Significance
Our work provides an insight into the complex network of basic helix–loop–helix (bHLH)/helix–loop–helix (HLH) transcription factors that regulates cell elongation. An unknown network motif (an incoherent feed-forward loop) has been discovered that was established by two negative regulators of brassinosteroid responses, namely the HLH transcription factor INCREASED LEAF INCLINATION1 BINDING bHLH1 (IBH1) and its unidentified homolog, IBH1-LIKE1 (IBL1). We also suggest that IBH1 and IBL1 coexist with PHYTOCHROME-INTERACTING FACTOR 4 (another key regulator of cell elongation) in transcriptional complexes.
Keywords: development, plant, growth
Abstract
Cell elongation is promoted by different environmental and hormonal signals, involving light, temperature, brassinosteroid (BR), and gibberellin, that inhibit the atypical basic helix–loop–helix (bHLH) transcription factor INCREASED LEAF INCLINATION1 BINDING bHLH1 (IBH1). Ectopic accumulation of IBH1 causes a severe dwarf phenotype, but the cell elongation suppression mechanism is still not well understood. Here, we identified a close homolog of IBH1, IBH1-LIKE1 (IBL1), that also antagonized BR responses and cell elongation. Genome-wide expression analyses showed that IBH1 and IBL1 act interdependently downstream of the BRASSINAZOLE-RESISTANT1 (BZR1)–PHYTOCHROME-INTERACTING FACTOR 4 (PIF4)–DELLA module. Although characterized as non-DNA binding, IBH1 repressed direct IBL1 transcription, and they both acted in tandem to suppress the expression of a common downstream helix–loop–helix (HLH)/bHLH network, thus forming an incoherent feed-forward loop. IBH1 and IBL1 together repressed the expression of PIF4, known to stimulate skotomorphogenesis synergistically with BZR1. Strikingly, PIF4 bound all direct and down-regulated HLH/bHLH targets of IBH1 and IBL1. Additional genome-wide comparisons suggested a model in which IBH1 antagonized PIF4 but not the PIF4–BZR1 dimer.
The plant hormones brassinosteroids (BRs), gibberellins (GAs), and auxin, together with environmental conditions and signals (such as shade avoidance, far-red light, and temperature), positively regulate cell elongation. This regulatory mechanism involves activation of members of the PACLOBUTRAZOL RESISTANCE (PRE) family of atypical basic helix–loop–helix (bHLH) transcription factors (1–6). PRE members are helix–loop–helix (HLH) proteins that lack the basic domain required for DNA binding and antagonize their interacting HLH or bHLH proteins, of which most act as cell elongation suppressors, thus forming triantagonistic bHLH systems (5, 6). An example of such a system is the Arabidopsis thaliana PRE1 and its interactor, the HLH factor INCREASED LEAF INCLINATION1 BINDING bHLH1 (IBH1). PRE1 expression is induced by GAs, whereas IBH1 functions in BR signaling. When ectopically expressed, IBH1 induces dwarfism and reduces BR sensitivity. BRs are perceived by the BR receptor BR-INSENSITIVE1 (BRI1) at the cell surface to initiate signaling cascades, activating two canonical BR transcription factors [BRASSINAZOLE-RESISTANT1 (BZR1) and the BZR2/BRI1-EMS-SUPPRESSOR1 (BES1) (7, 8)], which directly regulate BR-responsive gene expression and plant development (9). Both BZR1 and BZR2/BES1 bind the IBH1 promoter to repress it (10, 11). IBH1 does not bind DNA directly but interacts with the DNA binding bHLH factors HOMOLOG OF BR-ENHANCED EXPRESSION2 (BEE2), INTERACTING WITH IBH1 (HBI1), ACTIVATOR FOR CELL ELONGATION1 (ACE1), ACE2, ACE3, and CRYPTOCHROME INTERACTING bHLH1 (CIB1) (5, 6). HBI1, ACE1, ACE2, ACE3, and CIB1 positively regulate cell elongation downstream of BRs, GAs, temperature, and light signaling. ACE1 and HBI1 activate directly elongation-promoting genes, such as EXPANSIN and XYLOGLUCAN ENDOTRANSGLYCOSYLASE/HYDROLASE (5, 6). Whereas IBH1 interacts with HBI1 and ACE1 to inhibit their binding to DNA, PRE1 binds to IBH1 to prevent its repressive effect on HBI1 or ACE1, thus activating it. Hence, the ratio of positive (PRE1, HBI1, ACE1, ACE2, ACE3, and CIB1) and negative (IBH1) regulators determines the cell elongation outcome. Similar to IBH1, the putative non-DNA–binding HLH proteins ACTIVATION-TAGGED-BRI1-SUPPRESSOR1–INTERACTING FACTOR1 (AIF1), AIF2, AIF3, and AIF4 negatively regulate BR signaling and cell elongation and are antagonized by their interacting partners PRE1 and PRE3/ACTIVATION-TAGGED-BRI1-SUPPRESSOR1 (12, 13).
The mechanism of cell elongation in Arabidopsis had been further elucidated by the interplay between DELLA, BZR1, and PHYTOCHROME-INTERACTING FACTOR4 (PIF4) proteins that mediate GA, BR, and light and temperature responses, respectively (4, 14, 15). Cell elongation is promoted synergistically by the BRZ1–PIF4 dimer through its direct binding of the DNA-regulatory regions of common target genes but is inhibited by DELLA. Among the BRZ1–PIF4 targets are genes encoding HLH proteins with both positive effects on cell elongation (the PRE family) and negative effects [LONG HYPOCOTYL IN FAR-RED REDUCED PHYTOCHROME SIGNALING1 (HFR1) and PHYTOCHROME RAPIDLY REGULATED1 (PAR1)], forming together a module that controls skotomorphogenesis and shade avoidance (4, 14, 16, 17). Comparable with the IBH1/PRE1 regulatory pair, HFR1/PRE6 and PAR1/PRE1 act through sequestration of the inhibitory protein by interaction with PIF4 that is released to bind its target genes (18, 19).
Here, we characterized an unknown Arabidopsis HLH/bHLH transcription factor, designated IBH1-LIKE1 (IBL1), that acts as a negative regulator of the BR responses and cell elongation similarly to its close homolog IBH1. IBH1 and IBL1 showed largely overlapping transcriptional responses and inhibited the expression of several HLH/bHLH proteins, including PIF4. Although IBH1 had been characterized as a non-DNA–binding protein, it repressed IBL1 transcription directly and acted in tandem to suppress the expression of a common downstream HLH/bHLH network, thus forming the transcriptional regulation node known as the incoherent feed-forward loop (FFL). DNA-binding analysis revealed that all direct targets commonly suppressed by IBH1 and IBL1 were recognized by PIF4. Comparison of genomic colocalization between IBH1, PIF4, and BZR1 suggests a model in which IBH1 shares binding sites with PIF4 but not BZR1.
Results
IBL1 Negatively Regulates BR Signaling and Cell Elongation.
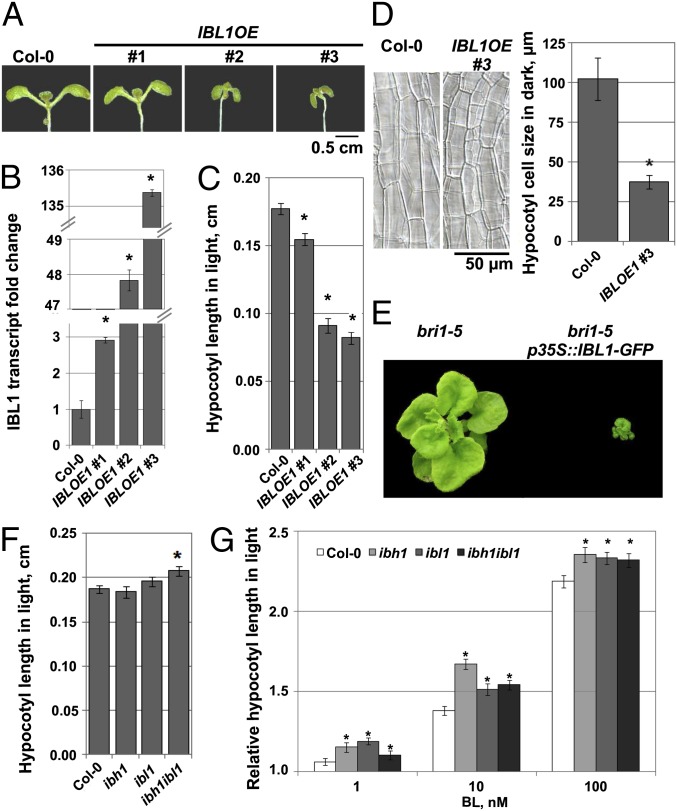
In a search for bHLH-type proteins that regulate BR responses, we screened genes previously identified by microarray analysis by comparing the actions of brassinolide (BL), the most active BR, with bikinin, a specific inhibitor of the negative BR signaling regulator, the BR-INSENSITIVE 2 kinase (20). In total, seven HLH/bHLH factors (At1g68810, At2g42280, At2g43060, At3g25710, At4g30410, At5g08130, and At5g57780) were significantly down-regulated by BL and bikinin. Next, we checked whether the seven HLH/bHLH genes were among the direct targets of the canonical BR transcriptional regulators BZR1 and BZR2/BES1 (10, 11). Four of them [FLOWERING BHLH4 (At2g42280) (21), BES-INTERACTING MYC-LIKE PROTEIN1 (At5g08130) (22), IBH1 (At2g43060) (3), and At4g30410] were targets of BZR1, but only IBH1 was a target of both. We focused on At4g30410, which had previously been identified as an IBH1 homolog (3, 23) (Fig. S1), and designated it IBL1. To determine whether IBL1 functioned in BR responses, we generated transgenic Arabidopsis plants that overexpressed (OE) IBL1 fused to the green fluorescent protein (GFP) under the 35S promoter [p35S::IBL1-GFP (IBL1OE)]. Of the T1 plants, 50% showed dwarfism and had shorter petioles and dark green round leaves. Three homozygous transgenic lines with different IBL1 transcript levels (Fig. 1 A and B) were selected for further analysis. They had short hypocotyls in light and dark because of impaired cell elongation (Fig. 1 C and D). Similar to IBH1 (3), overexpression of IBL1 enhanced the weak phenotype of bri1-5 (Fig. 1E), supporting its role in BR signaling downstream of BRI1.
Fig. 1.
IBL1 is a negative regulator of BR signaling and cell elongation. (A) Phenotypes of light-grown and homozygous IBL1OE (p35S::IBL1-GFP) seedlings compared with Columbia-0 (Col-0). (B) IBL1 transcript level in the IBL1OE lines compared with Col-0 (three quantitative RT-PCR experiments). *P < 0.05 relative to Col-0 (t test). (C) Hypocotyl measurement of light-grown IBL1OE lines compared with Col-0 (number of hypocotyls analyzed > 20). *P < 0.001 relative to Col-0 (t test). (D) Images of epidermal cells in hypocotyls from dark-grown IBL1OE line and Col-0 at 5 d after sowing (DAS; number of cells analyzed > 20). *P < 0.001 relative to Col-0 (t test). (E) Overexpression of IBL1-GFP enhanced the bri1-5 mutation. Hypocotyl measurement of light-grown ibh1, ibl1, and ibh1ibl1 mutants compared with Col-0 in the absence (F) and presence (G) of BL (number of hypocotyls analyzed > 20). (F) *P < 0.005 relative to Col-0 (t test). (G) *P < 0.05 relative to Col-0. Error bars indicate SE. Where not indicated, seedlings were analyzed at 8 DAS.
Next, we examined the loss-of-function mutant of IBL1. One bacterial transferred DNA (T-DNA) insertion line was identified in the exon of the IBL1 gene (ibl1; SALK 119457) (Fig. S2A). Although the ibl1 mutant completely lacked the IBL1 transcript (Fig. S2B), it had no obvious phenotype (Fig. S2E). Because we hypothesized that IBL1 functions redundantly with IBH1, we also characterized a knockdown line for IBH1 (ibh1; SALK 049177) (Fig. S2C). The ibh1 mutant contained a T-DNA insertion in the IBH1 promoter, and its transcript was reduced up to 80% (Fig. S2D), again without an obvious phenotype (Fig. S2E). Detailed examination of the double ibh1ibl1 seedlings revealed only a slight increase in hypocotyl length in the light (Fig. 1F). In addition, hypocotyls of single and double mutants were slightly hypersensitive to BL when grown in the light (Fig. 1G). These results indicate that IBL1 functions as a negative regulator of BR signaling and cell elongation.
IBL1 and IBH1 Act Interdependently.
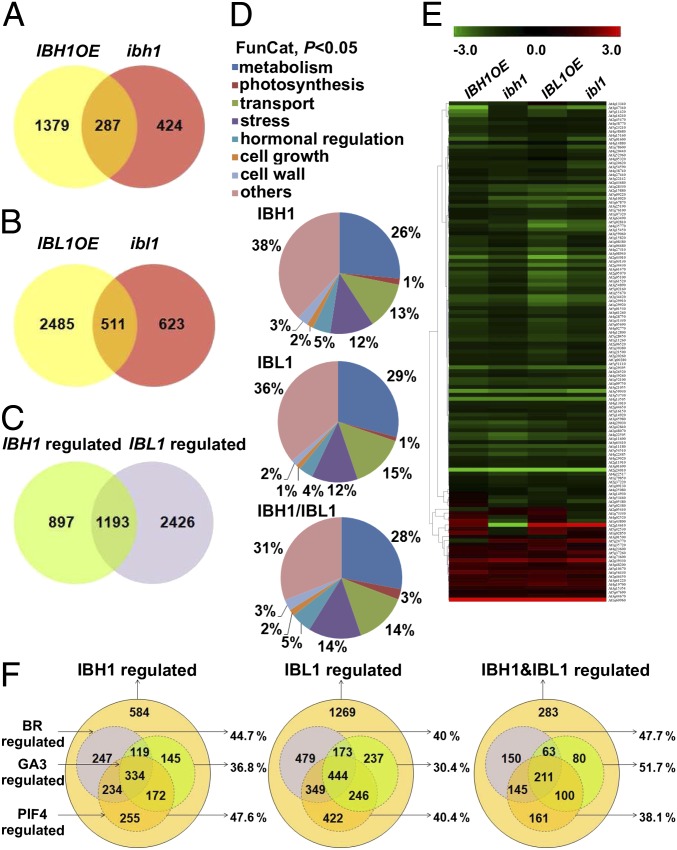
To understand how IBH1 and IBL1 interact to regulate cell elongation, we conducted genome-wide RNA analysis followed by sequencing (RNA-Seq) expression analyses of IBH1 [p35S::IBH1-GFP (IBH1OE)] and IBL1 gain-of-function (IBL1OE) mutants and loss-of-function mutants ibh1 and ibl1. The IBH1OE line showed a strong dwarf phenotype and enhanced the weak bri1-5 mutant, as reported previously (Fig. S2 F–H) (3). RNA-Seq analysis identified 2,090 IBH1-regulated genes (Dataset S1), of which 1,666 genes were differentially expressed in IBH1OE (870 and 796 down- and up-regulated, respectively), 711 genes were differentially expressed in ibh1 (431 and 280 down- and up-regulated, respectively), and 287 genes were affected in both mutants, which was significant compared with a random control (P = 1.3 × 10−188, Fisher exact test) (Fig. 2A and Dataset S2). Parallel RNA-Seq analyses identified 3,619 genes regulated by IBL1 (Datasets S1 and S2), of which 2,996 genes (769 and 2,227 down- and up-regulated, respectively) and 1,134 (915 and 219 down- and up-regulated, respectively) genes were affected in IBL1OE and ibl1, respectively, and 511 genes were affected in both mutants (P = 3.8 × 10−242; Fisher exact test) (Fig. 2B and Dataset S2). Of 2,090 and 3,619 genes regulated by IBH1 and IBL1, respectively, 1,193 genes were shared, corresponding to 57% and 33% of all of the genes, respectively (P < 1 × 10−242) (Fig. 2C). Gene ontology analysis showed that the common responsive IBH1 and IBL1 genes were enriched in similar functional categories, including cell metabolism, photosynthesis, hormone signaling, cell wall, and cell growth as well as responses to stress factors (Fig. 2D and Dataset S3).
Fig. 2.
Target gene expression regulated by IBH1 and IBL1. (A) Overlap of differentially expressed genes in RNA-Seq experiments performed on RNA from IBH1OE and ibh1 seedlings. (B) Overlap of differentially expressed genes in RNA-Seq performed on RNA from IBL1OE and ibl1 seedlings. (C) Overlap of genes regulated by IBH1 and IBL1. (D) Functional categories of IBH1, IBL1, and commonly regulated genes (P < 0.05). (E) Heat map representation of the expression profiles of 127 genes differentially expressed in IBH1OE, ibh1, IBL1OE, and ibl1 plants. The color scale indicates the fold change. (F) Comparison of IBH1, IBL1, and their commonly regulated target genes with BR, GA3, and PIF4 expression datasets available from the literature. Seedlings were analyzed at 10 DAS.
Comparison of genes up- and down-regulated by either IBH1 or IBL1 indicated that most of the overlapping genes between gain- and loss-of-function IBH1 and IBL1 plants had similar rather than opposite expression trends (Fig. S3 A and C and Dataset S2). The largest overlap occurred between genes down-regulated in both mutant backgrounds (168 genes for IBH1 and 321 genes for IBL1) and enriched in categories with possible functions in photosynthesis, light, and hormonal and temperature responses (Fig. S3 B and D and Dataset S3). The heat map of 127 genes, commonly regulated in IBH1OE, IBL1OE, ibh1, and ibl1 plants (Fig. 2E and Dataset S2), revealed that 98 (77%) of the coregulated genes were down-regulated, suggesting that IBH1 and IBL1 repress a common set of genes. RNA-Seq results were validated by quantitative (q)RT-PCR analysis in IBH1 and IBL1 gain- and loss-of-function plants. The expression of genes encoding the BR biosynthetic enzyme CONSTITUTIVE PHOTOMORPHOGENIC DWARF (24) was down-regulated in all four backgrounds, consistent with the RNA-Seq results (Fig. S4A).
Because IBH1 had been identified as a negative regulator of cell elongation that functioned downstream of the PIF4–BZR1–DELLA module integrating signals from BRs, GAs, light, and temperature (4, 14, 15), we compared the genes regulated by IBH1 and IBL1 with previously identified gene sets regulated by BRs (10, 11), PIF4 (14), and GAs (4). Like IBH1, genes regulated by IBL1 overlapped with genes regulated by BRs, PIF4, or GAs with similar percentages (Fig. 2F and Dataset S4). The genes commonly regulated by IBH1 and IBL1 were slightly more enriched in GA-regulated genes. Comparison of the expression data suggests that IBH1 and IBL1 act interdependently downstream of the PIF4–BZR1–DELLA module to regulate BR, light, and temperature responses.
IBH1 and IBL1 Act Through an Incoherent FFL to Control Several Downstream bHLH Transcription Factors.
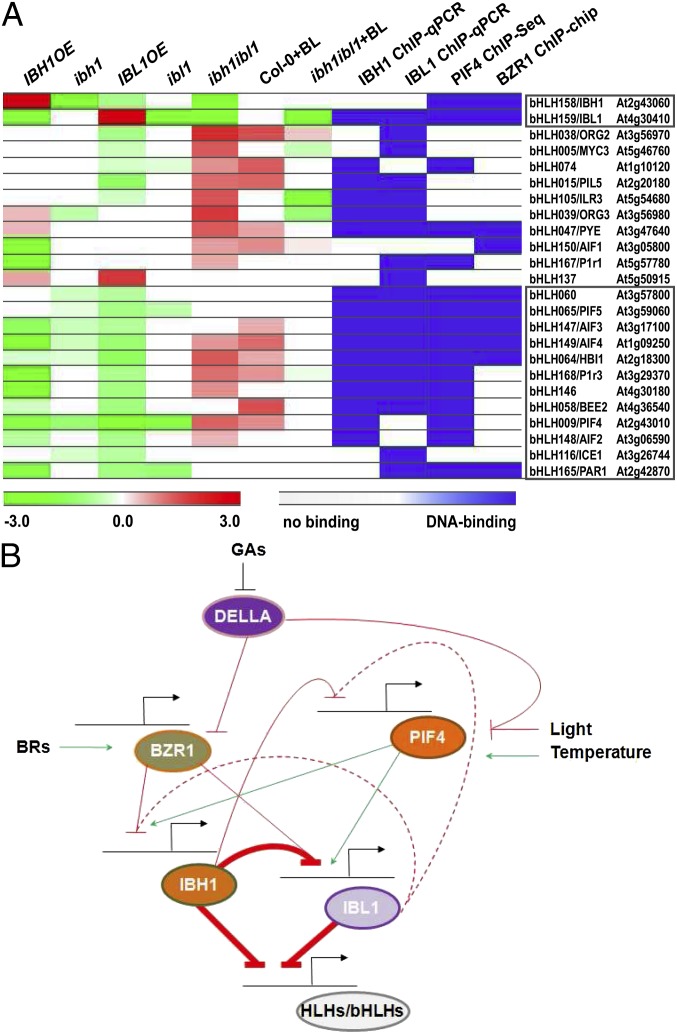
The large number of overlapping genes regulated by IBH1 and IBL1 prompted us to investigate whether IBH1 and IBL1 control the expression of other bHLH transcription factors. We compared the genes regulated by IBH1 and IBL1 identified by RNA-Seq with a combined list of HLH/bHLH transcription factors provided by the Arabidopsis Gene Regulatory Information Server (25) and the work by Carretero-Paulet et al. (26) (Dataset S5). In total, 28 HLH/bHLH transcription factors, including IBH1 and IBL1, were regulated by IBH1, IBL1, or both (Fig. S4B and Dataset S5). To verify this hypothetical bHLH transcription network, we analyzed all 28 differentially expressed HLH/bHLH genes (including IBH1 and IBL1) by qRT-PCR in IBH1OE, IBL1OE, ibh1, and ibl1 mutants (Fig. S4C and Dataset S5). For 24 of 28 HLH/bHLH transcription factors, the differential expression was validated (Fig. 3A). Interestingly, the expression of IBL1 and IBH1 was down-regulated in IBH1OE and IBL1OE plants, respectively. In addition, the expression of 13 HLH/bHLH transcription factors was regulated by both IBH1 and IBL1 (Dataset S5), of which 12 were down-regulated in both overexpression and knockdown backgrounds (Fig. 3A). AIF2, AIF3, AIF4 (12, 13), HBI1 (5), BEE2 (27), PIF4 (4, 14) and PIF5 (28), PAR1 (17), and INDUCER OF CBP EXPRESSION1 (29) (Dataset S5) were among the 12 HLH/bHLH factors, consistent with the observed down-regulation of target genes by IBH1 and IBL1 and the signaling cross-talk between BRs, GAs, temperature, and light in the cell elongation regulation (4, 14, 15).
Fig. 3.
Incoherent FFL formed by IBH1 and IBL1. (A) Heat map representation of 24 bHLH/HLH proteins differentially regulated by IBH1 and IBL1. From left to right, bHLH/HLH target gene expression in IBH1OE, ibh1, IBL1OE, ibl1, ibh1ibl1, and BL-treated Col-0 and ibh1ibl1 seedlings compared with controls at 10 DAS validated by qRT-PCR; direct binding of IBH1, IBL1, PIF4, and BZR1 to the target bHLH/HLH genes. The color scales indicate the fold change and direct binding. Highlighted in frames are the IBH1 and IBL1 genes and 12 down-regulated HLH/bHLH target genes. (B) Incoherent FFL model. The GA-negative regulator DELLA interacts with BZR1 and PIF4 and suppresses their DNA-binding activity. BZR1 forms a heterodimer with PIF4, and this protein complex regulates the transcription of IBH1 and IBL1 on signals, such as BRs, GAs, light, and temperature. BR inhibits the expression of IBH1 and IBL1 through direct binding by the BZR1. PIF4 also directly regulates IBH1 and IBL1 expression but oppositely to BZR1. IBH1 and IBL1 act downstream of the PIF4–BZR1–DELLA module by forming a type 2 incoherent FFL transcriptional node, where IBH1 down-regulates directly the expression of IBL1, and both repress the expression of downstream HLH/bHLH factors. Black lines and arrows, gene regulatory regions that control gene expression; blunt-ended red arrows, inhibition; dotted lines, indirect regulation; green arrows, induction; ovals, proteins; straight lines, direct regulation; thick lines, the incoherent FFL node.
Although IBH1 had recently been characterized as a putative non-DNA–binding transcription factor (5, 6), direct targets for another HLH/bHLH transcription factor, UP-BEAT1 (UPB1), also lacking the DNA binding domain were identified by chromatin immunoprecipitation (ChIP) followed by microarray analysis (30), showing that non-DNA–binding transcription factors could still be part of transcriptional complexes. Therefore, to test whether the differentially expressed HLH/bHLH transcription factors, including IBH1 and IBL1, were direct targets of IBH1 and IBL1, we used plants expressing GFP-tagged IBL1 and IBH1 under the corresponding endogenous promoters (pIBH1::IBH1-GFP and pIBL1::IBL1-GFP) for the ChIP assays followed by qPCR (ChiP-qPCR). The ChiP-qPCR revealed that IBH1 and IBL1 bound to the regulatory regions of 16 and 19 HLH/bHLH transcription factors, respectively (Fig. 3A, Fig. S5A, and Dataset S5). Whereas IBH1 was part of the transcriptional IBL1-regulating complex, IBL1 had no direct effect on the IBH1 expression; 8 of 12 HLH/bHLH transcription factors commonly down-regulated by IBL1 and IBH1 were also their direct targets (Dataset S5). The observation that IBH1 and IBL1 suppressed the expression of a common downstream HLH/bHLH network suggested that these proteins might act as a transcription regulation node, known as FFL. Such a node consists of two transcription factors—one transcription factor controlling the other one and jointly regulating a target gene (31, 32). Because IBH1 inhibited IBL1 and both repressed the target gene expression, the FFL was an incoherent type 2, in which the direct and indirect paths are in opposite direction; however, both result in target gene repression (Fig. 3B) (31, 32). In accordance with the suggested incoherent type 2 FFL model, the expression of 12 HLH/bHLH transcription factors was either up-regulated or not significantly modified when both repressors were absent in the double ibh1ibl1 mutant (Fig. 3A and Dataset S5). The expression tendency was similar in wild-type plants treated with BL that decreased the IBL1 and IBH1 levels (Fig. 3A and Dataset S5). In contrast, BL addition did not change significantly the expression of 12 HLH/bHLH targets in ibh1ibl1 plants. In summary, our results imply that a putative incoherent FFL network motif might control the transcriptional regulation of BRs, light, and/or temperature responses to ultimately, as shown previously, regulate cell elongation (14, 15).
IBH1 Is Part of the PIF4 Transcription Complexes.
Among the HLH/bHLH transcription factors regulated by IBH1 and IBL1 was the key light response regulator PIF4 (14), of which the expression was down-regulated in both IBH1 and IBL1 gain- and loss-of-function mutants (Fig. 3A and Dataset S5). Previously, PIF4 had been shown to bind directly to IBH1 and IBL1 and stimulate their expression (14). Remarkably, all HLH/bHLH transcription factors directly regulated by IBH1 and IBL1 were direct targets of PIF4 (14) but not all of BZR1 (10) (Fig. 3A).
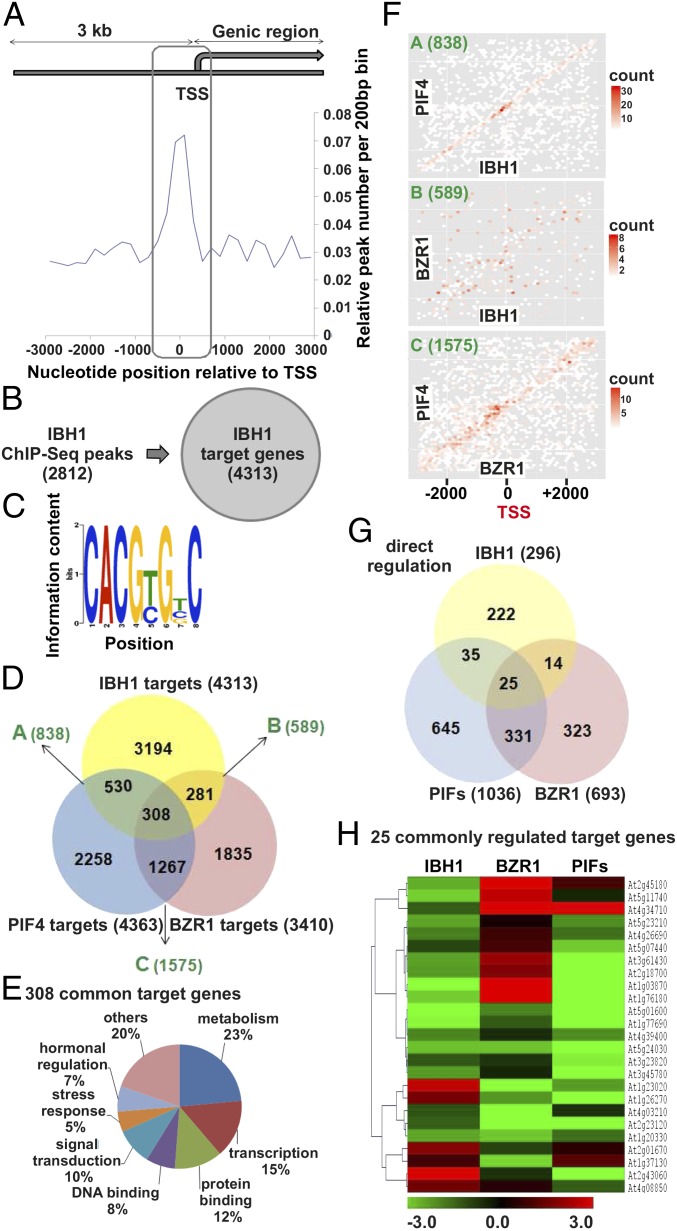
To investigate the possible link between PIF4, IBH1, and BZR1, we conducted a ChIP analysis followed by sequencing (ChIP-Seq) on the IBH1OE line and a control line p35S::GFP. Antibodies against GFP in the GFP-tagged IBH1 were used for immunoprecipitation of the endogenous protein–DNA complex, and the obtained DNA was subjected to next generation sequencing. The IBH1 ChIP-Seq provided satisfactory coverage of an approximate total of 11 million aligned reads. The alignment of 2–4 million reads was unique in the genome, representing ∼36% coverage, which is considered sufficient (33). Accordingly, the ChIP-Seq on the control p35S::GFP plants did not precipitate a significant amount of DNA because of the lack of DNA-binding capacity of the free GFP. Only uniquely aligned sequences were further analyzed, resulting in 2,945 statistically significant IBH1-binding peaks (P < 10−5). The screen was further restricted within 3 kb upstream of the transcriptional start site (TSS) and/or on the genic region, including untranslated regions, exons, and introns (Fig. 4A). In these genome segments, 2,812 (95.5%) of 2,945 IBH1 binding peaks had a prevalent distribution around 1 kb from the TSS (Fig. 4A). ChIP-qPCR amplification of random IBH1-binding peaks resulted in 87% successful validation of the ChIP-Seq results (Fig. S5B and Dataset S5). The binding of IBH1 to the regulatory region of BRI1 in plants with enhanced or endogenous IBH1 expression was confirmed as well (Fig. S5C). Because more than 50% of 2,812 peaks were located within 3 kb upstream of the TSS and/or on the genic region of at least two gene loci, they corresponded to a total of 4,313 nonredundant gene loci (Fig. 4B and Dataset S6), suggesting that IBH1 preferentially binds to high-gene density regions. In accordance with the binding of bHLH regulators to the hexanucleotide E-box DNA motif CANNTG (with N any nucleotide), the CACGTG (E-box type) G-box motif was found as the most enriched cis element among the IBH1-binding sites (Fig. 4C). Comparison of the gene sets bound directly by IBH1, PIF4 (14), and BZR1 (10) (Fig. 4D) showed that IBH1 shared targets with PIF4 (838 genes) and BZR1 (589 genes) and that, in total, 308 genes were common direct targets of IBH1, PIF4, and BZR1. This overlap was enriched in genes with functions in metabolism, transcription, protein and DNA binding, signal transduction, stress response, and hormonal regulation (Fig. 4E and Dataset S6). To better understand the possible mechanisms underlying gene regulation by these three transcription factors, we checked the distribution of the binding sites of IBH1, PIF4, and BZR1 in the genomic regions of the shared target genes. We found that IBH1 and PIF4 predominantly bound to overlapping DNA regions in their common targets but that IBH1 and BZR1 did not share common binding sites in the targeted genes (Fig. 4F). Because PIF4 and BZR1 had been reported to colocalize and interact at the regulatory regions of their shared targets (14), our results suggest that IBH1 can repress PIF4 targets, possibly by forming a complex with PIF4, which in turn, recognizes cis-regulatory elements that differ from the elements bound by the PIF4–BZR1 complex. To test this hypothesis, we examined how IBH1, PIF4, and BZR1 regulated the expression of direct common targets. Therefore, we considered genes that were differentially expressed in the IBH1OE line, the quadruple pifq mutant, and the dominant bzr1-D line (14) that had been identified as direct targets of IBH1, PIF4, and BZR1, respectively (Dataset S6). The comparison of genes directly regulated by IBH1, PIF4, and BZR1 resulted in a minimum of 25 genes (Fig. 4 G and H and Dataset S6). The expression profile of this gene set revealed that IBH1 and PIF4 regulate transcription in the opposite direction from BZR1, consistent with the model in which IBH1 suppresses primarily PIF4 but not PIF4–BZR1 targets.
Fig. 4.
IBH1 as part of transcriptional complexes. (A) Distribution of IBH1 binding peaks (frequency) relative to gene structure (−3 kb to the coding sequence). (B) Total IBH1-binding peaks in the region shown in A and corresponding target genes identified by the ChIP-Seq analysis. (C) Frequency of cis elements around the IBH1-binding sites. The sequence logo shows the most enriched motifs in the IBH1-binding regions. (D) Overlap between the direct target genes of IBH1, PIF4, and BZR1. (E) Functional categories in the 308 direct target genes shown in D (P < 0.05). (F) Genome colocalization of binding sites of IBH1, PIF4, and BZR1 along the promoter 3,000 bp upstream of the TSS and in the downstream genic region. The frequency of peak pairs is represented by a color scale. Counts are numbers of peak pairs in a certain area. (G) Overlap between the directly regulated targets of IBH1 (IBH1OE), PIF4 (pifq), and BZR1 (bzr1-D). (H) Heat map representation of the expression profiles of the 25 directly regulated genes shown in G. The color scale indicates the fold change.
Discussion
Here, we identified another negative regulator of BR signaling and cell elongation, the IBH1 homolog IBL1, that regulates downstream genes common with IBH1. The expression of IBH1 and IBL1 was inhibited by BRs through direct DNA binding of BZR1 to their promoters (10) and directly regulated by PIF4 but in a fashion opposite to BZR1 (14, 28). To clarify the role of IBH1 and IBL1 in cell elongation, we searched for gene regulatory networks through which these transcriptional regulators act. We found that IBH1 and IBL1 predominantly down-regulated genes with enriched functions in photosynthesis, light, hormonal, and temperature responses, which is in line with the IBH1 and IBL1 action downstream from the PIF4–BZR1–DELLA node (4, 14, 15). To elucidate the relevance of this type of regulation, we focused on genes differentially regulated by IBH1 and IBL1 that encode HLH/bHLH proteins, because this transcription factor family had been shown to control responses triggered by hormones, light, and stress factors in plants (1–6). Members of the plant bHLH family are important for organ formation, hormonal responses, stomata patterning, and flavonoid biosynthesis, and in analogy to the mammals, their action mechanism involves HLH/bHLH interactions as well as formation of complexes with other interacting transcription factors (34, 35). The analyses highlighted a type 2 incoherent FFL network motif, in which IBH1 down-regulates directly the IBL1 expression, whereas together, they repress the expression of common downstream HLH/bHLH factors. This type of negative regulation was confirmed computationally and experimentally to be essential for either accelerating target gene responses or generating transient transcription pulses (31, 32, 36). Although this incoherent FFL model awaits quantitative validation, we can speculate that such a mechanism is necessary to maintain homeostasis of plant responses after perception of signals related to hormones, light, temperature, and/or stress conditions that trigger IBH1 and IBL1 (31, 32). It remains to be determined whether, other than to BR hormones, IBH1 and IBL1 respond to different stimuli to execute either an interdependent action on shared targets or an independent regulation of unique targets.
Although IBH1 has been defined as non-DNA binding (5, 6) and the DNA-binding properties of IBL1 are unknown, the ChIP assay showed that both IBH1 and IBL1 are part of gene-regulating transcriptional complexes. In support, UPB1, another HLH type of transcription factor that lacks a typical DNA-binding domain, has been shown to bind target genes (30). Similarly, the recruitment of the non-DNA–binding MYELOBLASTOSIS FAMILY TRANSCRIPTION FACTOR-LIKE 2 to target genes occurred seemingly through its direct interaction with BZR2/BES1 (37). Moreover, despite a mutation preventing the protein–DNA binding of the bHLH protein R of maize (Zea mays), the mutated R has been shown to be part of the transcriptional complex occupying the target promoter (38).
In agreement with their roles as negative cell elongation regulators, IBH1 and IBL1 repress the expression of HLH/bHLH transcription factors that stimulate hypocotyl elongation, such as the BR-regulated HBI1, BEE2, and the light-related PIF4 and PIF5 (4, 14, 28). However, among the suppressed targets were also HLH factors encoding suppressors of cell elongation, including the AIF protein family, and PAR1 (16, 17), suggesting that IBH1 and IBL1 exert a feedback control on cell elongation by driving the expression of positive and negative regulators, thus integrating signals, such as BRs, GAs, light, temperature, and aging.
Our genome colocalization analysis showed that the IBH1-binding sites overlap with PIF4 but not BZR1. Accordingly, IBH1 and PIF4 regulated a subset of genes in the opposite direction of BZR1. Most of the IBH1-binding peaks were located in the promoter regions within 1 kb from the TSS, consistent with the mode in which PIF4 recognizes its target genes (14). In agreement with our hypothesis that IBH1 and PIF4 recognize similar binding sites, the typical bHLH G-box motif was the most enriched cis element in binding sites of both IBH1 and PIF4. Although IBH1 does not bind DNA directly (5, 6), it is possible that IBH1 is part of the transcriptional PIF4-repressing complex but not the PIF4–BZR1 dimer (Fig. 3B). However, neither yeast two-hybrid nor bimolecular fluorescent complementation assay showed a direct interaction between PIF4 and IBH1 (Fig. S6 and Dataset S7), suggesting that this putative interaction might require other proteins.
The fact that PIF4 transcription is repressed also by IBH1 and IBL1 hints at their control of the PIF4 de novo synthesis, indirectly affecting the respective ratio of the PIF4–BZR1 complexes. Based on our data, two distinct feedback regulatory mechanisms that impact on the PIF4 and BZR1 levels could be highlighted. In one case, IBH1 represses PIF4, whereas PIF4 induces expression of IBH1 and IBL1, which might be an additional mechanism to repress its own transcription. As an alternative scenario, which needs additional experimentation, IBH1 might block the DNA-binding activity of PIF4 at a posttranslational level, similar to HFR1 and PAR1 (18, 19). The results suggest that IBH1 and IBL1 are part of the PIF4 hub and play a role in fine tuning the BR-mediated cell elongation.
Methods
A. thaliana (L.) Heyhn plants (accession Columbia-0) were grown at 22 °C for a 16-h photoperiod (65 μE m−2 s−1) on half-strength Murashige and Skoog (MS) medium with 10 g L−1 sucrose. For ChIP-Seq and RNA-Seq analyses and validation, plants were grown on half-strength MS medium under continuous light. Whole seedlings were collected 10 d after sowing. For treatments, seeds were germinated on MS medium containing 10 nM BL or dimethyl sulfoxide as mock treatment. Genotyping of mutants was performed with the primers listed in Dataset S8.
Additional technical details are presented in SI Methods.
Supplementary Material
Acknowledgments
We thank D. Weijers, B. De Rybel, D. Inzé, and S. Prat for providing materials and B. Wittler, J. Boruc, K. Mejia-Guerra, W. Dejonghe, E. Mylle, and C. Betti for technical assistance. M.K.Z. thanks the Agency for Innovation by Science and Technology for a postdoctoral fellowship.
Footnotes
The authors declare no conflict of interest.
Data deposition: The data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (accession no. GSE51121).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1400203111/-/DCSupplemental.
References
- 1.Lee S, et al. Overexpression of PRE1 and its homologous genes activates gibberellin-dependent responses in Arabidopsis thaliana. Plant Cell Physiol. 2006;47(5):591–600. doi: 10.1093/pcp/pcj026. [DOI] [PubMed] [Google Scholar]
- 2.Hyun Y, Lee I. KIDARI, encoding a non-DNA binding bHLH protein, represses light signal transduction in Arabidopsis thaliana. Plant Mol Biol. 2006;61(1–2):283–296. doi: 10.1007/s11103-006-0010-2. [DOI] [PubMed] [Google Scholar]
- 3.Zhang L-Y, et al. Antagonistic HLH/bHLH transcription factors mediate brassinosteroid regulation of cell elongation and plant development in rice and Arabidopsis. Plant Cell. 2009;21(12):3767–3780. doi: 10.1105/tpc.109.070441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bai M-Y, et al. Brassinosteroid, gibberellin and phytochrome impinge on a common transcription module in Arabidopsis. Nat Cell Biol. 2012;14(8):810–817. doi: 10.1038/ncb2546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bai M-Y, Fan M, Oh E, Wang Z-Y. A triple helix-loop-helix/basic helix-loop-helix cascade controls cell elongation downstream of multiple hormonal and environmental signaling pathways in Arabidopsis. Plant Cell. 2012;24(12):4917–4929. doi: 10.1105/tpc.112.105163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ikeda M, Fujiwara S, Mitsuda N, Ohme-Takagi M. A triantagonistic basic helix-loop-helix system regulates cell elongation in Arabidopsis. Plant Cell. 2012;24(11):4483–4497. doi: 10.1105/tpc.112.105023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang Z-Y, et al. Nuclear-localized BZR1 mediates brassinosteroid-induced growth and feedback suppression of brassinosteroid biosynthesis. Dev Cell. 2002;2(4):505–513. doi: 10.1016/s1534-5807(02)00153-3. [DOI] [PubMed] [Google Scholar]
- 8.Yin Y, et al. BES1 accumulates in the nucleus in response to brassinosteroids to regulate gene expression and promote stem elongation. Cell. 2002;109(2):181–191. doi: 10.1016/s0092-8674(02)00721-3. [DOI] [PubMed] [Google Scholar]
- 9.Zhu J-Y, Sae-Seaw J, Wang Z-Y. Brassinosteroid signalling. Development. 2013;140(8):1615–1620. doi: 10.1242/dev.060590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sun Y, et al. Integration of brassinosteroid signal transduction with the transcription network for plant growth regulation in Arabidopsis. Dev Cell. 2010;19(5):765–777. doi: 10.1016/j.devcel.2010.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yu X, et al. A brassinosteroid transcriptional network revealed by genome-wide identification of BESI target genes in Arabidopsis thaliana. Plant J. 2011;65(4):634–646. doi: 10.1111/j.1365-313X.2010.04449.x. [DOI] [PubMed] [Google Scholar]
- 12.Wang H, et al. Regulation of Arabidopsis brassinosteroid signaling by atypical basic helix-loop-helix proteins. Plant Cell. 2009;21(12):3781–3791. doi: 10.1105/tpc.109.072504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ikeda M, Mitsuda N, Ohme-Takagi M. ATBS1 INTERACTING FACTORs negatively regulate Arabidopsis cell elongation in the triantagonistic bHLH system. Plant Signal Behav. 2013;8(3):e23448. doi: 10.4161/psb.23448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Oh E, Zhu J-Y, Wang Z-Y. Interaction between BZR1 and PIF4 integrates brassinosteroid and environmental responses. Nat Cell Biol. 2012;14(8):802–809. doi: 10.1038/ncb2545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gallego-Bartolomé J, et al. Molecular mechanism for the interaction between gibberellin and brassinosteroid signaling pathways in Arabidopsis. Proc Natl Acad Sci USA. 2012;109(33):13446–13451. doi: 10.1073/pnas.1119992109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hornitschek P, Lorrain S, Zoete V, Michielin O, Fankhauser C. Inhibition of the shade avoidance response by formation of non-DNA binding bHLH heterodimers. EMBO J. 2009;28(24):3893–3902. doi: 10.1038/emboj.2009.306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Roig-Villanova I, et al. Interaction of shade avoidance and auxin responses: A role for two novel atypical bHLH proteins. EMBO J. 2007;26(22):4756–4767. doi: 10.1038/sj.emboj.7601890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hao Y, Oh E, Choi G, Liang Z, Wang Z-Y. Interactions between HLH and bHLH factors modulate light-regulated plant development. Mol Plant. 2012;5(3):688–697. doi: 10.1093/mp/sss011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hong S-Y, et al. A competitive peptide inhibitor KIDARI negatively regulates HFR1 by forming nonfunctional heterodimers in Arabidopsis photomorphogenesis. Mol Cells. 2013;35(1):25–31. doi: 10.1007/s10059-013-2159-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.De Rybel B, et al. Chemical inhibition of a subset of Arabidopsis thaliana GSK3-like kinases activates brassinosteroid signaling. Chem Biol. 2009;16(6):594–604. doi: 10.1016/j.chembiol.2009.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ito S, et al. FLOWERING BHLH transcriptional activators control expression of the photoperiodic flowering regulator CONSTANS in Arabidopsis. Proc Natl Acad Sci USA. 2012;109(9):3582–3587. doi: 10.1073/pnas.1118876109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yin Y, et al. A new class of transcription factors mediates brassinosteroid-regulated gene expression in Arabidopsis. Cell. 2005;120(2):249–259. doi: 10.1016/j.cell.2004.11.044. [DOI] [PubMed] [Google Scholar]
- 23.De Rybel B, et al. A versatile set of ligation-independent cloning vectors for functional studies in plants. Plant Physiol. 2011;156(3):1292–1299. doi: 10.1104/pp.111.177337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Szekeres M, et al. Brassinosteroids rescue the deficiency of CYP90, a cytochrome P450, controlling cell elongation and de-etiolation in Arabidopsis. Cell. 1996;85(2):171–182. doi: 10.1016/s0092-8674(00)81094-6. [DOI] [PubMed] [Google Scholar]
- 25.Yilmaz A, et al. AGRIS: The Arabidopsis Gene Regulatory Information Server, an update. Nucleic Acids Res. 2011;39(Database Issue):D1118–D1122. doi: 10.1093/nar/gkq1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Carretero-Paulet L, et al. Genome-wide classification and evolutionary analysis of the bHLH family of transcription factors in Arabidopsis, poplar, rice, moss, and algae. Plant Physiol. 2010;153(3):1398–1412. doi: 10.1104/pp.110.153593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Friedrichsen DM, et al. Three redundant brassinosteroid early response genes encode putative bHLH transcription factors required for normal growth. Genetics. 2002;162(3):1445–1456. doi: 10.1093/genetics/162.3.1445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hornitschek P, et al. Phytochrome interacting factors 4 and 5 control seedling growth in changing light conditions by directly controlling auxin signaling. Plant J. 2012;71(5):699–711. doi: 10.1111/j.1365-313X.2012.05033.x. [DOI] [PubMed] [Google Scholar]
- 29.Lee B-h, Henderson DA, Zhu J-K. The Arabidopsis cold-responsive transcriptome and its regulation by ICE1. Plant Cell. 2005;17(11):3155–3175. doi: 10.1105/tpc.105.035568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tsukagoshi H, Busch W, Benfey PN. Transcriptional regulation of ROS controls transition from proliferation to differentiation in the root. Cell. 2010;143(4):606–616. doi: 10.1016/j.cell.2010.10.020. [DOI] [PubMed] [Google Scholar]
- 31.Mangan S, Alon U. Structure and function of the feed-forward loop network motif. Proc Natl Acad Sci USA. 2003;100(21):11980–11985. doi: 10.1073/pnas.2133841100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Alon U. Network motifs: Theory and experimental approaches. Nat Rev Genet. 2007;8(6):450–461. doi: 10.1038/nrg2102. [DOI] [PubMed] [Google Scholar]
- 33.Kaufmann K, et al. Chromatin immunoprecipitation (ChIP) of plant transcription factors followed by sequencing (ChIP-SEQ) or hybridization to whole genome arrays (ChIP-CHIP) Nat Protoc. 2010;5(3):457–472. doi: 10.1038/nprot.2009.244. [DOI] [PubMed] [Google Scholar]
- 34.Wang Z-Y, Bai M-Y, Oh E, Zhu J-Y. Brassinosteroid signaling network and regulation of photomorphogenesis. Annu Rev Genet. 2012;46:701–724. doi: 10.1146/annurev-genet-102209-163450. [DOI] [PubMed] [Google Scholar]
- 35.Feller A, Machemer K, Braun EL, Grotewold E. Evolutionary and comparative analysis of MYB and bHLH plant transcription factors. Plant J. 2011;66(1):94–116. doi: 10.1111/j.1365-313X.2010.04459.x. [DOI] [PubMed] [Google Scholar]
- 36.Kuttykrishnan S, Sabina J, Langton LL, Johnston M, Brent MR. A quantitative model of glucose signaling in yeast reveals an incoherent feed forward loop leading to a specific, transient pulse of transcription. Proc Natl Acad Sci USA. 2010;107(38):16743–16748. doi: 10.1073/pnas.0912483107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ye H, Li L, Guo H, Yin Y. MYBL2 is a substrate of GSK3-like kinase BIN2 and acts as a corepressor of BES1 in brassinosteroid signaling pathway in Arabidopsis. Proc Natl Acad Sci USA. 2012;109(49):20142–20147. doi: 10.1073/pnas.1205232109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kong Q, et al. Regulatory switch enforced by basic helix-loop-helix and ACT-domain mediated dimerizations of the maize transcription factor R. Proc Natl Acad Sci USA. 2012;109(30):E2091–E2097. doi: 10.1073/pnas.1205513109. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.