Significance
Streptococcus pneumoniae is a naturally competent organism, which can take up extracellular DNA by natural transformation. However, the mechanism by which DNA traverses the capsule and cell well layer is not understood. Here we describe a pilus structure in S. pneumoniae that is initially assembled on the bacterial surface when competence is induced, but subsequently secreted into the medium. We propose a mechanism for DNA uptake whereby the assembling pilus locally disrupts the rigid cell wall, creating a channel upon its release, creating an entry port for exogenous DNA. As DNA uptake coincides with pilus secretion, we suggest that, rather than acting as a retractile apparatus dragging DNA inside, the pilus acts as a cell-wall channel “drilling device.”
Abstract
Streptococcus pneumoniae is a major human pathogen that successfully adapts to the host environment via an efficient uptake system for free DNA liberated from other organisms in the upper respiratory tract, facilitating immune evasion and drug resistance. Although the initial signaling events leading to pneumococcal competence for DNA transformation and the fate of DNA when it has been taken up have been extensively studied, the actual mechanism by which DNA in the environment may traverse the thick capsular and cell wall layers remains unknown. Here we visualize that induction of competence results in the formation of a native morphologically distinct pilus structure on the bacterial surface. This plaited pilus is encoded by the competence (com)G locus, and, after assembly, it is rapidly released into the surrounding medium. Heterologous pneumococcal pilus expression in Escherichia coli was obtained by replacing the pulE-K putative pilin genes of the Klebsiella oxytoca type II secretion system with the complete comG locus. In the pneumococcus, the coordinated secretion of pili from the cells correlates to DNA transformation. A model for DNA transformation is proposed whereby pilus assembly “drills” a channel across the thick cell wall that becomes transiently open by secretion of the pilus, providing the entry port for exogenous DNA to gain access to DNA receptors associated with the cytoplasmic membrane.
The respiratory commensal pathogen Streptococcus pneumoniae or pneumococci is capable of transferring genetic information through a process known as transformation (1). The subsequent discovery by Avery et al. that the transforming agent is DNA initiated the biological revolution (2). However, we know little about the initial process that allows free DNA to be taken up by competent pneumococci.
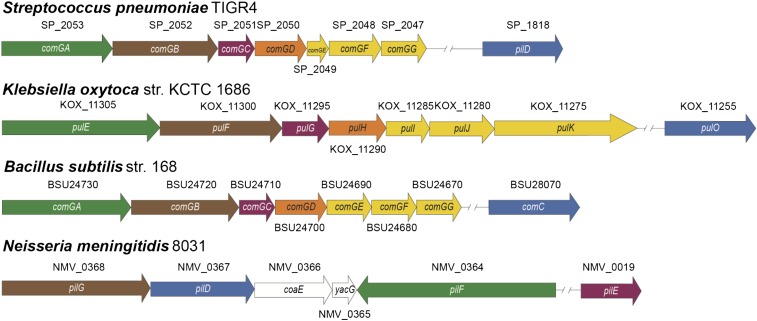
Competence is a highly regulated process in Gram-positive bacteria. It is initiated through quorum sensing by a competence stimulating peptide (CSP) and is shut off quickly after DNA uptake. Approximately 100 genes are regulated by the competence state (3). Among these genes, a locus with homology to the type II secretion system (T2SS) and the type IV pilus (T4P) of Gram-negative organisms is found in naturally competent Gram-positive organisms like Bacillus subtilis and S. pneumoniae, called comG (Fig. 1) (4, 5). The comG locus consists of seven genes and a pseudogene. Homology studies suggest that ComGA is the ATPase responsible for assembly of pilin subunits and that ComGB forms a base structure for assembly (6, 7). Although ComGC is thought to be the major pilin subunit, the ComGD-G proteins are homologous to minor pilins found in other species (8). The hydrophobic stretch located at the N terminus of the pilin subunits makes them stay on the membrane, and assembly can initiate from the membrane with the help of ComGA AAA+ ATPase on the ComGB base structure. The genomic organization of comG resembles that of the T4P and T2SS of Neisseria meningitidis and Klebsiella oxytoca and other Gram-negative organisms, with the major pilin placed after the gene for the base structure and followed by genes for the minor pilins (Fig. 1) (9, 10). In contrast to the Gram-positive systems, the T2SS and T4P in Gram-negative organisms possess a retraction ATPase in addition to the assembly ATPase that is homologous to ComGA (11). The retraction ATPase, along with the assembly ATPase, is thought to generate the piston-like movement of the putative T2SS pilus and the twitching motility in the T4P by cycles of assembly and retraction (11). Interestingly, in T2S and T4P systems in Gram-negative bacteria, the pilus structure acts as a physical tool to relay force to move the bacteria (T4P twitching motility) or to push the substrates to be secreted (T2S piston like movement). It remains unknown whether a similar role might be present in Gram-positive pilus structures associated with competence.
Fig. 1.
Genomic organization of the putative competence pilus of S. pneumoniae and B. subtilis and the type II secretion system of K. oxytoca and the type IV pilus of N. meningitides. The putative S. pneumoniae competence pilus is encoded by the comG operon. The operon consists of comGA AAA+ ATPase, comGB base, comGC major pilin, and comGD-G minor pilins. The putative functions are assessed from work performed in homologous systems like the N. meningitidis type IV pilus (T4P) and type two secretion system (T2SS) of K. oxytoca. The homologous genes in each system are represented by the same color. S. pneumoniae has an additional gene required for the assembly of the putative pilus, prepilin peptidase pilD, which recognizes special N-terminal motifs and cleaves the signal peptide in an authentic manner, leaving a hydrophobic stretch, and also modifying N terminus by methylation (18).
Previous structural studies of Gram-negative bacteria have been done in a recombinant E. coli system expressing K. oxytoca T2SS on a plasmid. These studies suggest that the putative major pilin pullulanase (Pul)G makes up a pseudopilus structure and that the minor pilins are involved in the initiation of polymerization (12). In support of this model, PulG was found to be assembled into long, surface-exposed pseudopili when overproduced in E. coli (13). The comG locus of Gram-positive B. subtilis has been shown to be required for transformation. The putative major pilin encoded by comGC, homologous to S. pneumoniae ComGC, has been shown biochemically to polymerize through cysteine mediated disulfide bridges, which are lacking in S. pneumoniae ComGC and in other secreted pneumococcal proteins, but no native pili structures have been observed in B. subtilis (8, 14).
Here we identify a native pilus dedicated to DNA transformation in pneumococci. We provide visual evidence that a morphologically distinct short pilus structure, encoded by the comG locus, is present on the bacteria early after competence induction, and show that it is rapidly secreted into the medium. Pilus secretion correlates with DNA uptake. Heterologous expression in E. coli of similar pili structures was obtained by replacing the T2SS pilin genes of K. oxytoca with the pneumococcal comG locus. The data represent visualized native T2SS pilus in bacteria.
Results
The Major Pneumococcal Pilin ComGC, but Not the Putative Minor Pilin ComGG, Is Released into the Medium via an Active Process.
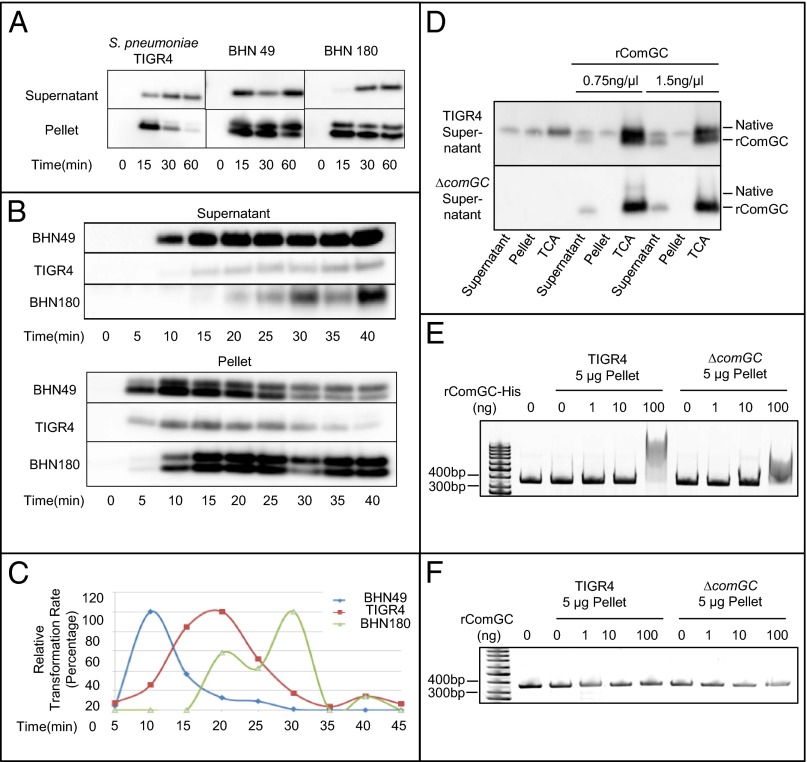
The window of DNA uptake in pneumococci is rather short and peaks at 20 min after induction by CSP in the clinical isolate TIGR4 (15). Because of the lack of a retraction ATPase and the short period allowed for DNA uptake in DNA transformation, we looked at time course expression of the putative major pilin ComGC in WT TIGR4 after induction with CSP. Fig. 2A shows that the ComGC protein level peaked at 15 min but then rapidly decreased from the bacterial fraction. Supernatant samples taken from the same culture revealed that ComGC was detected in the supernatant fraction for at least 120 min, which indicated that the protein was stable, but released from the bacterial cells to the supernatant. The last putative minor pilin in the comG locus, ComGG, was also analyzed by Western blot and found to be induced by CSP induction (Fig. 2B). ComGG disappeared from the pellet fraction after 30 min, but, unlike ComGC, did not appear in the supernatant. As previously reported, competence induces lysis of pneumococci through an autolysin-mediated mechanism (16). Hence, we looked at the appearance of ComGC in the supernatant also in an autolysin-defective (∆lytA) mutant in TIGR4 (Table S1) (17). The release of ComGC was similar in the WT and ∆lytA mutant (Fig. 2 C and D), suggesting that the release of ComGC into the medium is an active process and not a result of bacterial lysis. Because ComGG is not released, we propose that the gradual decrease of this minor pilin in the pellet fraction is a result of protein degradation after ComGC has been released.
Fig. 2.
ComGC is secreted into the medium and the comG genes are required for this secretion and transformation. (A) The putative major pilin subunit ComGC is secreted into the supernatant after competence induction. Secretion was detected by TCA precipitation followed by Western blot for the ComGC protein (Materials and Methods). ComGC peaked at 15 min after CSP induction, then rapidly released to the supernatant and stayed in the supernatant for a long time. (B) The minor pilin subunit ComGG was also induced by CSP induction. The expression and disappearance of ComGG from the pellet fraction was also rapid, but ComGG was not detected in the pneumococcal supernatant. This showed that the occurrence of ComGC in the supernatant was not caused by nonspecific lysis, (C and D) supported by the finding of ComGC release also in an autolysis-deficient mutant ∆lytA. (E) The ComG secretion phenotypes of comG mutants (∆comGB, ∆comGD, ∆comGE, ∆comGF, ∆comGG, ∆comGD-G, and ∆pilD) were assessed by using Western blot analyses. All tested mutants were capable of producing ComGC and processing it to the mature, cleaved form, but no comG mutant released ComGC to the supernatant. The ∆pilD mutant lacked the ability to cleave ComGC and nonprocessed ComGC was not found in the supernatant. The control strains of comGB, comGD, comGE, and ComGF were constructed to test if the erythromycin resistance (erm) cassette insertion had polar effects. In these control mutants, the cassette was inserted after the indicated gene, and no sequence was removed from the genome of the pneumococcus. The control strains produced and released ComGC at WT levels, showing no evidence of polar effects of the erm cassette insertion. (F) The prepilin peptidase motif was mutated to pinpoint which residues are important for cleavage of the signal peptide and release. Mutants comGCF16A and comGCE20A were fully capable of cleaving the signal peptide, but these mutants could not release ComGC. The comGCA15L F16A double mutant lacked the secretion phenotype and the comGCE20A mutant had a diminished ability to cleave the signal peptide and could not release ComGC. (G) The comG mutants were checked for transformability by transforming the mutants with chromosomal DNA from a SmR pneumococcal strain. The data indicate that these genes are required for release of ComGC as well as for transformation of pneumococci. Also, the control mutants with the erm cassette in between the comG genes were shown to be competent for DNA transformation. (H) The comGC mutants that lacked the ability to secrete ComGC were also nontransformable, as shown by transformation assays performed by using chromosomal DNA from a SmR pneumococcal strain.
Secretion of ComGC Requires Presence of Minor Pilins and Cleavage of ComGC by the Prepilin Peptidase PilD.
To identify factors required for the secretion of ComGC, we generated and analyzed a series of nonpolar mutants in the comG locus (Tables S1 and S2). All comG mutants expressed ComGC but they could not release ComGC into the medium (Fig. 2E). Hence, the minor pilins seem to be involved in assembly and/or secretion of ComGC. In homologous systems of T2SS and T4P, the pilins are processed by the prepilin peptidase to which pneumococcal PilD is homologous (Fig. 1). PilD cleaves in the middle of the signal peptide by recognizing a specific motif, thereby leaving a hydrophobic helical structure linked to the pilin proteins, allowing the cleaved proteins to stay on the membrane for further assembly by the ATPase (homolog in pneumococci, ComGA) and the base (homolog in pneumococci, ComGB) proteins (18). An AFTLVE motif is the predicted cleavage site, with cleavage taking place between A15 and F16 (Fig. S1) (19). Western blot analysis showed that a comGCA15L F16A double mutant was noncleavable and not secreted to the supernatant, whereas comGCE20A had a reduced capacity to be cleaved and completely lacked the secretion phenotype (Fig. 2F). The comGCF16A and comGCE20D mutants, interestingly, were cleavable by PilD but nevertheless did not secrete ComGC. These data suggest that cleavage by the prepilin peptidase requires the ComGC15A residue. After cleavage of ComGC, additional steps are required for its secretion, as indicated by the presence of mutants that are cleaved but not secreted.
Pneumococcal Transformation Requires Presence of Minor Pilins, ComGC Cleavage, and Secretion of Pili.
In addition to the inability to secrete ComGC, all mutants in the comG locus were completely nontransformable (Fig. 2G). Interestingly, the comGC mutants that were cleavable by PilD, but not secreted to the supernatant, were also not transformable, showing a correlation between ComGC secretion and DNA transformation (Fig. 2H).
We next examined the dynamics of ComGC expression in clinical isolates available in our collection. Among the tested isolates, all isolates that were highly transformable expressed a CSP-inducible ComGC that was released into the supernatant after it was expressed (Table S3). From this collection, we identified three strains with different release times of the putative major pilin protein ComGC (BHN49, TIGR4, and BHN180) and studied potential correlations to transformability. The CSP-induced cultures were sampled every 15 min (Fig. 3A) and 5 min (Fig. 3B). Fig. 3 A and B show their phenotype for expression and secretion of ComGC after CSP induction, revealing that BHN49 is an early, TIGR4 an intermediate, and BHN180 a late secretor. Strain BHN49 started to secrete ComGC at 10 min, whereas TIGR4 and BHN180 started secretion at 15 min. To compare ComGC secretion with DNA transformation, bacterial strains were coincubated with DNA for only 5 min, after which extracellular DNA was removed by addition of excess amounts of DNase I. The transformation profiles of these strains correlated with the peak of secretion (Fig. 3 A–C), suggesting that entry of DNA into the bacteria requires secretion of ComGC.
Fig. 3.
Clinical isolates of pneumococci show a differential release of ComGC that correlates with transformation. (A) In addition to TIGR4, two clinical isolates were selected from a collection of transformable pneumococcal strains to study release of ComGC. Strains were categorized by the time points at which they reached peak secretion levels. BHN49 reached the maximal ComGC signal in the supernatant at 15 min, making it an early secreting strain. BHN180 and TIGR4 strains reached maximal ComGC in the supernatant fraction at 30 min. (B) To dissect the ComGC release phenotype, samples were taken at 5-min intervals. This revealed that BHN180 peaked for the ComGC signal in the supernatant at 30 min, and TIGR4 between 20 and 25 min. (C) A transformation assay was performed in a 5-min-resolution experimental setup to find the dynamic correlation between the release of ComGC and DNA uptake. The bacteria were induced with CSP and incubated with DNA for 5 min before addition of DNase at the indicated time points. Also, a sample for Western blot to detect ComGC release to the supernatant was taken before addition of DNA to the induced samples. DNA used in this experiment was a 414-bp PCR product carrying the SmR rpsL gene, which can be transported through the CelA–CelB inner membrane DNA channel quicker than randomly sized SmR chromosomal DNA fragments (23, 30). This modification was done to enhance the time resolution of the experimental setup. The results demonstrate that the highest level of DNA uptake was observed when most of the ComGC was already present in the supernatant and show that ComGC release correlates with transformation. (D) Western blot for ComGC in the supernatant of untreated, ultracentrifuged, and TCA-precipitated samples. Two different concentrations of soluble rComGC∆1–39 (labeled “rComGC”) were added to the induced culture supernatants as controls. The native and soluble ComGC forms were detectable in the supernatant and TCA precipitated samples, but soluble ComGC was not detectable in the pellet fraction after ultracentrifugation. A total of 5 µL of each sample is loaded to the gel. (E) Electromobility shift assays using ultracentrifuged samples of induced culture supernatants. Recombinant ComGC proteins with a aminoterminal 6×His tag was added at different concentrations to 5 µg ultracentrifuged pellets of WT TIGR4 and its ∆comGC mutant. Addition of the His-tagged rComGC∆1–39 (labeled “rComGC-His”) protein caused a DNA retardation when added at 100 μg to both pelleted fractions. (F) The same experiment as in E except for the addition of soluble rComGC∆1–39 lacking a positively charged His tag (labeled “rComGC”) to the two pelleted fractions. No retardation of DNA was found in the electromobility shift assay.
We suggest that ComGC is released to the supernatant as a polymer, as it is pelleted by ultracentrifugation of CSP-induced pneumococcal supernatants (Fig. 3D). As further support for the polymeric nature of secreted ComGC, we added recombinant ComGC∆1–39 (which lacks the hydrophobic stretch at the N terminus) exogenously to the culture supernatant. The hydrophobic stretch is thought to be the polymerization domain, and hence rComGC∆1–39 should be monomeric. After ultracentrifugation of the supernatant of the TIGR4 strain with added rComGC∆1–39, the recombinant protein was found only in the supernatant fraction and not detected in the pellet fraction in WT or TIGR4∆comGC-derived culture supernatants (Fig. 3D). This suggests that the pelleting of native ComGC by ultracentrifugation is a result of polymerization that is dependent on the hydrophobic N-terminal stretch of the protein.
As release of the ComGC polymer correlated to bacterial DNA transformation, we next tested whether released ComGC could bind DNA. Concentrated supernatants from WT and ΔcomGC mutant strains were used in a gel-shift assay with a comGC PCR product as a DNA probe. No DNA-binding activity was detected in these pelleted fractions. As a positive control, we added the highly charged 6×His-tagged rComGC∆1–39 protein to the concentrated supernatants and obtained a shift in the DNA mobility assay at higher protein concentrations in the presence and absence of native ComGC polymers (Fig. 3E). This DNA binding was dependent on the positively charged His tag, as no DNA binding was found by addition of rComGC∆1–39 where the His-tag had been removed by thrombin treatment (Fig. 3F). Hence, neither the native ComGC polymer nor soluble ComGC monomer bind DNA under the conditions used.
A Distinct Plaited Pilus Is Observed on the Pneumococcal Surface Early After Competence Induction That Subsequently Appears in the Medium.
We looked for a polymerized surface structure on the bacteria early after competence induction, as well as for a similar structure in the medium at later time points. Interestingly, a short pilus structure was observed on the clinical isolate BHN49 at 7.5 min after induction with CSP on EM (Fig. 4A). The plaited, 8- to 10-nm-wide, and approximately 200-nm-long pilus protruded from the bacterium (Fig. 4 A–E). Morphologically similar pilus structures were observed in the supernatant of strain BHN49 from the same culture at later time points (30 min; Fig. 4C). Similar secreted pilus structures were also seen in the supernatant of CSP-induced TIGR4, but were not detected in the supernatant or on the surface of uninduced bacteria or induced comG mutants despite extensive observation by EM of these negative controls. Plaited pilus structures were significantly enriched by ultracentrifugation of a CSP-induced BHN49 supernatant (Fig. 4 F and G). The distribution of pilus length was determined by using ultracentrifuged enriched supernatants and found to range between 40 and 200 nm with an average of 106 nm (Fig. 4H).
Fig. 4.
A distinct pilus is visualized after induction of pneumococcal competence. (A) Strain BHN49, shown to express more ComGC than the other strains tested, was studied by using transmission EM. The samples were applied to a grid 7.5 min after CSP induction and stained with 2% uranyl acetate. A polymerized plaited pilus-like structure was observed on the bacteria. The image shows a sample of pilus-bearing bacteria. (B) A zoomed-in image in the dashed box A. The special plaited structure of the competence pilus is clearly visible. The pilus length is close to 200 nm, and 8 to 10 nm thick. (C–E) A morphologically similar plaited pilus structure was found in the supernatants of induced pneumococcal cultures after 30 min of CSP induction. (F and G) There is a higher abundance of pilus structures in the ultracentrifuge-enriched supernatant. (H) Distribution of the length of pili in the ultracentrifuge-enriched supernatant. The sample was imaged by transmission EM, and 56 randomly selected pili are measured and categorized according to their length. The average pilus length is 106.9 ± 5.2 nm (SEM).
Heterologous Expression of Pneumococcal Competence Pili in E. coli.
We attempted to label the competence-induced pilus structures with an immunogold procedure by using anti-ComGC primary antibody and secondary gold-labeled anti-rabbit antibody without success, probably because our ComGC antibodies generated against soluble recombinant ComGC do not recognize polymerized ComGC. To confirm our findings, we therefore reconstituted pneumococcal competence pilus expression in E. coli. The T2SS pseudopilus of K. oxytoca could be observed by overexpressing it in E. coli using a plasmid that encoded the structural elements of the type II secretion system (pCHAP231) (10). The comGA-G homologs, pulE-K, were removed from pCHAP231 (pMB93), and comGA-G were inserted instead (pSMT01; Fig. S2 and Table S4). pCHAP231 expressed the K. oxytoca pseudopilus in a maltose-dependent manner. In our system, the expression of comG genes from plasmid pSMT01 was constitutive, as shown by Western blots of ComGC and ComGG (Fig. 5A). Western blot of ComGC revealed the presence of two specific bands, which correlate to cleaved and noncleaved ComGC, suggesting that PulO, the PilD prepilin peptidase homolog, from pCHAP231 is functional in processing ComGC to its mature form (Fig. 5A). E. coli carrying pSMT01 were observed with transmission EM, and short pilus-like structures were observed on the bacteria as well as in the environment (Fig. 5 B–E), whereas no such structures were present in strain MB93 lacking the comGA-G genes (Fig. 5F). When these structures were observed at higher magnification, they showed a similar plaited structure to that found in competence-induced S. pneumoniae, a structure not shared by other E. coli pilus polymers (Fig. 5 B–E). The presence of a similar plaited structure present in E. coli expressing the comG operon suggests that the structures observed on pneumococci after CSP induction represent the pneumococcal competence pilus encoded by the comG locus.
Fig. 5.
Heterologous expression of the comG operon in E. coli. (A) A Western blot was performed to detect production of ComGC in the SMT01 K. oxytoca–S. pneumoniae hybrid plasmid carrier. The plasmid backbone carrier MB93 was used as a negative control. SMT01 produced two bands corresponding to uncleaved and cleaved ComGC. The occurrence of cleavage suggests that ComGC is processed in the reconstituted E. coli system and cleaved by the K. oxytoca PulO prepilin peptidase. The presence of noncleaved ComGC might be a result of overexpression and saturation of PulO leading to accumulation of uncleaved proteins at the membrane. (B) When comGA-G was inserted into pMB93, creating the SMT01 strain, short pilus-like curved structures were observed on the bacteria as well as released into the medium. (C–E) The high magnification of pilus structures from SMT01 revealed the same length and width and a similar plaited structure as the pilus observed in S. pneumoniae. (F) E. coli carrying a derivative of the pCHAP231 plasmid lacking the K. oxytoca pilins (∆pulE-K), MB93, did not have any pilus-like structures on the surface or free in the medium. (G) E. coli strain expressing the comGA-G locus by an IPTG-inducible vector (pT7.7). The bacteria are grown in Luria–Bertani broth and induced for 3 h at 37 °C with 1 mM IPTG. The bacteria are fixed and observed with transmission EM, and the arrow shows a bulge appearing on the bacterium.
In the K. oxytoca type II secretion system, the outer membrane-spanning secretin PulD forms a channel through which its pseudopilus can traverse. The extracellular presentation of the pneumococcal competence pilus in E. coli argues that also this Gram-positive structure uses the K. oxytoca secretin as an outer membrane channel. When E. coli carries a construct that expresses comGC-G on an IPTG-inducible pT7.7 vector, in the absence of pulD, bacteria do not express surface-located or secreted competence pili, but demonstrate bulges on the cells (Fig. 5G).
Discussion
Transformation in Gram-positive organisms is dependent on a conserved T2SS/T4P locus that also includes the comG operon in S. pneumoniae. The similarity of this locus to that of T4 pili of Gram-negative bacteria involved in DNA transformation and the identification of a macromolecular complex containing the putative major pilin ComGC on the surface of B. subtilis have led to the idea that transformable Gram-positive bacteria may form a T4-like pilus required for transformation. However, no native competence pilus structures have so far been observed on the surface of any Gram-positive organisms. The best-studied T2S system is encoded by K. oxytoca and dedicated to the secretion of pullulanase from the periplasm across the outer membrane into the medium. This secretion system is dependent on the assembly of a putative pilus initiated by minor pilin subunits preassembled in the membrane, allowing the assembly platform for the major pilin subunit PulG to produce pili of a definite length (12). However, only when PulG is overproduced in E. coli are long pseudopili assembled on the surface driven by self-polymerized PulG. These long PulG pseudopili are furthermore only formed in the presence of the outer membrane PulD secretin (20).
We argue that the native structure we observe in S. pneumoniae, upon competence induction, represents a self-secreting type II pilus whereby the pilus is shed into the supernatant quickly and does not stay on the surface of the bacteria for an extended period. It remains to be shown whether other factors are secreted along with the T2SS pilus. One possibility is that the pilus pushes a DNA-binding protein to the surface, thus secreting it through the cell wall and leaving it behind as the pilus itself is being shed into the supernatant. Further studies are however needed to find out the details of initial DNA binding to the surface of the bacterium.
The reasons why these pili structures have not been seen before are, besides their shortness, their low abundance on the cell surface, and, above all, their rapid release into the medium when they have been assembled. The native pneumococcal competence pili do not look like traditional T4 pili, T2SS PulG overexpressed pseudopili, or Gram-positive adhesion-mediating pili, as they are thicker (8–10 nm) and have a distinct plaited or wired appearance that might mean that each native pilus structure consists of two polymers. Indeed, in a very recent publication, micrometer-long thin pili (5–6 nm thick) were observed on S. pneumoniae cells when ComGC was overproduced relative to the other ComG components showing no resemblance to the short, plaited, and thicker native structures identified in the present study (21). In the study performed by Laurencau et al. (21), the observed ComGC pili were produced by a strain that had one extra copy of comGC in addition to the native locus. The increase in copy number of comGC might result in increased protein levels for ComGC, resulting in elongated pili. Also, the increase in the major pilin protein levels disturbs the stoichiometric balance between major and minor pilins. Minor pilins might play a role controlling the assembly, length, shape, and function of the pilus. Thus, the long ComGC pili observed might be similar in nature to the nonfunctional long pseudopili produced when the T2SS major pilin PulG was overproduced. In our studies, no micrometer-long pili were detected. If the pili we observe were fragments broken off from several micrometer-long structures, we would expect to see a normal distribution of pili between 0 and 1 to 2 µm, which is not the case. Instead, we see a distribution ranging mostly between 40 and 200 nm with an average of 106 nm. The longest pilus found were approximately 400 to 500 nm long.
By replacing the Klebsiella pulE-K genes for the corresponding pneumococcal comGA-G genes, we succeeded to express short plaited structures in E. coli with similar length and width as the native structures seen in pneumococci. In this reconstituted system, pneumococcal T2SS pili were observed on cells and in the medium only when the Klebsiella PulD outer membrane secretin was present. In its absence, we observed bulges on E. coli, suggesting periplasmic accumulation of ComGC and other ComG components, like the reconstituted T2SS Pul system in the absence of PulD (20). This suggests that the pneumococcal competence pilus may traverse the PulD channel in the outer membrane. Also, heterologous expression of E. coli T4 pili via the Klebsiella T2SS assembly system reveals a high degree of interchangeability of these systems (22). Nevertheless, our findings demonstrate heterologous expression of a Gram-positive pilus in a Gram-negative background. This will facilitate further studies on the assembly and function of pneumococcal competence pili.
The pneumococcal comG operon is rapidly induced after addition of CSP, and mutant analyses reveal each gene in the comG operon to be required for DNA transformation. A major finding here is that ComGC, presumably as part of the assembled T2SS pilus, is rapidly released into the medium. As inhibition of autolysis did not affect ComGC secretion and because one of the minor pilins, ComGG, was not released, the data suggest that pilus release is an ordered process and is not a result of bacterial lysis. All transformable pneumococcal isolates studied released ComGC shortly after competence stimulation, suggesting that pilus release is a general feature of competent pneumococci. No ComGC major pilin release was found, and no pilus structures were observed after each of the putative minor pilin genes (comGD-G) had been inactivated, suggesting that appropriate assembly of ComGC into a pilus structure is required for secretion. At present it is not clear whether or not the observed native pilus only consists of ComGC or whether it also has other ComG proteins incorporated.
In the recent publication by Laurencau et al. (21), exogenously added DNA was found to associate to the long, thin T4-like pili observed after overexpressing ComGC in S. pneumoniae. This finding led the authors to conclude that this pilus mediates DNA binding during DNA transformation. The findings provided in our manuscript argue that DNA uptake correlates to secretion of the native ComGC pilus rather than to the presence of the structure on the cell surface. Thus, clinical isolates that secrete the pilus early also reach maximal transformation earlier than isolates that secrete the pilus late. Also, in a late secreting isolate, abundant cell-associated and processed ComGC is seen almost 20 min before maximal transformation is reached. In a gel-shift experiment using ComGC and T2SS pili enriched supernatants (by ultracentrifugation), no DNA binding activity was found. Addition of 6×His-tagged ComGC at high concentrations to these enriched concentration resulted in DNA binding as a result of electrostatic charge provided by the His-tag, as addition of soluble non–His-tagged rComGC gave no DNA binding activity. Thus, our data suggest that the native secreted pneumococcal competence pilus is not a DNA-binding structure.
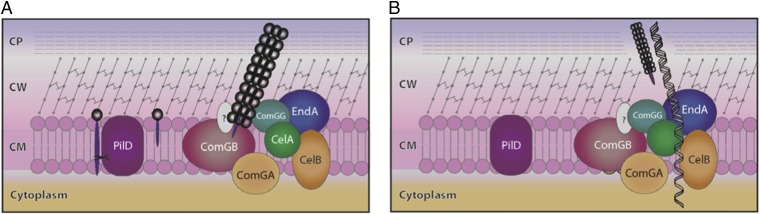
Based on the findings described here, we propose a new model for DNA transformation in S. pneumoniae (Fig. 6) whereby competence pilus secretion creates a transient 8- to 10-nm-wide channel across the peptidoglycan and capsular layers, allowing access of exogenous DNA (2 nm in diameter) to the cytoplasmic membrane-associated ComEA DNA receptor. This hypothesis is supported by a study performed by Provvedi and Dubnau demonstrating that protoplasts of B. subtilis lacking cell wall are able to bind DNA through the ComEA DNA receptor in the absence of ComGC, whereas comGC mutants were not able to bind in the presence of cell wall, suggesting that ComGC pilus might be involved in making membrane bound ComEA DNA receptor accessible to DNA (23). Furthermore, in Gram-negative bacteria, no T4pilin has ever been reported to bind DNA; the same is true in the Gram-positive organism B. subtilis (8).
Fig. 6.
Model showing the role of the T2SS competence pilus assembly and secretion for DNA uptake in pneumococci. (A and B) A new model describing the potential role of the pneumococcal competence pilus in DNA uptake is shown. (A) Competence induces ComGC to be translated and recruited to the inner membrane, probably at the cell pole and processed by the PilD peptidase. By using energy generated by the ComGA ATPase, ComGC is polymerized over the ComGB base, and as it polymerizes with the assistance of minor ComG pilins it breaks through the bacterial cell wall. (B) With the secretion of the pilus into the medium, a channel is created that traverses the cell wall and capsule that allows exogenous DNA to access the CelA/CelB (homologs of ComEA/ComEC of B. subtilis) DNA receptor/channel in the inner membrane and to become internalized. As the minor pilin ComGG is not released with the pilus, it is depicted to remain at the base of the access tunnel for DNA. Because a channel across the bacterial cell wall is potentially harmful for the bacteria, it is probably rapidly repaired, creating a narrow time window for DNA to be taken up by pneumococci (CM, cellular membrane; CP, capsule; CW, cell wall).
In nature, the T2SS and T4P act as physical relaying tools. Thus, twitching motility by T4P in Neisseria creates a pulling force and K. oxytoca T2SS creates a pushing force. We hypothesize that the polymerizing pilus structure relays a strong physical force powered by the ComGA AAA+ ATPase to “drill” a tunnel across the cell wall (24, 25). We expect such a pilus-generated DNA tunnel to be refilled by the cell wall synthetic machinery, and therefore only to exist over a short period. At least one of the minor pilins, ComGG, is not shed with the pilus. It is therefore possible that ComGG, together with other minor ComG pilins, remain at the base, or line the channel that has been opened by the release of the competence pilus. One or more minor pilins may bind DNA to increase the efficiency of the system. The recent finding that the minor T4 pilin ComP of N. meningitidis has DNA-binding properties is interesting in this respect (26).
It was recently demonstrated that a mutation in the dpr gene leads to an extended period of competence gene expression. Interestingly, this evokes an increased bacterial lysis (27), that we suggest could be a result of excessive cell wall damage generated by overexpression of the pilus. Similarly, the E. coli strain that carries the comG locus (SMT01) has a slight growth defect compared with the strain that lacks it (Fig. S3). These findings are consistent with our working model (Fig. 6), implying a cell wall hole generated upon pilus secretion. If pilus expression happens over an extended period, the holes generated by the pilus would undermine the osmotic stability of the bacterium and would cause lysis upon induction.
In Gram-negative bacteria, DNA binding to T4 pili has been proposed to be followed by pilus retraction via depolymerization mediated by a retraction ATPase dragging the DNA into the cell. However, there is no direct evidence for this process of DNA import. Actually, T4 pilus retraction in Neisseria gonorrhoeae occurs at a much higher speed than DNA uptake in B. subtilis (28, 29). Furthermore, in Gram-positive bacteria, there are no homologs to the PilT retraction ATPase. The model proposed here for transformation in pneumococci might also be applicable to Gram-negative T4 pili if pilus secretion and/or pilus retraction opens the outer membrane secretin channel and underlying peptidoglycan for DNA import.
Materials and Methods
Time-Course Induction of Pneumococcal Strains and Trichloroacetic Acid Precipitation of Supernatants.
S. pneumoniae was grown on blood plates overnight and transferred to liquid casitone and yeast extract (C+Y) containing cultures with 5% (wt/vol) dextrose and serum (DS)-containing medium, inoculated at an optical density at 620 nm of 0.05. When the cultures reached an optical density at 620 nm of 0.15, they were induced by CSP1/2 with a final concentration of 20 ng/mL A 1 mL culture was centrifuged in a microfuge tube for 1 min at 16,000 × g, and the supernatant was filtered by using a 0.22-µm syringe filter. The pellet was resuspended in 50 µL 1× NuPAGE-LDS sample buffer (Invitrogen) with 5% (wt/vol) β-mercaptoethanol. A total of 200 µL of Trichloroacetic Acid (TCA) was added to 1 mL of supernatant and incubated for 1 h on ice. The TCA-precipitated samples were centrifuged for 1 h at 4 °C and washed once with 500 µL ice-cold acetone without disturbing the pellet. The samples were air-dried and resuspended in 50 µL 1× NuPAGE-LDS sample buffer with 5% (wt/vol) β-mercaptoethanol for SDS/PAGE.
Transmission EM of E. coli and S. pneumoniae Samples.
A 20-µL droplet of CSP-induced or uninduced pneumococcal culture, over night plate-grown E. coli resuspended in PBS solution, or filtered pneumococcal culture supernatant after 30 min of induction was placed on a Parafilm. A carbon-coated Formvar copper EM grid was placed on the sample droplet for 5 min. For early time point samples, the CSP-induced culture of BHN49 was placed on a grid after 2.5 min and for 5 min at 37 °C, resulting in a total of 7.5 min of growth. Excess liquid was blotted on filter paper, and the grid was incubated with 2% (wt/vol) uranyl acetate for 30 s. The samples were examined by using a Tecnai Biotwin G2 at 100 kV.
Time-Course Transformation Assay.
S. pneumoniae was grown in C+Y with 5% (wt/vol) DS broth to an optical density at 620 nm of 0.15 and induced with 20 ng/mL CSP. A total of 500 µL of the bacterial culture was incubated with 100 ng of PCR-amplified rpsL from streptomycin resistant S. pneumoniae TIGR4 for 5 min at 37 °C. Then DNase I at a final concentration of 16 µg/mL was added and bacteria were incubated for an additional 90 min for recovery. After recovery, 100 µL of the culture were plated on blood–agar plates with 100 µg/mL streptomycin. Also, serial dilutions were plated on blood plates without antibiotics to assess the number of total bacteria. Relative transformation rate was determined by dividing the number of streptomycin resistant colonies to the number of colonies grown on nonselective plates. The maximum value was set to 100 and plotted against time.
Supplementary Material
Acknowledgments
We thank Dr. Olivera Francetic (Institut Pasteur, Paris, France) and Dr. Deborah Ribardo (University of Texas Southwestern Medical Center at Dallas, Dallas, TX) for providing the pCHAP231 and pT7.7 plasmids, respectively. This work was supported by research grants from the Swedish Research Council, the Knut and Alice Wallenberg Foundation, the Swedish Foundation for Strategic Research, and ALF-bidrag from Stockholm City Council.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1313860111/-/DCSupplemental.
References
- 1.Griffith F. The significance of pneumococcal types. J Hyg (Lond) 1928;27(2):113–159. doi: 10.1017/s0022172400031879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Avery OT, Macleod CM, McCarty M. Studies on the chemical nature of the substance inducing transformation of pneumococcal types: Induction of transformation by a desoxyribonucleic acid fraction isolated from pneumococcus type III. J Exp Med. 1944;79(2):137–158. doi: 10.1084/jem.79.2.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Claverys JP, Martin B, Polard P. The genetic transformation machinery: Composition, localization, and mechanism. FEMS Microbiol Rev. 2009;33(3):643–656. doi: 10.1111/j.1574-6976.2009.00164.x. [DOI] [PubMed] [Google Scholar]
- 4.Chung YS, Dubnau D. ComC is required for the processing and translocation of comGC, a pilin-like competence protein of Bacillus subtilis. Mol Microbiol. 1995;15(3):543–551. doi: 10.1111/j.1365-2958.1995.tb02267.x. [DOI] [PubMed] [Google Scholar]
- 5.Bergé M, Moscoso M, Prudhomme M, Martin B, Claverys JP. Uptake of transforming DNA in Gram-positive bacteria: A view from Streptococcus pneumoniae. Mol Microbiol. 2002;45(2):411–421. doi: 10.1046/j.1365-2958.2002.03013.x. [DOI] [PubMed] [Google Scholar]
- 6.Possot O, Pugsley AP. Molecular characterization of PulE, a protein required for pullulanase secretion. Mol Microbiol. 1994;12(2):287–299. doi: 10.1111/j.1365-2958.1994.tb01017.x. [DOI] [PubMed] [Google Scholar]
- 7.Possot O, d’Enfert C, Reyss I, Pugsley AP. Pullulanase secretion in Escherichia coli K-12 requires a cytoplasmic protein and a putative polytopic cytoplasmic membrane protein. Mol Microbiol. 1992;6(1):95–105. doi: 10.1111/j.1365-2958.1992.tb00841.x. [DOI] [PubMed] [Google Scholar]
- 8.Chen I, Provvedi R, Dubnau D. A macromolecular complex formed by a pilin-like protein in competent Bacillus subtilis. J Biol Chem. 2006;281(31):21720–21727. doi: 10.1074/jbc.M604071200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tønjum T, Koomey M. The pilus colonization factor of pathogenic neisserial species: Organelle biogenesis and structure/function relationships—a review. Gene. 1997;192(1):155–163. doi: 10.1016/s0378-1119(97)00018-8. [DOI] [PubMed] [Google Scholar]
- 10.d’Enfert C, Ryter A, Pugsley AP. Cloning and expression in Escherichia coli of the Klebsiella pneumoniae genes for production, surface localization and secretion of the lipoprotein pullulanase. EMBO J. 1987;6(11):3531–3538. doi: 10.1002/j.1460-2075.1987.tb02679.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Johnson TL, Abendroth J, Hol WG, Sandkvist M. Type II secretion: From structure to function. FEMS Microbiol Lett. 2006;255(2):175–186. doi: 10.1111/j.1574-6968.2006.00102.x. [DOI] [PubMed] [Google Scholar]
- 12.Cisneros DA, Bond PJ, Pugsley AP, Campos M, Francetic O. Minor pseudopilin self-assembly primes type II secretion pseudopilus elongation. EMBO J. 2012;31(4):1041–1053. doi: 10.1038/emboj.2011.454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sauvonnet N, Vignon G, Pugsley AP, Gounon P. Pilus formation and protein secretion by the same machinery in Escherichia coli. EMBO J. 2000;19(10):2221–2228. doi: 10.1093/emboj/19.10.2221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Daniels R, et al. Disulfide bond formation and cysteine exclusion in gram-positive bacteria. J Biol Chem. 2010;285(5):3300–3309. doi: 10.1074/jbc.M109.081398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Peterson SN, et al. Identification of competence pheromone responsive genes in Streptococcus pneumoniae by use of DNA microarrays. Mol Microbiol. 2004;51(4):1051–1070. doi: 10.1046/j.1365-2958.2003.03907.x. [DOI] [PubMed] [Google Scholar]
- 16.Steinmoen H, Teigen A, Håvarstein LS. Competence-induced cells of Streptococcus pneumoniae lyse competence-deficient cells of the same strain during cocultivation. J Bacteriol. 2003;185(24):7176–7183. doi: 10.1128/JB.185.24.7176-7183.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mellroth P, et al. LytA, major autolysin of Streptococcus pneumoniae, requires access to nascent peptidoglycan. J Biol Chem. 2012;287(14):11018–11029. doi: 10.1074/jbc.M111.318584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Strom MS, Nunn DN, Lory S. A single bifunctional enzyme, PilD, catalyzes cleavage and N-methylation of proteins belonging to the type IV pilin family. Proc Natl Acad Sci USA. 1993;90(6):2404–2408. doi: 10.1073/pnas.90.6.2404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pugsley AP. Processing and methylation of PuIG, a pilin-like component of the general secretory pathway of Klebsiella oxytoca. Mol Microbiol. 1993;9(2):295–308. doi: 10.1111/j.1365-2958.1993.tb01691.x. [DOI] [PubMed] [Google Scholar]
- 20.Vignon G, et al. Type IV-like pili formed by the type II secreton: Specificity, composition, bundling, polar localization, and surface presentation of peptides. J Bacteriol. 2003;185(11):3416–3428. doi: 10.1128/JB.185.11.3416-3428.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Laurenceau R, et al. A type IV pilus mediates DNA binding during natural transformation in Streptococcus pneumoniae. PLoS Pathog. 2013;9(6):e1003473. doi: 10.1371/journal.ppat.1003473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cisneros DA, Pehau-Arnaudet G, Francetic O. Heterologous assembly of type IV pili by a type II secretion system reveals the role of minor pilins in assembly initiation. Mol Microbiol. 2012;86(4):805–818. doi: 10.1111/mmi.12033. [DOI] [PubMed] [Google Scholar]
- 23.Provvedi R, Dubnau D. ComEA is a DNA receptor for transformation of competent Bacillus subtilis. Mol Microbiol. 1999;31(1):271–280. doi: 10.1046/j.1365-2958.1999.01170.x. [DOI] [PubMed] [Google Scholar]
- 24.Hanson PI, Whiteheart SW. AAA+ proteins: Have engine, will work. Nat Rev Mol Cell Biol. 2005;6(7):519–529. doi: 10.1038/nrm1684. [DOI] [PubMed] [Google Scholar]
- 25.Numata N, et al. Molecular mechanism of force generation by dynein, a molecular motor belonging to the AAA+ family. Biochem Soc Trans. 2008;36(pt 1):131–135. doi: 10.1042/BST0360131. [DOI] [PubMed] [Google Scholar]
- 26.Cehovin A, et al. Specific DNA recognition mediated by a type IV pilin. Proc Natl Acad Sci USA. 2013;110(8):3065–3070. doi: 10.1073/pnas.1218832110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mirouze N, et al. Direct involvement of DprA, the transformation-dedicated RecA loader, in the shut-off of pneumococcal competence. Proc Natl Acad Sci USA. 2013;110(11):E1035–E1044. doi: 10.1073/pnas.1219868110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kurre R, Maier B. Oxygen depletion triggers switching between discrete speed modes of gonococcal type IV pili. Biophys J. 2012;102(11):2556–2563. doi: 10.1016/j.bpj.2012.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Maier B, Chen I, Dubnau D, Sheetz MP. DNA transport into Bacillus subtilis requires proton motive force to generate large molecular forces. Nat Struct Mol Biol. 2004;11(7):643–649. doi: 10.1038/nsmb783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Draskovic I, Dubnau D. Biogenesis of a putative channel protein, ComEC, required for DNA uptake: membrane topology, oligomerization and formation of disulphide bonds. Mol Microbiol. 2005;55(3):881–896. doi: 10.1111/j.1365-2958.2004.04430.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.