Significance
During an immune response, B lymphocytes generate different classes of antibodies better suited to protect against particular pathogens by making two chromosomal cuts that are joined to replace one type of antibody gene with a different one. These cuts happen in widely separated segments of the chromosome that must be physically adjacent to be joined. We have asked how this happens. The surprising answer is that genes and gene segments lying certain distances apart on any chromosome may actually be packaged such that both are frequently touching or nearly touching and, if broken, can be efficiently joined by general processes that repair breaks in all our genes. The joining mechanisms we describe also may contribute to genetic deletions in cancers.
Keywords: intrachromosomal joining, topological domains, double-strand break synapsis
Abstract
Antibody class switch recombination (CSR) in B lymphocytes joins two DNA double-strand breaks (DSBs) lying 100–200 kb apart within switch (S) regions in the immunoglobulin heavy-chain locus (IgH). CSR-activated B lymphocytes generate multiple S-region DSBs in the donor Sμ and in a downstream acceptor S region, with a DSB in Sμ being joined to a DSB in the acceptor S region at sufficient frequency to drive CSR in a large fraction of activated B cells. Such frequent joining of widely separated CSR DSBs could be promoted by IgH-specific or B-cell–specific processes or by general aspects of chromosome architecture and DSB repair. Previously, we found that B cells with two yeast I-SceI endonuclease targets in place of Sγ1 undergo I-SceI–dependent class switching from IgM to IgG1 at 5–10% of normal levels. Now, we report that B cells in which Sγ1 is replaced with a 28 I-SceI target array, designed to increase I-SceI DSB frequency, undergo I-SceI–dependent class switching at almost normal levels. High-throughput genome-wide translocation sequencing revealed that I-SceI–generated DSBs introduced in cis at Sμ and Sγ1 sites are joined together in T cells at levels similar to those of B cells. Such high joining levels also occurred between I-SceI–generated DSBs within c-myc and I-SceI– or CRISPR/Cas9-generated DSBs 100 kb downstream within Pvt1 in B cells or fibroblasts, respectively. We suggest that CSR exploits a general propensity of intrachromosomal DSBs separated by several hundred kilobases to be frequently joined together and discuss the relevance of this finding for recurrent interstitial deletions in cancer.
Immunoglobulin heavy-chain locus (IgH) class switch recombination (CSR) is a deletional recombination process that allows antigen-activated B lymphocytes to change the type of IgH constant region expressed in association with a given variable region and, thereby, to produce different antibody classes. In the mouse, the downstream 200 kb of the IgH locus contain eight sets of constant-region exons (“CHs”) that each encodes a distinct IgH constant region. Each CH is organized into an independent transcriptional unit, with an “I” promoter and noncoding exon followed by a long (1–10 kb), repetitive S region and a set of CH exons encoding a particular constant region (e.g., Cμ, Cγs, Cε, Cα) (1). Mature B cells first express a productively assembled V(D)J exon in conjunction with the adjacent Cμ exons and, thereby, produce IgM. During CSR, double-strand breaks (DSBs) are introduced into the donor S region preceding Cμ (Sμ) and also into a downstream acceptor S region. These DSBs are initiated by activation-induced cytidine deaminase (AID), which is targeted to a specific acceptor S region by transcription initiated from the associated I promoter (1). To complete CSR, DSB ends in the donor Sμ are fused to DSB ends in the acceptor S region to delete intervening sequences, including Cμ, and juxtapose the downstream CH to the V(D)J exon. The joining phase of CSR is carried out largely by the classical nonhomologous end-joining pathway (2).
As CHs that undergo CSR lie 100–200 kb downstream of Sμ, CSR DSB joining usually occurs in cis over relatively long chromosomal distances. AID activity results in multiple DSBs within targeted S regions, many of which are rejoined or joined to other DSBs within the same S region (2). However, stimulation of purified B cells with bacterial lipopolysaccharide (LPS) or αCD40 plus interleukin-4 (IL-4) over a 4-day period leads to as much as 50% or more of the cells to undergo IgG1 CSR by joining Sμ to Sγ1 DSBs, which are separated by about 100 kb (1, 3). In this context, it has been proposed that the level of AID-initiated DSBs within donor and acceptor S regions is high enough to drive physiological CSR levels via the joining of a fraction of the total Sμ DSBs to a fraction of the total DSBs in the downstream target S regions (4, 5).
CSR requires DSBs in S regions separated by 100 kb or more to be physically juxtaposed (“synapsed”) for fusion by end joining. Such long-range synapsis of S-region DSBs might occur by several, not mutually exclusive, mechanisms. One possibility is that AID and/or S regions, potentially due to their ability to form higher order structures, may promote S-region synapsis before or after DSB initiation by AID (1). In addition, the ataxia teleangectasia mutated (ATM)-dependent DNA DSB response has been proposed to contribute to DSB synapsis, given that it can generate foci that spread in chromatin over distances of 100 kb or more flanking sites of DSBs (2). In the latter context, 53BP1, which is activated downstream of ATM, is required for CSR (6, 7), and one proposed 53BP1 role in this context is active synapsis of AID-initiated S-region DSBs (8). The 3D organization of the IgH locus also has been implicated as potentially contributing to CSR synapsis, based on detection of chromatin loops between intronic or 3′IgH enhancer regions that flank the two ends of the CH locus and I-region promoters upstream of given S regions in CSR-activated B cells (9). Finally, it has been hypothesized that 3D organization of chromatin more generally across the genome (10–12) may promote frequent joining of DSBs that lie within megabase-size or smaller domains and that CSR may have evolved to exploit this property (4, 5, 13).
As an approach to begin to distinguish among potential mechanisms of DSB synapsis during CSR, we previously asked whether DSBs generated at yeast I-SceI meganuclease target sites introduced in place of Sγ1 or in place of both Sμ and Sγ1 could support I-SceI–dependent recombinational IgM-to-IgG1 class switching in B cells. When the 10-kb Sγ1 was replaced with two I-SceI sites, CSR to Sγ1 was abrogated; however, class switching to IgG1 could be restored to about 10% of normal levels by ectopic I-SceI expression in activated B cells (5). In this case, recombinational class switching was mediated by joining of I-SceI–generated DSBs at the Sγ1 site to AID-initiated DSBs in Sμ. Indeed, when both the 4-kb Sμ and the 10-kb Sγ1 were each replaced with two I-SceI sites, CSR was abrogated but could again be restored to 10% of normal levels by ectopic I-SceI expression in the absence of AID (5). Here, we address the question of whether more physiological levels of IgH class switching can be achieved via introduction of more frequent I-SceI DSBs at sites of S regions and whether joining of DSBs over 100-kb distances at frequencies comparable to those that promote physiological IgH class switching occurs in cell types other than B lymphocytes and at a genomic site in addition to IgH.
Results
Increased Number of DSB Target Sites Promotes Physiological Levels of I-SceI–Dependent IgG1 Class Switching.
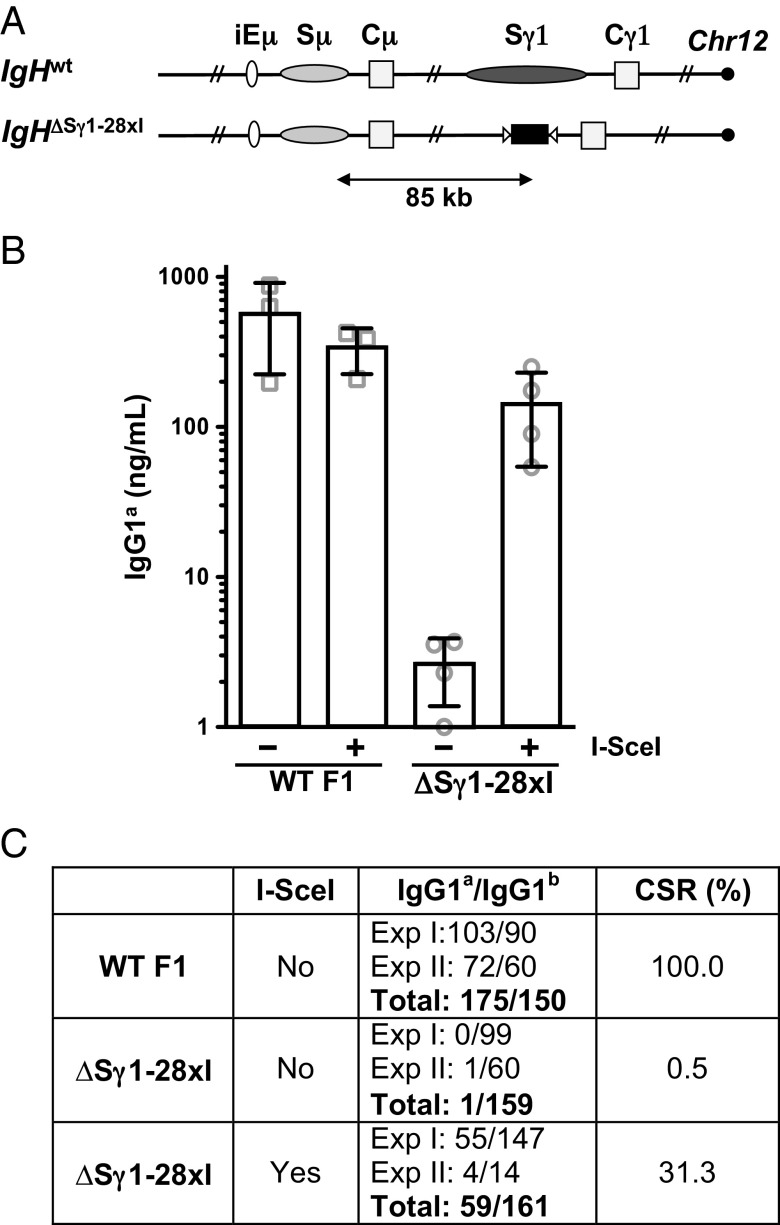
In prior studies, the level of I-SceI–dependent IgG1 class switching from a cassette with just two I-SceI target sites was only 10% of that of normal CSR, potentially due to much greater numbers of AID-initiated DSBs within the 10-kb Sγ1 region driving the reaction (5). To test this hypothesis, we used an approach based on EF1 embryonic stem (ES) cells, which derive from 129/Sv-C57BL/6 F1 mice and are heterozygous for the IgHa and IgHb alleles (5). We generated an EF1 ES cell line with a modified IgHa allele that contained an array of 28 I-SceI sites in place of Sγ1 (referred to as the IgHΔSγ1-28×I allele; Fig. 1A and Fig. S1). These ES cells were then used for RAG-2–deficient blastocyst complementation (“RDBC”) (14) to generate chimeric mice in which all mature lymphocytes, including splenic B cells, harbor the ES cell-derived mutant IgHa allele. Purified splenic B cells from WT F1 and IgHΔSγ1-28×I mice were stimulated in culture with LPS and IL-4 to induce CSR to IgG1 and also infected with either control or I-SceI–expressing retrovirus on day 1 of stimulation. Efficiency of infection, as evaluated by IRES-mediated GFP expression, ranged from 50% to 80%. On day 6, culture supernatants were collected and IgG1a secretion was measured by ELISA (Fig. 1B). Hybridomas were also generated from day 4 cultures to confirm recombinational class switching (Fig. 1C).
Fig. 1.
I-SceI–mediated IgH class switching is dependent on DSB frequency. (A) Schematic representation of the WT IgH locus and the IgHΔSγ1-28×I modified allele (not to scale). Black rectangle, I-SceI target cassette; white triangles, LoxP sites. (B) IgG1a class switching as measured by ELISA on supernatants from day 6 LPS/IL-4 B-cell cultures of the indicated genotypes, infected with either control (−) or I-SceI-expressing (+) retrovirus. Error bars represent SD. (C) Ratio of IgG1a/IgG1b in hybridomas obtained from WT or IgHΔSγ1-28×I B cells stimulated for 4 days as in A. Relative CSR frequency is defined by the ratio of IgG1a- to IgG1b-producing hybridomas and is arbitrarily set as 100% for WT cells. Part of the WT data are adopted from our previous study (5).
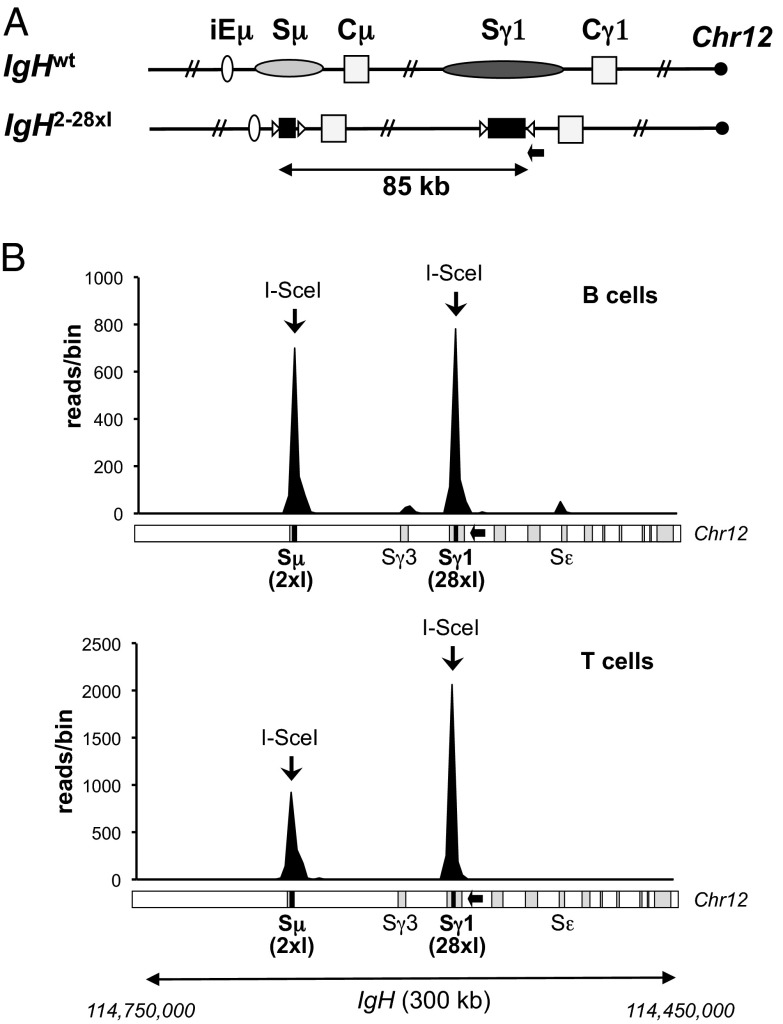
Infection with the I-SceI–expressing retrovirus had no obvious effect on IgG1a secretion in WT B cells; in addition, control-infected IgHΔSγ1-28×I B cells showed only very low background levels of IgG1a secretion (Fig. 1B). However, I-SceI–mediated switching to IgG1a in IgHΔSγ1-28×I B cells reached levels that were, on average, 40% of WT levels, within a range that overlapped with that of WT (Fig. 1B). Similar results (Fig. S2) were obtained with day 4 LPS/IL-4–stimulated F1 B cells carrying an IgHa modified allele in which two I-SceI sites replaced Sμ and 28 I-SceI sites replaced Sγ1 (“IgH2-28×I”; Fig. 2A). These results suggest that frequent DSBs at I-SceI sites in place of S regions can mediate IgG1 class switching at levels approaching those of AID-initiated CSR to IgG1 in WT B cells.
Fig. 2.
I-SceI–mediated IgH long-range joining is frequent in B and T cells. (A) Schematic of the WT IgH locus and the IgH2-28×I modified allele (not to scale); symbols are as in Fig. 1. The position of primers used for HTGTS is indicated by a black arrow. (B) Linear plots representing junctions mapping to a 300-kb region spanning the IgH locus, obtained in HTGTS libraries from IgH2-28×I activated B (Upper) and T (Lower) cells. Data are combined from three independent libraries per cell type. A schematic of the IgH locus is shown at the bottom for reference. The 300-kb region comprises Chr12:114,450,000–114,750,000 with I-SceI site cassettes at positions 114,568,854 (Sγ1) and 114,661,018 (Sμ), indicated by black bars. Bin size is 3 kb.
Following generation of hybridomas from control and mutant B-cell cultures, we identified those that expressed IgG1 by ELISA. As B cells undergo productive V(D)J recombination on one of their two IgH alleles and the process is random, about one-half of the IgM-positive B cells in an F1 population express IgMa and the other half expresses IgMb. Correspondingly, ELISA studies revealed that IgG1-positive hybridomas from WT F1 B cells were almost evenly distributed between IgG1a- and IgG1b-positive cells (Fig. 1C). However, of 160 IgHΔSγ1-28×I hybridomas generated from control virus infected cells, all but one were IgG1b-positive, confirming the requirement for I-SceI DSBs to support IgG1 switching on the mutant allele. In contrast, about 30% (59 of 220) of IgHΔSγ1-28×I hybridomas generated subsequent to ectopic I-SceI expression were IgG1a-positive (Fig. 1C). DNA sequences of the rearranged IgHa alleles from a series of IgG1a-expressing IgHΔSγ1-28×I hybridomas confirmed that each had joined DSBs in the 28× I-SceI cassette to DSBs in Sμ (Fig. S3). Thus, the hybridoma findings directly demonstrate that increasing the number of I-SceI–generated DSBs in place of the Sγ1 acceptor S region supports increased I-SceI–dependent IgG1 recombinational class switching.
Robust Joining of I-SceI–Generated IgH Locus DSBs at Sμ and Sγ1 in T Lymphocytes.
The ability of I-SceI–dependent DSBs in place of Sμ and Sγ1 to promote substantial IgH recombinational class switching could be promoted, all or in part, by a B-cell–specific conformation of the IgH locus that leads to their synapsis (9). To test such possibilities, we used high-throughput genome-wide translocation sequencing (HTGTS) to compare joining levels of I-SceI–generated DSBs at the Sμ location to I-SceI–generated DSBs at the Sγ1 location in activated IgH2-28×I B and T cells. The HTGTS approach was developed to follow the joining of a “bait” I-SceI DSB to other “prey” DSBs genome-wide (15) based on the nucleotide sequence of recovered bait-to-prey DBS junctions. Although a genome-wide method, HTGTS can also be used to assess joins between prey DSB ends and bait I-SceI DSB ends more focally, as demonstrated for joining of an I-SceI break in Sγ1 to AID-initiated DSBs in other S regions (15). For these experiments, IgH2-28×I purified splenic B were activated with αCD40 plus IL-4 to induce CSR to IgG1 and IgE, and splenic T cells were activated with Con A plus interleukin-2 (IL-2) for 4 days, with retroviral I-SceI expression introduced on day 1. HTGTS then was performed on activated B- or T-cell DNA by using PCR primers located on the centromeric side of the 28× I-SceI cassette inserted in place of Sγ1. This strategy captures junctions from the 3′ end of the bait I-SceI DSB (“3′-Sγ128×I DSB ends”) to other DSBs genome-wide, including those introduced into the 2× I-SceI cassette replacing Sμ (Fig. 2A).
From three independent HTGTS B-cell libraries, we obtained more than 2,000 genome-wide junctions (Fig. S4 A and B). However, most junctions occurred within a 300-kb region containing the CH portion of the IgH locus and the two sets of I-SceI target sites (Fig. 2B, Upper, and Fig. S4). Of these, over 40% occurred near the bait I-SceI site and predominantly reflected rejoining of I-SceI breaks in the 28× cassette following their resection, as expected (Fig. 2B, Upper, and Figs. S4 and S5; also see ref. 15). This estimate of break site junctions does not account for perfectly rejoined DSBs, and, thus, the relative frequency of break site joins could be higher. Notably, joining of 3′-Sγ128×I DSB ends to DSBs generated from the 2× I-SceI cassette in place of Sμ was substantial, accounting for over 40% of the total genome-wide junctions recovered (Fig. 2B, Upper, and Fig. S4). Of these junctions, only about 50% were within the 220-bp 2× I-SceI cassette, with less than 1% deriving from “perfect” joining events between two I-SceI sites, whereas the remaining 50% extended up to 10 kb from the break site (Fig. S4D). These results indicate that I-SceI–induced DSBs undergo extensive end processing before joining. Based on our IgG1 class-switching assays of the same cells (Fig. S2), the frequency of HTGTS joins between the I-SceI sites inserted in place of Sγ1 and Sμ corresponds to IgH class-switching levels of up to 30–40% of those of WT. The Sε region, which is a target of AID-initiated DSBs following αCD40/IL-4 stimulation, and the Sγ1-proximal Sγ3 region, which may be a lower-level AID target, were the other most frequent hot spots (Fig. 2B, Upper). Finally, as expected, a small percentage of reads also mapped to the WT Sγ1 and Sμ regions, representing translocations of the I-SceI DSBs to AID-initiated DSBs on the other copy of chromosome 12 (Fig. S5, Upper) (15).
From three independent activated T-cell HTGTS libraries, we obtained over 4,000 junctions (Fig. S4 A and B). Similarly to B cells, a substantial fraction of these junctions, mostly representing resection events, occurred at the Sγ1 I-SceI break site (Fig. 2B, Lower, and Figs. S4 and S5). As expected, given that AID is not expressed in activated T cells, junctions of 3′-Sγ128×I DSB ends to endogenous S regions did not occur at levels above background (Fig. 2B, Lower, and Fig. S5, Lower). However, junctions of 3′-Sγ128×I DSB ends to DSBs originating from the Sμ 2× I-SceI cassette occurred in T cells at frequencies approaching those observed in B cells (Fig. 2B and Fig. S4C), indicating that frequent joining between DSBs in Sγ1 and Sμ locations also occurs in T cells; indeed, by comparison with B cells, at levels high enough to support substantial CSR.
Frequent Joining Between DSBs Separated by 100 kb at the c-myc Locus.
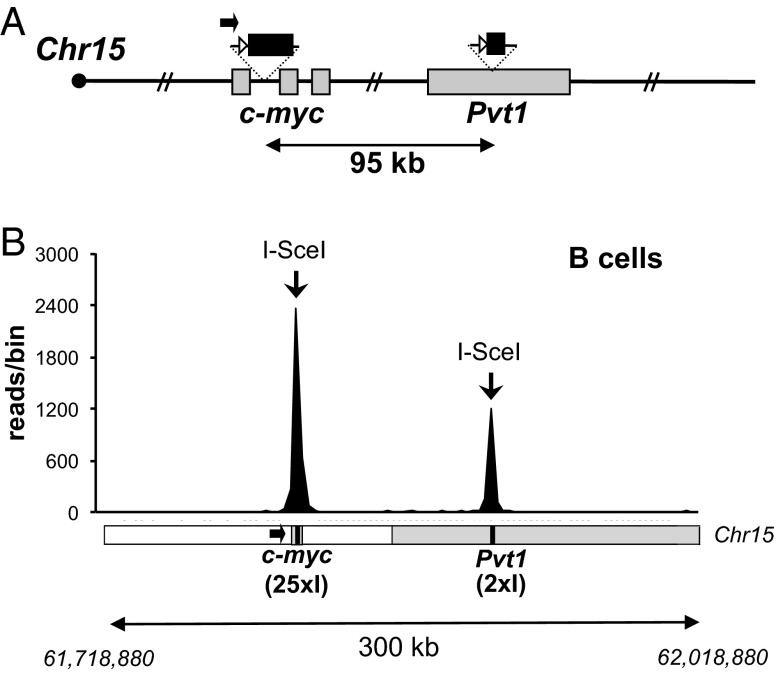
Although frequent joining between I-SceI–generated DSBs at Sγ1 and Sμ occurs in both B and T lymphocytes, it remains possible that this is due to specific IgH locus features. To ask whether similar, high-frequency joining is observed for widely separated DSBs outside of IgH, we generated murine ES cells with a modified chromosome 15 carrying a 25× I-SceI cassette inserted into intron 1 of c-myc (15) and a 2× I-SceI site cassette inserted into the Pvt1 locus, 95.7 kb downstream; we refer to this modified allele as “c-myc25-2×I” (Fig. 3A and Fig. S6). We chose to insert the 2× I-SceI cassette into the Pvt1 locus, which is a frequent target of translocations in B-cell lymphomas, because the distance from the c-myc I-SceI cassette is similar to that separating the two I-SceI cassettes in place of Sμ and Sγ1 in IgH. The c-myc25-2×I ES cells were used for RDBC, and purified splenic B cells from the resulting chimeras were activated and infected with I-SceI–expressing retrovirus as outlined above. HTGTS was used to isolate junctions between the 5′ end of the I-SceI DSBs in c-myc (“5′-c-myc25×I DSB ends”) and other cellular DSBs. The majority of more than 6,000 recovered junctions from three independent B-cell libraries (Fig. S7) mapped to a 300-kb region encompassing the I-SceI target sites (Fig. 3B and Fig. S7). Joining events within 30 kb of the 25× c-myc cassette, mostly representing resections (Fig. S8A), accounted for about 50% of total junctions (Fig. S7C). Strikingly, however, ∼25% of total recovered junctions corresponded to “long-range” joins of 5′-c-myc25×I DSB ends to DSBs at or near the 2× I-SceI cassette in Pvt1 (Fig. 3B and Fig. S7). End processing of these junctions was also substantial; only 2% corresponded to “perfect” joins, whereas 51% mapped outside the 180-bp 2× I-SceI cassette (Fig. S7D). As expected (15), the Sμ, Sγ1, and Sε regions of the IgH locus, which undergo AID-mediated DSBs during CSR, were the other most frequent hot spots in these libraries (Figs. S7C and S8B). These results show that high-frequency joining of DSBs in cis over 100-kb distances in lymphocytes occurs in loci other than IgH.
Fig. 3.
Efficient I-SceI–mediated long-range joining at the c-myc locus. (A) Schematic of the modified c-myc25-2×I allele (not to scale), as in Fig. 2. (B) Linear plots representing junctions mapping to a 300-kb region spanning the c-myc and Pvt1 loci, obtained in HTGTS libraries from c-myc25-2×I activated B cells. Data are combined from three independent libraries. (Lower) Schematic of the 300-kb region spanning Chr15:61,718,880–62,018,880; the c-myc and Pvt1 break sites are at positions 61,818,876 and 61,914,629, respectively. Bin size is 3 kb.
Frequent Long-Range Joining of I-SceI–Mediated DSBs to CRISPR/Cas9-Mediated DSBs in Fibroblasts.
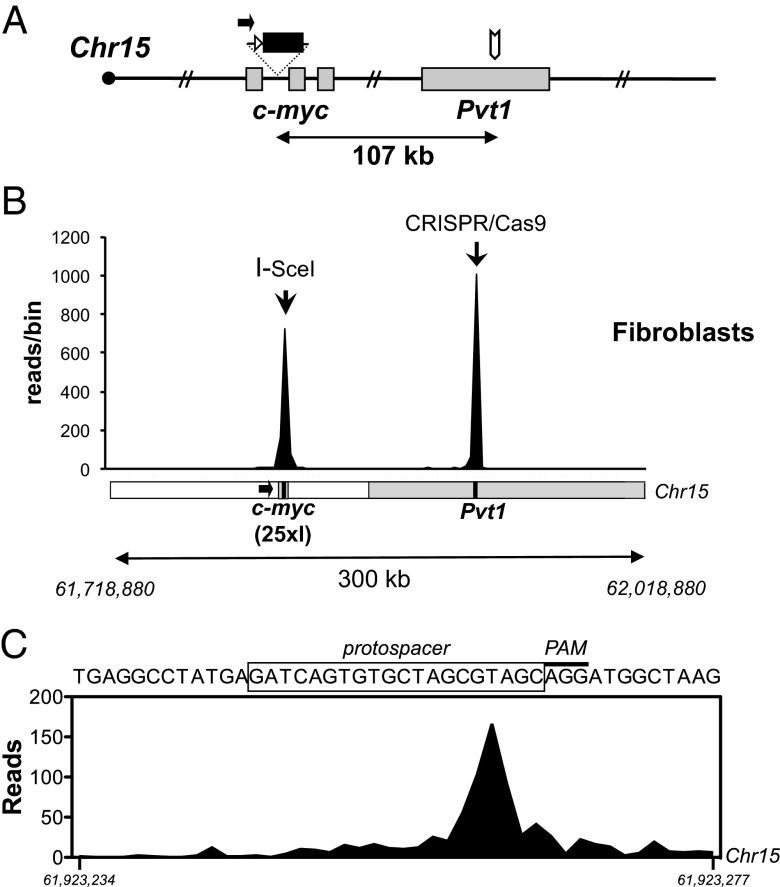
To investigate whether high-level joining of I-SceI–generated DSBs over 100-kb distances occurs in nonlymphoid cells and whether it could be observed with two different types of DSBs, we performed HTGTS on murine tail fibroblasts in which bait DSBs were the 5′-c-myc25×I DSB ends and additional targeted DSBs were generated 107 kb downstream in the Pvt1 locus (Fig. 4 A and B) via the CRISPR/Cas9 system (16, 17). For this purpose, Cas9 nuclease and RNA components required for Cas9 targeting to a 20-bp genomic target sequence (“protospacer”; Fig. 4C) in the Pvt1 locus were coexpressed from a plasmid termed “pX-Pvt1” (16, 17). To induce simultaneous DSBs in c-myc and Pvt1, fibroblasts homozygous for the 25× I-SceI cassette in c-myc and hemizygous for a Rosa26-targeted I-SceI–glucocorticoid receptor (GR) fusion transgene (“myc25×I;ROSAI-SceI-GR fibroblasts”) were transfected with pX-Pvt1, followed by treatment with the GR analog TA to activate the I-SceI–GR fusion protein (15). Genome-wide translocations to I-SceI–mediated c-myc bait DSBs then were identified by HTGTS as outlined above.
Fig. 4.
Long-range joining between I-SceI–mediated c-myc and CRISPR/Cas9-mediated Pvt1 breaks in fibroblasts. (A) Schematic of the modified c-myc25×I allele, as in Fig. 3, showing the CRISPR/Cas9 target site in the Pvt1 locus (white arrow). (B) Linear plot showing junctions mapping to a 300-kb region spanning the c-myc and Pvt1 loci (as in Fig. 3), obtained in HTGTS libraries from c-myc25×I;ROSAI-SceI-GR fibroblasts expressing the Pvt1-specific CRISPR/Cas9 and treated with TA. Data are combined from four independent experiments. A schematic of the locus (as in Fig. 3) is shown for reference. Bin size is 3 kb. (C) Identified c-myc/Pvt1 joining events peak 4 bp 5′ of the PAM. The protospacer sequence (black box) corresponds to position Chr15:61,923,246–61,923,265.
In four independent HTGTS experiments, we obtained almost 3,000 junctions between 5′-c-myc25×I DSB ends and other DSB ends genome-wide, nearly 40% of which represented joins between I-SceI– and CRISPR/Cas9-induced DSBs (Fig. 4B and Fig. S9). Indeed, the majority (71%) of these I-SceI to CRISPR/Cas9 junctions occurred within the 20-bp Pvt1 protospacer target sequence (Fig. 4C and Fig. S9E), consistent with 5′-c-myc25×I DSB ends joining to the blunt DSB introduced by Cas9 three base pairs 5′ of the protospacer-adjacent motif (PAM) (16, 18–20). As about 80% of the recovered junctions in this region occurred within 200 bp of the expected Cas9 break site in Pvt1 (Fig. S9E), CRISPR/Cas9-induced breaks that joined to 5′-c-myc25×I DSB ends, on average, showed less end processing than I-SceI–induced breaks that joined to either 5′-c-myc25×I or 3′-Sγ128×I DSB ends (Figs. S4D, S7D, and S9E). Whether such apparent difference in the degree of DSB end processing before joining reflects the type of breaks being joined (e.g., blunt CRISPR/Cas9-generated ends versus 3′-overhang I-SceI–generated ends) (18, 21), location of the breaks, or other factors, will require further investigation. Overall, these findings demonstrate that high-frequency intrachromosomal joining over distances of ∼100 kb occurs in cells other than lymphoid cells and between different types of DSBs.
Discussion
We previously found that I-SceI–generated DSBs at a pair of I-SceI target sites that replaces Sμ and Sγ1, respectively, support I-SceI–dependent IgM-to-IgG1 recombinational class switching at about 5% of WT B-cell levels and that such I-SceI–dependent IgH class switching occurs in the absence of S regions and AID (5). These findings led us to hypothesize that AID functions in CSR primarily to initiate DSBs and that S regions mainly serve as substrates to promote sufficient levels of AID-initiated DSBs to drive physiological CSR DSB joining over 100- to 200-kb distances via general DNA DSB response and repair mechanisms (5). This model predicted that increasing the numbers of I-SceI DSBs in this system should drive higher levels of IgH class switching. We now have confirmed this prediction by showing that inserting a cassette of 28 I-SceI sites in place of Sγ1 supports I-SceI–dependent IgM-to-IgG1 recombinational class switching in activated B cells at levels of 30% or more of those of normal cells. To put this in perspective, CSR in appropriately activated normal B cells can result in 50% or more being driven to undergo AID-dependent IgG1 class switching over a 4- to 5-day period (3). These findings support the model that introduction of sufficiently frequent DSBs at the positions of Sμ and Sγ1 can provide physiological CSR levels in the absence of any specialized “long-range” synapsis functions of S regions or AID (5). Our current findings are also consistent with the hypothesis that the S-region–based mechanism of CSR evolved from an earlier SHM process, with primitive S regions using numerous palindromic SHM motifs that allow AID to generate a sufficient number of DSBs to drive physiological CSR via general DSB synapsis and joining mechanisms (1).
The frequency at which the ends of two separate DSBs are joined within the genome of cells is influenced by several factors, including the frequency of DSBs at each site and the frequency at which they are physically juxtaposed (“synapsed”) for joining (4, 13). In this regard, the question arises as to how I-SceI DSBs separated by 100 kb in place of Sγ1 and Sμ within IgH are synapsed for such frequent joining. Our current findings rule out a requisite role for AID or S-region sequences in this process. Moreover, our findings of high-frequency joining of IgH DSBs separated by 100 kb in cell types other than B cells and the similarly frequent joining of DSBs over equivalent distances in the c-myc and Pvt1 loci now demonstrate that this phenomenon is neither IgH locus nor B-cell specific. In this regard, our studies of the translocation of I-SceI–generated DSBs to IR-induced genome-wide DSBs in G1-arrested pro-B cells showed that DSBs lying in cis on the same chromosome have a much higher probability of translocating to each other than to other genomic DSBs (13), and that, within a chromosome, the highest frequency joining occurs for two DSBs within a megabase or less (4). Thus, our current and prior studies indicate that two DSBs lying within several hundred kilobases are synapsed frequently enough to support such high-frequency joining.
There are several potential mechanisms, not mutually exclusive, that may contribute to high-frequency synapsis of sequences separated in cis by several hundred kilobases. We have previously suggested the possibility that the ATM-dependent DNA DSB response, which modifies chromatin on either side of a DSB over hundreds of kilobases, may, beyond tethering DSBs for end joining, also facilitate synapsis over 100-kb distances during CSR (22). In this regard, the 53BP1 DSB response factor has been implicated in the movement of telomeric “breaks” (23). Beyond this, DSBs can rapidly diffuse over distances of 1 μm (equivalent to a few hundred kilobases) via Brownian motion (24); such local movement could potentially contribute to frequent synapsis of sequences separated by such distances (13). In addition, various recent studies indicate that chromatin is organized into megabase or submegabase topological domains that contribute to increased frequency of interactions between sequences within them (10, 12, 25, 26). Our current studies provide further support for our proposed model that CSR evolved to exploit general mechanisms that promote frequent DSB synapsis over submegabase distances (5). More specifically, we suggest that one or more of the general mechanisms outlined above lead to frequent synapsis of S regions, potentially enhanced by DSBs, in activated B cells. Moreover, we suggest that AID induces sufficiently high levels of DSBs in target S regions such that the probability of DSBs being present in these synapsed S regions is high enough to drive physiological CSR in a large fraction of activated B cells over a several-day period (5).
Our current findings suggest that the general mechanisms we outline above could support recombinational class switching in many different chromosomal locations separated by several hundred kilobases if appropriate IgH coding sequences were present and DSBs were introduced in the appropriate locations. More generally, our findings have implications for the interstitial deletions that frequently occur in cancer. In this regard, the processes we now describe could be mechanistically relevant to a number of recurrent interstitial oncogenic deletions associated with T- and B-cell acute lymphoblastic leukemias (27, 28) and with early T-cell precursor acute lymphoblastic leukemias (29). Our findings may also be relevant for mechanisms underlying some of the focal chromosomal deletions found in many different cancers (30) and that have been implicated in oncogenesis (31).
Materials and Methods
Generation of IgHΔSγ1–28×I, IgH2-28×I, and c-myc25-2×I Mice.
To generate the 0.5-kb 28× I-SceI repeat, an oligonucleotide containing the I-SceI site was oligomerized by sequential cloning into the BamHI site of the S85 vector (32). After each cloning step, correct insert orientation was confirmed by sequencing and restriction analysis. I-SceI target site repeats were confirmed to be unidirectional and ligated into the targeting vector, as previously described (32, 33). To generate the IgHΔSγ1-28×I and IgH2-28×I alleles, the targeting construct was transfected into EF1 (129/Sv × C57BL/6) ES cells in which the Sγ1a was deleted, with either WT Sμ or with Sμ replaced by a 2× I-SceI cassette (5). Targeted clones were identified by Southern blotting as described (5) (Fig. S1). To generate the c-myc25-2×I replacement allele, c-myc25×I ES cells (15) were used to insert a 2× I-SceI cassette at position Chr15:61,914,629 in Pvt1 by gene targeting, using a 4.3-kb 5′- and a 4-kb 3′-homology arm, and targeted clones were identified by Southern blotting (Fig. S7). Targeted ES cells were infected with Cre-recombinase–expressing adenovirus to remove the neomycin resistance gene and used for RDBC. Chimeric mice were analyzed at 8–16 wk of age. Animal experiments were performed under protocols approved by the Institutional Animal Care and Use Committee of Boston Children’s Hospital (Protocol 11-11-2074R).
B- and T-Cell Culture and FACS Analysis.
Mature B and T cells were separated from total spleen cell suspensions using α-CD43 magnetic microbeads (Miltenyi). The CD43-negative fraction (B cells) was cultured with either α-CD40 (1 μg/mL; eBioscience) or LPS (25 ng/mL; Sigma) plus IL-4 (20 ng/mL; PeproTech); the CD43-positive fraction (T cells) was cultured with Con A (2.5 μg/mL; Sigma) and IL-2 (R&D Systems) for 4 days. Infection with I-SceI–expressing or control retrovirus was performed at day 1 as previously described (15). At day 4, infection efficiency was evaluated by flow cytometry as the percentage of cells expressing the retroviral IRES-GFP and ranged from 40% to 80%.
ELISA and Hybridoma Generation.
IgG1a secretion levels were determined by ELISA in day 4 or 6 B-cell culture supernatants by using a monoclonal α-mouse IgG1a antibody (BD Pharmingen) followed by alkaline phosphatase-conjugated goat α-mouse IgG1 antibody (Southern Biotech) for detection. For day 4 assays, levels of Ig light-chain expression were measured in the same supernatants with goat α-mouse κ and λ antibodies (Southern Biotech) and IgG1a values were normalized based on Ig light-chain amounts to account for variability in cell number across different cultures. Hybridomas were generated as previously described (5) from day 4-stimulated B cells and assayed by ELISA, as above. Genomic DNA from selected hybridomas was subjected to nested PCR to isolate and sequence Sμ to 28×I cassette junctions (5).
HTGTS.
Libraries were prepared by the adapter-PCR method from 30 to 50 μg of B- or T-cell genomic DNA as previously described (15), with the following modifications. All libraries, excluding one c-myc25-2×I B-cell library, were prepared using sonication to fragment DNA to a 500- to 3,000-bp size range. Fragmented DNA was then end repaired with T4 DNA polymerase, T4 polynucleotide kinase, and DNA polymerase I, large Klenow fragment (NEB) according to published protocols, followed by A-tailing with 3′–5′ exo- Klenow polymerase and linker ligation. Blocking digestion was carried out with XbaI for c-myc25-2×I libraries and with NotI for IgH28-2×I libraries. The following locus-specific primers were used for nested PCR amplification: biotinylated mycH (5′-AGCAGCTGCTAGTCCGACGA-3′) followed by mycPreLox (5′-ACCGCCGCTAATTCCGATCATATTC-3′) for the c-myc locus; biotinylated Tel-Sg1 (5′-TAGAAGGCCGCTCTTTTGC-3′) followed by Tel-Sg1-6F (5′-GCAGGAATTCGATATCAAGCTA-3′) for the IgH locus. Libraries were barcoded and sequenced on the Illumina MiSeq platform. Data analysis was performed as previously described (15), using modified reference genomes (based on the National Center for Biotechnology Information Build37/mm9 assembly of the mouse genome) in which the sequence of the 2× I-SceI site cassette was inserted either in place of Sμ in the IgH locus or in the Pvt1 locus. Junctions within the 2× I-SceI cassette were further inspected to rule out and remove potential artifacts. Combined libraries were further processed for removal of duplicate reads.
For I-SceI/CRISPR/Cas9 HTGTS, 44–50 μg of genomic DNA were used for library preparation, as described above, with the following modifications: after A-tailing and linker ligation, two rounds of PCR were performed; a first-round PCR was carried out using a biotinylated 5′ primer (MycL; 5′-CGAGCGTCACTGATAGTAGGGAGT-3′) and standard 3′ primer (AP1; 5′-GTAATACGACTCACTATAGGGC-3′). Subsequent to capture of biotinylated amplification products on streptavidin-coated magnetic beads, libraries were further amplified in a second, nested, emulsion-based PCR by using primers containing Illumina flow cell adapter sequences with (MiSeq-MID-MycPreLox; 5′-AAT GAT ACG GCG ACC ACC GAG ATC TAC ACT CTT TCC CTA CAC GAC GCT CTT CCG ATC T-MID-ACCGCCGCTAATTCCGATCATATTC-3′) or without (MiSeq-AP2; 5′-CAA GCA GAA GAC GGC ATA CGA GAT CGG TCT CGG CAT TCC TGC TGA ACC GCT CTT CCG ATC TAC TAT AGG GCA CGC GTG GT-3′) multiplex identifiers. Samples were ether-extracted, subjected to PCR purification, and library size selection, followed by sequencing.
CRISPR/Cas9-Mediated DSB Induction.
To induce CRISPR/Cas9-mediated DSBs in the Pvt1 locus, oligonucleotides (5′-CACCGATCAGTGTGCTAGCGTAGC-3′; 5′-AAACGCTACGCTAGCACACTGATC-3′) were annealed and ligated into the BbsI-digested pSpCas9(BB) expression construct [Addgene plasmid 42230 (16)] to generate pX-Pvt1.
SV40 large T-antigen–transformed 25×-I-SceI-c-mycI/I;ROSA-GR-I-SceI+/w (c-myc25×I;ROSAI-SceI-GR) murine tail fibroblasts were grown in a 50% (vol/vol) mix of DMEM/Ham’s F-10 medium supplemented with 19% (vol/vol) heat-inactivated FCS, 1× MEM nonessential amino acids solution (Gibco), 1.12 mM sodium pyruvate, 2.25 mM glutamine, 18.7 mM Hepes, penicillin–streptomycin (112 U/mL), and amphotericin B (0.25 μg/mL). Twelve to 16 h after pX-Pvt1 transfection, TA (10 μM) was added, and fibroblasts were harvested 3.5 days later and processed for HTGTS.
Supplementary Material
Acknowledgments
This work was supported by grants from the National Institute of Allergy and Infectious Diseases (Grant R01AI077595) and the National Cancer Institute (Grants R01CA098285 and P01CA109901) of the National Institutes of Health (to F.W.A.). F.W.A. is an Investigator of the Howard Hughes Medical Institute. M.G. was a V Foundation Scholar and is supported by an Eleanor and Miles Shore/Boston Children’s Hospital Career Development Fellowship Award. R.C. was supported by Grants FP7 ERC-2009-StG (Proposal 242965, “Lunely”), AIRC IG-12023, and AICR 12-0216.
Footnotes
The authors declare no conflict of interest.
Data deposition: The HTGTS data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (accession no. GSE53755).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1324176111/-/DCSupplemental.
References
- 1.Chaudhuri J, et al. Evolution of the immunoglobulin heavy chain class switch recombination mechanism. Adv Immunol. 2007;94:157–214. doi: 10.1016/S0065-2776(06)94006-1. [DOI] [PubMed] [Google Scholar]
- 2.Boboila C, Alt FW, Schwer B. Classical and alternative end-joining pathways for repair of lymphocyte-specific and general DNA double-strand breaks. Adv Immunol. 2012;116:1–49. doi: 10.1016/B978-0-12-394300-2.00001-6. [DOI] [PubMed] [Google Scholar]
- 3.Cheng HL, et al. Integrity of the AID serine-38 phosphorylation site is critical for class switch recombination and somatic hypermutation in mice. Proc Natl Acad Sci USA. 2009;106(8):2717–2722. doi: 10.1073/pnas.0812304106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Alt FW, Zhang Y, Meng FL, Guo C, Schwer B. Mechanisms of programmed DNA lesions and genomic instability in the immune system. Cell. 2013;152(3):417–429. doi: 10.1016/j.cell.2013.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zarrin AA, et al. Antibody class switching mediated by yeast endonuclease-generated DNA breaks. Science. 2007;315(5810):377–381. doi: 10.1126/science.1136386. [DOI] [PubMed] [Google Scholar]
- 6.Manis JP, et al. 53BP1 links DNA damage-response pathways to immunoglobulin heavy chain class-switch recombination. Nat Immunol. 2004;5(5):481–487. doi: 10.1038/ni1067. [DOI] [PubMed] [Google Scholar]
- 7.Ward IM, et al. 53BP1 is required for class switch recombination. J Cell Biol. 2004;165(4):459–464. doi: 10.1083/jcb.200403021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Reina-San-Martin B, Chen J, Nussenzweig A, Nussenzweig MC. Enhanced intra-switch region recombination during immunoglobulin class switch recombination in 53BP1-/- B cells. Eur J Immunol. 2007;37(1):235–239. doi: 10.1002/eji.200636789. [DOI] [PubMed] [Google Scholar]
- 9.Kenter AL, et al. Three-dimensional architecture of the IgH locus facilitates class switch recombination. Ann N Y Acad Sci. 2012;1267:86–94. doi: 10.1111/j.1749-6632.2012.06604.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dixon JR, et al. Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature. 2012;485(7398):376–380. doi: 10.1038/nature11082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lieberman-Aiden E, et al. Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science. 2009;326(5950):289–293. doi: 10.1126/science.1181369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nagano T, et al. Single-cell Hi-C reveals cell-to-cell variability in chromosome structure. Nature. 2013;502(7469):59–64. doi: 10.1038/nature12593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang Y, et al. Spatial organization of the mouse genome and its role in recurrent chromosomal translocations. Cell. 2012;148(5):908–921. doi: 10.1016/j.cell.2012.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen J, Lansford R, Stewart V, Young F, Alt FW. RAG-2-deficient blastocyst complementation: An assay of gene function in lymphocyte development. Proc Natl Acad Sci USA. 1993;90(10):4528–4532. doi: 10.1073/pnas.90.10.4528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chiarle R, et al. Genome-wide translocation sequencing reveals mechanisms of chromosome breaks and rearrangements in B cells. Cell. 2011;147(1):107–119. doi: 10.1016/j.cell.2011.07.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cong L, et al. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339(6121):819–823. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ran FA, et al. Genome engineering using the CRISPR-Cas9 system. Nat Protoc. 2013;8(11):2281–2308. doi: 10.1038/nprot.2013.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gasiunas G, Barrangou R, Horvath P, Siksnys V. Cas9-crRNA ribonucleoprotein complex mediates specific DNA cleavage for adaptive immunity in bacteria. Proc Natl Acad Sci USA. 2012;109(39):E2579–E2586. doi: 10.1073/pnas.1208507109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jinek M, et al. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. 2012;337(6096):816–821. doi: 10.1126/science.1225829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mali P, et al. RNA-guided human genome engineering via Cas9. Science. 2013;339(6121):823–826. doi: 10.1126/science.1232033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Richardson C, Elliott B, Jasin M. Chromosomal double-strand breaks introduced in mammalian cells by expression of I-Sce I endonuclease. Methods Mol Biol. 1999;113:453–463. doi: 10.1385/1-59259-675-4:453. [DOI] [PubMed] [Google Scholar]
- 22.Bassing CH, Alt FW. H2AX may function as an anchor to hold broken chromosomal DNA ends in close proximity. Cell Cycle. 2004;3(2):149–153. doi: 10.4161/cc.3.2.689. [DOI] [PubMed] [Google Scholar]
- 23.Dimitrova N, Chen Y-CM, Spector DL, de Lange T. 53BP1 promotes non-homologous end joining of telomeres by increasing chromatin mobility. Nature. 2008;456(7221):524–528. doi: 10.1038/nature07433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Marshall WF, et al. Interphase chromosomes undergo constrained diffusional motion in living cells. Curr Biol. 1997;7(12):930–939. doi: 10.1016/s0960-9822(06)00412-x. [DOI] [PubMed] [Google Scholar]
- 25.Naumova N, et al. Organization of the mitotic chromosome. Science. 2013;342(6161):948–953. doi: 10.1126/science.1236083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nora EP, et al. Spatial partitioning of the regulatory landscape of the X-inactivation centre. Nature. 2012;485(7398):381–385. doi: 10.1038/nature11049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Larmonie NS, et al. Breakpoint sites disclose the role of the V(D)J recombination machinery in the formation of T-cell receptor (TCR) and non-TCR associated aberrations in T-cell acute lymphoblastic leukemia. Haematologica. 2013;98(8):1173–1184. doi: 10.3324/haematol.2012.082156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Onozawa M, Aplan PD. Illegitimate V(D)J recombination involving nonantigen receptor loci in lymphoid malignancy. Genes Chromosomes Cancer. 2012;51(6):525–535. doi: 10.1002/gcc.21942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang J, et al. The genetic basis of early T-cell precursor acute lymphoblastic leukaemia. Nature. 2012;481(7380):157–163. doi: 10.1038/nature10725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Beroukhim R, et al. The landscape of somatic copy-number alteration across human cancers. Nature. 2010;463(7283):899–905. doi: 10.1038/nature08822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Solimini NL, et al. Recurrent hemizygous deletions in cancers may optimize proliferative potential. Science. 2012;337(6090):104–109. doi: 10.1126/science.1219580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zarrin AA, Tian M, Wang J, Borjeson T, Alt FW. Influence of switch region length on immunoglobulin class switch recombination. Proc Natl Acad Sci USA. 2005;102(7):2466–2470. doi: 10.1073/pnas.0409847102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zarrin AA, et al. An evolutionarily conserved target motif for immunoglobulin class-switch recombination. Nat Immunol. 2004;5(12):1275–1281. doi: 10.1038/ni1137. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.