Significance
Although knowledge about the human interactome is increasing in coverage because of the development of high-throughput technologies, fundamental gaps remain. In particular, interactions mediated by short linear motifs are of great importance for signaling, but systematic experimental approaches for their detection are missing. We fill this important gap by developing a dedicated approach that combines bioinformatics, custom oligonucleotide arrays and peptide-phage display. We computationally design a library of all possible motifs in a given proteome, print representatives of these on custom oligonucleotide arrays, and identify natural peptide binders for a given protein using phage display. Our approach is scalable and has broad application. Here, we present a proof-of-concept study using both designed human and viral peptide libraries.
Abstract
The human proteome contains a plethora of short linear motifs (SLiMs) that serve as binding interfaces for modular protein domains. Such interactions are crucial for signaling and other cellular processes, but are difficult to detect because of their low to moderate affinities. Here we developed a dedicated approach, proteomic peptide-phage display (ProP-PD), to identify domain–SLiM interactions. Specifically, we generated phage libraries containing all human and viral C-terminal peptides using custom oligonucleotide microarrays. With these libraries we screened the nine PSD-95/Dlg/ZO-1 (PDZ) domains of human Densin-180, Erbin, Scribble, and Disks large homolog 1 for peptide ligands. We identified several known and putative interactions potentially relevant to cellular signaling pathways and confirmed interactions between full-length Scribble and the target proteins β-PIX, plakophilin-4, and guanylate cyclase soluble subunit α-2 using colocalization and coimmunoprecipitation experiments. The affinities of recombinant Scribble PDZ domains and the synthetic peptides representing the C termini of these proteins were in the 1- to 40-μM range. Furthermore, we identified several well-established host–virus protein–protein interactions, and confirmed that PDZ domains of Scribble interact with the C terminus of Tax-1 of human T-cell leukemia virus with micromolar affinity. Previously unknown putative viral protein ligands for the PDZ domains of Scribble and Erbin were also identified. Thus, we demonstrate that our ProP-PD libraries are useful tools for probing PDZ domain interactions. The method can be extended to interrogate all potential eukaryotic, bacterial, and viral SLiMs and we suggest it will be a highly valuable approach for studying cellular and pathogen–host protein–protein interactions.
There are an estimated 650,000 protein–protein interactions in a human cell (1). These interactions are integral to cellular function and mediate signaling pathways that are often misregulated in cancer (2) and may be hijacked by viral proteins (3). Commonly, signaling pathways involve moderate affinity interactions between modular domains and short linear motifs (SLiMs; conserved 2- to 10-aa stretches in disordered regions) (4) that are difficult to capture using high-throughput methods, such as yeast two-hybrid (Y2H) or affinity-purification mass spectrometry (AP/MS) but can be identified using peptide arrays, split-protein systems (5, 6), or peptide-phage display (7–10). A major limitation of peptide arrays is coverage, because the number of potential binding peptides in the proteome is orders of magnitude larger than what can be printed on an array. Conventional phage libraries display combinatorially generated peptide sequences that can identify biophysically optimal ligands of modular domains but this approach can exhibit a hydrophobic bias and may not be ideal for detecting natural binders (11). Thus, there is a need for alternative approaches for identification of relevant domain–SLiMs interactions.
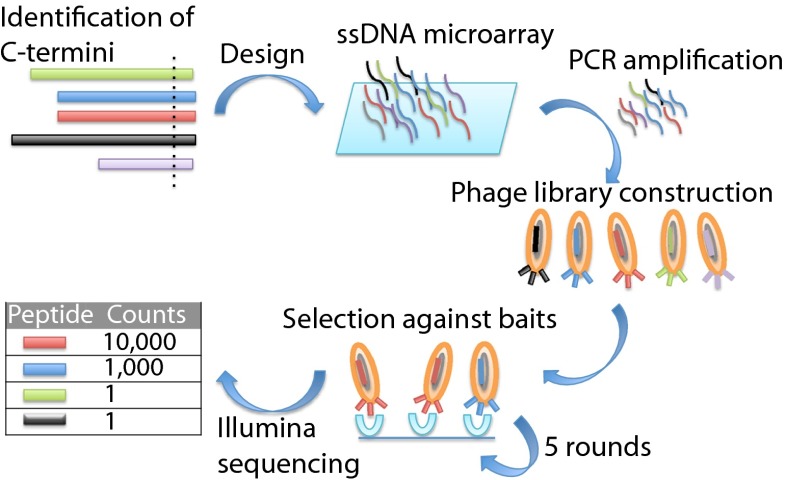
Here, we report an approach that solves both the problem of coverage and the problem of artificial binders. We take advantage of microarray-based oligonucleotide synthesis to construct custom-made peptide-phage libraries for screening peptide–protein interactions, an approach we call proteomic peptide-phage display (ProP-PD) (Fig. 1). This process is similar in concept to the method for autoantigen discovery recently proposed by Larman et al. (12). In this earlier work, a T7 phage display library comprising 36-residue overlapping peptides covering all ORFs in the human genome was used to develop a phage immunoprecipitation sequencing methodology for the identification of autoantigens. A more general application of the library for the identification of protein–peptide interactions was introduced, but not explored in depth. We here establish that ProP-PD is a straightforward method for the identification of potentially relevant ligands of peptide binding domains. Our approach is based on the filamentous M13 phage, which is highly suited for efficient screening of peptide binding domains (13). The main advantage of our display system is that it is nonlytic and highly validated; random M13 phage-displayed peptide libraries have been used to map binding specificities of hundreds of diverse modular domains (7, 8, 14–16). We showcase our approach by identifying interactions of PSD-95/Dlg/ZO-1 (PDZ) domains.
Fig. 1.
Overview of the ProP-PD. The human and viral ProP-PD libraries were designed to contain over 50,000 or 10,000 C-terminal heptapeptides, respectively. Oligonucleotides encoding the sequences were printed on microarray slides, PCR-amplified, and cloned into a phagemid designed for the display of peptides fused to the C terminus of the M13 major coat protein P8. The libraries were used in binding selections with PDZ domains and the selected pools were analyzed by next-generation sequencing on the Illumina platform.
The PDZ family is one of the largest domain families in the human proteome, with about 270 members that typically interact with C-terminal peptides (class I binding motif: x-S/T-x-Φ-COO-, class II: x-Φ-x-Φ-COO-) (17) but also with internal peptide stretches and phosphoinositides (18, 19). PDZ–peptide interactions have been extensively analyzed by distinct experimental efforts, such as peptide-phage display (7, 20), peptide arrays (9, 21, 22), and split-ubiquitin membrane Y2H (23), as well as by computational approaches (24–28). Furthermore, the PDZ family has been shown to be the target of viral hijacking, whereby virus proteins mimic the C termini of human proteins to exploit these interactions (29). Thus, the PDZ family offers an excellent model system for validation of the ProP-PD approach.
We created ProP-PD libraries displaying all known human and viral C-terminal peptide sequences and used these to identify binding partners for the nine PDZ domains of Densin-180, Erbin, Scribble, and disk large homolog 1 (DLG1) (Fig. 1). These proteins have crucial roles in the postsynaptic density of excitatory neuronal synapses, in the establishment of adherens and tight junctions in epithelial cells, and in the regulation of cell polarity and migration (30–32). Additionally, both Scribble and DLG1 are known targets of viral proteins (33, 34). Using the ProP-PD libraries we identified known and novel human and viral ligands and validated candidates in vivo and in vitro. Our results demonstrate that ProP-PD is a powerful approach for the proteomic screening of human and viral targets. Future studies with larger libraries tiling the complete disordered regions of any proteome can be envisioned, as the technology is highly scalable.
Results
Library Design and Construction.
We designed a human peptide library containing 50,549 heptamer C-terminal sequences, corresponding to 75,797 proteins, including isoforms and cleaved sequences (Dataset S1), reported in the RefSeq, TopFind, and ENSEMBL databases (Status December 2011) (Fig. 2A). The peptides only listed in TopFind represent experimentally validated alternative C termini resulting from proteolytic cleavage events (35). Four percent of the entries map to more than one protein because they have identical C-terminal peptide sequences. In addition, we designed a library of all known viral protein C termini, containing the 10,394 distinct viral protein C termini found in Swissprot corresponding to 15,995 viral proteins (Fig. S1 and Dataset S2). Oligonucleotides encoding the peptides flanked by annealing sites were printed on custom microarrays, PCR-amplified, and used in combinatorial mutagenesis reactions to create libraries of genes encoding for peptides fused to the C terminus of the M13 major coat protein P8 in a phagemid vector (Fig. 1) (36). In our hybrid M13 phage systems, the phage particle contains all of the wild-type coat proteins with the addition of the fusion protein for display. The system has previously been optimized for efficient display of C-terminal peptides (37). The display level of the fusion protein is expected to be between 5% and 40% of the about 2,700 copies of the P8 protein on the phage particle (38). The avidity of the displayed peptides ensures the capture of transient domain–SLiMs interactions.
Fig. 2.
Library design and quality. (A) Histogram showing the number of entries taken from distinct databases to design the human C-terminal ProP-PD library. (B) Pie chart showing the composition of the libraries as determined by deep sequencing.
From each obtained oligonucleotide microarray we constructed two distinct phage libraries that were used in replicate screens against the target domain. Deep sequencing of the naïve libraries confirmed the presence of more than 80% and 90% of the designed human and viral sequences, respectively. The majority of the incorporated sequences were designed wild-type peptides but about 30% of the sequences had mutations (Fig. 2B). The mutations may arise from the oligonucleotide synthesis, the copying of the oligonucleotides of the microarray surface, the PCR amplification of the oligonucleotide library, or during the phage library construction and amplification. Indeed, the M13 phage has a mutation rate of 0.0046 per genome per replication event (39). The percentage of mutations in our libraries is lower than what was observed in the previous study by Larman et al. (12). Moreover, each library contained 108 to 109 unique members, which far exceeded the number of unique C-terminal peptides encoded by the DNA arrays, and thus, the mutations did not compromise coverage of our designed library sequences.
Analysis of the ProP-PD Selection Data.
The replicate ProP-PD libraries were used to capture binders for nine recombinant GST-tagged PDZ domains (Densin-180 PDZ; Erbin PDZ; Scribble PDZ1, PDZ2, PDZ3, and PDZ4; and DLG1 PDZ1, PDZ2, and PDZ3) following five rounds of selection. The selections were successful as judged by pooled phage ELISA, except for Scribble PDZ4, which has previously been found to fail in conventional C-terminal peptide-phage display, suggesting that this domain may not recognize C-terminal peptide ligands or that it is not functional when immobilized on the plastic surface (7, 40). Resultant phage pools were analyzed by next-generation sequencing. To define a high interest set of peptides that interact with the PDZ domains, we filtered as follows: (i) discarded mutated sequences, (ii) required a minimum threshold of read count (as indicated in Fig. 3A), and (iii) selected peptides found in either Uniprot/Swissprot or RefSeq (April 2013).
Fig. 3.
Analysis of the ProP-PD selection data. (A) Assignment of cut-off values. The histogram shows the deep-sequencing data of the phage pool selected for DLG1 PDZ2 from the human ProP-PD library. The gray dotted line indicates the assigned cut-off value, which is after the peak of the nonspecific peptides. (B) Correlation between selections against replicate libraries using all sequencing data when applicable (Tables S1 and S2). The data from the selections against the human libraries are in black and the data from the viral libraries are in gray. Most of the points are in the low count range and clustered in the lower left corner. (C) Comparisons between ProP-PD data and predictions based on PWMs derived from conventional phage display for domains with more than two ProP-PD ligands. The datapoints are shown as red circles, except the outliers (defined as PWM rank > 1,000) that are shown as black dots. The blue line represent is the linear fit of the data, excluding outliers. (D) Overlaps between identified ligands and interactions reported in the domino and BioGRID databases. For Scribble and DLG1 we pooled the results for the ProP-PD selections for their respective PDZ1, PDZ2, and PDZ3 domains.
For the replicate libraries, the overall correlation between the selected peptides for all domains was high (Fig. 3B) (r2 = 0.8 for all data), providing an estimate of the reproducibility of the procedure. Looking at individual domains, we found that the correlations between the replicate selections were lower in some cases (Scribble PDZ2 and PDZ3, DLG1 PDZ2 0.5 < r2 < 0.7) than in others (Scribble PDZ1, Erbin PDZ, and DLG1 PDZ3, r2 = 0.99). It thus appears to be good practice to construct more than one library for each design to ensure good coverage of the sequence space.
Comparison with Conventional Peptide-Phage Display.
To compare the data obtained from the ProP-PD selections with results from conventional peptide-phage display, we derived position weight matrices (PWMs) based on the ProP-PD data and found good overall agreement with PWMs derived from random peptide-phage display libraries of a previous study (7) (Fig. S2). The ProP-PD–based PWMs were generally less hydrophobic, as evidenced by calculation of their accumulated hydrophobicity values. We further investigated if conventional phage display would have identified proteins containing the C-terminal sequences obtained from ProP-PD (Fig. 3C and Table S1). There is good agreement between the two systems for Erbin, DLG1 PDZ2, and PDZ3; however, clear differences were observed for Scribble PDZ1, PDZ2, and PDZ3 targets (Fig. 3C). For Erbin PDZ there is one notable outlier (YYDYTDV) that lacks the C-terminal [T/S]WV motif, which is otherwise the hallmark of the ligands of this domain. For Scribble PDZ1 the three highest ranked ProP-PD ligands are captured by the PWM predictions, but not the lower ranked peptides.
There are several discrepancies between the PWM-based predictions and the ProP-PD data for Scribble PDZ2 and PDZ3. For example, for Scribble PDZ2, the first (GSPDSWV) and fifth (ASPDSWV) highest ProP-PD ligand score badly in the PWM-based predictions, which may in part be explained by the S at position −2 that is not represented in the PWM used for predictions. Among the outliers of Scribble PDZ3 we note the IRETHLW peptide, which appears to contain a cryptic PDZ class I motif with a shift of one amino acid, as previously suggested for other PDZ ligands (25). Other outliers (ASFWETS, GDLFSTD, and THWRETI) do not contain typical class I binding motifs and are therefore missed by the PWM-based predictions.
Comparison Between Human ProP-PD Data and Known Ligands.
We compared the overlap between our identified putative human ligands with the physical interactions reported in the BioGRID and DOMINO databases (excluding high-throughput AP/MS data to avoid comparing binary interactions with complexes). The overlaps (Fig. 3D) are rather low, and there are two likely reasons for this. First, BioGRID (and other related databases) do not yet annotate the domains/motifs mediating the interactions. Hence, the interactions reported therein may be mediated by other parts of the protein not represented in this study. Second, the coverage of DOMINO is known to be relatively low (41). A more extensive literature search provided support for about 50% of the interactions for the PDZ domains of Erbin, DLG1, and Densin-180, suggesting that a high proportion of the ligands identified by ProP-PD are relevant (Fig. 3A and Table S1). Curiously, we found support for only 5 of the 36 ligands identified for the Scribble PDZ domains and therefore attempted to validate some of these new interactions using in vitro affinity determination and cell-based assays.
Validation of Human Scribble Ligands in Vitro.
We determined in vitro affinities using fluorescence polarization assays (Table 1). We synthesized fluorescein-labeled peptides for the first ranked ligands for each of the Scribble PDZ domains (PDZ1: RFLETKL and AWDETNL, PDZ2: GSPDSWV and VQRHTWL, PDZ3: VQRHTWL and AWDETNL). The affinities (Table 1) were in the low micromolar range (1–40 μM), which is typical for PDZ domain-mediated interactions (42) and similar to what have been observed for synthetic ligands derived from combinatorial phage libraries (7, 20).
Table 1.
Dissociation constants of the PDZ domains of Scribble with selected peptides as determined using synthetic fluorescein-labeled peptides
|
KD (μM) |
||||||||||
| Protein | Peptide |
PDZ1 | PDZ2 | PDZ3 | ||||||
| Human | −6 | −5 | −4 | −3 | −2 | −1 | 0 | |||
| B7Z2Y1 | R | F | L | E | T | K | L | 2.1 ± 0.2 | 29 ± 5 | 5.8 ± 0.6 |
| ARHG7 | A | W | D | E | T | N | L | 2.3 ± 0.3 | 17 ± 2 | 3.5 ± 0.2 |
| NXPE2 | V | Q | R | H | T | W | L | NA | 5 ± 1 | 7 ± 2 |
| PKP4 | G | S | P | D | S | W | V | NB | 37 ± 7 | 22 ± 5 |
| DNM1L | I | R | E | T | H | L | W | NB | NA | 1.1 ± 0.4 |
| MK12 | V | S | K | E | T | P | L | NA | NA | 5.0 ± 0.5 |
| GCYA2 | F | L | R | E | T | S | L | NA | NA | 10 ± 2 |
| CTNB1 | A | W | F | D | T | D | L | NA | NA | 8.5 ± 2 |
| MET | A | S | F | W | E | T | S | NA | NA | NB |
| Viral | ||||||||||
| TAX HTL1L | H | F | H | E | T | E | V | NA | 7 ± 2 | 2.5 ± 0.7 |
NA, not available as the dissociation constants were not determined; NB, no binding under conditions used. No binding was observed with the scrambled NATWLED peptide used as negative control.
Furthermore, we measured affinities for additional Scribble PDZ3 interactions to investigate if there was a correlation between affinities and the sequencing counts (covering a range of 0–10,000 counts). The peptides (Table 1) conform to a class I binding motif (x-S/T-x-Φ-COO-), with the exceptions of the IRETHLW and the ASFWETS peptides, as discussed previously. There is a weak correlation (r2 = 0.36) between the logarithm of the sequencing counts and the affinities (Fig. S3), suggesting that ProP-PD data can be used in a semiquantitative manner, similar to intensities from peptide arrays. The observed counts can be influenced by factors other than affinities—such as phage growth rates (43), different display levels, and biases in amplicon PCR (44)—but such confounding effects can be minimized by exceedingly high library coverage during selections, using a display system with minimal growth bias for different clones and optimizing PCR conditions for linear amplification. From the linear fit we estimate that peptides with affinities weaker than 20 μM may be lost, and the GSPDSWV peptide (Kd = 22 μM) was indeed not retrieved in the sequencing data from this selection. We failed to detect an interaction between Scribble PDZ3 and the ASFWETS peptide in the concentration range used, indicating that it is a false-positive hit.
Validation of Scribble Ligands in Vivo.
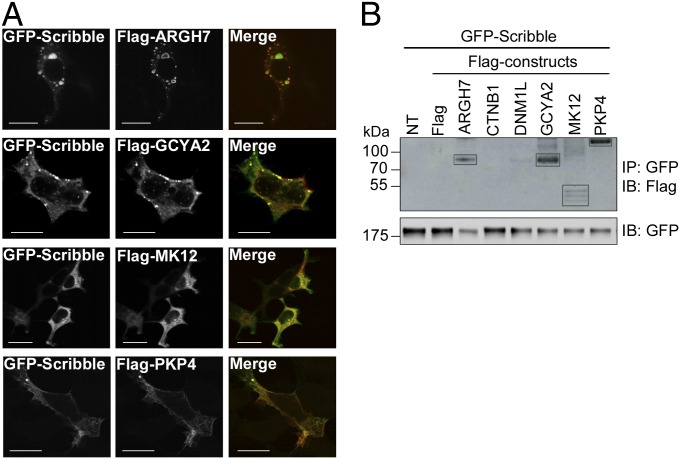
For additional validations we performed colocalization and coimmunoprecipitation (Co-IP) experiments using N-terminally GFP-tagged Scribble and N-terminally Flag-tagged full-length target proteins containing six of the peptides used for affinity determinations, namely β-PIX (ARGH7, positive control), PKP4, β-catenin (CTNB1), mitogen-activated kinase 12 (MK12), guanylate cyclase soluble subunit α-2 (GCYA2), and dynamin-1-like protein (DNM1L). Upon transient overexpression in HEK293T cells, Scribble clearly colocalized with ARGH7, GCYA2, and PKP4 (Fig. 4A) at distinct subcellular sites. Notably, Scribble was targeted to distinct vesicular structures when coexpressed with ARGH7 and GCYA2 but enriched at filamentous structures when expressed with PKP4. These interactions were further supported by Co-IP experiments (Fig. 4). Some colocalization was noted between CTNB1 and Scribble (Fig. S4), but we failed to confirm an interaction between the two proteins through Co-IP. CTNB1 and Scribble have previously been shown to colocalize in hippocampal neurons and have been coimmunoprecipitated from neuronal lysates (45), and may thus interact under other cellular contexts. Coexpressed MK12 and Scribble were found diffused in the cytoplasm, but weak yet consistent bands were observed from their Co-IP supporting an interaction (Fig. 4B). In contrast, when Scribble was coexpressed with DNM1L, it was targeted to vesicular structures, whereas DNM1L was found to be diffused in the cells. Furthermore, the Co-IP between the two proteins was largely negative. Colocalizations and Co-IPs thus support the interactions between full-length Scribble and ARGH7, GCYA2, PKP4, and MK12 but not with DNM1L.
Fig. 4.
Scribble interacts with ARGH7, GCAY2, MK12, and PKP4. (A) Colocalization of Flag-tagged ARGH7, GCYA2, MK12 and PKP4 with GFP-tagged full length Scribble as shown by confocal micrographs taken 48 h after cotransfection in HEK293T cells. (Scale bars, 15 μm.) (B) Coimmunoprecipitation of GFP-Scribble and Flag-tagged proteins in HEK293T cells upon transient overexpression. IP: GFP indicates that the Co-IPs were made using an anti-GFP antibody, and IB: Flag indicates that the Western blot detection was performed using an anti-Flag antibody. Controls: NT, nontransfected; Flag, only the Flag-tag. Lanes with protein names show the immunoblots of the single proteins, an immunoblot with GFP is shown as control (see Methods for details).
Overview of Human Targets.
We created a protein–protein interaction network of the four PDZ-containing proteins with their 78 putative binding partners for a comprehensive overview of the data (Fig. S5). Consistent with previous studies and roles in cell polarity and adhesion, the network of the LAP proteins Densin-180, Erbin, and Scribble contains interactions with the catenin family members PKP4, δ-catenin, and ARVCF (40, 46–48), whereas the DLG1 part of the network contains previously known interactions with anion transporters, potassium channels, and G protein-coupled receptors (see SI Methods for a detailed discussion of the network and biological relevance of the previously unknown interactions).
Host–Virus Protein–Protein Interactions.
The viral ProP-PD library was created to identify putative interactions between viral proteins and human PDZ domains. For the PDZ domains of Scribble and DLG1, we retrieved mainly previously known interactors (SI Methods and Table S2) (29). We determined the affinities of the Tax-1 C-terminal peptide (HFETEV) for Scribble PDZ2 and PDZ3 and found them to be in the low micromolar range (Table 1), similar to the affinity for the human ligands.
The viral ProP-PD further suggested a set of novel host–virus protein–protein interactions listed in Table S2, including an interaction between Scribble and the rabies virus glycoprotein G, which has previously been shown to bind other PDZ proteins (41). In addition, we revealed interactions between DLG1 PDZ2 and the C termini of the cytomegalovirus protein HHRF7 and the glycoprotein U47 of human herpes virus 6A. Finally, the ProP-PD data suggest several new ligands for Erbin PDZ, such as the Vpu protein of HIV and the Bat coronavirus envelope small membrane protein. These results show how the ProP-PD approach can be used to identify novel putative host–virus protein–protein interactions.
Discussion
We made use of custom oligonucleotide arrays to construct defined phage display libraries comprising the entire human and viral C-terminomes found in Swissprot. We demonstrated the power of such customized peptide-phage libraries in identifying ligands of potential biological relevance using PDZ domains as model proteins. Compared with conventional phage display, the main strength of our approach is the defined search space encompassing biological ligands, which obviates the need for predictions. Next-generation sequencing of the phage pools provides a list of selected peptide sequences that are directly associated with target proteins of potential biological relevance. We identified interactions between PDZ domains and C-termini of human proteins, and expanded the ProP-PD approach to screen for host–virus protein–protein interactions. Future studies with more extensive viral libraries can be envisioned. For example, it is possible to generate comprehensive libraries of viral species, including extensive sequence variations from strain sequencing, for the rapid screening of interactions between host proteins and virus proteins and for potential subtyping of viral strains based on their binding preferences. The method could also be extended to pathogenic bacteria that have been shown to exploit modular domains (41).
The PDZ ligands retrieved from the ProP-PD appear generally less hydrophobic than ligands derived using combinatorial phage libraries, although the affinities for the bait proteins are in the same range (7, 20). The hydrophobic bias might be explained by a bias in the M13 phage display system toward displaying hydrophobic peptides (49). Because such hydrophobic peptides are less abundant in the ProP-PD libraries, this issue is circumvented. However, the ProP-PD method has other limitations. First, it does not account for spatial or temporal separation of the ligands within cells, although it can be envisioned to filter the data for such factors. Second, ProP-PD is not suitable for tackling posttranslational modifications, which are common regulatory mechanisms of domain–SLiMs interactions (50).
ProP-PD can be compared with other methods for detection of protein–peptide interactions, such as SPOT microarrays, where defined peptides are synthesized on a cellulose membrane (10, 51). The SPOT array technique has the key advantage of allowing for studies of modifications, such as phosphorylation and acetylation, but has several disadvantages. First, the number of peptides that can be printed on a SPOT microarray is still smaller than necessary. By contrast, ProP-PD libraries scale easily and could contain all potential human binding motifs. Second, SPOT microarrays have relatively high false-positive rates, which does not appear to be the case for Prop-PD. The approach can also be compared with Y2H. Although Y2H has the advantage of screening full proteins (rather than only peptides), it has generally had both lower sensitivity and specificity for detecting domain–SLiM interactions (52). Another advantage of the ProP-PD approach over Y2H is that it is not limited to proteins that can be translocated to the nucleus. Finally, ProP-PD can be compared with AP/MS, which has the advantage of probing interactions in a cellular context. However, elusive SLiMs interactions are often not detected in these experiments. Thus, ProP-PD can be used as to complement AP/MS derived networks.
Over the last decade there has been increasing interest in intrinsically disordered regions, which are present in about 30% of human proteins (53) and are enriched in SLiMs that may serve as binding sites for target proteins. Although there are more than 100,000 SLiMs instances in the human proteome (4), the function is only known for a fraction (54). By creating ProP-PD libraries that represent all of the disordered regions of target proteomes, it will be possible to rapidly and comprehensively screen for SLiMs–domain interactions. A library of the complete human proteome has indeed already been constructed using the T7 display system, and it was validated for protein–peptide interaction screening by the identification of a known ligand for GST-tagged replication protein A2. However, other binding partners were not picked up as the target motifs were at the breakpoints between peptides, highlighting the importance of the initial design of the libraries.
As outlined by Larman et al., the ProP-PD approach can also be used for the identification of antibody epitopes, and the peptides may to some extent retain some secondary structures when expressed on the coat protein (12). This aspect is reminiscent of other studies where libraries of highly structured natural peptides have been used to identify inhibitors of protein–protein interactions (55). Folded peptides from proteomes distinct from the target organism may be used for identification of inhibitors of specific human protein–protein interactions. The design of folded rather than disordered peptide libraries could be a possible extension of our ProP-PD approach.
We believe that the ProP-PD technology can be scaled to any proteome of interest and will become a widely applicable method for the rapid proteome-wide profiling of peptide-binding modules. It will enable the unbiased search for potential biologically relevant targets for network analysis and comparative studies.
Methods
Design of Human and Viral ProP-PD Libraries.
The human ProP-PD library (Dataset S1) was designed by retrieving information from Ensembl62 (version GRCh37.6, built 64), RefSeq and TopFind (downloaded December 2011). The viral C-terminal library contained the nonredundant C-terminal heptapeptides (Dataset S2) retrieved from Swissprot with an overview of host specificities in Fig. S1. The C-terminal peptide sequences were reverse translated using the most frequent Escherichia coli codons (56) and the coding sequences were flanked by primer annealing sites for PCR amplification and site-directed mutagenesis reactions.
Oligonucleotide Pool from Microarray Chip.
The designed oligonucleotides were obtained on 244k microarray chips (Agilent) and copied from the microarray chips through hybridization of primers designed to anneal to the single stranded templates. The primer (GCCTTAATTGTATCGGTTTA) complementary to the 3′ end of the designed oligonucleotides was dissolved (30 μM) in hybridization buffer [1 M NaCl, 10 mM Tris⋅HCl pH 7.5, 0.5% Trition-X100, 1 mM dithiothritol (DTT)] and allowed to hybridize to the templates for 4 h at 30 °C under rotation. Unbound primer was removed by washing with 50 mL of low-stringency wash buffer (890 mM phosphate buffer, pH 7.4, 60 mM NaCl, 6 mM EDTA, 0.5% Triton-X100) followed by 50 mL of high-stringency wash buffer (8.9 mM phosphate buffer pH 7.4, 0.6 mM NaCl, 0.06 mM EDTA, 0.5% Triton-X100). A complementary strand was synthesized through a polymerase reaction [900 μL reaction: 1× NEB buffer 2 (10 mM Tris⋅HCl, 10 mM MgCl2, 50 mM NaCl, 1 mM DTT, pH 7.9), 90 μg BSA, 0.1 mM of each dNTP, 54 units of T4 DNA polymerase, 75 units of Klenow Fragment (3′-5′ exo-; New England Biolabs)] at 30 °C for 30 min. The newly synthesized oligonucleotides were removed from the microarray chip by incubation with 1 mL 20 mM NaOH at 65 °C for 20 min. The eluted single-stranded oligonucleotides were precipitated in Eppendorf tubes at −80 °C for 2 h by addition of 3 M sodium acetate, molecular grade glycogen, and 100% (vol/vol) ethanol [final concentrations 85 mM sodium acetate, 0.7% glycogen, 70% (vol/vol) ethanol]. The DNA was pelleted by centrifugation at 16,100 × g at 4 °C for 30 min, the supernatant removed, and the pellets washed by addition of cold 70% (vol/vol) ethanol and centrifugation at 16,100 × g at 4 °C for 5 min. The DNA pellets were allowed to dry at room temperature for 30 min and resuspended in a total volume of 40 μL water. The single-stranded oligonucleotides (1 μL for 50-μL reaction) were used as template for a PCR using Taq polymerase to amplify the library (24 cycles of 55 °C annealing, 72 °C elongation, and 98 °C denaturation). To improve coverage, the template was amplified in 16 separate reactions. The PCR products were confirmed by gel electrophoresis [2.5% (wt/vol) agarose] with SYBR Safe (Invitrogen) staining, purified on four columns of the QIAgen nucleotide removal kit and eluted in 40 μL water from each column. The concentration of the dsDNA was estimated using PicoGreen dye (Invitrogen) and using a twofold dilution series (100–0.8 μg/μL) of λ-phage double-stranded DNA (dsDNA, Invitrogen) as a standard. The PicoGreen dye was diluted 1:400 in TE buffer and mixed with 1 μL of dsDNA standard or PCR product in a low-fluorescence 96-well plate (Bio-Rad). The fluorescence was read in a quantitative PCR machine (Bio-Rad) (excitation 480 nm, emission 520 nm) and the sample DNA concentration was determined from the standard curve.
Library Construction and Amplification
ProP-PD libraries were constructed following a modified version of a published procedure (57, 58). The PCR-amplified dsDNA (0.6 μg) was used as primers for oligonucleotide-directed mutagenesis after removal of residual single-stranded (ssDNA) by ExoI treatment (0.2 units/μL, 37 °C for 30 min, 85 °C for 15 min) followed by flash cooling on ice. The dsDNA was then directly 5′ phosphorylated for 1 h at 37 °C in TM buffer (10 mM MgCl2, 50 mM Tris, pH 7.5) and 1 mM ATP, 5 mM DTT using a T4 polynucleotide kinase (1 unit/μL; New England Biolabs). The phosphorylated dsDNA was denatured and annealed (95 °C for 3 min, 50 °C for 3 min and 20 °C for 5 min) to ssDNA template [10 μg ssDNA encoding the M13 major coat protein P8 (36) prepared as described elsewhere (57)] in TM buffer in a total volume of 250 μL. dsDNA was synthesized overnight at 20 °C by addition of 10 μL 10 mM ATP, 10 μL 10 mM dNTP mixture, 15 μL 100 mM DTT, 30 Weiss units T4 DNA ligase, and 30 units T7 DNA. The DNA was purified using a QIAquick DNA purification kit and eluted with 35 μL water. The phagemid library was converted into a ProP-PD library by electroporation into E. coli SS320 cells preinfected with M13KO7 helper phage (58). The transformation efficiency was 108 to 109 transformants per reaction thus exceeding the theoretical diversity of the library by more than 1,000-fold. The phage-producing bacteria were grown over night in 500 mL 2YT (16 g Bacto tryptone, 10 g Bacto yeast extract, 5 g NaCl, per liter water) medium at 37 °C and then pelleted by centrifugation (10 min at 11,270 × g). The supernatant was transferred to a new tube and phages were precipitated by adding one-fifth volume polyethylene glycol⋅NaCl, [20% PEG-8000 (wt/vol), 2.5 M NaCl], incubating for 5 min at 4 °C and centrifuging at 28,880 × g at 4 °C for 20 min. The phage pellet was resuspended in 20 mL PBT (PBS, 0.05% Tween-20, 0.2% BSA), insoluble debris was removed by centrifugation and the library was stored at −20 °C in 20% (vol/vol) glycerol. The naïve libraries were deep sequenced using the Illumina platform (SI Methods and Fig. S6). The library was reamplified in E. coli SS320 cells in presence of 0.4 M IPTG.
ProP-PD Selections and Validation Experiments.
Selections and analyses were carried out at 4 °C essentially as described by Ernst et al. (59). Similarly, peptide synthesis, affinity measurements, and Co-IPs were carried out using standard protocols. Detailed descriptions are given in SI Methods.
Supplementary Material
Acknowledgments
This work was supported in part by an Ontario Genomics Institute SPARK grant (to P.M.K.), and Canadian Institutes of Health Research Grants MOP-123526 and MOP-93684 (to P.M.K. and S.S.S.).
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1312296111/-/DCSupplemental.
References
- 1.Stumpf MP, et al. Estimating the size of the human interactome. Proc Natl Acad Sci USA. 2008;105(19):6959–6964. doi: 10.1073/pnas.0708078105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arkin MR, Wells JA. Small-molecule inhibitors of protein-protein interactions: Progressing towards the dream. Nat Rev Drug Discov. 2004;3(4):301–317. doi: 10.1038/nrd1343. [DOI] [PubMed] [Google Scholar]
- 3.Davey NE, Travé G, Gibson TJ. How viruses hijack cell regulation. Trends Biochem Sci. 2011;36(3):159–169. doi: 10.1016/j.tibs.2010.10.002. [DOI] [PubMed] [Google Scholar]
- 4.Davey NE, et al. Attributes of short linear motifs. Mol Biosyst. 2012;8(1):268–281. doi: 10.1039/c1mb05231d. [DOI] [PubMed] [Google Scholar]
- 5.Shekhawat SS, Ghosh I. Split-protein systems: Beyond binary protein-protein interactions. Curr Opin Chem Biol. 2011;15(6):789–797. doi: 10.1016/j.cbpa.2011.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lam MH, Stagljar I. Strategies for membrane interaction proteomics: no mass spectrometry required. Proteomics. 2012;12(10):1519–1526. doi: 10.1002/pmic.201100471. [DOI] [PubMed] [Google Scholar]
- 7.Tonikian R, et al. A specificity map for the PDZ domain family. PLoS Biol. 2008;6(9):e239. doi: 10.1371/journal.pbio.0060239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tonikian R, Zhang Y, Boone C, Sidhu SS. Identifying specificity profiles for peptide recognition modules from phage-displayed peptide libraries. Nat Protoc. 2007;2(6):1368–1386. doi: 10.1038/nprot.2007.151. [DOI] [PubMed] [Google Scholar]
- 9.Stiffler MA, et al. PDZ domain binding selectivity is optimized across the mouse proteome. Science. 2007;317(5836):364–369. doi: 10.1126/science.1144592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Breitling F, Nesterov A, Stadler V, Felgenhauer T, Bischoff FR. High-density peptide arrays. Mol Biosyst. 2009;5(3):224–234. doi: 10.1039/b819850k. [DOI] [PubMed] [Google Scholar]
- 11.Luck K, Travé G. Phage display can select over-hydrophobic sequences that may impair prediction of natural domain-peptide interactions. Bioinformatics. 2011;27(7):899–902. doi: 10.1093/bioinformatics/btr060. [DOI] [PubMed] [Google Scholar]
- 12.Larman HB, et al. Autoantigen discovery with a synthetic human peptidome. Nat Biotechnol. 2011;29(6):535–541. doi: 10.1038/nbt.1856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.McLaughlin ME, Sidhu SS. Engineering and analysis of peptide-recognition domain specificities by phage display and deep sequencing. Methods Enzymol. 2013;523:327–349. doi: 10.1016/B978-0-12-394292-0.00015-1. [DOI] [PubMed] [Google Scholar]
- 14.Xin X, et al. SH3 interactome conserves general function over specific form. Mol Syst Biol. 2013;9:652. doi: 10.1038/msb.2013.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vetter SW. Phage display selection of peptides that target calcium-binding proteins. Methods Mol Biol. 2013;963:215–235. doi: 10.1007/978-1-62703-230-8_14. [DOI] [PubMed] [Google Scholar]
- 16.Pande J, Szewczyk MM, Grover AK. Phage display: Concept, innovations, applications and future. Biotechnol Adv. 2010;28(6):849–858. doi: 10.1016/j.biotechadv.2010.07.004. [DOI] [PubMed] [Google Scholar]
- 17.Luck K, Charbonnier S, Travé G. The emerging contribution of sequence context to the specificity of protein interactions mediated by PDZ domains. FEBS Lett. 2012;586(17):2648–2661. doi: 10.1016/j.febslet.2012.03.056. [DOI] [PubMed] [Google Scholar]
- 18.Ivarsson Y, et al. Prevalence, specificity and determinants of lipid-interacting PDZ domains from an in-cell screen and in vitro binding experiments. PLoS ONE. 2013;8(2):e54581. doi: 10.1371/journal.pone.0054581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wawrzyniak AM, Vermeiren E, Zimmermann P, Ivarsson Y. Extensions of PSD-95/discs large/ZO-1 (PDZ) domains influence lipid binding and membrane targeting of syntenin-1. FEBS Lett. 2012;586(10):1445–1451. doi: 10.1016/j.febslet.2012.04.024. [DOI] [PubMed] [Google Scholar]
- 20.Sharma SC, Memic A, Rupasinghe CN, Duc AC, Spaller MR. T7 phage display as a method of peptide ligand discovery for PDZ domain proteins. Biopolymers. 2009;92(3):183–193. doi: 10.1002/bip.21172. [DOI] [PubMed] [Google Scholar]
- 21.Boisguerin P, et al. An improved method for the synthesis of cellulose membrane-bound peptides with free C termini is useful for PDZ domain binding studies. Chem Biol. 2004;11(4):449–459. doi: 10.1016/j.chembiol.2004.03.010. [DOI] [PubMed] [Google Scholar]
- 22.Wiedemann U, et al. Quantification of PDZ domain specificity, prediction of ligand affinity and rational design of super-binding peptides. J Mol Biol. 2004;343(3):703–718. doi: 10.1016/j.jmb.2004.08.064. [DOI] [PubMed] [Google Scholar]
- 23.Gisler SM, et al. Monitoring protein-protein interactions between the mammalian integral membrane transporters and PDZ-interacting partners using a modified split-ubiquitin membrane yeast two-hybrid system. Mol Cell Proteomics. 2008;7(7):1362–1377. doi: 10.1074/mcp.M800079-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen JR, Chang BH, Allen JE, Stiffler MA, MacBeath G. Predicting PDZ domain-peptide interactions from primary sequences. Nat Biotechnol. 2008;26(9):1041–1045. doi: 10.1038/nbt.1489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gfeller D, et al. The multiple-specificity landscape of modular peptide recognition domains. Mol Syst Biol. 2011;7:484. doi: 10.1038/msb.2011.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hui S, Xing X, Bader GD. Predicting PDZ domain mediated protein interactions from structure. BMC Bioinformatics. 2013;14:27. doi: 10.1186/1471-2105-14-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kim J, et al. Rewiring of PDZ domain-ligand interaction network contributed to eukaryotic evolution. PLoS Genet. 2012;8(2):e1002510. doi: 10.1371/journal.pgen.1002510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Smith CA, Kortemme T. Structure-based prediction of the peptide sequence space recognized by natural and synthetic PDZ domains. J Mol Biol. 2010;402(2):460–474. doi: 10.1016/j.jmb.2010.07.032. [DOI] [PubMed] [Google Scholar]
- 29.Javier RT, Rice AP. Emerging theme: cellular PDZ proteins as common targets of pathogenic viruses. J Virol. 2011;85(22):11544–11556. doi: 10.1128/JVI.05410-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Feng W, Zhang M. Organization and dynamics of PDZ-domain-related supramodules in the postsynaptic density. Nat Rev Neurosci. 2009;10(2):87–99. doi: 10.1038/nrn2540. [DOI] [PubMed] [Google Scholar]
- 31.Harris BZ, Lim WA. Mechanism and role of PDZ domains in signaling complex assembly. J Cell Sci. 2001;114(Pt 18):3219–3231. doi: 10.1242/jcs.114.18.3219. [DOI] [PubMed] [Google Scholar]
- 32.Hatzfeld M. The p120 family of cell adhesion molecules. Eur J Cell Biol. 2005;84(2–3):205–214. doi: 10.1016/j.ejcb.2004.12.016. [DOI] [PubMed] [Google Scholar]
- 33.Lee SS, Weiss RS, Javier RT. Binding of human virus oncoproteins to hDlg/SAP97, a mammalian homolog of the Drosophila discs large tumor suppressor protein. Proc Natl Acad Sci USA. 1997;94(13):6670–6675. doi: 10.1073/pnas.94.13.6670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nakagawa S, Huibregtse JM. Human scribble (Vartul) is targeted for ubiquitin-mediated degradation by the high-risk papillomavirus E6 proteins and the E6AP ubiquitin-protein ligase. Mol Cell Biol. 2000;20(21):8244–8253. doi: 10.1128/mcb.20.21.8244-8253.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lange PF, Overall CM. TopFIND, a knowledgebase linking protein termini with function. Nat Methods. 2011;8(9):703–704. doi: 10.1038/nmeth.1669. [DOI] [PubMed] [Google Scholar]
- 36.Fuh G, et al. Analysis of PDZ domain-ligand interactions using carboxyl-terminal phage display. J Biol Chem. 2000;275(28):21486–21491. doi: 10.1074/jbc.275.28.21486. [DOI] [PubMed] [Google Scholar]
- 37.Held HA, Sidhu SS. Comprehensive mutational analysis of the M13 major coat protein: Improved scaffolds for C-terminal phage display. J Mol Biol. 2004;340(3):587–597. doi: 10.1016/j.jmb.2004.04.060. [DOI] [PubMed] [Google Scholar]
- 38.Malik P, et al. Role of capsid structure and membrane protein processing in determining the size and copy number of peptides displayed on the major coat protein of filamentous bacteriophage. J Mol Biol. 1996;260(1):9–21. doi: 10.1006/jmbi.1996.0378. [DOI] [PubMed] [Google Scholar]
- 39.Drake JW. A constant rate of spontaneous mutation in DNA-based microbes. Proc Natl Acad Sci USA. 1991;88(16):7160–7164. doi: 10.1073/pnas.88.16.7160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhang Y, et al. Convergent and divergent ligand specificity among PDZ domains of the LAP and zonula occludens (ZO) families. J Biol Chem. 2006;281(31):22299–22311. doi: 10.1074/jbc.M602902200. [DOI] [PubMed] [Google Scholar]
- 41.Ceol A, et al. DOMINO: A database of domain-peptide interactions. Nucleic Acids Res. 2007;35(Database issue):D557–D560. doi: 10.1093/nar/gkl961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jemth P, Gianni S. PDZ domains: Folding and binding. Biochemistry. 2007;46(30):8701–8708. doi: 10.1021/bi7008618. [DOI] [PubMed] [Google Scholar]
- 43.Thomas WD, Golomb M, Smith GP. Corruption of phage display libraries by target-unrelated clones: Diagnosis and countermeasures. Anal Biochem. 2010;407(2):237–240. doi: 10.1016/j.ab.2010.07.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Aird D, et al. Analyzing and minimizing PCR amplification bias in Illumina sequencing libraries. Genome Biol. 2011;12(2):R18. doi: 10.1186/gb-2011-12-2-r18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sun Y, Aiga M, Yoshida E, Humbert PO, Bamji SX. Scribble interacts with beta-catenin to localize synaptic vesicles to synapses. Mol Biol Cell. 2009;20(14):3390–3400. doi: 10.1091/mbc.E08-12-1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Laura RP, et al. The Erbin PDZ domain binds with high affinity and specificity to the carboxyl termini of delta-catenin and ARVCF. J Biol Chem. 2002;277(15):12906–12914. doi: 10.1074/jbc.M200818200. [DOI] [PubMed] [Google Scholar]
- 47.Izawa I, et al. ERBIN associates with p0071, an armadillo protein, at cell-cell junctions of epithelial cells. Genes Cells. 2002;7(5):475–485. doi: 10.1046/j.1365-2443.2002.00533.x. [DOI] [PubMed] [Google Scholar]
- 48.Appleton BA, et al. Comparative structural analysis of the Erbin PDZ domain and the first PDZ domain of ZO-1. Insights into determinants of PDZ domain specificity. J Biol Chem. 2006;281(31):22312–22320. doi: 10.1074/jbc.M602901200. [DOI] [PubMed] [Google Scholar]
- 49.Krumpe LR, et al. T7 lytic phage-displayed peptide libraries exhibit less sequence bias than M13 filamentous phage-displayed peptide libraries. Proteomics. 2006;6(15):4210–4222. doi: 10.1002/pmic.200500606. [DOI] [PubMed] [Google Scholar]
- 50.Van Roey K, Dinkel H, Weatheritt RJ, Gibson TJ, Davey NE. The switches.ELM resource: A compendium of conditional regulatory interaction interfaces. Sci Signal. 2013;6(269):rs7. doi: 10.1126/scisignal.2003345. [DOI] [PubMed] [Google Scholar]
- 51.Volkmer R. Synthesis and application of peptide arrays: Quo vadis SPOT technology. ChemBioChem. 2009;10(9):1431–1442. doi: 10.1002/cbic.200900078. [DOI] [PubMed] [Google Scholar]
- 52.Davey NE, Edwards RJ, Shields DC. Computational identification and analysis of protein short linear motifs. Front Biosci. 2010;15:801–825. doi: 10.2741/3647. [DOI] [PubMed] [Google Scholar]
- 53.Dunker AK, et al. The unfoldomics decade: An update on intrinsically disordered proteins. BMC Genomics. 2008;9(Suppl 2):S1. doi: 10.1186/1471-2164-9-S2-S1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Dinkel H, et al. ELM—The database of eukaryotic linear motifs. Nucleic Acids Res. 2012;40(Database issue):D242–D251. doi: 10.1093/nar/gkr1064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Watt PM. Screening for peptide drugs from the natural repertoire of biodiverse protein folds. Nat Biotechnol. 2006;24(2):177–183. doi: 10.1038/nbt1190. [DOI] [PubMed] [Google Scholar]
- 56.Nakamura Y, Gojobori T, Ikemura T. Codon usage tabulated from international DNA sequence databases: Status for the year 2000. Nucleic Acids Res. 2000;28(1):292. doi: 10.1093/nar/28.1.292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Rajan S, Sidhu SS. Simplified synthetic antibody libraries. Methods Enzymol. 2012;502:3–23. doi: 10.1016/B978-0-12-416039-2.00001-X. [DOI] [PubMed] [Google Scholar]
- 58.Sidhu SS. Phage display in pharmaceutical biotechnology. Curr Opin Biotechnol. 2000;11(6):610–616. doi: 10.1016/s0958-1669(00)00152-x. [DOI] [PubMed] [Google Scholar]
- 59.Ernst A, et al. Coevolution of PDZ domain-ligand interactions analyzed by high-throughput phage display and deep sequencing. Mol Biosyst. 2010;6(10):1782–1790. doi: 10.1039/c0mb00061b. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.