were aligned against National Center for Biotechnology Information (NCBI) build 37 (hg19) of the human genome using the NCBI RefSeq transcript annotation with TopHat version 2 (31, 32). The assembly and quantification of transcripts were performed with Cufflinks (32) without a reference transcriptome. To determine the relative expression of identified BRAF isoforms, Cufflinks analysis was repeated with a reference transcriptome to which the identified BRAF p61 variant was added. The supervised analysis of differential expression was performed using gene-level–aligned read counts in which a gene-wise exact test assessed the difference in the means between two groups (VR1-VR2 vs. VR3–5) of negative-binomially distributed counts using edgeR (33). Significant genes were those with a q-value <10−6. Heatmaps were drawn from the set of genes differentially expressed between the VR1-VR2 and VR3–5 sublines in which the expression values were calculated as the log2 of read count plus a pseudo count of 1. These values then were converted to Z-scores before visualization. Functional enrichment analyses were performed with the molecular signatures database (MSigDB) (34) and were reviewed manually. To summarize the significance of functional enrichment across multiple, independent enriched signatures that fell into one of five distinct categories (as indicated in Fig. 1D), multiple independent P values were combined using Fisher’s method.
Cell-Viability Assay.
Twenty-four hours before drug treatment, 3,000–5,000 cells were plated in each well of 96-well plates. Viable cells were determined 72 h after drug treatment using CellTiter-Glo luminescent cell-viability reagent according to the manufacturer’s protocols (Promega). Each assay consisted of four replicate wells and was repeated at least three times. Data were expressed as the percentage of the cell viability of control cells. The data were displayed graphically using GraphPad Prism version 5.0 for Mac (GraphPad Software Inc.).
Clonal Outgrowth Assay.
Clonal outgrowth studies were conducted in 96-well format using 100 wells for each condition and were conducted in triplicate. Resistant wells were assayed after 2 mo of treatment and are plotted relative to vemurafenib-treated cells.
Combination Drug Screen.
Resistant cells were plated in two 384-well plates and 1 d later were exposed to a library of 94 compounds at a defined concentration along with DMSO or 1 μM vemurafenib. Cells were allowed to proliferate for 72 h, nuclei were stained with Hoechst dye, and cell number was quantified using high-content microscopy. The screen was repeated three times using varying library concentrations of 5 μg/mL, 500 ng/mL, and 50 ng/mL. Each combination was measured in quadruplicate. Raw cell numbers were median normalized on a per-plate basis. For each compound in the library, the relative cell number in the DMSO plate was compared with the number in the vemurafenib plate using a t test. A synergy score was developed based on the −log10 of the P value of the t test and was signed to indicate synergistic inhibition of growth (positive score) or enhanced growth (negative score). The final synergy score is based on the analysis of three different library concentrations.
Immunohistochemical Analyses.
All specimens were acquired from individuals with NSCLC under the auspices of institutional review board-approved clinical protocols at each hospital; informed consent was obtained. Immunohistochemistry for pAKT and pS6 was conducted on matched-pair formalin-fixed paraffin-embedded tumor sections as previously described (35).
Supplementary Material
Acknowledgments
We thank Paul Lin, Martin McMahon, Jonathan Weissman, and members of the T.G.B. laboratory for critical review of the manuscript. T.G.B. received funding from a National Institutes of Health Director's New Innovator Award, the Howard Hughes Medical Institute, the Doris Duke Charitable Foundation, American Lung Association, National Lung Cancer Partnership, the Sidney Kimmel Foundation for Cancer Research, and the Searle Scholars Program. E.A.C. was supported by National Cancer Institute Grant NCI- CA137153.
Footnotes
The authors declare a conflict of interest. Gideon Bollag is an employee of Plexxikon Inc., which owns and markets vemurafenib.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1320956111/-/DCSupplemental.
References
- 1.Sawyers CL. The cancer biomarker problem. Nature. 2008;452(7187):548–552. doi: 10.1038/nature06913. [DOI] [PubMed] [Google Scholar]
- 2.Garraway LA, Jänne PA. Circumventing cancer drug resistance in the era of personalized medicine. Cancer Discov. 2012;2(3):214–226. doi: 10.1158/2159-8290.CD-12-0012. [DOI] [PubMed] [Google Scholar]
- 3.Glickman MS, Sawyers CL. Converting cancer therapies into cures: Lessons from infectious diseases. Cell. 2012;148(6):1089–1098. doi: 10.1016/j.cell.2012.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhang Z, et al. Activation of the AXL kinase causes resistance to EGFR-targeted therapy in lung cancer. Nat Genet. 2012;44(8):852–860. doi: 10.1038/ng.2330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bivona TG, et al. FAS and NF-κB signalling modulate dependence of lung cancers on mutant EGFR. Nature. 2011;471(7339):523–526. doi: 10.1038/nature09870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dhomen N, Marais R. New insight into BRAF mutations in cancer. Curr Opin Genet Dev. 2007;17(1):31–39. doi: 10.1016/j.gde.2006.12.005. [DOI] [PubMed] [Google Scholar]
- 7.Davies H, et al. Mutations of the BRAF gene in human cancer. Nature. 2002;417(6892):949–954. doi: 10.1038/nature00766. [DOI] [PubMed] [Google Scholar]
- 8.Bollag G, et al. Clinical efficacy of a RAF inhibitor needs broad target blockade in BRAF-mutant melanoma. Nature. 2010;467(7315):596–599. doi: 10.1038/nature09454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chapman PB, et al. BRIM-3 Study Group Improved survival with vemurafenib in melanoma with BRAF V600E mutation. N Engl J Med. 2011;364(26):2507–2516. doi: 10.1056/NEJMoa1103782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hauschild A, et al. Dabrafenib in BRAF-mutated metastatic melanoma: Amulticentre, open-label, phase 3 randomised controlled trial. Lancet. 2012;380(9839):358–365. doi: 10.1016/S0140-6736(12)60868-X. [DOI] [PubMed] [Google Scholar]
- 11.Corcoran RB, et al. EGFR-mediated re-activation of MAPK signaling contributes to insensitivity of BRAF mutant colorectal cancers to RAF inhibition with vemurafenib. Cancer Discov. 2012;2(3):227–235. doi: 10.1158/2159-8290.CD-11-0341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Prahallad A, et al. Unresponsiveness of colon cancer to BRAF(V600E) inhibition through feedback activation of EGFR. Nature. 2012;483(7387):100–103. doi: 10.1038/nature10868. [DOI] [PubMed] [Google Scholar]
- 13.Montero-Conde C, et al. Relief of feedback inhibition of HER3 transcription by RAF and MEK inhibitors attenuates their antitumor effects in BRAF-mutant thyroid carcinomas. Cancer Discov. 2013;3(5):520–533. doi: 10.1158/2159-8290.CD-12-0531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Paik PK, et al. Clinical characteristics of patients with lung adenocarcinomas harboring BRAF mutations. J Clin Oncol. 2011;29(15):2046–2051. doi: 10.1200/JCO.2010.33.1280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Brose MS, et al. BRAF and RAS mutations in human lung cancer and melanoma. Cancer Res. 2002;62(23):6997–7000. [PubMed] [Google Scholar]
- 16.Marchetti A, et al. Clinical features and outcome of patients with non-small-cell lung cancer harboring BRAF mutations. J Clin Oncol. 2011;29(26):3574–3579. doi: 10.1200/JCO.2011.35.9638. [DOI] [PubMed] [Google Scholar]
- 17.Imielinski M, et al. Mapping the hallmarks of lung adenocarcinoma with massively parallel sequencing. Cell. 2012;150(6):1107–1120. doi: 10.1016/j.cell.2012.08.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Joseph EW, et al. The RAF inhibitor PLX4032 inhibits ERK signaling and tumor cell proliferation in a V600E BRAF-selective manner. Proc Natl Acad Sci USA. 2010;107(33):14903–14908. doi: 10.1073/pnas.1008990107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Engelman JA, et al. MET amplification leads to gefitinib resistance in lung cancer by activating ERBB3 signaling. Science. 2007;316(5827):1039–1043. doi: 10.1126/science.1141478. [DOI] [PubMed] [Google Scholar]
- 20.Katayama R, et al. Mechanisms of acquired crizotinib resistance in ALK-rearranged lung Cancers. Sci Transl Med. 2012;4(120):20ra17. doi: 10.1126/scitranslmed.3003316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Poulikakos PI, et al. RAF inhibitor resistance is mediated by dimerization of aberrantly spliced BRAF(V600E) Nature. 2011;480(7377):387–390. doi: 10.1038/nature10662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lemmon MA, Schlessinger J. Cell signaling by receptor tyrosine kinases. Cell. 2010;141(7):1117–1134. doi: 10.1016/j.cell.2010.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lito P, et al. Relief of profound feedback inhibition of mitogenic signaling by RAF inhibitors attenuates their activity in BRAFV600E melanomas. Cancer Cell. 2012;22(5):668–682. doi: 10.1016/j.ccr.2012.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chandarlapaty S, et al. AKT inhibition relieves feedback suppression of receptor tyrosine kinase expression and activity. Cancer Cell. 2011;19(1):58–71. doi: 10.1016/j.ccr.2010.10.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schulze A, Lehmann K, Jefferies HB, McMahon M, Downward J. Analysis of the transcriptional program induced by Raf in epithelial cells. Genes Dev. 2001;15(8):981–994. doi: 10.1101/gad.191101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lopez-Bergami P, et al. Rewired ERK-JNK signaling pathways in melanoma. Cancer Cell. 2007;11(5):447–460. doi: 10.1016/j.ccr.2007.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.McCarthy SA, et al. Rapid phosphorylation of Ets-2 accompanies mitogen-activated protein kinase activation and the induction of heparin-binding epidermal growth factor gene expression by oncogenic Raf-1. Mol Cell Biol. 1997;17(5):2401–2412. doi: 10.1128/mcb.17.5.2401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rudin CM, Hong K, Streit M. Molecular characterization of acquired resistance to the BRAF inhibitor dabrafenib in a patient with BRAF-mutant non-small-cell lung cancer. J Thorac Oncol. 2013;8(5):e41–e42. doi: 10.1097/JTO.0b013e31828bb1b3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gautschi O, et al. A patient with BRAF V600E lung adenocarcinoma responding to vemurafenib. J Thorac Oncol. 2012;7(10):e23–e24. doi: 10.1097/JTO.0b013e3182629903. [DOI] [PubMed] [Google Scholar]
- 30.Schulze A, Nicke B, Warne PH, Tomlinson S, Downward J. The transcriptional response to Raf activation is almost completely dependent on Mitogen-activated Protein Kinase Kinase activity and shows a major autocrine component. Mol Biol Cell. 2004;15(7):3450–3463. doi: 10.1091/mbc.E03-11-0807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Trapnell C, Pachter L, Salzberg SL. TopHat: Discovering splice junctions with RNA-Seq. Bioinformatics. 2009;25(9):1105–1111. doi: 10.1093/bioinformatics/btp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Trapnell C, et al. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat Protoc. 2012;7(3):562–578. doi: 10.1038/nprot.2012.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Robinson MD, McCarthy DJ, Smyth GK. edgeR: A bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26(1):139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Subramanian A, et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA. 2005;102(43):15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Engelman JA, et al. Effective use of PI3K and MEK inhibitors to treat mutant Kras G12D and PIK3CA H1047R murine lung cancers. Nat Med. 2008;14(12):1351–1356. doi: 10.1038/nm.1890. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.