Abstract
For decades, it has been hypothesized that gene regulation has had central role in human evolution, yet much remains unknown about the genome-wide impact of regulatory mutations. Here we use whole-genome sequences and genome-wide chromatin immunoprecipitation and sequencing data to demonstrate that natural selection has profoundly influenced human transcription factor binding sites since the divergence of humans from chimpanzees 4–6 million years ago. Our analysis uses a new probabilistic method, called INSIGHT, for measuring the influence of selection on collections of short, interspersed noncoding elements. We find that, on average, transcription factor binding sites have experienced somewhat weaker selection than protein-coding genes. However, the binding sites of several transcription factors show clear evidence of adaptation. Several measures of selection are strongly correlated with predicted binding affinity. Overall, regulatory elements seem to contribute substantially to both adaptive substitutions and deleterious polymorphisms with key implications for human evolution and disease.
It has long been argued that mutations affecting mechanisms of gene regulation must have had a prominent role in the evolution of humans and other mammals1–3. For several theoretical reasons, transcription factor binding sites and other cis-regulatory sequences may be particularly important in evolutionary adaptation4–7. For example, mutations in such sequences might help to minimize the functional tradeoffs associated with evolutionary changes because these elements often primarily influence the expression of a single gene in a particular cell type or under a particular condition, whereas proteins tend to have broader effects. In addition, cis-regulatory mutations are often co-dominant (neither allele dominates), which may allow natural selection to act on them more efficiently than on protein-coding mutations. Accordingly, several examples of phenotypic changes driven by cis-regulatory mutations have been identified in recent years, including pigmentation and bristle patterns in Drosophila melanogaster, skeletal reduction in stickleback fish and lactose persistence in humans7,8. In addition, some genome-wide analyses have found bulk statistical evidence of natural selection in noncoding regions near genes, presumably due to cis-regulatory elements9–12.
Nevertheless, evidence in support of the overall prominence of cis-regulatory mutations in evolutionary adaptation remains largely anecdotal and indirect, and there is continuing controversy about the relative roles of regulatory and protein-coding sequences in evolution8. Large-scale genomic studies of the evolution of transcription factor binding sites have the potential to advance this debate, but a major limitation of such studies so far has been a lack of precisely annotated binding sites across the genome. The analysis of conserved noncoding sequences and/or promoter regions rather than experimentally identified transcription factor binding sites tends to dilute the signature of natural selection and makes it more difficult to connect DNA mutations with fitness-influencing phenotypes. In addition, because polymorphisms are sparse and provide weak information at individual binding sites, most studies have either pooled polymorphism counts across genomic regions9,13, which can produce significant statistical biases14, or they have relied on divergence patterns over longer evolutionary time scales10,12,15, which can be influenced by binding site turnover, alignment errors or misidentifications of orthology. As a result, many questions remain about the manner in which transcription factor binding sites evolve and the general functional consequences of noncoding mutations.
We sought to address these questions using two types of data that have recently become available: whole-genome sequences for dozens of human individuals16,17 and genome-wide chromatin immunoprecipitation and sequencing (ChIP-seq) data identifying binding sites for dozens of transcription factors in multiple human cell types18. We also made use of whole-genome sequences for several nonhuman primates, which enable patterns of human variation to be contrasted with patterns of molecular evolution since the divergence of humans and their closest living relatives 4–6 million years ago. To interpret these data, we made use of a new probabilistic method, called Inference of Natural Selection from Interspersed Genomically coHerent elemenTs (INSIGHT), that characterizes the effects of natural selection on collections of short transcription factor binding sites. Our analysis of these data using INSIGHT sheds new light on the evolution of transcription factor binding sites in the human lineage.
RESULTS
Probabilistic model
Full mathematical details of the INSIGHT model and inference procedure are presented separately19, but we summarize the approach here to aid in the interpretation of our results. Similar to McDonald-Kreitman–based methods for identifying departures from neutrality9,20–22, INSIGHT obtains information about natural selection by contrasting patterns of polymorphism and divergence in transcription factor binding sites with those in flanking neutral regions, thereby mitigating biases from demography, variation in mutation rate and differences in coalescence time. However, INSIGHT improves substantially on these methods by making use of a full generative probabilistic model, directly accommodating weak negative selection23, allowing information from many short (~10-bp) elements to be combined in a rigorous manner and integrating phylogenetic information from multiple outgroup species with genome-wide population genetic data.
The INSIGHT model (Supplementary Fig. 1) assumes that nucleotides within a transcription factor binding site evolve by a mixture of four selective modes: (i) neutral drift, (ii) weak negative selection, (iii) strong negative selection and (iv) positive selection. All flanking nucleotides are assumed to evolve neutrally. This coarse-grained, categorical approach to modeling the distribution of fitness effects (DFE) is inspired by a simpler method developed for protein-coding genes in Drosophila24 and by observations indicating that the data contain only limited information about a full, continuous DFE25,26. Three key assumptions allow these selective modes to be disentangled. First, strong selection (positive or negative) is assumed to eliminate polymorphism because it causes mutations to rapidly reach fixation or be lost24. Second, weak negative selection permits polymorphism, but does not allow derived alleles to reach high frequencies27. Third, positive selection tends to favor the fixation of derived alleles (such events are denoted ‘adaptive substitutions’), whereas negative selection (strong or weak) guarantees that derived alleles will eventually be lost. Consequently, information about the overall prevalence of selection derives primarily from rates of high-frequency derived alleles, information about positive selection comes from levels of divergence, and information about weak negative selection comes from relative rates of low- and high-frequency derived alleles, all in transcription factor binding sites relative to flanking neutral regions.
Maximum-likelihood estimates of the model parameters yield three quantities of particular interest: (i) the fraction of sites under selection (ρ) and the expected numbers of (ii) adaptive substitutions (E[A]) and (iii) weakly deleterious polymorphisms (E[W]) (Table 1 and Online Methods). The model allows for likelihood ratio tests for negative and/or positive selection and the straightforward estimation of confidence intervals for all parameters. Model parameters can also be used to obtain estimates of two ratios that have been of interest in previous studies: the fraction of fixed differences driven by positive selection (α)22,27 and the fraction of polymorphisms subject to weak negative selection (here denoted τ)28,29. However, because the denominators of α and τ are strongly influenced by both negative and positive selection, we focus on the more readily interpretable quantities E[A] and E[W] in our analysis.
Table 1.
summary of model parameters and expected values
| Locus-specific model parameters | |
| θ i | Population-scaled mutation rate at locus i |
| λ i | Scale factor for neutral divergence at locus i |
| Global model parameters | |
| ρ | Probability that each transcription factor binding site nucleotide is under selection |
| β1, β2, β3, | Fractions of neutral polymorphic sites with low-, intermediate- and high-frequency derived alleles |
| γ | Scale factor for θi in selected sites |
| η | Scale factor for λi in selected sites |
| Derived parameters | |
| α | Fraction of fixed differences due to positive selection |
| τ | Fraction of polymorphic sites subject to weak negative selection |
| Posterior expected values | |
| E[A] | Number of fixed differences due to positive selection (adaptive substitutions) |
| E[W] | Number of polymorphic sites subject to weak negative selection |
| E[D] | Number of weakly deleterious mutations per haploid genome |
Simulation study
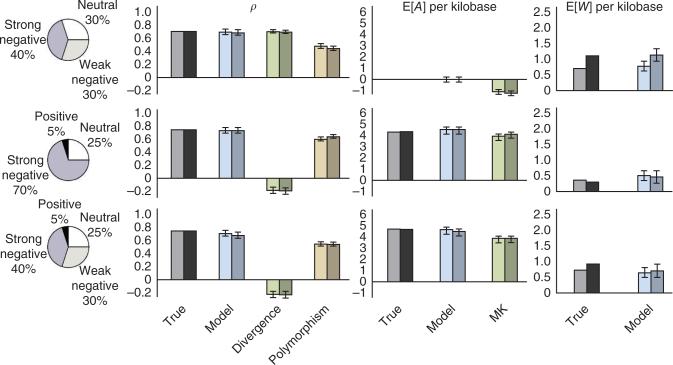
To test our methods, we generated synthetic data sets consisting of 10,000 instances of a 10-bp transcription factor binding site with various mixtures of selective effects, flanked by 5,000 neutral bases on each side (Supplementary Note). We then estimated all model parameters (Table 1 and Online Methods) from each data set and compared our estimates with the ‘true’ values used in the simulation (Supplementary Note). We found that our model-based estimates substantially improved on estimators based on divergence alone, polymorphism alone or the McDonald-Kreitman framework22. In particular, divergence-based estimators tended to be distorted by combinations of positive and negative selection (which have opposing influences on substitution rates; Fig. 1, rows 2 and 3), whereas polymorphism- and McDonald-Kreitman–based estimators were biased in the presence of weak negative selection23,27 (Fig. 1, rows 1 and 3). Our model-based estimates, by contrast, could accommodate combinations of selective effects with minimal bias. In addition, our estimates were robust to the assumption of a realistically complex demographic history, unlike methods that estimate a full DFE from the site frequency spectrum25,30–32, which typically require corrections for demography when applied to real data. Our methods did slightly underestimate ρ in the presence of moderate positive selection because many positively selected mutations are lost to drift and leave no signature in the data. Nevertheless, the effect on estimates of adaptive substitutions (E[A] and α) was small.
Figure 1.
Results for data sets simulated under three different mixtures of selective modes. Four selective modes (pie charts) are considered: neutral evolution (2Nes = 0), weak negative selection (2Nes = –10), strong negative selection (2Nes = –100) and positive selection (2Nes = 10). Bars represent the fraction of nucleotides under selection (ρ) and the expected numbers of adaptive substitutions (E[A]) and weakly deleterious polymorphisms (E[W]) per kilobase of transcription factor binding site sequence analyzed. Separate bars are shown for true values in the simulations and model-based estimates. Estimates of ρ are additionally compared with simple estimators based on divergence and polymorphism rates, and estimates for E[A] per kilobase are compared with a McDonald-Kreitman–based estimator (MK). The first bar in each pair represents simulations with constant population sizes, and the second bar represents a realistically complex demographic scenario for human populations. The nonzero values of E[W] per kilobase in the absence of weak negative selection (second row) reflect residual polymorphism in strongly selected sites. Error bars, 1 s.e.m. (additional results are shown in supplementary Figs. 12 and 13, and further details are given in the supplementary Note).
Patterns of selection on transcription factor binding sites
Next, we applied our methods to real data describing transcription factor binding sites, human polymorphism and patterns of divergence in primate genomes. First, we developed a pipeline for identifying high-confidence binding sites using ChIP-seq data from the Encyclopedia of DNA Elements (ENCODE) Project18. Briefly, this pipeline involved de novo motif discovery, manual inspection of motifs and binding-site prediction at ChIP-seq peaks, followed by filtering (Supplementary Note). Data were combined across cell types. Our pipeline yielded a total of 1.4 million binding sites for 78 transcription factors, with 582–106,113 binding sites per transcription factor (median of 9,748; Supplementary Table 1). Each transcription factor binding site was associated with a collection of putatively neutral nucleotide positions in a 20-kb block containing the binding site. These neutral positions excluded known protein-coding and RNA genes, conserved non-coding elements, their immediate flanking sites and the predicted transcription factor binding sites, leaving an average of 7,315 neutral sites per block. Finally, we summarized patterns of polymorphism and divergence within transcription factor binding sites and flanking neutral sites using high-coverage human genome sequence data for 54 unrelated individuals of diverse ancestry from Complete Genomics17 and synteny-based alignments of the chimpanzee33, orangutan34 and rhesus macaque35 genomes (Supplementary Note).
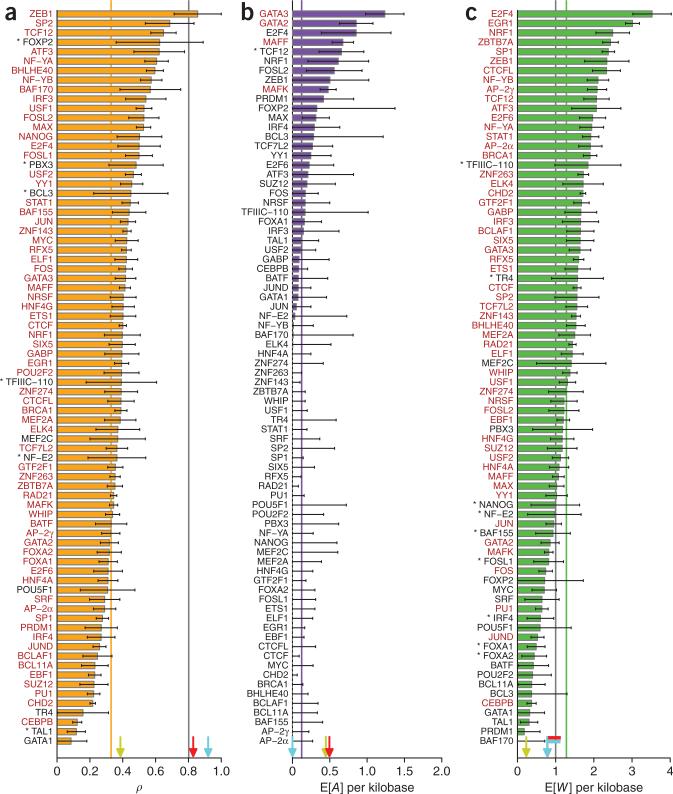
We applied our model and inference procedure to the complete set of binding sites for each transcription factor and obtained transcription factor–specific estimates of all parameters and expected values (Fig. 2). For comparison, we also applied our methods to second codon positions (CDS2 sites, at which all mutations cause amino acid substitutions) in 15,864 protein-coding genes that were carefully filtered to avoid errors from alignment, orthology identification and gene annotation (Supplementary Note). We obtained an average estimate across transcription factors (weighted by the number of nucleotides considered) of ρ = 0.33 (Fig. 2a and Supplementary Table 2). In comparison, our estimate for CDS2 sites was ρ = 0.80 (Supplementary Table 3), in reasonable agreement with estimates of 0.70–0.76 from comparative genomic analyses15,36. Thus, we detect a strong signature of natural selection in transcription factor binding sites, with about a third of nucleotides estimated to be under selection, but the fraction is considerably lower than at CDS2 sites. Notably, our comparison excluded synonymous sites in protein-coding genes but included analogous positions in transcription factor binding sites at which transcription factors exhibit, at most, weak base preferences.
Figure 2.
Estimates of key parameters for the binding sites of each transcription factor in our study. (a–c) Shown are estimates of the fraction of nucleotides under selection (ρ) (a), the expected number of adaptive substitutions per kilobase (E[A] per kilobase) (b) and the expected number of deleterious mutations per kilobase (E[W] per kilobase) (c). Weighted averages are indicated by lines in matching colors. Estimates for second codon positions in protein-coding sequences (CDS2) are shown for comparison (dark-gray lines). Arrows indicate estimates for CDS2 sites in subsets of genes identified as being under positive selection in mammalian phylogenies39 (yellow) or human populations38 (red) or denoted as housekeeping genes on the basis of gene expression patterns (light blue) (supplementary note). Flags in c indicate overlapping arrows. Transcription factor names in red indicate statistical significance after a correction for multiple tests61 (adjusted P < 0.05). Asterisks indicate nominal P < 0.05. Error bars, 1 s.e.m. (additional results are shown in supplementary Fig. 2 and supplementary Table 2). Notably, these estimates are fairly insensitive to the threshold for low-frequency derived alleles (supplementary Fig. 14).
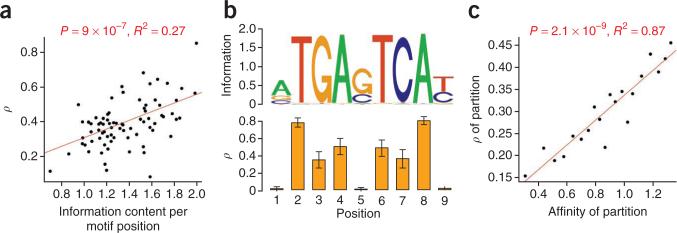
We observed considerable variation across transcription factors in the estimates of key parameters (Fig. 2). Not unexpectedly, many of the transcription factors showing the strongest evidence of natural selection in their binding sites, such as ZEB1, SP2, FOXP2, ATF3 and BAF170, have fairly short binding sites (6–9 bp) with strong base preferences and few degenerate positions. Many of these transcription factors have relatively few binding sites in our set (700–2,000). Basic helix-loop-helix (bHLH) proteins were significantly over- represented among the transcription factors associated with strong selection, with five of six bHLH proteins ranking in the top third by ρ (P = 0.014, one-sided Fisher's exact test). Transcription factors having roles in apoptosis and development were slightly underrepresented (Supplementary Tables 4–6). A substantial fraction of the variation in estimates of ρ was explained by the information content of the associated motifs (R2 = 0.27; Fig. 3a), a measure of the average binding affinity of the transcription factor for its binding sites. This finding suggests that the same forces that constrain the sequences of many binding sites across a genome also influence patterns of evolution at each individual transcription factor binding site (as has been observed over longer evolutionary time scales15,37). For transcription factors that have many binding sites, it was possible to estimate a separate ρ value for each position within the motif, and, in several of these cases, we observed a clear correlation between position-specific estimates of ρ and information content (Fig. 3b).
Figure 3.
Information content, binding affinity and selection. (a) Information content per motif position versus estimates of ρ (the fraction of sites under selection) for the 78 transcription factors analyzed in our study. (b) Motif logo for JUND (top) and position-specific estimates of ρ (bottom). Error bars, 1 s.e.m. Notice that positions with high information content tend to be under selection, and positions with low information content tend not to be under selection. This relationship holds for some but not all transcription factors. (c) Predicted binding affinity versus ρ. All binding sites were partitioned into 20 equally sized bins by predicted binding affinity, and ρ was estimated separately for each partition using INSIGHT. Additional details are given in the supplementary Note.
Notably, a substantial minority of transcription factors showed significant evidence of positive selection in their binding sites (Fig. 2). We estimated that, on average, an adaptive substitution occurred about once for every ~8,300 nucleotides in transcription factor binding sites (E[A] per kilobase = 0.12) (Fig. 2b), and about 1 in 20 recent nucleotide substitutions in binding sites had been driven by positive selection (α = 0.05). By contrast, CDS2 sites showed little evidence of adaptive evolution on average (E[A] per kilobase ≈0, α ≈0 for all sites pooled). Classes of genes previously described as being under positive selection38,39 showed evidence of adaptation by our methods, but seven transcription factors had estimated values of E[A] per kilobase that exceeded those for positively selected genes (Fig. 2b and Supplementary Fig. 2). We also found substantial evidence of weak negative selection in transcription factor binding sites, with an average estimate of E[W] per kilobase of 1.3, 30% greater than the estimate for CDS2 sites. Overall, our results support previous findings that negative selection is dominant in the evolution of human protein-coding sequences, with positive selection primarily influencing a relatively small subset of genes25,32,38, but they indicate that positive selection has had a somewhat more pronounced influence on binding-site evolution, at least for some transcription factors.
The two transcription factors whose binding sites showed the strongest evidence of positive selection, as measured by E[A] per kilobase, were the GATA-binding zinc-finger proteins GATA2 and GATA3, both key regulators of gene expression in hematopoietic cells. GATA2 and GATA3 are unusual among the transcription factors associated with strong selection in having fairly large numbers of binding sites (27,475 and 15,617, respectively), which together contribute an expected 312 adaptive substitutions, 19% of the total from all transcription factor binding sites in our study. By contrast, binding sites for the third member of this family, GATA1, showed much weaker evidence of selection. The 50,389 binding sites for MAFF, a basic leucine-zipper protein best known for enhancing expression of the oxytocin receptor gene (OTR) but also thought to have broad roles in the cellular stress response, contributed an additional 286 adaptive substitutions (18% of the total). Also of interest was the presence of the forkhead-box protein P2 (FOXP2) among transcription factors whose binding sites are under strong selection, given apparent positive selection in this gene's protein-coding sequence and its possible role in the development of human speech40. However, FOXP2 has relatively few binding sites in our set (743), and evidence for selection was not statistically significant.
Correlation with binding affinity
To allow for variability across transcription factor binding sites, we estimated the local binding affinity of each binding site in our set (Supplementary Note) and then partitioned all binding sites by binding affinity and estimated ρ values separately for each group. We observed a strong correlation between binding affinity and ρ (R2 = 0.87; Fig. 3c), indicating that the strength of natural selection at individual binding sites is well predicted by local binding affinity. We also compared the signatures of selection at transcription factor binding sites that have experienced recent affinity-increasing and affinity-decreasing mutations and found that affinity-increasing mutations showed an enrichment for adaptive substitutions, whereas affinity-decreasing mutations showed an enrichment for weakly deleterious polymorphisms (Supplementary Fig. 3).
Additional correlates of selection
We examined several other possible covariates of natural selection on transcription factor binding sites and identified a few other trends of interest. The fraction of sites under selection, ρ, was positively correlated with the expression levels of associated genes, consistent with observations in protein-coding sequences39 (R2 = 0.36, P = 0.002; Supplementary Fig. 4a). We also observed a positive correlation between ρ and the number of cell types in which each transcription factor binding site was bound as determined from our ChIP-seq data (R2 = 0.22, P = 0.0004; Supplementary Fig. 4b) and a negative correlation between the prevalence of weak negative selection (E[W] per kilobase) and distance to the nearest coding exon (Supplementary Fig. 5). We found no significant correlations with the tissue specificity of gene expression or distance to the transcription start site. In tests for elevated or reduced values of ρ, E[A] per kilobase and E[W] per kilobase near subsets of transcription factor binding sites associated with putative target genes of particular Gene Ontology (GO)41 categories, several transcription factors showed elevated values of E[A] per kilobase (indicating adaptive evolution) near genes involved in neural processes, consistent with a recent analysis of the promoter regions of primate genomes10. Other transcription factors showed elevated values of ρ near genes involved in development or cellular differentiation (Supplementary Tables 7–9 and Supplementary Note). To enable other researchers to explore this data set further, we created a UCSC Genome Browser track that displays all analyzed transcription factor binding sites and summarizes all relevant parameter estimates (Supplementary Fig. 6).
Total numbers of adaptive and deleterious mutations
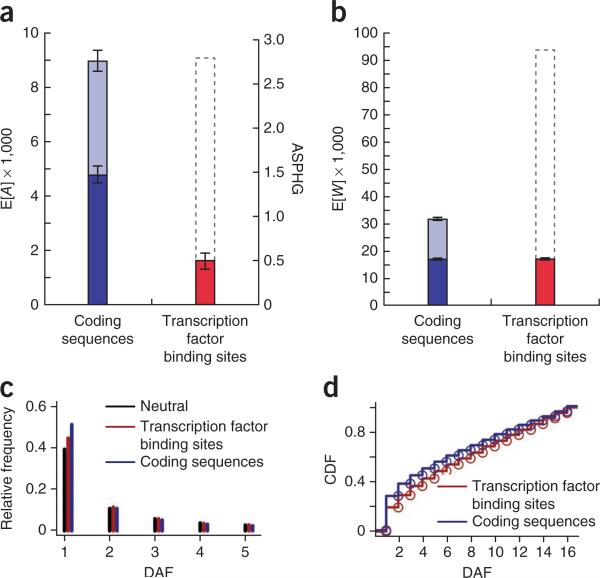
We applied our model to the 15,864 filtered protein-coding genes described, treating each coding exon as a ‘locus’ and drawing neutral sites from flanking regions. We then compared estimates of the total expected number of adaptive substitutions (E[A]) for these coding sequences and our 1.4 million transcription factor binding sites. (We experimented with several approaches for fitting the model to coding sequences and here report results for the approach that was most sensitive to adaptation; Supplementary Note.) This analysis produced cumulative estimates of E[A] = 4,786 for coding sequences and E[A] = 1,635 for transcription factor binding sites (Fig. 4a and Supplementary Table 3) or about 1.5 and 0.5 adaptive substitutions per hundred generations (ASPHG), respectively, assuming an average genomic divergence time of 6.5 million years42 and an average generation time of 20 years43. Our estimate for coding sequences implies that the fraction of fixed differences driven by adaptation in coding regions is ~20%, in reasonable agreement with estimates of 10–35% from previous studies25,27,44. When we extrapolated to all annotated protein-coding genes, our coding sequence estimate rose to E[A] = 8,954 or 2.8 ASPHG (Supplementary Note). Although our filtered gene set may not be representative of genome-wide coding sequences in all respects, it provides an approximate benchmark against which to compare the estimate for our 1.4 million transcription factor binding sites. Despite the incompleteness of our transcription factor binding site annotations, their estimated cumulative contribution to adaptive substitutions is nearly one-fifth that estimated for all protein-coding sequences (1,635 versus 8,954 adaptive substitutions). We also compared total expected numbers of weakly deleterious polymorphisms, obtaining estimates of E[W] = 16,937 for coding sequences and E[W] = 17,024 for transcription factor binding sites (Fig. 4b). Extrapolating to a full set of genes, as above, yielded E[W] = 31,687 for coding sequences, suggesting that the contribution of weakly deleterious polymorphisms from our transcription factor binding sites is more than half that from all coding sequences.
Figure 4.
Genome-wide analyses of adaptive and deleterious mutations in protein-coding sequences and transcription factor binding sites. (a) Expected numbers of adaptive substitutions on the human lineage (E[A]). The analysis was performed on a subset of genes that passed rigorous data quality filters (dark blue), and results were extrapolated to a full set of genes (light blue) (supplementary Note). The gray dashed outline for transcription factor binding sites indicates a crude extrapolation to the entire genome, assuming that two nucleotides function in gene regulation for every one that encodes proteins. The alternative y axis (right) shows estimated adaptive substitutions per hundred generations (ASPHG). Error bars indicate 1 s.e.m. above and below the mean (supplementary Note). (b) Plot as in a showing expected numbers of weakly deleterious polymorphisms (E[W]). (c) Site frequency spectra (SFS) for polymorphic sites in transcription factor binding sites, coding sequences and neutral flanking sequences. The first 5 derived allele frequencies (DAFs) are shown as counts out of 108 chromosomes (complete results in supplementary Fig. 15). (d) Cumulative distribution function (CDF) for expected weakly deleterious mutations per haploid genome (E[D]) in transcription factor binding site and coding sequences. Notice that the distribution is shifted toward more common alleles in transcription factor binding sites. Results are similar with alternative thresholds for low-frequency alleles.
Using the minor allele frequency at each site, we could further calculate the expected numbers of weakly deleterious mutations per haploid genome (E[D]) for coding sequences and transcription factor binding sites. Here a deficiency of rare alleles and a slight enrichment for more common low-frequency alleles in transcription factor binding sites relative to coding sequences (Fig. 4c) disproportionally increased the estimates for transcription factor binding sites, yielding E[D] = 386.1 and E[D] = 431.1 for coding sequences and transcription factor binding sites, respectively (Supplementary Fig. 7). The extrapolated estimate for a full set of genes was E[D] = 722.3, which is fairly similar to previous estimates of 500 (ref. 45) and 674 (ref. 46) (Supplementary Note). Thus, we estimated the contribution of deleterious mutations per haploid genome from our transcription factor binding sites to be well over half the total contribution from coding sequences. Notably, common low-frequency alleles accounted for a substantially larger fraction of deleterious mutations in transcription factor binding sites than in coding sequences (Fig. 4d), which may have implications for human disease.
DISCUSSION
Since the discovery by Jacob and Monod of the cis-regulatory control of transcription more than 50 years ago47, there has been a great deal of speculation about the role of cis-regulatory elements in evolution1–7. Complete genome sequences and genome-wide ChIP-seq data make it possible to begin to examine these issues empirically on a genomic scale. Using a novel statistical approach designed to exploit these new data, we have shown that natural selection has indeed exerted substantial influence on transcription factor binding sites in the human genome. The transcription factor binding sites we have analyzed have some limitations—for example, some may represent sites of nonfunctional protein binding or may be present only in the immortalized cell lines examined by ENCODE—but our use of experimentally defined binding sites leads to a much clearer signature of selection than has been observed in studies based on less precisely defined noncoding regions proximal to genes9–11. Notably, we obtain qualitatively similar results when applying our methods to alternative genome-wide sets of transcription factor binding sites (Supplementary Note).
We estimate that, overall, the transcription factor binding sites analyzed in this study have contributed nearly a fifth as many adaptive substitutions (E[A]) and more than half as many weakly deleterious polymorphisms (E[W]) and weakly deleterious mutations per haploid genome (E[D]) as coding sequences. However, our analysis considers only a relatively small subset of all transcription factor binding sites, corresponding to perhaps 5% of all transcription factors48, a limited set of cell types and conditions, and fairly conservative predictions of binding sites. In addition, we have not considered regulatory elements involved in splicing, post-transcriptional regulation and other forms of gene regulation. It is not possible, at present, to estimate with any accuracy the total number of bases that function in gene regulation. However, if we assume that noncoding functional elements outnumber coding bases by at least 2:1, as suggested by patterns of long-term evolutionary conservation49–51, that these elements predominately function in gene regulation52 and, further, that mutations in these elements have similar distributions of fitness effects as in our transcription factor binding sites, then we can obtain rough estimates of the genome-wide contributions of regulatory sequences. This extrapolation yields a genome-wide estimate of E[A] = 9,017 (2.8 ASPHG), roughly equal to the extrapolated estimate for coding sequences (Fig. 4a and Supplementary Note). The projections for weakly deleterious polymorphisms and mutations per haploid genome from transcription factor binding sites rise to values 3.0 and 3.3 times as large as the corresponding estimates for coding sequences. Although these calculations are crude, they nevertheless highlight the substantial genome-wide contribution from regulatory sequences at both the positive and negative ends of the distribution of fitness effects.
Our observations raise the question of what impact regulatory mutations have on the genetic load associated with segregating deleterious mutations53,54. This question cannot be addressed directly using our methods because they do not yield estimates of the selection coefficient s. However, simulations suggest that the weakly deleterious mutations detectable by our methods have average population-scaled selection coefficients (2Nes, where Ne is the effective population size and s is the selection coefficient) of between about –16.7 and –7.7 (Supplementary Table 10 and Supplementary Note). Assuming Ne = 10,000, the average reduction in fitness per haploid genome due to our estimates for coding sequences–or, equivalently, the expected number of lethal equivalents per gamete–would therefore be 0.3–0.6 (Supplementary Note). This estimate is somewhat lower than the estimates of 0.7–2.5 lethal equivalents per gamete obtained from reductions in survival in the presence of human inbreeding54,55. However, if transcription factor binding sites are included and we extrapolate to a larger set of regulatory elements, our estimates rise to 1.2–2.6, in better agreement with inbreeding studies. These calculations are also crude, but they hint that most deleterious mutations segregating in human populations may be regulatory in nature. Furthermore, our finding that these regulatory mutations occur at higher frequencies, on average, than those in coding sequences (Fig. 4d) suggests somewhat different genetic architectures for regulatory and coding deleterious variations. For example, elevated derived allele frequencies most likely correspond to increased average allele ages and greater sharing of deleterious alleles across populations, differences that may have implications for the roles of these mutations in human diseases and their detectability in association studies.
Our results may be influenced in some respects by the simplifying assumptions underlying our model. Our estimates of the fraction of nucleotides under selection (ρ) depend on the assumption that a negligible fraction of high-frequency derived alleles is under selection. If modes of selection that cause alleles to remain at elevated frequencies or produce a steady influx of high-frequency derived alleles–such as balancing selection or recurrent positive selection–are common, the parameter ρ will be underestimated, and other parameters could also be influenced. To have a substantial influence on our results, however, these phenomena would have to be more prevalent than suggested by current evidence39,56,57. Another possible concern is that our estimates of ρ (and derived quantities) could be artificially elevated by reduced diversity in flanking neutral sites due to background selection or hitchhiking. Similarly, it is conceivable that fine-scale differences between binding sites and flanking neutral regions in mutation, fixation or SNP detection rates could lead to biased parameter estimates. However, follow-up experiments indicate that our approach adequately controls for these effects (Supplementary Figs. 5 and 8–10 and Supplementary Note). Evolutionary turnover of transcription factor binding sites58,59 is another possible confounding factor in our analysis. For example, losses of transcription factor binding sites in chimpanzee could lead to the overcounting of substitutions in the human lineage. However, simulations indicate that our model is effective in mitigating this problem by making use of outgroup sequences only to infer ancestral alleles (Supplementary Fig. 11 and Supplementary Note). Finally, our analysis ignores structural variants, focusing on point mutations, because they are most common, easiest to detect and easiest to model. Structural variants and point mutations generally show qualitatively similar patterns of selection51,60, but it will be of interest in future work to consider broader classes of mutation.
ONLINE METHODS
Probabilistic model
The INSIGHT model is detailed elsewhere19 and outlined in the Supplementary Note. Briefly, it incorporates a statistical phylogenetic model for outgroup genomes, a Jukes-Cantor62 model for divergence along the human branch and simple Bernoulli models for the occurrence of low- and high-frequency polymorphisms in human samples. It assumes a mixture of selected and neutral positions in binding sites and assumes that all flanking sequences evolve neutrally. The parameter ρ is the coefficient for the mixture model. The other two key global free parameters are η, which scales the rate of divergence, and γ, which scales the rate of low-frequency polymorphisms, both in selected sites relative to neutral flanking sites (Table 1). The phylogenetic parameters are pre-estimated using RPHAST63, and the neutral parameters (β1, β2 and β3, λi and θi for each locus i) are pre-estimated from the flanking sites. The remaining parameters are estimated by maximum likelihood using an expectation maximization algorithm.
Simulation study
Synthetic data were generated by forward simulation using SFS CODE64. We considered two demographic scenarios: one with a single human population of constant size (Ne = 10,000) and another with separate African, East Asian and European populations and recently estimated demographic parameters65. For each of the assumed distributions of fitness effects and each demographic scenario (Fig. 1 and Supplementary Figs. 12 and 13), we generated 10,000 independent loci consisting of a transcription factor binding site of 10 bp in size with 5,000 neutral flanking sites on each side. Nucleotides within the transcription factor binding sites were assigned selective modes by sampling from a multinomial distribution corresponding to each assumed mixture of fitness effects. We assumed mutation and recombination rates of μ = 1.8 × 10–8 and ρ = 1.1 × 10–8 events per nucleotide per generation, respectively. The recombination rate was held constant, but mutation rates were allowed to vary across loci by sampling from a normal distribution with the given mean and a standard deviation equal to one-tenth of the mean. Data were generated for 50 individuals (100 chromosomes). The true values of all parameters were based on the selective modes assigned during simulation. For comparison, we used the simple divergence-based estimator of Kondrashov and Crow66, an analogous polymorphism-based estimator and the McDonald-Kreitman–based estimator of Smith and Eyre-Walker22. Complete details appear in the Supplementary Note.
Pipeline for transcription factor binding site identification
The transcription factor binding site identification pipeline is detailed in the Supplementary Note. Briefly, precomputed ChIP-seq peaks for 122 transcription factors from the Hudson Alpha Institute for Biotechnology and the Stanford-Yale-USC-Harvard consortium in the ENCODE Project were obtained from the UCSC Genome Browser (see URLs), excluding time-course experiments, chemically treated cell types, controls and data sets with release dates after June 2012. Motifs were identified for each transcription factor in multiple rounds of analysis with MEME67 using subsampling strategies similar to those used in MEME-ChIP68. Motifs were manually inspected and compared with those in motif databases, and a single best motif was selected for each transcription factor. Transcription factors were discarded if a high-quality motif could not be identified. Binding sites at ChIP-seq peaks were then identified using MAST (P < 0.0001, E value < 10). Binding sites were merged across cell lines and, where applicable, from the two data providers. Transcription factors having fewer than 500 binding sites were discarded, leaving 78 transcription factors. Motifs and corresponding binding sites were trimmed by eliminating edge positions with information content of <0.5.
Genome sequence data
Information about human polymorphisms came from the 69 Genomes data set from Complete Genomics (see URLs). Although larger data sets are available16, this one was selected for its high coverage, which allows singleton variants to be characterized with fairly high confidence. Our simulation results indicate that the sample size is large enough for our purposes. We eliminated data from the child in each of 2 trios and all but the 4 grandparents in the 17-member CEPH (Centre d'Etude de Polymorphisme Humain) pedigree, leaving 54 individuals. Genotype calls were extracted from the masterVar files. Outgroup data were obtained from the alignments in the UCSC Genome Browser of the chimpanzee (panTro2), orangutan (ponAbe2) and rhesus macaque (rheMac2) genomes with the human reference genome (hg19). Filters were applied to eliminate repetitive sequences, recent duplications, CpG sites and regions not showing conserved synteny with outgroup genomes. Our analysis considered only the autosomes (chromosomes 1–22). Complete details appear in the Supplementary Note.
Analysis of real data sets
The INSIGHT program was run separately on the full set of binding sites for each of the 78 transcription factors analyzed. Binding sites with fewer than 100 flanking nucleotides (after filtering) were excluded. To avoid overfitting due to sparse data, we added a single small ‘pseudolocus’ to each data set. We assumed a low frequency threshold of f = 15% for most analyses but also experimented with f = 10% and 20% (Supplementary Fig. 14). After parameter estimation, the expected values E[A] and E[W] were obtained by summing over the appropriate conditional distributions across all non-filtered nucleotide positions in the set, given the estimated parameters and observed data. Site frequency spectra (Fig. 4 and Supplementary Fig. 15) represent counts of derived alleles out of 108 chromosomes. Ancestral alleles were determined by parsimony at sites where the chimpanzee allele was shared by at least one of the other outgroups (orangutan or macaque). Details appear in the Supplementary Note.
Variances in parameter estimates
Variances in parameter estimates of ρ, η and γ were obtained by calculating the 3 × 3 negative Hessian matrix for the log-likelihood function at its maximum (an approximation of the Fisher information matrix) and then inverting this matrix. The square roots of the diagonal elements of the resulting matrix were used as approximate standard errors for these parameters. These variances were propagated to the quantities E[A] and E[W] using an approximation. The error bars in the figures extend one standard error above and below the maximum-likelihood estimates of the parameters. Complete details appear in the Supplementary Note.
Likelihood ratio tests
Likelihood ratio tests (LRTs) were used to assess the significance that parameters of interest have values greater than zero (Fig. 2) and that the parameters differ significantly between two sets of elements belonging to different classes (GO analysis for ρ). The first type of test was carried out by fitting the model to the data with all parameters free, fitting it again with a parameter of interest fixed at zero and then comparing twice the difference in the maximized log likelihoods to an appropriate asymptotic null distribution (all variants of χ2 distributions). The second type of test was accomplished by fitting the model separately to two partitions of a data set, fitting it once to the complete data set and again comparing twice the difference in maximized log likelihoods to an appropriate null distribution. Complete details appear in the Supplementary Note.
Information content and binding affinity
Information content was calculated from the inferred motif models as
where k is the number of positions in the motif and is the probability of observing base b at position i of a binding site69,70.
The predicted binding affinity of a binding site having sequence X = (x1,..., xk) was calculated as
where is the probability of observing base xi at position i in a binding site and πxi is the background frequency of base xi (estimated across the genome)71,72. Additional details appear in the Supplementary Note.
Robustness to binding site turnover
The potential impact of binding site turnover was assessed by simulation. We simulated both losses of chimpanzee binding sites and gains of human binding sites, at various rates, under various mixtures of selective effects. We observed almost no sensitivity to relaxation of constraint on the chimpanzee lineage, even at fairly high rates of binding site loss (Supplementary Fig. 11). The model also behaves reasonably when gains in humans involve genuine positive selection. We did observe some overestimation of adaptive substitutions in a scenario in which binding sites emerge fully formed and immediately come under negative selection, with no intermediate adaptive interval, but this scenario is unlikely to have a major impact on our results (Supplementary Note).
Supplementary Material
ACKNOWLEDGMENTS
We thank R. Blekhman, C. Danko, A. Boyko, K. Pollard, N. Goldman and A. Clark for comments on the manuscript. This research was supported by a Packard Fellowship, a Sloan Research Fellowship, US National Science Foundation grant DBI-0644111 and US National Institutes of Health (National Institute of General Medical Sciences, NIGMS) grant GM102192 (to A.S.). In addition, L.A. was supported in part by a postdoctoral fellowship award from the Cornell Center for Vertebrate Genomics, and B.A.A. was supported by US National Institutes of Health training grant T32-GM083937.
Footnotes
AUTHOR CONTRIBUTIONS
L.A., I.G. and A.S. conceived and designed the study. L.A., I.G., B.A.A., M.J.H., B.G. and A.S. analyzed the data. L.A., I.G., B.A.A. and M.J.H. contributed materials and analysis tools. A.S. and A.K. supervised the research. L.A., I.G. and A.S. wrote the manuscript with review and contributions from all authors.
URLs. UCSC Genome Browser, http://genome.ucsc.edu/; Complete Genomics human variation data, http://www.completegenomics.com/public-data/69-Genomes; INSIGHT source code and webserver, http://compgen.bscb.cornell.edu/INSIGHT/.
Supplementary information is available in the online version of the paper.
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interests.
References
- 1.Ohno S. An argument for the genetic simplicity of man and other mammals. J. Hum. Evol. 1972;1:651–662. [Google Scholar]
- 2.King MC, Wilson AC. Evolution at two levels in humans and chimpanzees. Science. 1975;188:107–116. doi: 10.1126/science.1090005. [DOI] [PubMed] [Google Scholar]
- 3.Wilson AC, Maxson LR, Sarich VM. Two types of molecular evolution. Evidence from studies of interspecific hybridization. Proc. Natl. Acad. Sci. USA. 1974;71:2843–2847. doi: 10.1073/pnas.71.7.2843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Britten RJ, Davidson EH. Gene regulation for higher cells: a theory. Science. 1969;165:349–357. doi: 10.1126/science.165.3891.349. [DOI] [PubMed] [Google Scholar]
- 5.Stern DL. Evolutionary developmental biology and the problem of variation. Evolution. 2000;54:1079–1091. doi: 10.1111/j.0014-3820.2000.tb00544.x. [DOI] [PubMed] [Google Scholar]
- 6.Carroll SB. Evolution at two levels: on genes and form. PLoS Biol. 2005;3:e245. doi: 10.1371/journal.pbio.0030245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wray GA. The evolutionary significance of cis-regulatory mutations. Nat. Rev. Genet. 2007;8:206–216. doi: 10.1038/nrg2063. [DOI] [PubMed] [Google Scholar]
- 8.Hoekstra HE, Coyne JA. The locus of evolution: evo devo and the genetics of adaptation. Evolution. 2007;61:995–1016. doi: 10.1111/j.1558-5646.2007.00105.x. [DOI] [PubMed] [Google Scholar]
- 9.Andolfatto P. Adaptive evolution of non-coding DNA in Drosophila. Nature. 2005;437:1149–1152. doi: 10.1038/nature04107. [DOI] [PubMed] [Google Scholar]
- 10.Haygood R, Fedrigo O, Hanson B, Yokoyama K-D, Wray GA. Promoter regions of many neural- and nutrition-related genes have experienced positive selection during human evolution. Nat. Genet. 2007;39:1140–1144. doi: 10.1038/ng2104. [DOI] [PubMed] [Google Scholar]
- 11.Torgerson DG, et al. Evolutionary processes acting on candidate cis-regulatory regions in humans inferred from patterns of polymorphism and divergence. PLoS Genet. 2009;5:e1000592. doi: 10.1371/journal.pgen.1000592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gaffney DJ, Blekhman R, Majewski J. Selective constraints in experimentally defined primate regulatory regions. PLoS Genet. 2008;4:e1000157. doi: 10.1371/journal.pgen.1000157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen K, Rajewsky N. Natural selection on human microRNA binding sites inferred from SNP data. Nat. Genet. 2006;38:1452–1456. doi: 10.1038/ng1910. [DOI] [PubMed] [Google Scholar]
- 14.Stoletzki N, Eyre-Walker A. Estimation of the neutrality index. Mol. Biol. Evol. 2011;28:63–70. doi: 10.1093/molbev/msq249. [DOI] [PubMed] [Google Scholar]
- 15.Pollard KS, Hubisz MJ, Rosenbloom KR, Siepel A. Detection of nonneutral substitution rates on mammalian phylogenies. Genome Res. 2010;20:110–121. doi: 10.1101/gr.097857.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.1000 Genomes Project Consortium A map of human genome variation from population-scale sequencing. Nature. 2010;467:1061–1073. doi: 10.1038/nature09534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Drmanac R, et al. Human genome sequencing using unchained base reads on self-assembling DNA nanoarrays. Science. 2010;327:78–81. doi: 10.1126/science.1181498. [DOI] [PubMed] [Google Scholar]
- 18.ENCODE Project Consortium An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gronau I, Arbiza L, Mohammed J, Siepel A. Inference of natural selection from interspersed genomic elements based on polymorphism and divergence. Mol. Biol. Evol. 2013;30:1159–1171. doi: 10.1093/molbev/mst019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McDonald JH, Kreitman M. Adaptive protein evolution at the Adh locus in Drosophila. Nature. 1991;351:652–654. doi: 10.1038/351652a0. [DOI] [PubMed] [Google Scholar]
- 21.Sawyer SA, Hartl DL. Population genetics of polymorphism and divergence. Genetics. 1992;132:1161–1176. doi: 10.1093/genetics/132.4.1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Smith NG, Eyre-Walker A. Adaptive protein evolution in Drosophila. Nature. 2002;415:1022–1024. doi: 10.1038/4151022a. [DOI] [PubMed] [Google Scholar]
- 23.Charlesworth J, Eyre-Walker A. The McDonald-Kreitman test and slightly deleterious mutations. Mol. Biol. Evol. 2008;25:1007–1015. doi: 10.1093/molbev/msn005. [DOI] [PubMed] [Google Scholar]
- 24.Bierne N, Eyre-Walker A. The genomic rate of adaptive amino acid substitution in Drosophila. Mol. Biol. Evol. 2004;21:1350–1360. doi: 10.1093/molbev/msh134. [DOI] [PubMed] [Google Scholar]
- 25.Boyko AR, et al. Assessing the evolutionary impact of amino acid mutations in the human genome. PLoS Genet. 2008;4:e1000083. doi: 10.1371/journal.pgen.1000083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wilson DJ, Hernandez RD, Andolfatto P, Przeworski M. A population genetics–phylogenetics approach to inferring natural selection in coding sequences. PLoS Genet. 2011;7:e1002395. doi: 10.1371/journal.pgen.1002395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fay JC, Wyckoff GJ, Wu CI. Positive and negative selection on the human genome. Genetics. 2001;158:1227–1234. doi: 10.1093/genetics/158.3.1227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ohta T. Slightly deleterious mutant substitutions in evolution. Nature. 1973;246:96–98. doi: 10.1038/246096a0. [DOI] [PubMed] [Google Scholar]
- 29.Kondrashov AS. Contamination of the genome by very slightly deleterious mutations: why have we not died 100 times over? J. Theor. Biol. 1995;175:583–594. doi: 10.1006/jtbi.1995.0167. [DOI] [PubMed] [Google Scholar]
- 30.Williamson SH, et al. Simultaneous inference of selection and population growth from patterns of variation in the human genome. Proc. Natl. Acad. Sci. USA. 2005;102:7882–7887. doi: 10.1073/pnas.0502300102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Eyre-Walker A, Woolfit M, Phelps T. The distribution of fitness effects of new deleterious amino acid mutations in humans. Genetics. 2006;173:891–900. doi: 10.1534/genetics.106.057570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Eyre-Walker A, Keightley PD. Estimating the rate of adaptive molecular evolution in the presence of slightly deleterious mutations and population size change. Mol. Biol. Evol. 2009;26:2097–2108. doi: 10.1093/molbev/msp119. [DOI] [PubMed] [Google Scholar]
- 33.Chimpanzee Sequencing and Analysis Consortium Initial sequence of the chimpanzee genome and comparison with the human genome. Nature. 2005;437:69–87. doi: 10.1038/nature04072. [DOI] [PubMed] [Google Scholar]
- 34.Locke DP, et al. Comparative and demographic analysis of orang-utan genomes. Nature. 2011;469:529–533. doi: 10.1038/nature09687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rhesus Macaque Genome Sequencing and Analysis Consortium Evolutionary and biomedical insights from the rhesus macaque genome. Science. 2007;316:222–234. doi: 10.1126/science.1139247. [DOI] [PubMed] [Google Scholar]
- 36.Eory L, Halligan DL, Keightley PD. Distributions of selectively constrained sites and deleterious mutation rates in the hominid and murid genomes. Mol. Biol. Evol. 2010;27:177–192. doi: 10.1093/molbev/msp219. [DOI] [PubMed] [Google Scholar]
- 37.Moses AM, Chiang DY, Kellis M, Lander ES, Eisen MB. Position specific variation in the rate of evolution in transcription factor binding sites. BMC Evol. Biol. 2003;3:19. doi: 10.1186/1471-2148-3-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bustamante CD, et al. Natural selection on protein-coding genes in the human genome. Nature. 2005;437:1153–1157. doi: 10.1038/nature04240. [DOI] [PubMed] [Google Scholar]
- 39.Kosiol C, et al. Patterns of positive selection in six mammalian genomes. PLoS Genet. 2008;4:e1000144. doi: 10.1371/journal.pgen.1000144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Enard W, et al. Molecular evolution of FOXP2, a gene involved in speech and language. Nature. 2002;418:869–872. doi: 10.1038/nature01025. [DOI] [PubMed] [Google Scholar]
- 41.Ashburner M, et al. Gene ontology: tool for the unification of biology. Nat. Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Green RE, et al. A draft sequence of the Neandertal genome. Science. 2010;328:710–722. doi: 10.1126/science.1188021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chen F-C, Li W-H. Genomic divergences between humans and other hominoids and the effective population size of the common ancestor of humans and chimpanzees. Am. J. Hum. Genet. 2001;68:444–456. doi: 10.1086/318206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gojobori J, Tang H, Akey JM, Wu CI. Adaptive evolution in humans revealed by the negative correlation between the polymorphism and fixation phases of evolution. Proc. Natl. Acad. Sci. USA. 2007;104:3907–3912. doi: 10.1073/pnas.0605565104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sunyaev S, et al. Prediction of deleterious human alleles. Hum. Mol. Genet. 2001;10:591–597. doi: 10.1093/hmg/10.6.591. [DOI] [PubMed] [Google Scholar]
- 46.Lohmueller KE, et al. Proportionally more deleterious genetic variation in European than in African populations. Nature. 2008;451:994–997. doi: 10.1038/nature06611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jacob F, Monod J. Genetic regulatory mechanisms in the synthesis of proteins. J. Mol. Biol. 1961;3:318–356. doi: 10.1016/s0022-2836(61)80072-7. [DOI] [PubMed] [Google Scholar]
- 48.Vaquerizas JM, Kummerfeld SK, Teichmann SA, Luscombe NM. A census of human transcription factors: function, expression and evolution. Nat. Rev. Genet. 2009;10:252–263. doi: 10.1038/nrg2538. [DOI] [PubMed] [Google Scholar]
- 49.Mouse Genome Sequencing Consortium Initial sequencing and comparative analysis of the mouse genome. Nature. 2002;420:520–562. doi: 10.1038/nature01262. [DOI] [PubMed] [Google Scholar]
- 50.Siepel A, et al. Evolutionarily conserved elements in vertebrate, insect, worm, and yeast genomes. Genome Res. 2005;15:1034–1050. doi: 10.1101/gr.3715005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lunter G, Ponting CP, Hein J. Genome-wide identification of human functional DNA using a neutral indel model. PLoS Comput. Biol. 2006;2:e5. doi: 10.1371/journal.pcbi.0020005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Shen Y, et al. A map of the cis-regulatory sequences in the mouse genome. Nature. 2012;488:116–120. doi: 10.1038/nature11243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Muller HJ. Our load of mutations. Am. J. Hum. Genet. 1950;2:111–176. [PMC free article] [PubMed] [Google Scholar]
- 54.Morton NE, Crow JF, Muller HJ. An estimate of the mutational damage in man from data on consanguineous marriages. Proc. Natl. Acad. Sci. USA. 1956;42:855–863. doi: 10.1073/pnas.42.11.855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bittles AH, Neel JV. The costs of human inbreeding and their implications for variations at the DNA level. Nat. Genet. 1994;8:117–121. doi: 10.1038/ng1094-117. [DOI] [PubMed] [Google Scholar]
- 56.Asthana S, Schmidt S, Sunyaev S. A limited role for balancing selection. Trends Genet. 2005;21:30–32. doi: 10.1016/j.tig.2004.11.001. [DOI] [PubMed] [Google Scholar]
- 57.Bubb KL, et al. Scan of human genome reveals no new loci under ancient balancing selection. Genetics. 2006;173:2165–2177. doi: 10.1534/genetics.106.055715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Schmidt D, et al. Five-vertebrate ChIP-seq reveals the evolutionary dynamics of transcription factor binding. Science. 2010;328:1036–1040. doi: 10.1126/science.1186176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kasowski M, et al. Variation in transcription factor binding among humans. Science. 2010;328:232–235. doi: 10.1126/science.1183621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Mu XJ, Lu ZJ, Kong Y, Lam HY, Gerstein MB. Analysis of genomic variation in non-coding elements using population-scale sequencing data from the 1000 Genomes Project. Nucleic Acids Res. 2011;39:7058–7076. doi: 10.1093/nar/gkr342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Holm S. A simple sequentially rejective multiple test procedure. Scand. J. Stat. 1979;6:65–70. [Google Scholar]
- 62.Jukes TH, Cantor CR. Evolution of protein molecules. In: Munro H, editor. Mammalian Protein Metabolism. Academic Press; New York: 1969. pp. 21–132. [Google Scholar]
- 63.Hubisz MJ, Pollard KS, Siepel A. PHAST and RPHAST: phylogenetic analysis with space/time models. Brief. Bioinform. 2011;12:41–51. doi: 10.1093/bib/bbq072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hernandez RD. A flexible forward simulator for populations subject to selection and demography. Bioinformatics. 2008;24:2786–2787. doi: 10.1093/bioinformatics/btn522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Gravel S, et al. Demographic history and rare allele sharing among human populations. Proc. Natl. Acad. Sci. USA. 2011;108:11983–11988. doi: 10.1073/pnas.1019276108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kondrashov AS, Crow JF. A molecular approach to estimating the human deleterious mutation rate. Hum. Mutat. 1993;2:229–234. doi: 10.1002/humu.1380020312. [DOI] [PubMed] [Google Scholar]
- 67.Bailey TL, Elkan C. Fitting a mixture model by expectation maximization to discover motifs in biopolymers. Proc. Int. Conf. Intell. Syst. Mol. Biol. 1994;2:28–36. [PubMed] [Google Scholar]
- 68.Machanick P, Bailey TL. MEME-ChIP: motif analysis of large DNA datasets. Bioinformatics. 2011;27:1696–1697. doi: 10.1093/bioinformatics/btr189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Schneider TD, Stormo GD, Gold L, Ehrenfeucht A. Information content of binding sites on nucleotide sequences. J. Mol. Biol. 1986;188:415–431. doi: 10.1016/0022-2836(86)90165-8. [DOI] [PubMed] [Google Scholar]
- 70.Wasserman WW, Sandelin A. Applied bioinformatics for the identification of regulatory elements. Nat. Rev. Genet. 2004;5:276–287. doi: 10.1038/nrg1315. [DOI] [PubMed] [Google Scholar]
- 71.Berg OG, von Hippel PH. Selection of DNA binding sites by regulatory proteins. Statistical-mechanical theory and application to operators and promoters. J. Mol. Biol. 1987;193:723–750. doi: 10.1016/0022-2836(87)90354-8. [DOI] [PubMed] [Google Scholar]
- 72.Stormo GD. DNA binding sites: representation and discovery. Bioinformatics. 2000;16:16–23. doi: 10.1093/bioinformatics/16.1.16. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.