Abstract
A single nucleotide polymorphism in the protein tyrosine phosphatase nonreceptor type 22 gene (PTPN22), that encodes the Lyp tyrosine phosphatase LypR620W, has been linked to a number of autoimmune diseases including type I diabetes, rheumatoid arthritis and systemic lupus erythematosus. Studies in PTPN22 KO mice and in mice expressing the mouse homolog of the pro-autoimmune allele, PepR619W, have reported increased germinal center activity and enhanced antibody production. Here we present findings that explain the basis for increased germinal center activity in PTPN22 mutant mice. As compared with their wild type equivalents, TFH cells from PTPN22 KO mice proliferate and accumulate to a greater extent, and exhibit enhanced production of IL-21. The follicular regulatory T cells (TFR) in PTPN22 KO mice do not expand to effectively regulate these TFH cells, resulting in an increase in B cell numbers and antibody production. This is evident in the KBxN mouse model of arthritis in which PTPN22 deficiency results in increased severity of disease. Our findings demonstrate the importance of cell type specific PTPN22 activity on regulation of antibody production.
Introduction
Genome wide association studies (GWAS) have identified a single nucleotide polymorphism (SNP) in the PTPN22 gene, R620W, to be strongly associated with a number of autoimmune diseases including type I diabetes (T1D), rheumatoid arthritis, systemic lupus erythematosus, Graves disease, and others (1–3). Of interest, it does not increase the frequency of Crohns disease or multiple sclerosis (4, 5). Since the protein is expressed in essentially all bone-marrow derived cells, such disease selectively is likely to reflect differences in the types of immune cells contributing to each disease and how the alternative alleles of the phosphatase affects the function of the various cell types. PTPN22 encodes the lymphoid tyrosine phosphatase (LYP) in humans and PEST-enriched protein phosphatase (PEP) in mice. The functional outcome of the disease associated allele is controversial. Originally it was thought to be a gain of function mutation (6–9) however this has been challenged in recent models with suggestions that the mutation causes a loss of function (10, 11) or an alteration of substrate specificity (12).
In order to learn about the effect of PTPN22 on immune cells, several labs have produced knockout (KO) mice (13, 14). When on the B6 background, mice deficient in Pep show no overt autoimmune disease, although they do exhibit splenomegaly and increased Teff/mem cells that accumulate over time. This T cell phenotype is attributable to the fact that PEP targets include the Src-family kinases including signaling molecules proximal to the TCR, including Lck, Fyn and ZAP70 (15, 16). In the absence of PEP, T cell signaling is increased (13) and mice exhibit greater numbers of GCs and have higher levels of IgG in their sera compared to WT mice. Despite this observation, B cell signaling was reported to be similar between WT and KO mice suggesting this is an indirect effect attributable to the enhanced activity of T cells (13).
PTPN22 also influences Treg number and function, which is important in the context of autoimmunity. We have shown previously that PTPN22 deficiency increases thymic development of Tregs leading to an increase in the numbers of peripheral Tregs (17). This increase has been shown by other groups in various PTPN22 models (9, 14). It has also been reported that Treg suppressive function is enhanced through an LFA-1 mediated mechanism in PTPN22 KO mice (14).
PTPN22 is expressed in B cells, although at a lower level than in T cells (18). The majority of studies on the effect of PTPN22 in B cells have been performed by comparing human samples that carry the R620W variant and the common allele. These studies have suggested the R620W allele impairs BCR signaling, leading to expansion of transitional and anergic B cells which exhibit reduced apoptosis upon BCR engagement (19, 20). Another study has also reported that the risk variant leads to escape of autoimmune B cells though a defect in central and peripheral tolerance (21). Mouse studies have shown that PTPN22 KO mice can develop a lupus like phenotype when bred to mice containing a mutation in CD45 resulting in increased B cell activation (11).
Recently, two groups have introduced the disease associated allele (PEP-R619W analogous to the human LYP R620W) and found the knock-in mice exhibit a phenotype similar to the KO mouse with increased GCs and increased serum IgG (10, 12). One of these studies suggested that on a mixed genetic background (129/SJL backcrossed several generations to B6) B cell tolerance was impaired in the PEPR619W mouse (12). Finally, in a recent report PTPN22 expression in the NOD mouse was knocked-down by RNAi resulting in enhanced B cell upregulation of CD25 and CD69 in response to anti-IgM and anti-CD40 ligation. In addition apoptosis was increased, overall suggesting the KO has the opposite effect of the R620W allele on human B cells and consistent with the hypothesis that R620W increases the phosphatase activity of Pep/Lyp (9). However due to the nature of the autoimmune prone backgrounds thus far reported, the B cell phenotypes described would be difficult to attribute solely to a mutation in PTPN22. Working with a non-autoimmune prone background such as the B6 PTPN22 KO mouse allows us to identify PTPN22 dependent phenotypes in the absence of genetic interactions with other pro-autoimmune alleles that can alter immune phenotype.
Efficient antibody generation by B cells relies on the formation of a germinal center (GC) in which optimal interaction between T and B cells occurs (22). Follicular helper T cells (TFH) are a subset of CD4+ T cells that express CXCR5 and migrate to the GC via a CXCL13 chemokine gradient to provide help to B cells (23–26). Help in this case can be through production of effector cytokines IL-21 (27, 28) and IL-4 (29, 30) as well as engagement of CD40 on B cells through CD40L expression, promoting activation of B cells and class switching, as well as formation of plasma cells (31, 32). Interaction with dendritic cells in the initial stages of activation of CD4+ T cells is thought to determine the fate and differentiation of Th0 cells into TFH, controlled by the transcription factor Bcl-6(33–35). Further interaction with B cells stabilizes this phenotype (22). Strength of signal during these early stages may favor TFH differentiation, a point of particular relevance in the case of PTPN22 KO mice (29). Regulation of the GC is crucial to maintaining peripheral tolerance. A recently described population of thymically derived Tregs that express CXCR5 and also localized to the GC, known as follicular regulatory T cells (TFR), can control the GC reaction (36, 37). It has been suggested that the balance of TFR:TFH is important in controlling the magnitude of the humoral immunity (38).
In this study we have used PTPN22 KO mice to investigate the mechanisms by which PTPN22 may alter formation and regulation of the GC. Overall understanding of how this critical disease susceptibility gene functions in humoral immunity will allow us to identify targets in a number of autoimmune conditions which may be beneficial for therapy. We have found that loss of PTPN22 increases GC activity predominantly by increasing proliferation, survival and cytokine secretion by TFH. Through adoptive transfer models we show that the T cell genotype is more important than the B cell genotype in terms of the contribution by PTPN22. We also find a decreased ratio of TFR:TFH in PTPN22 KO mice which we hypothesize may be due to an observed increase in IL-21 production in the GC. We propose the TFR cells are unable to control the expanded number of TFH present in PTPN22 KO mice, resulting in enhanced antibody production.
Materials and Methods
Mice
Experimental procedures were carried out according to the National Institutes of Health Guide for the care and use of laboratory animals. PTPN22 −/− mice were obtained from Dr. Andrew Chan (Genentech, San Francisco, CA) and have previously been described (13). PTPN22+/− mice were obtained by interbreeding PTPN22 −/− mice with C57BL/6 (Jackson Laboratories, Bar Harbor, ME). FoxP3 GFP and OT-II Thy 1.1+ mice were provided by Dr. Charles Surh (TSRI) and were bred to PTPN22 −/− mice. Nur-77 GFP knock-in mice were provided by Dr. Kristin Hogquist (University of Minnesota) and bred to PTPN22 −/− mice (39). KBxN mice were produced through breeding of KRN mice (provided by Dr. Kerri Mowen, TSRI) with B6.H2g7 mice (Jackson Laboratories). PTPN22−/− mice were bred to KRN and/or B6.H2g7 mice to produce PTPN22−/− and PTPN22+/− KBxN mice.
Immunizations
8–12 week old mice were immunized sub-cutaneously (sub.cut.) in the flank with 100μg of NP-KLH (Biosearch technologies, Novarto, CA) or Ovalbumin (Invivogen, San Diego, CA) in complete Freund’s adjuvant (CFA) (Difco, Detroit, MI). Secondary immunizations were carried out by injecting the same dose of antigen s.c. without CFA, 14 days following primary immunization. The draining lymph node refers to the inguinal lymph node on the side of the mouse being injected.
Flow Cytometry
Cells to be stained were resuspended in FACS buffer (HBSS containing 1% FCS) and incubated with the indicated antibodies for 15 minutes on ice. Cells were then washed in FACS buffer before acquisition on an LSR-II flow cytometer (BD Bioscience, Franklin Lakes, NJ) and analysis using Flowjo (Treestar). Antibodies (all Biolegend, San Diego, CA unless otherwise stated) used were anti-mouse CD4 PerCP Cy5.5, CD8 Pacific Blue/APC Cy7, PD-1 FITC/PE, CXCR5-biotin (BD Bioscience), CD44 Pacific Blue, GL-7 FITC, FAS PE, CD138 APC, CD19 PerCP Cy5.5, CD23 PE, CD21 PerCP Cy5.5, streptavidin APC. For intracellular staining of markers, an intracellular staining kit (Fix/Perm, eBioscience, San Diego CA) was used together with anti-mouse Foxp3 PE (Ebioscience) and Ki-67 APC (Biolegend). Intracellular cytokine staining was carried out using Cytofix/Cytoperm buffers (BD) and IL-21 PE (Ebioscience).
For cell sorting, splenocyte and thymocyte single cell suspensions were antibody labeled as described above and the desired populations were sorted using FACS Aria (BD Bioscience) or Mo Flo XDP (Beckman Coulter, Indianapolis, IN) by TSRI Flow cytometry core facility.
Calcium Flux
Splenocytes were rested in cRPMI for 20 mins at 37°C, before staining. Splenocytes from one genotype (5×106) were labeled with 1μg Cy5 dye in cRPMI (GE healthcare, Chalfont, UK) for 5 mins at room temperature and the other genotype was left unstained. Cy5 labeled cells were washed and mixed with unlabeled cells. These cells were then loaded with INDO-1 AM (Invitrogen) in RPMI media for 30 minutes at 37°C. Following washing the cells were stained with CD19 FITC, CD23 PE and CD21 PerCP Cy5.5 (all Biolegend) and resuspended in HBSS (Gibco) and kept on ice. Cells were then warmed to 37°C prior to running the sample and ran for 30 seconds to establish a baseline, 10μg/ml anti-IgM Fab2 (Jackson Immunoresearch, WestGrove, PA) was added at 30 seconds to stimulate B cells. At 2 minutes, 10mM CaCl2 (Sigma Aldrich, St. Louis, MO) was added and the cells run until 5 minutes. At 5 minutes 1μg/ml of ionomycin (EMD Biosciences, La Jolla, CA) was added and the sample run until 7 minutes.
Lymphocyte Purification
FACS sorting was carried out as described in the above method. Magnetic separation of CD4+ T cells was carried out using a CD4 negative depletion kit according to manufacturers instructions (BD). B cells were isolated from splenocytes using a B220 positive selection kit according to manufacturers instructions (Miltenyi Biotec Inc. Auburn, CA).
ELISA
Serum was collected from mice at the stated time points. Maxisorp plates (Nunc, Rochester, NY) were coated with 5μg/ml ovalbumin (Invivogen) or NP-BSA (Biosearch technologies) (NP4 for high affinity Abs or NP32 for broad affinity Abs) overnight at 4°C. Plates were blocked in 5% BSA (Sigma Aldrich) for an hour at 37°C. Plates were washed three times with wash buffer (HBSS with 0.1% Tween-20 (Sigma Aldrich)). Sera were diluted accordingly following optimization for each experiment in reagent buffer (HBSS containing 1% BSA, 0.1% Tween-20) and incubated on the plate in triplicate for 1 hour at 37°C. Plates were washed three times. Anti-mouse IgG alkaline phosphatase (AP) or anti-mouse IgG1 horse radish peroxidase (HRP) were then diluted and added to the wells for a further hour at 37°C (Both Jackson Immunoresearch). Plates were washed and then incubated with either pNPP AP substrate or TMB HRP substrate (both Sigma Aldrich). Plates were read using a Versamax plate reader (Molecular devices, Sunnyvale, CA) at 405 or 450nm respectively.
Q-PCR
NP-KLH immunized dLN were collected from FoxP3-GFP PTPN22 WT and KO mice, 7 days post immunization. The cells were stained for CXCR5, CD44, CD4 and PD-1 and FACS sorted for TFH (CD4+ CD44hi CXCR5+ PD-1+ FoxP3−) and TFR (CD4+ CD44hi CXCR5+ PD-1+ FoxP3−). mRNA was then extracted by Trizol (Life Technologies, Carlsbad, CA) and cDNA was produced using the High capacity cDNA RT kit (Applied Biosystems, Carlsbad, CA) according to manufacturers instructions. Q-PCR, using a fixed amount of cDNA produced from comparable numbers of cells from WT and KO mice was performed to measure levels of IL-21, IL-4, IL-10, Bcl2, Mcl-1, Bim L, Bim EL and Bax (sequences available on request) using a 7900HT Real time PCR system (Applied Biosystems). Ct values were normalized to β2M using the 2−ΔΔCt method. KO values were normalized to WT.
In Vitro B cell help assay
PTPN22 KO and WT mice were immunized with NP-KLH as described. 8 days later dLN were extracted and CD4 T cells were purified as described above. Unimmunized PTPN22 WT and KO mouse spleens were also collected at this time point and B cells were purified as described above. CD4 T cells (5×104/well) and B cells (5×105/well) were co-cultured in the combinations stated in the results section with 20ng/ml IL-2, β-mercaptoethanol and NP-KLH (Biosearch technologies) for a further 7 days at 37°C. Supernatant was collected and analysed for NP-specific IgG by ELISA as described above.
Confocal Microscopy
Lymph nodes were collected at the stated time points post immunization, immersion fixed in 1% v/v paraformaldehyde (Electron Micoscopy Sciences, Hatfield, PA). LN were then transferred to 15% sucrose (Sigma Aldrich) until the organs sank, and then frozen in Tissue-Tek OCT compound (Sakura Finetek, Torrance, CA). LN sections were cut at 10μM using a cryostat (which one). Sections were acetone (Sigma Aldrich) treated for 10 mins at room temperature and then washed in PBS. Antibodies used to stain for markers are as follows; anti-mouse CD4 (Ebioscience), PNA-biotin (Biosearch technologies), anti-rat AF647, streptavidin AF555 (both Life Technologies, Carlsbad, CA). Images were acquired on a Leica 780 confocal microscope (Leica Microsystems, Buffalo Grove, IL) and analyzed with Imaris (Bitplane, South Windsor, CT) and Image Pro Plus software (Media Cybernetics, Rockville, MD).
Scoring of KBxN arthritis
KBxN mice were weaned at 3 weeks of age and paw thickness (front and hind) was measured by calipers. Clinical scoring was based on previously described scoring criteria (40). Serum transferred arthritis was induced in PTPN22 KO and WT mice through i.p. injection of 150μl of serum collected from 6–10 week old KBxN mice.
Statistics
Graphs were assembled and analyzed using Prism 5 software (Graphpad, San Diego, CA). For multiple group analyses, one-way ANOVA with Tukeys post-test was carried out. For comparison of two group data-sets Students t-test was used.
Results
PTPN22 KO mice have increased follicular B cells
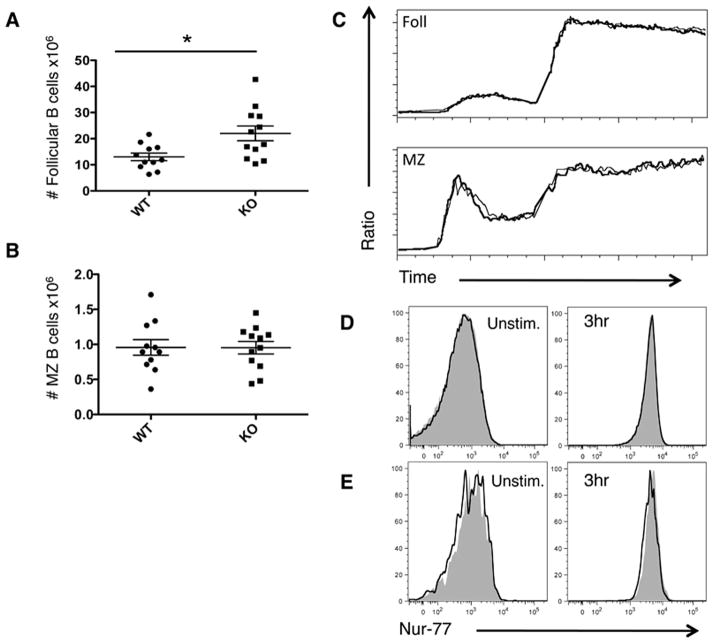
Following the observation that PTPN22 KO mice had increased numbers of GCs, we analyzed spleens of unimmunized mice for the distribution of cells among the various B cell subsets by flow cytometry (Figure 1). PTPN22 KO mice have significantly more follicular B cells compared to WT mice, however marginal zone B cell numbers were similar in both types of mice (Figure 1 A and B). Calcium flux in response to anti-IgM stimulation was also similar between PTPN22 WT and KO B cells of both follicular and marginal zone subsets (Figure 1C). We utilized Nur-77 GFP mice as a reporter of strength of BCR signaling, to measure the response to anti-IgM stimulation (41). Follicular (Figure 1D) and MZ (Figure 1E) subsets from PTPN22 WT and KO mice show no difference in resting GFP levels or in response to IgM stimulation. Overall these data suggest an accumulation of follicular B cells occurs in PTPN22 KO mice but intrinsic signaling differences were not detectable by calcium flux or activation of Nur-77.
Figure 1. Numbers and intracellular signaling of B cells in PTPN22 KO mice.
Spleens were taken from WT and KO mice and stained for follicular (CD19+ CD21+ CD23+) (A) and marginal zone B cells (CD19+ CD21hi CD23lo) (B) and absolute numbers in the spleen are shown (graphs show mean + S.E.M each dot represents 1 mouse). Splenocytes were labeled with Indo-1 and calcium flux was measured for both follicular and marginal zone B cells in response to anti-IgM at 30 seconds and Ionomycin at 3 minutes (C) (thick line=WT thin line=KO). Nur-77 GFP PTPN22 WT (filled grey) and KO (unfilled black line) spleens were analysed for GFP expression at ex vivo or following 3hr anti-IgM stimulation. GFP expression of follicular B cells is shown in (D) and MZ in (E). C, D and E are representative of 2 independent experiments. *p<0.05
PTPN22 KO demonstrate increased responses to T-dependent antigen
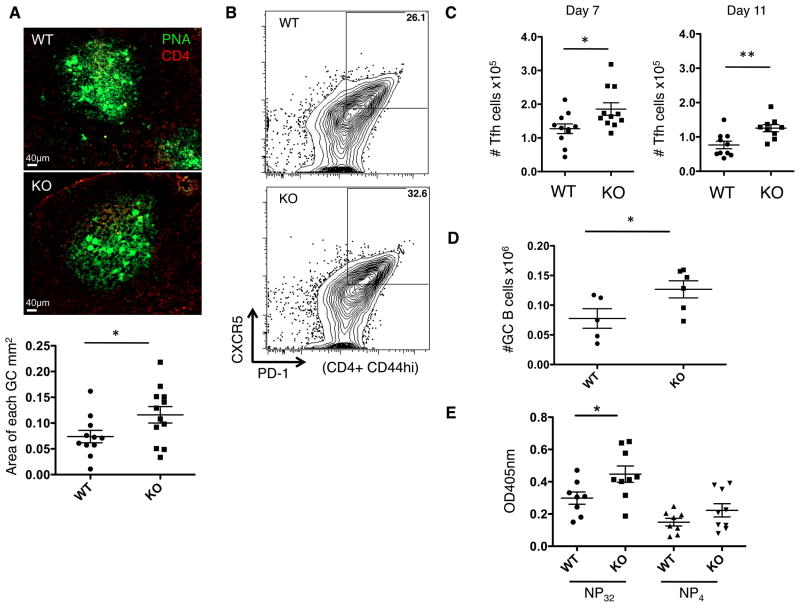
To assess the influence of PTPN22 on GC activity, mice were immunized with NP-KLH in CFA (Figure 2) and cells from the draining lymph nodes (dLN) analyzed at various time points following immunization. In agreement with previous reports (13), we observed larger and more numerous GCs in KO mice compared to WT mice (Figure 2A). We also observed that both the percentage and absolute number of TFH cells in the dLN was increased in KO mice. This peaked 7 days post-immunization and reached statistical significance (Figure 2 B and C). In a similar manner, numbers of GC B cells were significantly increased in dLN of PTPN22 KO mice compared to WT mice (Figure 2 D). NP-specific antibody levels in the sera of these mice 14 days post-immunization was quantified by ELISA (Figure 2E). There was a significantly higher level of broad affinity anti-NP IgG in KO mice compared to WT, as well as slightly higher, yet not statistically significant levels of high affinity anti-NP IgG.
Figure 2. T-dependent antigen responses in PTPN22 KO mice.
WT and KO mice were immunized with NP-KLH in CFA s.c. 7 days later dLN were isolated and analyzed. A) frozen lymph node sections, 7 days post immunization were stained with PNA (green) and CD4 (red) to visualize germinal centers using confocal microscopy (20x). Imaris software was used to calculate the area of each germinal center, the graph shows GCs measured from 3 independent experiments, each data point is 1 GC B) representative flow cytometry dot plots showing TFH (CD4+ CD44hi CXCR5+ PD-1+) percentage in the dLN 7 days post immunization. C) absolute number of TFH cells in the dLN over the timecourse of the immunization. D) shows GC B cell numbers (CD19+ FAS+ GL-7+) in the dLN of WT and KO mice 11 days post immunization. E) serum anti-NP IgG antibody levels 14 days post immunization were measured by ELISA. *P<0.05; **P<0.01 (graphs show mean ± S.E.M and each data point represents 1 mouse in panels C, D and E)
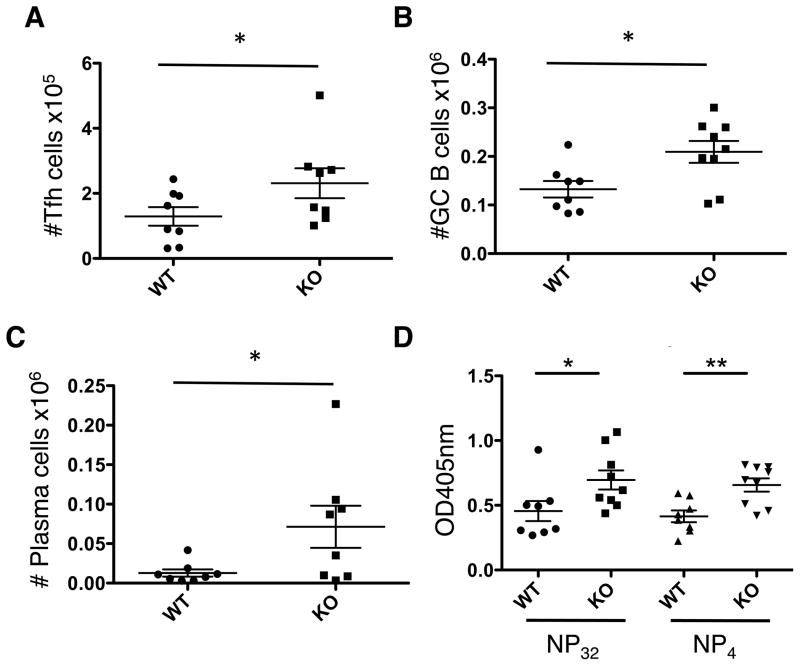
We next assessed the recall response to antigen. Mice were immunized with NP-KLH in CFA, and then re-challenged with NP-KLH. Analysis of the dLN was carried out 7 days after re-challenge. Similar to the primary response, PTPN22 KO mice show significantly higher numbers of TFH cells and GC B cells (Figure 3A and B). Numbers of plasma cells were also significantly increased in KO dLN compared to WT (Figure 3C). Surprisingly the numbers of plasma cells in the WT dLN were low. We hypothesize that the majority of these may have migrated to the bone marrow by this time. Anti-NP specific IgG, of both broad and high affinity, were significantly increased in the sera of PTPN22 KO mice compared to WT mice (Figure 3D). The significant increase in the KO of high affinity antibodies in the secondary response compared to the primary response suggests the effect of the KO increases over time.
Figure 3. Secondary T-dependent antigen responses in PTPN22 KO mice.
WT and KO mice were immunized with NP-KLH in CFA s.c. 14 days later mice were re-challenged with NP-KLH. After a further 7 days dLN were isolated and analyzed. A) TFH (CD4+ CD44hi CXCR5+ PD-1+) numbers in the dLN 7 days post re-challenge. B) shows GC B cell numbers (CD19+ FAS+ GL-7+) in the dLN of WT and KO mice. C) Plasma cell numbers (CD19lo CD138+) D) serum anti-NP IgG antibody levels 7 days post re-challenge were measured by ELISA. *P<0.05; **P<0.01 (graphs show mean ± S.E.M. and each data point represents 1 mouse)
Overall these data are consistent with increased GC activity in PTPN22 KO mice resulting in higher levels of antibody production.
Follicular regulatory cells (TFR) are not upregulated in PTPN22 KO mice
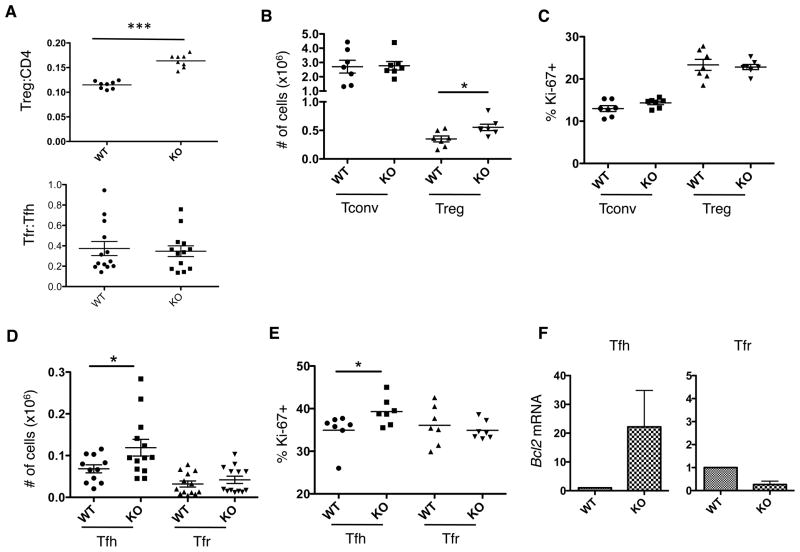
We, as well as others, have previously shown that PTPN22 deficient mice have increased numbers of Tregs (9, 14, 17). This may help compensate for the increased activity of Tconv cells. Recently, a subset of thymically derived Tregs has been described that specifically regulates the GCs (36, 37). These TFR express CXCR5, which allows them to migrate to the follicle in the same manner as TFH cells. In an attempt to reconcile the fact that PTPN22 KO mice have increased Treg but also increased GC activity, we assessed the ratio of TFR and TFH cells in dLN 7 days post-immunization (Figure 4). As previously shown, the Treg:Tconv ratio is increased in PTPN22 KO mice compared to WT mice, however the TFR:TFH ratio was found to be similar in both WT and PTPN22 KO mice (Figure 4A). The spleen in these same mice showed a reduced TFR:TFH ratio in KO mice compared to WT (supplementary figure 1). Analyzing both total cell numbers and cell proliferation (by Ki-67 staining), we found that conventional CD4 T cells (Tconv) were similar in number in both WT and KO mice whereas the Treg numbers were significantly higher in KO mice (Figure 4B). Numbers of dividing (Ki-67+) cells were similar between WT and KO mice in both the CD4+ Tconv population as well as the Treg population (Figure 4C).
Figure 4. Regulation of the Germinal Center in PTPN22 KO mice.
WT and KO mice were immunized with NP-KLH in CFA s.c. 7 days later dLN were isolated and stained for flow cytometric analysis. A) Treg:CD4 ratio and TFR:TFH ratio in the dLN. B) Absolute numbers and Ki-67 staining (C) of Tconv (CD4+ FoxP3− CXCR5− PD-1−) and Treg (CD4+ FoxP3+ CXCR5− PD-1−). D) TFH (CD4+ CXCR5+ PD-1+ FoxP3−) and TFR (CD4+ CXCR5+ PD-1+ FoxP3+) numbers were calculated by flow cytometry and Ki-67 staining is shown in (E). F) TFH and TFR were sorted from the dLN 7 days post immunization and mRNA was isolated. Bcl2 mRNA was quantified by Q-PCR (F is the result of 3 independent sorts, 4 mice per genotype were pooled per sort) *p<0.05; ***p<0.001 (graphs show mean ± S.E.M and each data point represents 1 mouse).
When analyzing the numbers of CD4+ cells within the follicular subset (CXCR5+ PD-1+), TFH numbers were found to be significantly higher and more of them are in the cell cycle (higher % of Ki-67+ cells) in the KO mouse as compared to the WT (Figure 4D and E). In contrast, the absolute number and the number of Ki-67+ TFR cells was the same in WT and KO mice (Figure 4D and E).
In addition to a proliferative advantage by PTPN22 KO TFH cells, mRNA levels of Bcl2 were higher in this population compared to WT (Figure 4F), suggesting a survival advantage. Bcl2 mRNA in TFR cells was similar in both genotypes. Analysis of further apoptosis associated markers was carried out by Q-PCR and is shown in supplementary figure 2. Amongst TFH cells, the pro-survival molecule Mcl-1 and the pro-apoptotic Bim L and Bax were unchanged between WT and KO. The pro-apoptotic Bim EL splice variant was upregulated slightly in KO TFH cells, although not to the levels observed with Bcl-2. KO TFR cells showed a slight increase in Mcl-1 transcript, but the other apoptotic markers were similarly expressed in WT and KO. Overall, due to the large increase in Bcl-2 and relatively small changes to other molecules these data suggest that PTPN22 KO TFH cells are more protected from apoptosis than WT, which could contribute to the increased numbers observed in the LN.
PTPN22 KO TFH produce more IL-21 and IL-4 than WT TFH
The disparity between Treg and TFR in the KO mouse may arise from differences in their cytokine environments. TFH cells can produce high levels of IL-21 that destabilize FoxP3 and promotes Bcl-6 transcription, the master regulator of TFH differentiation (33–35). Analysis of total mRNA from the dLN showed a significantly higher level of IL-21 transcript in KO mice compared to WT (Figure 5A). Indeed, IL-21+ TFH cells were present in greater numbers (Figure 5B), and we observed higher IL-21 transcript levels in a sorted population of PTPN22 KO TFH cells (Figure 5C) compared to WT TFH cells. KO TFH cells also expressed more IL-4 mRNA (Figure 5D) than WT cells, a cytokine that provides B cell help. In contrast, IL-10 was similarly expressed in WT and KO TFR. To date, this is the only cytokine to be associated with their function (37) (Figure 5E).
Figure 5. Cytokine production in PTPN22 KO mice.
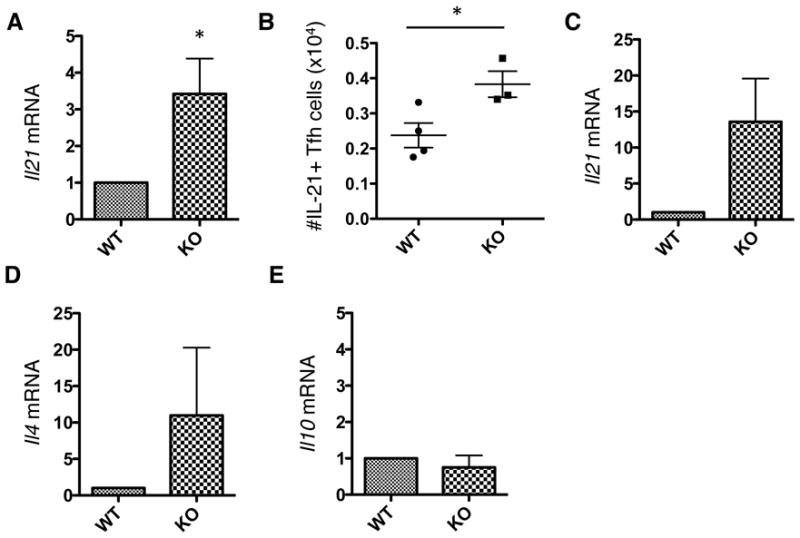
FoxP3-GFP PTPN22 WT and KO mice were immunized with NP-KLH in CFA for 7 days. Total mRNA from the dLN was extracted and the levels of IL-21 transcript were measured by Q-PCR (A). The number of IL-21+ TFH cells in the dLN 7 days post-immunization was measured by intracellular cytokine staining (B). Cells from the dLN were FACS sorted into TFH (CD4+ CXCR5+ PD-1+ FoxP3−) and TFR (CD4+ CXCR5+ PD-1+ FoxP3+) populations. mRNA was purified and cytokines were measured by Q-PCR. TFH mRNA levels of IL-21 (C) and IL-4 (D) were measured as well as IL-10 (E) mRNA levels in TFR cells. All Q-PCR data shown is normalized to WT levels. Panel A is a combination of n=3 mice per group. Panel B is representative of 3 independent experiments (each data point represents 1 mouse). Panels C, D and E are the result of 3 independent sorts combined with a total of 4 LN pooled per sort, per genotype. (graphs show mean ± S.E.M).
PTPN22 KO CD4 T cells provide increased help to B cells
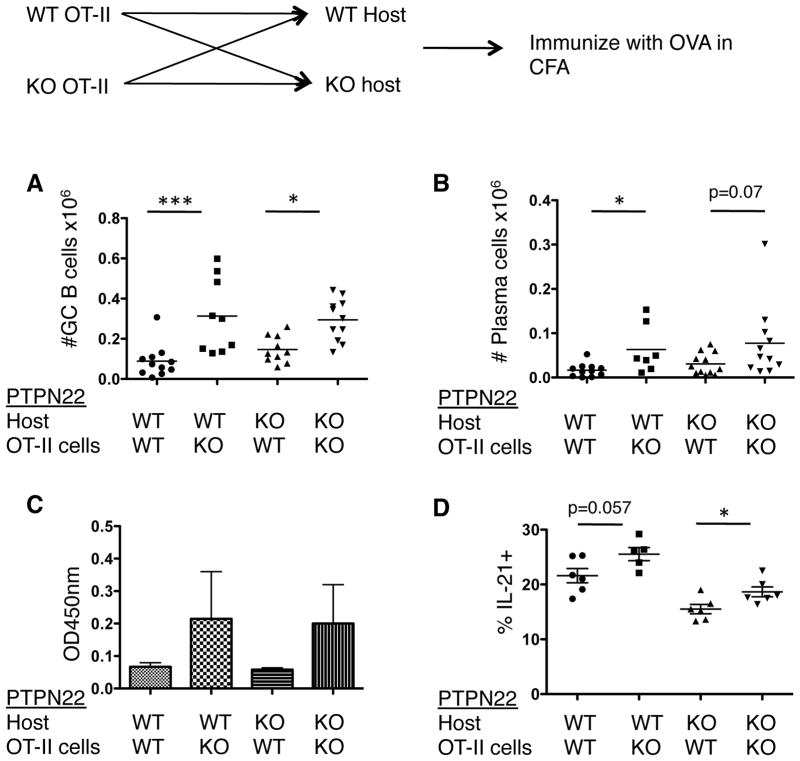
In order to further dissect the cellular basis for how PTPN22 affects the GCs, we utilized a TCR transgenic adoptive transfer model (Figure 6). Thy 1.1 labeled PTPN22 WT or KO OT-II CD4 T cells were purified and transferred into either WT or KO hosts. Two days later these mice were immunized with ovalbumin in CFA and the dLN and sera were collected at various timepoints. The numbers of GC B cells in the dLN were measured by flow cytometry at 11 days post-immunization (Figure 6A). In the presence of PTPN22 KO OT-II cells, GC B cell numbers were significantly higher than in the presence of WT OT-II cells, regardless of host genotype. In a similar manner, plasma cell numbers were increased in the presence of PTPN22 KO OT-II cells. This difference is significant in the WT host but not in the KO host (Figure 6B). This was also reflected in the level of anti-OVA IgG1 in the sera at day 14 (Figure 6C). In addition, there was a small increase in the percentage of IL-21+ OT-II T cells in the mice that received the PTPN22 KO OT-II cells as compared to WT OT-II T cells (Figure 6D). This was significant in the case of the KO host but did not reach statistical significance in the WT host. Overall, these data suggest that PTPN22 deficiency in CD4 T cells is sufficient to provide improved help to B cells.
Figure 6. PTPN22 KO CD4 T cells can provide improved help to B cells.
Thy 1.1+ OT-II+ PTPN22 WT and KO CD4+ T cells were isolated by MACS and (2×106) injected into WT or KO hosts. Mice were immunized with ovalbumin in CFA s.c. and the dLN and sera was analyzed 14 days later. A) Number of GC B cells (CD19+ FAS+ GL-7+). B) number of plasma cells (CD19lo CD138+). C) Anti-OVA IgG1 in the sera measured by ELISA. D) OT-II IL-21+ cells in the dLN 7 days post immunization as detected by intracellular cytokine staining. *p<0.05; **p<0.01 (graphs show mean. Error bars = S.E.M and each data point in panels A, B and C represent 1 mouse).
Increased GC activity in PTPN22 KO mice is influenced by T cells rather than B cells
The OT-II transfer model allowed us to examine how the loss of PTPN22 in the T cell would affect the GC, however we also wished to measure a polyclonal response by a conventional repertoire with a normal ratio of TFR:TFH. To this end, we used sub-lethally irradiated PTPN22 WT hosts and transferred CD4+ T cells and B cells purified from either PTPN22 WT or KO mice (Figure 7). Two days later mice were immunized with NP-KLH and the B cell response was analyzed 14 days later.
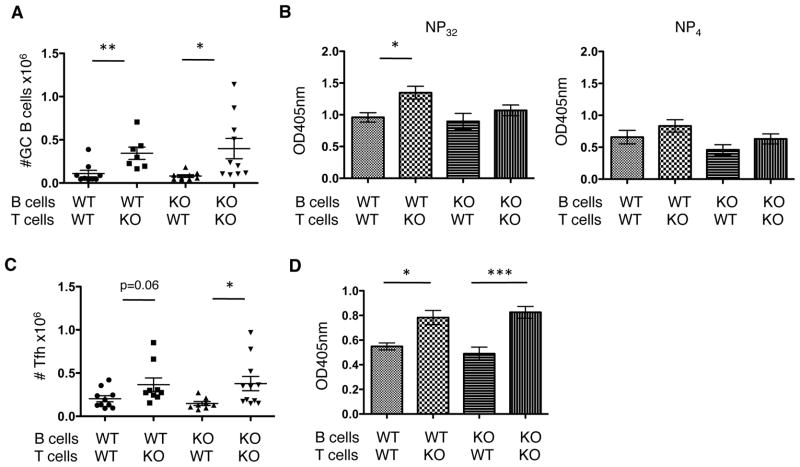
Figure 7. T cell PTPN22 genotype is dominant in controlling B cell responses.
WT and KO CD4+ T cells (5×106) and B cells (1×107) were isolated by MACS and injected into irradiated WT host mice. Mice were then immunized with NP-KLH in CFA s.c. and the sera and spleens were analyzed 14 days later. X-axis labels denote the PTPN22 genotype of the cells. A) shows GC B cell (CD19+ FAS+ GL-7+) numbers. B) shows sera anti-NP IgG of broad affinity (NP32) and high affinity (NP4) (n=5 mice per group). TFH cell numbers were counted in the spleens of mice (C). D) PTPN22 KO and WT mice were immunized with NP-KLH s.c. 7 days later dLN were removed and CD4 T cells were isolated and cultured with B cells isolated from either WT or KO spleens (unimmunized) for a further 8 days in vitro. NP-specific IgG was measured by ELISA (n=6 mice per group).*p<0.05; **p<0.01; ***p<0.001 (graphs show mean ± S.E.M. Data points in A and C represent 1 mouse).
GC B cells numbers and NP-specific antibodies titers (Figure 7A and B) were dependent on T cell rather than B cell genotype as the numbers of TFH in mice that received PTPN22 KO CD4 T cells were increased compared to those that received WT CD4 T cells (Figure 7C). Looking specifically at anti-NP titers there are significant increases in broad affinity (NP32) antibody if the mice received KO CD4 T cells compared to WT T cells. Similarly there are trends towards higher levels of high affinity antibody (NP4) in mice that received KO CD4 T cells compared to WT although not to significant levels. This may reflect the fact that the assay is measuring the primary response. As previously described, high affinity antibody levels were significantly increased in secondary responses in KO mice (Figure 3) whereas there was no statistical significance in the increase observed in the primary response of KO mice compared to WT (Figure 2). Overall the data in figure 7 show GC activity was increased in mice with PTPN22 KO CD4 T cells.
In addition to this in vivo model we utilized an in vitro approach to determine the influence of PTPN22 on T and B cells by adapting a model described by Yusuf et al. (30) (Figure 7D). WT and KO mice were immunized with NP-KLH in CFA and after 7 days the LN were removed and CD4 cells isolated. The purified cells were co-cultured with B cells from either WT or KO mice for a further 8 days and anti-NP IgG in the supernatant was measured. The presence of KO CD4 T cells significantly increased anti-NP IgG production regardless of the genotype of the B cells. This further supports the conclusion that T cell help in PTPN22 KO mice is increased compared to WT mice and is responsible for increased Ig.
KBxN arthritis severity is increased in PTPN22 deficient mice
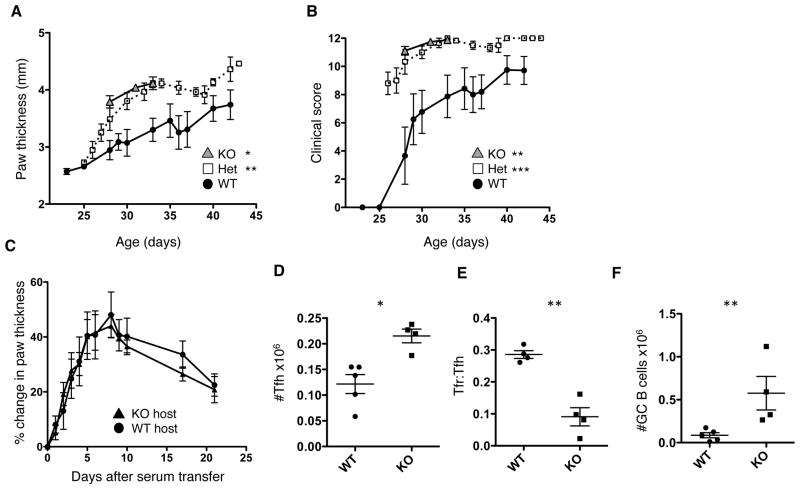
Mice expressing the KRN TCR and carrying the Ag7 MHC molecule (KBxN) develop severe arthritis. This arthritis is characterized by auto-antibodies against glucose-6-phosphate isomerase (GPI) (42). PTPN22 KO, Het and WT KBxN mice were monitored for arthritis by paw thickness measurements (Figure 8A) and clinical scoring (Figure 8B). PTPN22 deficiency significantly increased the severity of disease and PTPN22 KO KBxN mice either died early (around 32 days of age) or were euthanized due to complications as a result of severe arthritis (e.g. weight loss). Serum from KBxN mice can transfer disease to B6 mice (42). To assess the role of PTPN22 deficiency in events downstream of antibody production, serum from PTPN22 WT KBxN mice was transferred into PTPN22 WT and KO B6 mice (Figure 8C). There was no difference in either the incidence or severity of disease between these hosts, indicating that PTPN22 was not influencing events downstream of autoantibody production.
Figure 8. PTPN22 deficiency increases severity of KBxN arthritis.
PTPN22 KO, Het and WT KBxN mice were monitored for signs for disease onset from around 21 days of age by paw thickness measurements (A) and clinical score (B) (WT n=10 Het n=11 KO n=9). PTPN22 KO KBxN mice either died or were euthanized by 32 days due to severe disease. C) shows change in paw thickness following serum transfer from PTPN22 WT KBxN mice into PTPN22 WT or KO B6 hosts (WT n=4 KO n=5). Spleens were collected from PTPN22 WT and KO KBxN at 28 days for age and stained for TFH cells (D), TFR (E) and GC B cells (F). *p<0.05; **p<0.01; ***p<0.001 (graphs show mean ± S.E.M and each data point in panel D, E and F represent 1 mouse).
At 28 days of age, spleens were collected from PTPN22 KO and WT KBxN mice and analyzed for numbers of TFH, TFR and GC B cells. TFH cell numbers were significantly higher in PTPN22 KO mice compared to WT (Figure 8D) and the TFR:TFH ratio was also decreased (Figure 8E). GC B cell frequency was significantly increased in the spleens of PTPN22 KO KBxN mice compared to WT (Figure 8F). Taken together these results indicate that PTPN22 deficiency in the induction phase of KBxN arthritis, but not the later effector stage, can increase disease severity. This was associated with a greater expansion of TFH cells at a young age in PTPN22 KO KBxN mice compared to WT KBxN mice.
Discussion
Although autoimmunity is not associated with deficiency of PTPN22, there is a consistent increase in GC activity and serum levels of immunoglobulin in both PTPN22 KO and mice expressing the susceptibility allele. This study was undertaken to identify the cellular and molecular events by which PTPN22 contributes to GC activity and antibody production. PTPN22 KO mice show increased GC formation and accumulation of follicular B cells, however there appears to be little difference in the intrinsic signaling properties of B cells in the KO mouse compared to WT (Figure 1) (13). Through investigation of key germinal center cell types we found that the increased GC activity and antibody levels in PTPN22 KO mouse can largely be attributed to the T cell compartment. Following immunization, we observed significantly greater expansion of TFH cells in KO compared to WT mice. There were more Ki-67+ TFH cells and they expressed higher transcript levels of the pro-survival molecule, Bcl-2, in the KO than in the WT draining lymph node. Through adoptive transfer experiments of CD4 T cells we demonstrated that KO CD4 T cells, which include the TFH population, could provide improved help to B cells regardless of the B cell genotype. One possible reason for this improved help, aside from a larger expansion of this population, is that KO TFH cells produce higher levels of IL-21 compared to WT TFH cells. This cytokine is important in 2 ways, firstly it provides help to GC B cells allowing class switching and differentiation into plasma cells (27, 28) and secondly, in the environment of the GC, we hypothesize that this may have an inhibitory/destabilizing effect on TFR.
TFR cells are a recently described subset of thymically derived Tregs (36, 37). These cells express CXCR5 and PD-1, allowing them to migrate to the GC, and are important in controlling GC reactions. PTPN22 KO mice have increased numbers of Tregs in the periphery compared to WT mice, however in spite of their increased numbers we have shown that GC reactions are amplified suggesting lack of control specifically in the GC. Analysis of the TFR:TFH ratio in the dLN following immunization showed that PTPN22 KO and WT mice have similar (Figure 2), or possibly lower (supplementary figure 1) ratios of TFR:TFH. This is in contrast to the conventional Treg compartment, which is expanded in the KO mouse. There are significantly more Ki-67+ KO TFH cells than their WT counterparts, however, this is not the case in the TFR population and as such the compensatory increase in Tregs that occurs elsewhere in the KO mouse is absent in the GC. We hypothesize that this is due to the higher level of IL-21 in the lymph node, produced by overactive and greater numbers of TFH cells in the PTPN22 KO. IL-21 is inhibitory to Treg function and stability as it signals downstream through STAT3 rather than STAT5, resulting in FoxP3 instability (43–45). Due to the localized production of IL-21 by TFH cells in the follicle, this does not affect the stability of the conventional Treg population that is located in the T cell zone of the LN. Overall, TFH cells in the GCs of PTPN22 KO mice overwhelm the TFR cells and the balance is shifted in favor of B cell activation.
This unique alteration in the balance of Treg:Teff in different locations could possibly explain the association of certain diseases with PTPN22. A predominantly Th1/Th17 driven disease such as MS has no association with PTPN22, and in a mouse model of EAE the increased Treg:CD4 ratio was sufficient to protect the KO mice from disease (4, 17). However in an antibody-mediated disease, loss of PTPN22 may not prevent disease despite the overall increase in Treg numbers because the specific Treg subset that controls the GC, the TFR population, does not expand in the same manner.
The effect of PTPN22 on the GC has a clear effect on an autoimmune prone background. Disease in the KBxN model has been associated with many of the changes we find associated with PTPN22 deficiency. Loss of Tregs in KBxN scurfy mice leads to an increase in TFH cells and disease (46). Deletion of the IL-21 receptor on the KBxN background resulted in mice refractory to disease (47). We show that arthritis in the KBxN model was increased with PTPN22 deficiency and associated with increased frequency of TFH cells and a lower TFR:TFH ratio. PTPN22 is important in the induction phase of this model, however not the later stages downstream of antibody production as highlighted by our serum transfer data (Figure 8C) which confirms a recent report by Wang et al. (48)
The cell transfer studies presented in this paper provide the first clear evidence that a deficiency in PTPN22 in T cells can provide increased help to B cells regardless of B cell genotype. As previously suggested by Hasegawa et al.(13) we find that the increased numbers of GCs observed B6 PTPN22 KO mouse models is directly related to T cell hyperactivity rather than aberrations in B cell function. TFH differentiation is thought to be a multi-step process that involves firstly interaction with antigen presenting dendritic cells to induce Bcl-6 expression and then further interaction with B cells in the follicle to stabilize this phenotype (22). As the B cell genotype had no effect in the cell transfer models, we hypothesize that the difference between WT and KO TFH cells is initiated at the time of DC priming of the T cell. It has been reported that strength of signal between the T cell and DC can influence the differentiation of a T cell to the TFH lineage, with stronger interactions favoring TFH commitment and IL-21 production (29). It is known that in the absence of PTPN22 or in PTPN22R619W knock-in mice, proximal TCR signaling is increased, particularly in the CD44hi compartment (10, 12–14). We show PTPN22 KO T cells differentiate more readily into TFH compared to WT T cells and produce more IL-21 (figures 2, 3 and 5). In addition, monoclonal PTPN22 KO OT-II CD4+ T cells produce more IL-21 than WT OT-II cells, despite having equal affinity TCRs (figure 6). The strength of signal in the case of PTPN22 KO OT-II cells would be higher than WT OT-II cells. Once committed, further TCR signals received by the KO TFH will result in greater proliferation and cytokine production, much in the same way that has been reported for Teff/mem cells in this KO mouse (13, 14). Combined with a lack of TFR expansion to match the increased activity of the TFH cells in the KO mouse, this leads to greater GC activity.
The exact mechanism by which the R620W allele of Lyp enhances autoimmune diseases is not certain. Reports have argued a gain of function (6–9) as well as a loss of function in lymphocytes (11). Recently two mouse studies using the R619W PEP knock-in mice have reported conflicting hypotheses as to the impact of this allele (10, 12). The most recent of which demonstrates that the former report, suggesting R619W PEP is susceptible to increased protein degradation, was possibly an artifact of the antibody used to detect this protein. Nevertheless both studies show a phenotype closely resembling the PTPN22 KO mouse. Our study suggests that loss of PTPN22 results in increased GC activity, a phenotype which would expect to lead to an increase in disease in an antibody mediated autoimmune condition driven by other genetic loci, arguing that the R620W allele associated with disease would result in a loss of function. However, as hypothesized by Dai et al. the introduction of the R620W allele could lead to an alteration of substrate specificity for the protein and as such lead to a loss of function with some substrates and a gain of function with others. The “loss of function” forced through knockout of PTPN22 in the mice in this study would suggest that this affect is sufficient to trigger increased GC activity. However in the presence of the R620W allele this phenotype may be more subtle and the outcome could be influenced by other disease susceptibility loci which may also result in the loss of B cell tolerance through endogenous effects (10, 12).
Supplementary Material
Acknowledgments
These experiments were funded by NIH grants A1070351-05S1 and AI50864.
We would like to thank Dr. Kristin Hogquist for providing the Nur-77 GFP mice and Dr. Kerri Mowen for providing KRN mice and serum. We also thank Dr. David Nemazee for helpful discussions. This is manuscript #25021 from The Scripps Research Institute.
Footnotes
Conflict of Interest: The authors declare that no conflict of interest exists
References
- 1.Bottini N, Musumeci L, Alonso A, Rahmouni S, Nika K, Rostamkhani M, MacMurray J, Meloni GF, Lucarelli P, Pellecchia M, Eisenbarth GS, Comings D, Mustelin T. A functional variant of lymphoid tyrosine phosphatase is associated with type I diabetes. Nat Genet. 2004;36:337–338. doi: 10.1038/ng1323. [DOI] [PubMed] [Google Scholar]
- 2.Criswell LA, Pfeiffer KA, Lum RF, Gonzales B, Novitzke J, Kern M, Moser KL, Begovich AB, Carlton VE, Li W, Lee AT, Ortmann W, Behrens TW, Gregersen PK. Analysis of families in the multiple autoimmune disease genetics consortium (MADGC) collection: the PTPN22 620W allele associates with multiple autoimmune phenotypes. Am J Hum Genet. 2005;76:561–571. doi: 10.1086/429096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Todd JA, Walker NM, Cooper JD, Smyth DJ, Downes K, Plagnol V, Bailey R, Nejentsev S, Field SF, Payne F, Lowe CE, Szeszko JS, Hafler JP, Zeitels L, Yang JH, Vella A, Nutland S, Stevens HE, Schuilenburg H, Coleman G, Maisuria M, Meadows W, Smink LJ, Healy B, Burren OS, Lam AA, Ovington NR, Allen J, Adlem E, Leung HT, Wallace C, Howson JM, Guja C, Ionescu-Tirgoviste C, Simmonds MJ, Heward JM, Gough SC, Dunger DB, Wicker LS, Clayton DG. Robust associations of four new chromosome regions from genome-wide analyses of type 1 diabetes. Nat Genet. 2007;39:857–864. doi: 10.1038/ng2068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.De Jager PL, Sawcer S, Waliszewska A, Farwell L, Wild G, Cohen A, Langelier D, Bitton A, Compston A, Hafler DA, Rioux JD. Evaluating the role of the 620W allele of protein tyrosine phosphatase PTPN22 in Crohn’s disease and multiple sclerosis. Eur J Hum Genet. 2006;14:317–321. doi: 10.1038/sj.ejhg.5201548. [DOI] [PubMed] [Google Scholar]
- 5.Franke A, McGovern DP, Barrett JC, Wang K, Radford-Smith GL, Ahmad T, Lees CW, Balschun T, Lee J, Roberts R, Anderson CA, Bis JC, Bumpstead S, Ellinghaus D, Festen EM, Georges M, Green T, Haritunians T, Jostins L, Latiano A, Mathew CG, Montgomery GW, Prescott NJ, Raychaudhuri S, Rotter JI, Schumm P, Sharma Y, Simms LA, Taylor KD, Whiteman D, Wijmenga C, Baldassano RN, Barclay M, Bayless TM, Brand S, Buning C, Cohen A, Colombel JF, Cottone M, Stronati L, Denson T, De Vos M, D’Inca R, Dubinsky M, Edwards C, Florin T, Franchimont D, Gearry R, Glas J, Van Gossum A, Guthery SL, Halfvarson J, Verspaget HW, Hugot JP, Karban A, Laukens D, Lawrance I, Lemann M, Levine A, Libioulle C, Louis E, Mowat C, Newman W, Panes J, Phillips A, Proctor DD, Regueiro M, Russell R, Rutgeerts P, Sanderson J, Sans M, Seibold F, Steinhart AH, Stokkers PC, Torkvist L, Kullak-Ublick G, Wilson D, Walters T, Targan SR, Brant SR, Rioux JD, D’Amato M, Weersma RK, Kugathasan S, Griffiths AM, Mansfield JC, Vermeire S, Duerr RH, Silverberg MS, Satsangi J, Schreiber S, Cho JH, Annese V, Hakonarson H, Daly MJ, Parkes M. Genome-wide meta-analysis increases to 71 the number of confirmed Crohn’s disease susceptibility loci. Nat Genet. 2010;42:1118–1125. doi: 10.1038/ng.717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fiorillo E, Orru V, Stanford SM, Liu Y, Salek M, Rapini N, Schenone AD, Saccucci P, Delogu LG, Angelini F, Manca Bitti ML, Schmedt C, Chan AC, Acuto O, Bottini N. Autoimmune-associated PTPN22 R620W variation reduces phosphorylation of lymphoid phosphatase on an inhibitory tyrosine residue. J Biol Chem. 2010;285:26506–26518. doi: 10.1074/jbc.M110.111104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rieck M, Arechiga A, Onengut-Gumuscu S, Greenbaum C, Concannon P, Buckner JH. Genetic variation in PTPN22 corresponds to altered function of T and B lymphocytes. J Immunol. 2007;179:4704–4710. doi: 10.4049/jimmunol.179.7.4704. [DOI] [PubMed] [Google Scholar]
- 8.Vang T, Congia M, Macis MD, Musumeci L, Orru V, Zavattari P, Nika K, Tautz L, Tasken K, Cucca F, Mustelin T, Bottini N. Autoimmune-associated lymphoid tyrosine phosphatase is a gain-of-function variant. Nat Genet. 2005;37:1317–1319. doi: 10.1038/ng1673. [DOI] [PubMed] [Google Scholar]
- 9.Zheng P, Kissler S. PTPN22 silencing in the NOD model indicates the type 1 diabetes-associated allele is not a loss-of-function variant. Diabetes. 2013;62:896–904. doi: 10.2337/db12-0929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang J, Zahir N, Jiang Q, Miliotis H, Heyraud S, Meng X, Dong B, Xie G, Qiu F, Hao Z, McCulloch CA, Keystone EC, Peterson AC, Siminovitch KA. The autoimmune disease-associated PTPN22 variant promotes calpain-mediated Lyp/Pep degradation associated with lymphocyte and dendritic cell hyperresponsiveness. Nat Genet. 2011;43:902–907. doi: 10.1038/ng.904. [DOI] [PubMed] [Google Scholar]
- 11.Zikherman J, Hermiston M, Steiner D, Hasegawa K, Chan A, Weiss A. PTPN22 deficiency cooperates with the CD45 E613R allele to break tolerance on a non-autoimmune background. J Immunol. 2009;182:4093–4106. doi: 10.4049/jimmunol.0803317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dai X, James RG, Habib T, Singh S, Jackson S, Khim S, Moon RT, Liggitt D, Wolf-Yadlin A, Buckner JH, Rawlings DJ. A disease-associated PTPN22 variant promotes systemic autoimmunity in murine models. J Clin Invest. 2013;123:2024–2036. doi: 10.1172/JCI66963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hasegawa K, Martin F, Huang G, Tumas D, Diehl L, Chan AC. PEST domain-enriched tyrosine phosphatase (PEP) regulation of effector/memory T cells. Science. 2004;303:685–689. doi: 10.1126/science.1092138. [DOI] [PubMed] [Google Scholar]
- 14.Brownlie RJ, Miosge LA, Vassilakos D, Svensson LM, Cope A, Zamoyska R. Lack of the phosphatase PTPN22 increases adhesion of murine regulatory T cells to improve their immunosuppressive function. Sci Signal. 2012;5:ra87. doi: 10.1126/scisignal.2003365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cloutier JF, Veillette A. Cooperative inhibition of T-cell antigen receptor signaling by a complex between a kinase and a phosphatase. J Exp Med. 1999;189:111–121. doi: 10.1084/jem.189.1.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wu J, Katrekar A, Honigberg LA, Smith AM, Conn MT, Tang J, Jeffery D, Mortara K, Sampang J, Williams SR, Buggy J, Clark JM. Identification of substrates of human protein-tyrosine phosphatase PTPN22. J Biol Chem. 2006;281:11002–11010. doi: 10.1074/jbc.M600498200. [DOI] [PubMed] [Google Scholar]
- 17.Maine CJ, Hamilton-Williams EE, Cheung J, Stanford SM, Bottini N, Wicker LS, Sherman LA. PTPN22 Alters the Development of Regulatory T Cells in the Thymus. J Immunol. 2012;188:5267–5275. doi: 10.4049/jimmunol.1200150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Heng TS, Painter MW. The Immunological Genome Project: networks of gene expression in immune cells. Nat Immunol. 2008;9:1091–1094. doi: 10.1038/ni1008-1091. [DOI] [PubMed] [Google Scholar]
- 19.Arechiga AF, Habib T, He Y, Zhang X, Zhang ZY, Funk A, Buckner JH. Cutting edge: the PTPN22 allelic variant associated with autoimmunity impairs B cell signaling. J Immunol. 2009;182:3343–3347. doi: 10.4049/jimmunol.0713370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Habib T, Funk A, Rieck M, Brahmandam A, Dai X, Panigrahi AK, Luning Prak ET, Meyer-Bahlburg A, Sanda S, Greenbaum C, Rawlings DJ, Buckner JH. Altered B cell homeostasis is associated with type I diabetes and carriers of the PTPN22 allelic variant. J Immunol. 2012;188:487–496. doi: 10.4049/jimmunol.1102176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Menard L, Saadoun D, Isnardi I, Ng YS, Meyers G, Massad C, Price C, Abraham C, Motaghedi R, Buckner JH, Gregersen PK, Meffre E. The PTPN22 allele encoding an R620W variant interferes with the removal of developing autoreactive B cells in humans. J Clin Invest. 2011;121:3635–3644. doi: 10.1172/JCI45790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Crotty S. Follicular helper CD4 T cells (TFH) Annu Rev Immunol. 2011;29:621–663. doi: 10.1146/annurev-immunol-031210-101400. [DOI] [PubMed] [Google Scholar]
- 23.Breitfeld D, Ohl L, Kremmer E, Ellwart J, Sallusto F, Lipp M, Forster R. Follicular B helper T cells express CXC chemokine receptor 5, localize to B cell follicles, and support immunoglobulin production. J Exp Med. 2000;192:1545–1552. doi: 10.1084/jem.192.11.1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Forster R, Mattis AE, Kremmer E, Wolf E, Brem G, Lipp M. A putative chemokine receptor, BLR1, directs B cell migration to defined lymphoid organs and specific anatomic compartments of the spleen. Cell. 1996;87:1037–1047. doi: 10.1016/s0092-8674(00)81798-5. [DOI] [PubMed] [Google Scholar]
- 25.Kim CH, Rott LS, Clark-Lewis I, Campbell DJ, Wu L, Butcher EC. Subspecialization of CXCR5+ T cells: B helper activity is focused in a germinal center-localized subset of CXCR5+ T cells. J Exp Med. 2001;193:1373–1381. doi: 10.1084/jem.193.12.1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schaerli P, Willimann K, Lang AB, Lipp M, Loetscher P, Moser B. CXC chemokine receptor 5 expression defines follicular homing T cells with B cell helper function. J Exp Med. 2000;192:1553–1562. doi: 10.1084/jem.192.11.1553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Linterman MA, Beaton L, Yu D, Ramiscal RR, Srivastava M, Hogan JJ, Verma NK, Smyth MJ, Rigby RJ, Vinuesa CG. IL-21 acts directly on B cells to regulate Bcl-6 expression and germinal center responses. J Exp Med. 2010;207:353–363. doi: 10.1084/jem.20091738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nurieva RI, Chung Y, Hwang D, Yang XO, Kang HS, Ma L, Wang YH, Watowich SS, Jetten AM, Tian Q, Dong C. Generation of T follicular helper cells is mediated by interleukin-21 but independent of T helper 1, 2, or 17 cell lineages. Immunity. 2008;29:138–149. doi: 10.1016/j.immuni.2008.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fazilleau N, McHeyzer-Williams LJ, Rosen H, McHeyzer-Williams MG. The function of follicular helper T cells is regulated by the strength of T cell antigen receptor binding. Nat Immunol. 2009;10:375–384. doi: 10.1038/ni.1704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yusuf I, Kageyama R, Monticelli L, Johnston RJ, Ditoro D, Hansen K, Barnett B, Crotty S. Germinal center T follicular helper cell IL-4 production is dependent on signaling lymphocytic activation molecule receptor (CD150) J Immunol. 2010;185:190–202. doi: 10.4049/jimmunol.0903505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Foy TM, Laman JD, Ledbetter JA, Aruffo A, Claassen E, Noelle RJ. gp39-CD40 interactions are essential for germinal center formation and the development of B cell memory. J Exp Med. 1994;180:157–163. doi: 10.1084/jem.180.1.157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Han S, Hathcock K, Zheng B, Kepler TB, Hodes R, Kelsoe G. Cellular interaction in germinal centers. Roles of CD40 ligand and B7-2 in established germinal centers. J Immunol. 1995;155:556–567. [PubMed] [Google Scholar]
- 33.Johnston RJ, Poholek AC, DiToro D, Yusuf I, Eto D, Barnett B, Dent AL, Craft J, Crotty S. Bcl6 and Blimp-1 are reciprocal and antagonistic regulators of T follicular helper cell differentiation. Science. 2009;325:1006–1010. doi: 10.1126/science.1175870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nurieva RI, Chung Y, Martinez GJ, Yang XO, Tanaka S, Matskevitch TD, Wang YH, Dong C. Bcl6 mediates the development of T follicular helper cells. Science. 2009;325:1001–1005. doi: 10.1126/science.1176676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yu D, Rao S, Tsai LM, Lee SK, He Y, Sutcliffe EL, Srivastava M, Linterman M, Zheng L, Simpson N, Ellyard JI, Parish IA, Ma CS, Li QJ, Parish CR, Mackay CR, Vinuesa CG. The transcriptional repressor Bcl-6 directs T follicular helper cell lineage commitment. Immunity. 2009;31:457–468. doi: 10.1016/j.immuni.2009.07.002. [DOI] [PubMed] [Google Scholar]
- 36.Chung Y, Tanaka S, Chu F, Nurieva RI, Martinez GJ, Rawal S, Wang YH, Lim H, Reynolds JM, Zhou XH, Fan HM, Liu ZM, Neelapu SS, Dong C. Follicular regulatory T cells expressing Foxp3 and Bcl-6 suppress germinal center reactions. Nat Med. 2011;17:983–988. doi: 10.1038/nm.2426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Linterman MA, Pierson W, Lee SK, Kallies A, Kawamoto S, Rayner TF, Srivastava M, Divekar DP, Beaton L, Hogan JJ, Fagarasan S, Liston A, Smith KG, Vinuesa CG. Foxp3+ follicular regulatory T cells control the germinal center response. Nat Med. 2011;17:975–982. doi: 10.1038/nm.2425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sage PT, Francisco LM, Carman CV, Sharpe AH. The receptor PD-1 controls follicular regulatory T cells in the lymph nodes and blood. Nat Immunol. 2013;14:152–161. doi: 10.1038/ni.2496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Moran AE, Holzapfel KL, Xing Y, Cunningham NR, Maltzman JS, Punt J, Hogquist KA. T cell receptor signal strength in Treg and iNKT cell development demonstrated by a novel fluorescent reporter mouse. J Exp Med. 2011;208:1279–1289. doi: 10.1084/jem.20110308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang J, Fathman JW, Lugo-Villarino G, Scimone L, von Andrian U, Dorfman DM, Glimcher LH. Transcription factor T-bet regulates inflammatory arthritis through its function in dendritic cells. J Clin Invest. 2006;116:414–421. doi: 10.1172/JCI26631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zikherman J, Parameswaran R, Weiss A. Endogenous antigen tunes the responsiveness of naive B cells but not T cells. Nature. 2012;489:160–164. doi: 10.1038/nature11311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Monach PA, Mathis D, Benoist C. The K/BxN arthritis model. Curr Protoc Immunol. 2008;Chapter 15(Unit 15):22. doi: 10.1002/0471142735.im1522s81. [DOI] [PubMed] [Google Scholar]
- 43.Attridge K, Wang CJ, Wardzinski L, Kenefeck R, Chamberlain JL, Manzotti C, Kopf M, Walker LS. IL-21 inhibits T cell IL-2 production and impairs Treg homeostasis. Blood. 2012;119:4656–4664. doi: 10.1182/blood-2011-10-388546. [DOI] [PubMed] [Google Scholar]
- 44.Clough LE, Wang CJ, Schmidt EM, Booth G, Hou TZ, Ryan GA, Walker LS. Release from regulatory T cell-mediated suppression during the onset of tissue-specific autoimmunity is associated with elevated IL-21. J Immunol. 2008;180:5393–5401. doi: 10.4049/jimmunol.180.8.5393. [DOI] [PubMed] [Google Scholar]
- 45.Peluso I, Fantini MC, Fina D, Caruso R, Boirivant M, MacDonald TT, Pallone F, Monteleone G. IL-21 counteracts the regulatory T cell-mediated suppression of human CD4+ T lymphocytes. J Immunol. 2007;178:732–739. doi: 10.4049/jimmunol.178.2.732. [DOI] [PubMed] [Google Scholar]
- 46.Jang E, Cho WS, Cho ML, Park HJ, Oh HJ, Kang SM, Paik DJ, Youn J. Foxp3+ regulatory T cells control humoral autoimmunity by suppressing the development of long-lived plasma cells. J Immunol. 2011;186:1546–1553. doi: 10.4049/jimmunol.1002942. [DOI] [PubMed] [Google Scholar]
- 47.Jang E, Cho SH, Park H, Paik DJ, Kim JM, Youn J. A positive feedback loop of IL-21 signaling provoked by homeostatic CD4+CD25− T cell expansion is essential for the development of arthritis in autoimmune K/BxN mice. J Immunol. 2009;182:4649–4656. doi: 10.4049/jimmunol.0804350. [DOI] [PubMed] [Google Scholar]
- 48.Wang Y, Shaked I, Stanford SM, Zhou W, Curtsinger JM, Mikulski Z, Shaheen ZR, Cheng G, Sawatzke K, Campbell AM, Auger JL, Bilgic H, Shoyama FM, Schmeling DO, Balfour HH, Jr, Hasegawa K, Chan AC, Corbett JA, Binstadt BA, Mescher MF, Ley K, Bottini N, Peterson EJ. The Autoimmunity-Associated Gene PTPN22 Potentiates Toll-like Receptor-Driven, Type 1 Interferon-Dependent Immunity. Immunity. 2013 doi: 10.1016/j.immuni.2013.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.