Abstract
C-C Chemokine receptor five knockout (Ccr5-/-) mice develop fewer experimental pulmonary metastases than wild type (WT) mice. This phenomenon was explored by applying gene-expression profiling to the lungs of mice with these metastases. Consequently, Erythroid Differentiation Regulator 1 (Erdr1) was identified as upregulated in the WT mice. Though commonly associated with bone marrow stroma, Erdr1 was differentially expressed in WT pulmonary mesenchymal cells (PMCs) and murine embryonic fibroblasts (MEFs). Moreover, the Ccr5 ligand Ccl4 increased its expression by 3.36 ± 0.14 fold. Ccr5 signaling was dependent on the Map2k but not the Pi3k pathway since treatment with U0126 inhibited upregulation of Erdr1 but treatment with LY294002 increased the expression by 3.44 ± 0.92 fold (p < 0.05). Erdr1's effect on B16-F10 melanoma metastasis was verified by the adoptive transfer of WT MEFs into Ccr5-/- mice. In this model, MEFs that had been transduced with Erdr1 shRNA lowered metastasis by 33% compared to control transduced MEFs. The relevance of ERDR1 on human disease was assessed by co-culturing chronic lymphocytic leukemia (CLL) cells with M2-10B4 stromal cells that had been transfected with shRNA or control plasmids. After 96 hours of co-culture, the cell counts were higher with control cell lines compared with Erdr1 knockdown lines (OR 1.88 ± 0.27, 2.52 ± 0.66 respectively). This increase was associated with a decrease in apoptotic cells (OR 0.69 ± 0.18, 0.58 ± 0.12 respectively).
Implications
Therefore, ERDR1 is a stromal-derived factor that promotes cancer cell survival in vitro and in an experimental metastasis model.
Keywords: ERDR1, chemokines, tumor stroma, metastasis
Introduction
Over the past ten years, evidence has emerged highlighting the importance of stromal cells in cancer progression (1-4). During metastasis, the associated stroma is remodeled by the activation of fibroblasts and the recruitment of bone-marrow derived cells. While the mediators of this process continue to be elucidated, several are well established including PDGFR ligands (5), MMP-9 (6), IL-6 (7), S100A4 and S100A8 (8, 9). These mediators are of clinical importance because they provide therapeutic targets, which may be critical for the treatment of chemotherapy-resistant tumors.
We have previously shown that stromal cells expressing Ccr5 contribute to the formation of a metastasis-promoting stroma. Specifically, the activation of Ccr5 on pulmonary mesenchymal cells (PMCs) leads to the induction of Mmp-9 (10). These results have been extended by others studying the Ccr5 ligand Ccl3. In addition to Mmp9, tumors in Ccr5-/- and Ccl3-/- mice produce less Hgf and accumulate fewer monocytes and T cells than tumors in WT mice (11). Ccr5 expression by the host also contributes to the survival of cancer cells by inhibiting apoptosis (12).
Given the importance of CCR5, we decided to further investigate this mechanism by comparing gene expression in WT vs. Ccr5-/- lungs after the injection of B16-F10 melanoma cells. These experiments led to our characterization of the gene erythroid differentiation regulator 1 (Erdr1) as a metastasis-promoting signal.
ERDR1 was first isolated from the myelomonocytic cell line WEHI-3B as a cytokine that induced hemoglobin synthesis (13). However, a wider biologic function was suggested by its expression in other cell types including murine fibroblasts, lymphoma cells, and human peripheral blood mononuclear cells (14). In addition to hemoglobin synthesis, ERDR1 also acts as a survival signal during periods of cellular stress. This function has been established in erythroblasts, granulocyte/monocyte progenitors, and hematopoietic stem cells (14). A greater significance of this gene is also implied by its differential expression on a number of cDNA microarrays including those analyzing brain inflammation, neurodegeneration (15-17) and development (18, 19).
In this report, we show that ERDR1-expressing stroma promotes cancer cell survival in vitro and during cancer cell invasion. Furthermore, the gene is upregulated by chemokine ligand binding to CCR5. Therefore, ERDR1 may be an innovative therapeutic target either directly or indirectly by inhibiting CCR5.
Experimental Procedures
Mice
C57BL/6J (WT) mice were purchased from Jackson Laboratories (Bar Harbor, ME). The generation of B6.129P2-Ccr5tm1Kuz/J (Ccr5-/-) mice has been described previously (20). Males and females between eight and twelve weeks were used in these experiments. Controls and experimental groups were balanced by age and gender. All animals were housed in pathogenfree conditions and all experiments were conducted using protocols approved by the Institutional Animal Care and Use Committee of the University of North Carolina at Chapel Hill.
Cells
B16-F10 melanoma, A293T, and M2-10B4 cells were purchased from American Type Culture Collection (ATCC, Rockland, MD). Upon receipt, these cells were expanded by three to four passages and were frozen into aliquots to maintain their uniformity and veracity. B16-F10 cells were additionally verified by melanin production. Aliquots were passaged twice after thawing and before use.
Isolation of pulmonary mesenchymal cells (PMCs) was performed as previously described (10). In all experiments, PMCs were used within two passages of isolation. Murine embryonic fibroblasts (MEFs) were harvested from day E 13.5 embryos as described elsewhere (21) and were transduced after two to five ex vivo passages.
All murine cell types were cultured in DMEM supplemented with 10% FBS and 1% penicillin/streptomycin (Gibco, Grand Island, NY). Human CLL cells were cultured in complete RPMI (Gibco) with 10% FBS.
Flow cytometry and cell sorting
Identification of PMC populations was accomplished by incubating the cells with anti-Thy-1.2-PECy7 (eBioscience, San Diego, CA) and anti-CD45-eFluor450 (eBioscience) antibodies for 15 minutes at room temperature. CD45+ cells were sorted by the UNC Flow Cytometry Facility. Apoptotic cells were identified by staining the cells with propidium iodide and Annexin-V- APC (eBioscience) for 15 minutes at room temperature. Fluorescence was measured on the MACSQuant Analyzer (Miltenyi, Bergisch Gladbach, Germany) and was analyzed using Summit software (Beckman Coulter Brea, CA).
In Vitro Experiments
PMCs were maintained overnight in serum-free conditions prior to chemokine treatment. The following day, 50 ng/ml of Ccl4 (Peprotech Inc., Rocky Hill, NJ) was added. mRNA was harvested 24 and 48 hours after stimulation. Intracellular signaling pathways were investigated by adding 10 μM of the MAP2K inhibitor U0129 or 20 μM of the PI3K inhibitor LY294002 (Cell Signaling Technology, Danvers, MA). Both inhibitors were reconstituted in DMSO before being diluted with media. Control wells were treated with media containing equivalent amounts of DMSO.
Apoptosis was induced with staurosporine treatment. For these experiments, MEFs were seeded onto 6-well plates 48 hours after transduction. After resting overnight, the cells were incubated with 100 nM staurosporine plus 20 μM Z-VAD-FMK (inhibitor) or Z-FA-FMK (control peptide) (Sigma-Aldrich, St. Louis, MO). Apoptosis was measured 24 hours later using annexin V and PI as described above.
In Vivo Experiments
Cell transfer experiments were performed by tail vein injection in a total volume of 200μl of PBS. 4 × 105 MEFs were injected followed by 7.5 × 105 B16-F10 cells. The mice were given 48 hours rest between injections. Fourteen days after melanoma injection, the lungs were harvested and insufflated with Fekete's solution. Lung metastatic nodules were counted by an individual blinded to the experimental group (10).
eGFP expression by cells within the lung was measured using an eGFP ELISA (Cell BioLabs, San Diego, CA). To do so, the left upper lobe of the lung was harvested after perfusing the animal with PBS. These samples were then homogenized in the presence of PBS and a protease-inhibitor cocktail (Roche, Pleasanton, CA). Supernatants were added to the ELISA plate following two centrifugation steps. The plates were then processed according to the manufacturer's instructions.
RNA Analysis
Gene array experiments were performed on the lungs of WT and Ccr5-/- mice. These lungs were harvested at 6, 24, and 48 hours after intravenous injection with 7.5 × 105 B16-F10 melanoma cells. Passenger leukocytes were removed by perfusing the mice with PBS warmed to 37o C. The samples were the flash frozen and stored in liquid nitrogen prior to processing. mRNA isolation and cDNA formation have been previously described (10). The resulting cDNA was normalized by amount, pooled by time point, and hybridized to Affymetrix GeneChip® Genome 430 Arrays (six arrays in total) (Affymetrix, Santa Clara, CA). Gene expression was analyzed using GeneSpring software (Agilent Technologies, Santa Clara, CA). All microarray data are MIAME compliant and have been deposited at GEO (GEO ID: GSE51422). Confirmation of the microarray studies was performed by semi-quantitative RT-PCR as previously described (10). (See Supplementary Experimental Procedures for primer sequences).
PMC or MEF mRNA was amplified by RT-PCR using the reagents and primers given in the Supplementary Experimental Procedures. Real-time PCR was performed using SYBR Green Master Mix (Applied Biosystems, Foster City, CA). For analysis of PMC or MEFs, Erdr1 expression was calculated relative SDHα. This housekeeping gene was chosen from eight candidate genes based on its expression compared to Erdr1 and its stability under experimental conditions. For analysis of the whole lung, expression was normalized to the amount of mRNA as measured by the Qubit fluorometer (Invitrogen, Grand Island, NY).
Cloning and Manipulation of Erdr1 expression
Full-length Erdr1 was cloned from PMC and MEF cDNA and sequenced as described in the Supplementary Experimental Procedures. For expression by lentiviral vectors, Erdr1 cDNA was cloned into a pLenti7.3 plasmid (Invitrogen) containing either the EF1α or CMV promoter (see Supplementary Experimental Procedures). Lentiviral vectors were packaged in A293T cells according to the manufacturer's instructions.
Erdr1 expression was inhibited by transducing target cells with shRNA. Candidate shRNA sequences were confirmed by the UNC Genome Analysis facility. These sequences and a scrambled control sequence were cloned into pHSPG vectors (see Supplementary Experimental Procedures) (22, 23). HSPG viral vectors were packaged as described in the Supplementary Experimental Procedures. PMCs and MEFs were transduced by spin inoculation in six-well plates with polybrene (4 μg/mL) (Sigma) and virus (MOI=5). Transduction efficiency was assessed by measuring the percentage of eGFP positive cells by flow cytometry. Knockdown efficiency was determined by real time PCR. Of the candidate sequences tested, two were chosen for this study based on the degree of Erdr1 inhibition.
Over-expression of Erdr1 was accomplished using the GeneSwitch vector system (Invitrogen). After cloning, Erdr1 and LacZ control plasmids were linearized with SAPI (NEB, Ipswich, MA). One μg of plasmid DNA was transfected using Qiagen Attractene reagent according to the manufacturer's instructions (Qiagen, Germantown, MD). After an eight hour incubation, RU486 (one μM final concentration) was added to the cells. 72 hours later, expression of Erdr1 was measured by real time PCR in an aliquot of these cells. The remainder was injected into mice as described above.
Analysis of CLL Samples
Peripheral blood samples were obtained from CLL patients through UNC's Tissue Procurement Facility. Only patients with more than 95% lymphocytes on their complete blood count differential were included; all patients were consented according to policies of UNC Institutional Review Board. Peripheral blood monocytes (PBMCs) were isolated from EDTA samples using LSM (MP Biomedicals, Santa Ana, CA) according to the manufacturer's instructions. Cells were washed five times, resuspended in 10% RPMI, and maintained at 5% CO2 and 37o overnight. On the day of collection, 104 M2-10B4 cells in 10% DMEM were added to six-well plates and were treated with two μg/ml mitomycin C (Sigma). The following day, the wells were washed twice with warm PBS and 106 PBMCs were added in 10% RPMI.
Ninety-six hours after the initiation of the co-culture, peripheral blood cells were harvested and analyzed for apoptosis with Annexin V and propidium iodide. M2-10B4 cells were excluded by their eGFP expression.
Statistics
Unless otherwise stated, data are presented as the mean of measurements taken from three or more separate experiments. Statistical error for these means is presented as ±1 SEM. p values were determined by Mann-Whitney test; values < 0.05 were considered significant. The Bonferroni correction was applied to experiments with more than two experimental groups.
Results
Erdr1 has greater expression in the lungs and PMCs of WT than Ccr5-/- mice
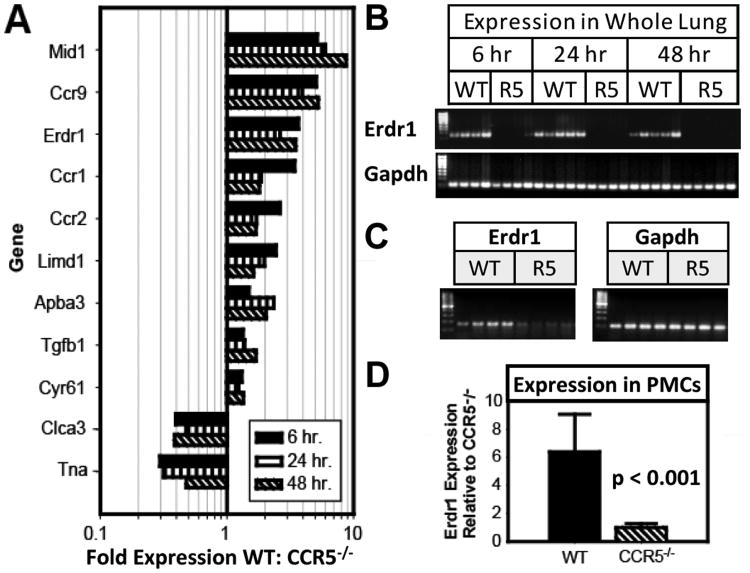
We searched for genes that might be critical for the CCR5-dependent promotion of metastasis by comparing mRNA from WT and Ccr5-/- mice that were intravenously injected with B16-F10 melanoma cells. Analysis revealed 11 candidate genes that were differentially expressed at 6, 12, and 24 hours after injection (Fig. 1A). Semi-quantitative RT-PCR was then performed on the unpooled cDNA samples using primers for each of these genes (Supplemental Fig. S1). Erdr1 expression was detected in 100% of WT mice (n=14); but in 0 (n=13) of the Ccr5-/- mice (Fig. 1B). This same pattern was also seen in control mice that did not receive tumor injections (Fig. 1C). The expression patterns for the remaining ten genes were much more heterogeneous. For example, Mid1 was detectable in 50% of the WT and 38.5% of the Ccr5-/- mice while Ccr9 was evident in 64.3% of the WT and 46.2% of the Ccr5-/- mice. As a result, we chose to further characterize the role of ERDR1 in the promotion of lung metastasis.
Figure 1. Erdr1 is upregulated in the lungs and PMCs of WT mice compared with Ccr5-/- mice.
(A) The bar graph shows relative expression of genes from WT mice compared with Ccr5-/- mice. Lungs were harvested 6, 24, and 48 hours after intravenous injection of B16-F10 melanoma cells. After mRNA extraction, cDNA was formed and pooled for application to an Affymetrix GeneChip® Genome 430 Arrays (n=27). (B) Semi-quantitative RT-PCR for Erdr1 applied to the unpooled mouse lung samples from the experiment described above. The gel shows detectable Erdr1 mRNA for each WT mouse. (C) Semi-quantitative PCR as applied to cDNA from four WT and four CCR5-/- mice without tumor injection. (D) Relative Erdr1 expression in PMCs of WT and Ccr5-/- mice by real-time PCR. Gene expression was standardized to β-actin and then was normalized to Erdr1 expression in Ccr5-/- PMCs (n=12).
Semi-quantitative RT-PCR was then performed on the unpooled cDNA samples using primers for each of these genes (Supplemental Fig. S1). Erdr1 expression was detected in all 14 WT mice; however, expression was near or below the threshold of detection for all 13 Ccr5-/- mice (Fig. 1B). This same pattern was also seen in control mice that did not receive tumor injections (Fig. 1C). The expression patterns for the remaining ten genes were much more heterogeneous. As a result, we chose to further characterize the role of ERDR1 in the promotion of lung metastasis.
We have previously shown metastases can be increased in Ccr5-/- mice with the adoptive transfer of WT pulmonary mesenchymal cells (PMCs) but not Ccr5-/- PMCs (24). Since this disparity might have been due to the increased expression of Erdr1 in the WT cells, gene expression was measured in WT and Ccr5-/- PMCs using real-time PCR. As was the case for the whole lung, Erdr1 expression was 6.4 ± 2.7 fold higher in the WT PMCS than the Ccr5-/- cells (p < 0.001) (Fig. 1D).
Given the limited published data on Erdr1, the sequence of the gene expressed by PMCs was compared with that described in WEHI-3B cell line (13). As shown in Supplemental Fig. S2, the open reading frame in the Erdr1 consensus sequence was 100% identical to the sequence found in the WEHI-3B cell line.
CCL4 induces the expression of Erdr1 in CCR5-expressing PMCs
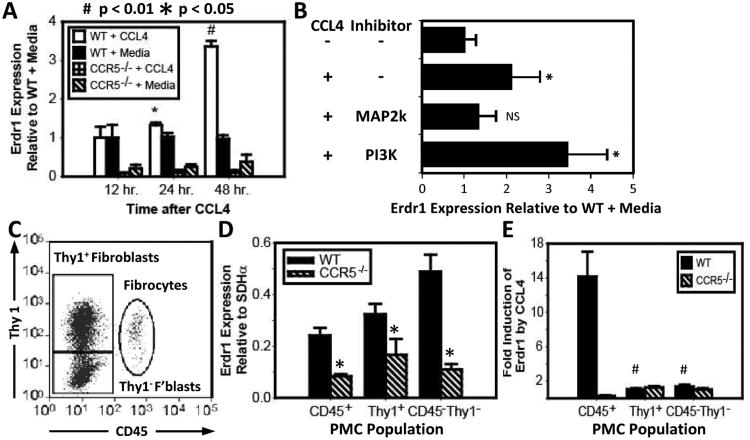
We established an in vitro association between ERDR1 and CCR5 by treating PMCs with 50 ng/ml of the Ccr5-specific chemokine Ccl4. As measured by real-time PCR, Erdr1 expression in WT PMCs was increased by 1.33 fold ± 0.06 (p < 0.05) after 24 hours of incubation with Ccl4. By 48 hours, expression had increased by 3.36 fold ± 0.14 (p < 0.001) (Fig. 2A). Ccl4 did not increase expression of Erdr1 in Ccr5-/- PMCs.
Figure 2. Stimulation of Ccr5 on PMCs induces expression ofErdr1.
(A) Erdr1 expression by real time PCR after Ccl4 treatment. WT and Ccr5-/- PMCs were stimulated with 50 ng/ml of Ccl4 after overnight culture in serum-free media. mRNA was harvested at 0, 24, and 48 hours. Bar graph shows expression of Erdr1 normalized to baseline expression in WT PMCs. Significant increases in Erdr1 expression were detectable in WT PMCs at 24 and 48 hours. No significant changes were noted in Ccr5-/- PMCs. (B) The response of Erdr1 to Ccl4 following treatment with 10μM of the MAP2K inhibitor U0129 and 20 μM of the PI3K inhibitor LY294002. Erdr1 expression is inhibited by U0129 (3rd bar) but not LY294002 (4th bar). (C) PMC subpopulations. The dot plot shows flow cytometric analysis of PMC populations as defined by CD45 and Thy-1. (D) Bar graph showing Erdr1 expression in PMCs sorted by CD45 and Thy-1. All subpopulations of WT cells expressed more Erdr1 than Ccr5-/- (p < 0.05). (E) Bar graph shows induction of Erdr1 after stimulation with Ccl4 in PMC subpopulations. PMCs were sorted 48 hours after treatment with 50 ng/ml of Ccl4. Only WT CD45+ PMCs exhibited a significant increase in Erdr1 expression (p < 0.01).
The relationship between ERDR1 and CCL4 was verified by inhibiting the intracellular signaling pathways induced by CCL4. To do so, WT PMCs were treated with the MAP2K inhibitor U0126 or the PI3K inhibitor LY294002. As shown in Fig 2B, Ccl4 increased expression of Erdr1 following treatment with LY294002 by 3.44 ± 0.92 fold (p < 0.05), but not with U0126 (Fig. 2B).
Cultured PMCs contain three different populations of stromal cells, which can be differentiated by their expression of CD45 and Thy-1 (10). Flow cytometric analysis identified fibrocytes as CD45 positive with intermediate expression of Thy-1. Fibroblasts, on the other hand, were CD45- and could be separated into two additional populations based on Thy-1 expression (Fig. 2C). All three of these WT populations expressed more Erdr1 than their Ccr5-/- counterparts (Fig. 2D).
Fibrocytes also express more CCR5 than fibroblasts (10). Given this observation, fibrocytes were expected to have a greater response to CCL4 than fibroblasts. This hypothesis was tested by treating PMCs with Ccl4 for 48 hours and then by sorting the cells by Thy-1 and CD45 expression. As predicted, WT CD45+ fibrocytes increased their expression of Erdr1 by 14.2 fold ± 2.9. Erdr1 expression was not induced in the Ccr5-/- fibrocytes or fibroblasts (p < 0.01) (Fig. 2E).
Experimental pulmonary metastasis is reduced after knockdown of Erdr1
The effect of ERDR1 on metastasis was directly tested by manipulating the amount of ERDR1 expressed by stromal cells within the lung prior to injection with B16 F10 cells. In the first series of experiments, metastasis was measured in Ccr5-/- mice injected with Erdr1 knockdown or control stromal cells. To this end, we selected two HSPG retroviral vectors carrying different Erdr1-specific shRNA sequences and one vector carrying scrambled shRNA control sequence (Supplemental Fig. 3A).
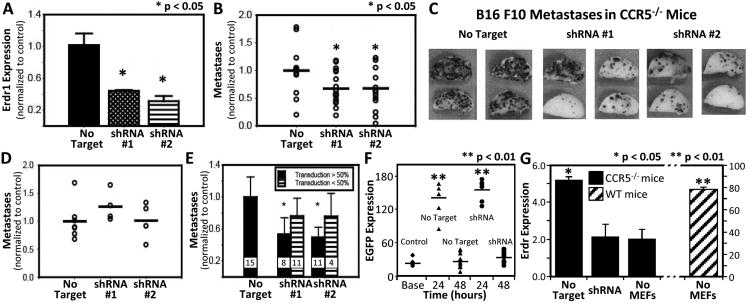
This strategy was applied to murine embryonic fibroblasts (MEFs) rather than PMCs for several reasons. First, MEFs expressed greater amounts of Erdr1 than PMCs (Supplemental Fig. 3B). As a result, shRNA treatment would generate a greater difference in Erdr1 expression and amplify effects of the gene on metastasis. Second, PMCs transduced with Erdr1 shRNAs expanded poorly (Supplemental Fig. 3C). This constraint limited the technical feasibility of these experiments and raised questions about the persistence of the knock-down cells in vivo. Despite these differences, unmanipulated WT MEFs did promote metastasis in Ccr5-/- mice similar to that found by injecting WT PMCs (Supplemental Fig. 3D). Transduction of MEFs also produced viable cells with a mean reduction in Erdr1 expression of 55% ± 1.0% for shRNA #1 and 70% ± 6.5% for shRNA #2 (Fig 3A).
Figure 3. Inhibition ofErdr1inhibits metastasis.
(A) Real-time PCR measurement of Erdr1 expression in MEFs 48 hours after transduction with shRNA retroviral constructs. Results normalized to MEFs transduced with control plasmid. (B) Graph showing number of B16-F10 metastases in Ccr5-/- mice injected with MEFs transduced with control (left), shRNA#1 (middle), or shRNA#2 (right). Mice were injected with 4×105 MEFs 24 hours before injection with 7.5 × 105 melanoma cells. Metastases are normalized to mice treated with control MEFs; results are pooled from three different experiments. (C) Photographs of the lungs (left upper lobe) of representative mice described in (B). (D) Graph showing metastases in WT mice injected with control or shRNA transduced MEFs. Results normalized to control MEF group. (E) Bar graph representing the number of B16-F10 metastases in Ccr5-/- mice. Black bars represent mice that received more than 50% transduced MEFs; striped bars are mice that received less than 50% transduced MEFs. The sample sizes are given just above the x-axis. (F) The graph shows the amount of eGFP detected from the lung of Ccr5-/- injected with MEFs that had been transduced with control or shRNA plasmids. eGFP was detected by ELISA. First column on the left is eGFP expression in mice injected with unmanipulated MEFs. (G) Real time PCR measurement of Erdr1 in the lung of Ccr5-/- mice 24 hours after injection with 4×105 MEFs transduced with control or shRNA. Columns on the right show Erdr1 expression of Ccr5-/- and WT mice without MEF injection.
Compared to the control MEFs, Erdr1 knockdown MEFs were associated with a 32.7% and 32.4% reduction in lung metastasis (p<0.05) (Fig. 3B, C). This result suggested that ERDR1 acts to promote metastasis or inhibition of ERDR1 reduces metastasis. The latter was ruled out by transferring Erdr1 knockdown MEFs into WT mice. There were no measurable differences in metastasis when comparing shRNA knockdown MEFs to control transduced MEFs (Fig. 3D).
The transduction efficiency varied from experiment to experiment. This observation prompted the comparison of mice that received greater than 50% transduced MEFs with those that received less than 50%. Ccr5-/- mice that received greater than 50% shRNA transduced MEFs had significantly fewer metastases (shRNA#1 = 53.3%, shRNA#2 = 50.4%) than those that received less than 50% transduced cells (shRNA#1 =23.8%, shRNA#2 =15.4%) (Fig. 3E). Thus, a greater number of shRNA transduced MEFs were associated with fewer metastases.
Our observation that inhibition of ERDR1 inhibits metastasis could also be explained by differences in the survival of the injected MEFs. The number of viable MEFs expressing eGFP in the lung was determined by measuring eGFP in the lung by ELISA. This assay revealed no differences in the lungs from animals injected with control or Erdr1 shRNA transduced MEFs. Animals receiving transduced MEFs had significantly more eGFP than animals injected with unmanipulated MEFs at 24 hours (p<0.01). Differences in eGFP amount were lost at 48 hours (Fig. 3F). Our technique was further confirmed by measuring Erdr1 expression in the lungs of Ccr5-/- mice after injection with shRNA or controlled transduced MEFs. 24 hours after injection with 4 × 105 control MEFs, Erdr1 expression was increased by 2.5 ± 0.8 fold over shRNA transduced MEFs (Fig. 3G).
ERDR1 promotes survival of chronic lymphocytic leukemia cells
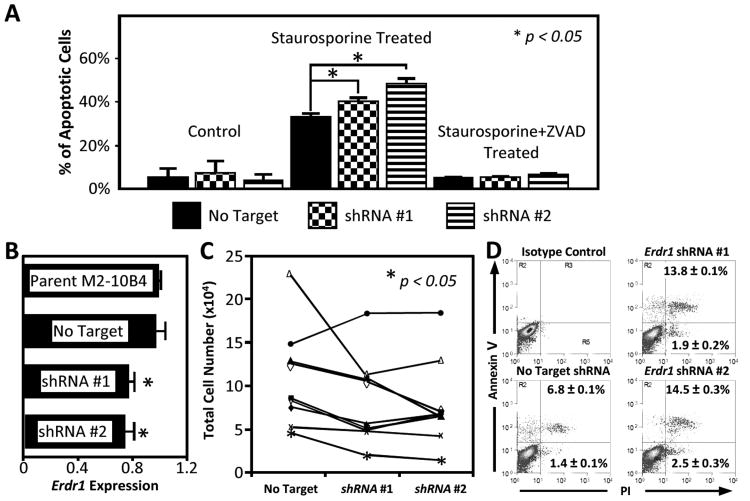
The ability of ERDR1 to promote experimental metastasis is not well explained by its principle function of hemoglobinization. Other functions have not been well described. Our experience with MEFs suggested ERDR1 could oppose apoptosis. Though no differences in apoptosis were noted at baseline, ERDR1 countered apoptosis when cells were challenged with staurosporine, an apoptotic stimulus. In these experiments, Erdr1 knockdown by shRNA #1 and #2 was associated with a greater percentage of apoptotic cells than control transduction (32.7% ± 1.8% vs. 40.2% ± 1.7% vs. 48.1% ± 2.1%, p < 0.05). These differences in apoptosis were lost when these cells were treated with the pan-caspase inhibitor Z-VAD (Fig.4A).
Figure 4. Inhibition ofErdr1promotes apoptosis of primary chronic lymphocytic leukemia cells.
(A) Effect of Erdr1 knockdown on staurosporine induced apoptosis. MEFs were transduced with Erdr1 knockdown or control shRNA and were treated with or without staurosporine (100 nM). The bar graph shows a disproportionate increase the apoptosis of MEFs treated with Erdr1 shRNA. The differences between control and knockdown shRNA transduced cells were lost when the caspase inhibitor Z-VAD-FMK was added. (B) Erdr1 expression in M2-10B4 cell lines. Results are expressed relative to the parent line (top). The bottom three lines are stable clones transduced with non-target control, shRNA#1, or shRNA#2 plasmids. (C) Total cell number of CLL cells co-cultured with M2-10B4 clones. Each line represents a sample from an individual patient. The points on the left are the cell number after culture with non-targeted control M2-10B4 cells. The points in the center and right are numbers after culture with shRNA knockdown lines. Cells were harvested after 96 hours of culture. (D) Representative sample dot plots of CLL samples co-cultured with M2-10B4 clones. The mean ± SEM is given for all 10 CLL samples.
ERDR1's effect on apoptosis was also tested by co-culturing primary chronic lymphocytic leukemia (CLL) cells with M2-10B4 stromal cells. Without stromal cell support, CLL cells undergo apoptosis within 48 to 72 hours. Therefore, ERDR1's contribution to this process could be examined by comparing CLL cell survival using stromal cells treated with Erdr1shRNAs. Accordingly, we selected stable control and knock-down clones based on similarities in proliferation and on expression of Erdr1. Compared with the non-targeted line, the Erdr1 expression was 76.8% ± 3.3% for shRNA #1 and 74.0% ± 6.5% for shRNA #2 (p<0.05) (Fig. 4B).
At 96 hours, the cell counts were higher when the CLL cells were co-cultured with control cell lines compared with Erdr1 knockdown lines (OR 1.88 ± 0.27, 2.52 ± 0.66 respectively, p < 0.05) (Fig. 4C). This increase in total cell number was associated with a decrease in the percentage of apoptotic cells (OR 0.69 ± 0.18, 0.58 ± 0.12 respectively) (Fig. 4D).
Discussion
Therapeutic strategies that disrupt the tumor stroma have been limited by the lack of suitable targets. Our group searched for such a target by comparing the gene expression from the lungs of WT and Ccr5-/- mice since Ccr5 signaling by the host stromal cells promotes metastasis (24). This search identified Erdr1 as overexpressed in the lungs of WT compared to Ccr5-/- mice injected with tumor cells. Though previously associated with the bone marrow stroma, the expression of this gene in the lung stroma was established by detecting it in PMCs, which are made up of both fibroblasts and fibrocytes. The reintroduction of ERDR1 into Ccr5-/- mice by the adoptive transfer of MEFs restored the pro-metastatic stroma. Furthermore, we found that ERDR1 inhibited apoptosis of human CLL cells. Therefore, we propose that ERDR1 expression by stromal cells promotes invasion and survival of cancer cells by inhibiting apoptosis of these cells.
We chose to study Erdr1 since it was one of 11 genes that were differentially expressed in the lung during the first 48 hours of tumor growth. Several of these 11 genes have already been associated with cancer progression. For example, Cyr61 has been associated with poor prognosis in squamous cell cancers of the head and neck (25) and esophagus (26). CCR2 has been linked to breast cancer metastasis (27). The inclusion of genes with known tumorpromoting properties substantiates the robustness of our approach. Nevertheless, of these 11 genes, only Erdr1 demonstrated differential expression in all 27 mice tested.
The identification of ERDR1 was somewhat unexpected since previous work had focused on its role in hemoglobin synthesis. However, a broader physiologic role for this gene is implied by its expression in a wide variety of developing and mature tissues. In addition to the bone marrow (13), gene expression has been demonstrated in the mammary gland, spleen, and lymph node (28). Others have found Erdr1 expression during retinal development (29), gonadal differentiation (18), liver development (30) and placental morphogenesis (31). The expression of Erdr1 during development is noteworthy because it suggests a potential role in malignancy. Cancer gene expression often recapitulates tissue-specific development programs (32, 33). Consequently, developmental genes are candidate oncogenes. In this context, the contribution of ERDR1 to cancer cell survival may not be surprising.
These data provide an additional mechanism by which CCR5 promotes cancer progression, namely by the induction of Erdr1. The capacity of Ccr5 to increase Erdr1 expression was demonstrated in PMCs following treatment with Ccl4, a ligand with specificity for Ccr5. This expression was the greatest in the migrating fibrocyte population, which was expected given their increased expression of Ccr5. This observation is notable since fibrocytes have the greater effect on metastasis than either Thy-1+ or Thy-1- fibroblasts (10).
ERDR1's role in cancer progression is complex. Jung et al have shown that recombinant ERDR1 suppresses the invasion of human gastric cancer cells (34). This observation follows reports that ERDR1 inhibits metastasis of melanoma cells (35). Our data can be harmonized with these apparently contradictory studies by noting major differences in the experimental methods. Our studies were conducted in Ccr5-/- mice that have a low basal expression of Erdr1. Furthermore, Erdr1 expression was manipulated through bystander cells rather than within the cancer cell. Therefore, our experiments highlight the effect of ERDR1 on the surrounding stroma whereas the preceding references focus on the effect of ERDR1 on the cancer cells.
The pro-metastatic and anti-metastatic properties of ERDR1 attest to the bimodal response induced by ERDR1 (14). When the baseline expression of Erdr1 is reduced, induction of the gene likely promotes cell survival by inhibiting apoptosis. This phenomenon was observed during treatment with staurosporine, and is similar to our CLL data. This relationship is also substantiated by the observation that apoptosis of melanoma cells is increased in the Ccr5-/- mouse (12). An inverse relationship between Erdr1 expression and apoptosis has also been demonstrated in a Nox2-/- mouse undergoing reperfusion injury (36). This model is consistent with our data showing inhibition of Erdr1 inhibits metastasis. However, when the transcription of Erdr1 reaches a threshold, apoptosis increases and cell viability diminishes. This phenomenon has been demonstrated in keratinocytes that are undergoing UVB treatment (35).
Our experiments looked at the effects of ERDR1 as it was expressed by several cell types including PMCs, MEFs, and M2-10B4 cells. We have seen similar results when comparing PMCs and fibrocytes from WT and Ccr5-/- mice (37). Like our genetically manipulated cells, cells from these animals differentially express Erdr1. In all of these experiments, greater expression of Erdr1 in the transferred cells led to a greater the tumor burden for the recipient.
CCR5 expression can have diverse implications for cancer prognosis depending on the tissues that express it. In mouse models, Ccr5 boosts CD4+-dependent, CD8+ T cell responses (38) although it may also lower infiltration by those T cells (12). Patients with CCR5+ infiltrating lymphocytes had a better outcome in colorectal cancer (39, 40) or when undergoing immunotherapy for melanoma (41). On the other hand, expression by the tumor tissue is considered a poor marker in melanoma (42), prostate cancer (43) and breast cancer (44, 45). CCR5 expression on osteosarcoma cells upregulates integrins and predisposes them to metastasis (46). Expression of Ccr5 by the tumor fibroblasts has been associated with the promotion of pulmonary metastasis in murine models (11, 24) and the progression of lung cancer in patients (47). Targeting ERDR1 may be more advantageous than targeting CCR5 since it may inhibit progression mediated by the stroma without inhibiting the anti-tumor immune response.
The study of ERDR1 in human disease is complicated by the lack of a known human homolog. Nevertheless, the gene has been sequenced from human tissues by several labs. ERDR1 has been detected in human keratinocytes (48), lymphoma (13), and HEK293 cells (15). Our lab has also detected ERDR1 in nurse-like cells (49). All of these investigators have found near sequence identity between the human and murine genes.
In summary, this paper identifies ERDR1 as a stromal-derived cytokine that promotes cancer progression. Furthermore, it establishes an additional means by which inflammatory chemokines contribute to cancer progression. The relationship between ERDR1 and CCR5 is clinically significant since CCR5 inhibitors are already in clinical trials (50). Further understanding of this interaction may provide a means of separating the beneficial anti-tumor immune responses from the harmful pro-tumor stromal support. Finally, our data suggest that this relatively unknown gene may be critical to understanding and manipulating tumor-stroma interactions.
Supplementary Material
Acknowledgments
We thank Peter Dormer for initial assistance obtaining antibody for western blots, and Deborah Taxman for advice on constructing shRNA vectors. This work was supported by NIH Grant P50CA058223 (JSS) and ACS Grant RSG 118750 (HVD). RLM is a member of the Medical Scientist Training Program T32 GM008719.
Grant Support: NIH Grant P50CA058223 (JSS) and ACS Grant RSG 118750 (HVD). RLM is a member of the Medical Scientist Training Program T32 GM008719.
Footnotes
Conflict of interest: None of the authors have current or former affiliations with entities that have personal or financial interest in this work. Specifically, the authors have no conflict of interest to declare surrounding the use of CCR5 inhibitors.
References
- 1.Zhou Y, You MJ, Young KH, Lin P, Lu G, Medeiros LJ, et al. Advances in the molecular pathobiology of B-lymphoblastic leukemia. Hum Pathol. 2012;43:1347–1362. doi: 10.1016/j.humpath.2012.02.004. [DOI] [PubMed] [Google Scholar]
- 2.Rasanen K, Vaheri A. Activation of fibroblasts in cancer stroma. Exp Cell Res. 2010;316:2713–2722. doi: 10.1016/j.yexcr.2010.04.032. [DOI] [PubMed] [Google Scholar]
- 3.Grivennikov SI, Greten FR, Karin M. Immunity, inflammation, and cancer. Cell. 2010;140:883–899. doi: 10.1016/j.cell.2010.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhang J, Liu J. Tumor stroma as targets for cancer therapy. Pharmacol Therapeut. 2013;137:200–215. doi: 10.1016/j.pharmthera.2012.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Steller EJ, Raats DA, Koster J, Rutten B, Govaert KM, Emmink BL, et al. PDGFRB promotes liver metastasis formation of mesenchymal-like colorectal tumor cells. Neoplasia. 2013;15:204–217. doi: 10.1593/neo.121726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hiratsuka S, Nakamura K, Iwai S, Murakami M, Itoh T, Kijima H, et al. MMP9 induction by vascular endothelial growth factor receptor-1 is involved in lung-specific metastasis. Cancer Cell. 2002;2:289–300. doi: 10.1016/s1535-6108(02)00153-8. [DOI] [PubMed] [Google Scholar]
- 7.Studebaker AW, Storci G, Werbeck JL, Sansone P, Sasser AK, Tavolari S, et al. Fibroblasts isolated from common sites of breast cancer metastasis enhance cancer cell growth rates and invasiveness in an interleukin-6-dependent manner. Cancer Res. 2008;68:9087–9095. doi: 10.1158/0008-5472.CAN-08-0400. [DOI] [PubMed] [Google Scholar]
- 8.Cabezón T, Skibshøj I, Klingelhöfer J, Grigorian M, Gromov P, Rank F, et al. Expression of S100A4 by a variety of cell types present in the tumor microenvironment of human breast cancer. Int J Cancer. 2007;121:1433–1444. doi: 10.1002/ijc.22850. [DOI] [PubMed] [Google Scholar]
- 9.Saha A, Lee YC, Zhang Z, Chandra G, Su SB, Mukherjee AB. Lack of an endogenous anti-inflammatory protein in mice enhances colonization of B16F10 melanoma cells in the lungs. J Biol Chem. 2010;285:10822–10831. doi: 10.1074/jbc.M109.083550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.van Deventer HW, Wu QP, Bergstralh DT, Davis BK, O'Connor BP, Ting JP, et al. C-C chemokine receptor 5 on pulmonary fibrocytes facilitates migration and promotes metastasis via matrix metalloproteinase 9. Am J Pathol. 2008;173:253–264. doi: 10.2353/ajpath.2008.070732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wu Y, Li YY, Matsushima K, Baba T, Mukaida N. CCL3-CCR5 axis regulates intratumoral accumulation of leukocytes and fibroblasts and promotes angiogenesis in murine lung metastasis process. J Immunol. 2008;181:6384–6393. doi: 10.4049/jimmunol.181.9.6384. [DOI] [PubMed] [Google Scholar]
- 12.Song JK, Park MH, Choi DY, Yoo HS, Han SB, Yoon do Y, et al. Deficiency of C-C chemokine receptor 5 suppresses tumor development via inactivation of NF-kappaB and upregulation of IL-1Ra in melanoma model. PLoS One. 2012;7:e33747. doi: 10.1371/journal.pone.0033747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dormer P, Spitzer E, Frankenberger M, Kremmer E. Erythroid differentiation regulator (EDR), a novel, highly conserved factor I. Induction of haemoglobin synthesis in erythroleukaemic cells. Cytokine. 2004;26:231–242. doi: 10.1016/j.cyto.2004.02.005. [DOI] [PubMed] [Google Scholar]
- 14.Dormer P, Spitzer E, Moller W. EDR is a stress-related survival factor from stroma and other tissues acting on early haematopoietic progenitors (E-Mix) Cytokine. 2004;27:47–57. doi: 10.1016/j.cyto.2004.03.014. [DOI] [PubMed] [Google Scholar]
- 15.Gillingwater TH, Wishart TM, Chen PE, Haley JE, Robertson K, MacDonald SHF, et al. The neuroprotective WldS gene regulates expression of PTTG1 and erythroid differentiation regulator 1-like gene in mice and human cells. Hum Mol Genet. 2006;15:625–635. doi: 10.1093/hmg/ddi478. [DOI] [PubMed] [Google Scholar]
- 16.Lee JH, Kim CH, Kim DG, Ahn YS. Microarray analysis of differentially expressed genes in the brains of tubby mice. Korean J Physiol Pharmacol. 2009;13:91–97. doi: 10.4196/kjpp.2009.13.2.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Brown AR, Rebus S, McKimmie CS, Robertson K, Williams A, Fazakerley JK. Gene expression profiling of the preclinical scrapie-infected hippocampus. Biochem Bioph Res Co. 2005;334:86–95. doi: 10.1016/j.bbrc.2005.06.060. [DOI] [PubMed] [Google Scholar]
- 18.Nef S, Schaad O, Stallings NR, Cederroth CR, Pitetti JL, Schaer G, et al. Gene expression during sex determination reveals a robust female genetic program at the onset of ovarian development. Dev Biol. 2005;287:361–377. doi: 10.1016/j.ydbio.2005.09.008. [DOI] [PubMed] [Google Scholar]
- 19.Anderson KC. Targeted therapy of multiple myeloma based upon tumormicroenvironmental interactions. Exp Hematol. 2007;35:155–162. doi: 10.1016/j.exphem.2007.01.024. [DOI] [PubMed] [Google Scholar]
- 20.Andres PG, Beck PL, Mizoguchi E, Mizoguchi A, Bhan AK, Dawson T, et al. Mice with a selective deletion of the CC chemokine receptors 5 or 2 are protected from dextran sodium sulfate-mediated colitis: Lack of CC chemokine receptor 5 expression results in a NK1.1+ lymphocyte-associated Th2-type immune response in the intestine. J Immunol. 2000;164:6303–6312. doi: 10.4049/jimmunol.164.12.6303. [DOI] [PubMed] [Google Scholar]
- 21.Sipkins DA, Wei X, Wu JW, Runnels JM, Cote D, Means TK, et al. In vivo imaging of specialized bone marrow endothelial microdomains for tumour engraftment. Nature. 2005;435:969–973. doi: 10.1038/nature03703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Coffield VM, Jiang Q, Su L. A genetic approach to inactivating chemokine receptors using a modified viral protein. Nat Biotechnol. 2003;21:1321–1327. doi: 10.1038/nbt889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Taxman DJ, Livingstone LR, Zhang J, Conti BJ, Iocca HA, Williams KL, et al. Criteria for effective design, construction, and gene knockdown by shRNA vectors. BMC Biotechnol. 2006;6:7. doi: 10.1186/1472-6750-6-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.van Deventer HW, O'Connor W, Jr, Brickey WJ, Aris RM, Ting JP, Serody JS. C-C chemokine receptor 5 on stromal cells promotes pulmonary metastasis. Cancer Res. 2005;65:3374–3379. doi: 10.1158/0008-5472.CAN-04-2616. [DOI] [PubMed] [Google Scholar]
- 25.Kok SH, Chang HH, Tsai JY, Hung HC, Lin CY, Chiang CP, et al. Expression of Cyr61 (CCN1) in human oral squamous cell carcinoma: An independent marker for poor prognosis. Head Neck. 2010;32:1665–1673. doi: 10.1002/hed.21381. [DOI] [PubMed] [Google Scholar]
- 26.Yang Z, Lei Z, Li B, Zhou Y, Zhang G-M, et al. Rapamycin inhibits lung metastasis of B16 melanoma cells through down-regulating alphav integrin expression and upregulating apoptosis signaling. Cancer Sci. 2009;101:494–500. doi: 10.1111/j.1349-7006.2009.01412.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lu Y, Chen Q, Corey E, Xie W, Fan J, Mizokami A, et al. Activation of MCP-1/CCR2 axis promotes prostate cancer growth in bone. Clin Exp Metastas. 2009;26:161–169. doi: 10.1007/s10585-008-9226-7. [DOI] [PubMed] [Google Scholar]
- 28.Lattin JE, Schroder K, Su AI, Walker JR, Zhang J, Wiltshire T, et al. Expression analysis of G Protein-Coupled Receptors in mouse macrophages. Immunome Res. 2008;4:5. doi: 10.1186/1745-7580-4-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Blackshaw S, Harpavat S, Trimarchi J, Cai L, Huang H, Kuo WP, et al. Genomic Analysis of Mouse Retinal Development. PLoS Biol. 2004;2:e247. doi: 10.1371/journal.pbio.0020247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Otu HH, Naxerova K, Ho K, Can H, Nesbitt N, Libermann TA, et al. Restoration of liver mass after injury requires proliferative and not embryonic transcriptional patterns. J Biol Chem. 2007;282:11197–11204. doi: 10.1074/jbc.M608441200. [DOI] [PubMed] [Google Scholar]
- 31.Sferruzzi-Perri AN, Macpherson AM, Roberts CT, Robertson SA. Csf2 null mutation alters placental gene expression and trophoblast glycogen cell and giant cell abundance in mice. Biol Reprod. 2009;81:207–221. doi: 10.1095/biolreprod.108.073312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kho AT, Zhao Q, Cai Z, Butte AJ, Kim JY, Pomeroy SL, et al. Conserved mechanisms across development and tumorigenesis revealed by a mouse development perspective of human cancers. Genes Dev. 2004;18:629–640. doi: 10.1101/gad.1182504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liu H, Kho AT, Kohane IS, Sun Y. Predicting survival within the lung cancer histopathological hierarchy using a multi-scale genomic model of development. PLoS Med. 2006;3:e232. doi: 10.1371/journal.pmed.0030232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jung MK, Houh YK, Ha S, Yang Y, Kim D, Kim TS, et al. Recombinant ERDR1 suppresses the migration and invasion ability of human gastric cancer cells, SNU-216, through the JNK pathway. Immunol Lett. 2013;150:145–151. doi: 10.1016/j.imlet.2013.01.012. [DOI] [PubMed] [Google Scholar]
- 35.Jung MK, Park Y, Song SB, Cheon SY, Park S, Houh Y, et al. Erythroid differentiation regulator 1, an interleukin 18-regulated gene, acts as a metastasis suppressor in melanoma. J Invest Dermatol. 2011;131:2096–2104. doi: 10.1038/jid.2011.170. [DOI] [PubMed] [Google Scholar]
- 36.Thirunavukkarasu M, Adluri R, Juhasz B, Samuel S, Zhan L, Kaur A, et al. Novel role of NADPH oxidase in ischemic myocardium: a study with Nox2 knockout mice. Funct Integr Genomics. 2012;12:501–514. doi: 10.1007/s10142-011-0256-x. [DOI] [PubMed] [Google Scholar]
- 37.van Deventer HW, Palmieri DA, Wu QP, McCook EC, Serody JS. circulating fibrocytes prepare the lung for cancer metastasis by recruiting Ly-6c+ monocytes via CCL2. J Immunol. 2013 doi: 10.4049/jimmunol.1202857. In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.González-Martín A, Gómez L, Lustgarten J, Mira E, Mañes S. Maximal T cell–mediated antitumor responses rely upon CCR5 expression in both CD4+ and CD8+ T cells. Cancer Res. 2011;71:5455–5466. doi: 10.1158/0008-5472.CAN-11-1687. [DOI] [PubMed] [Google Scholar]
- 39.Musha H, Ohtani H, Mizoi T, Kinouchi M, Nakayama T, Shiiba K, et al. Selective infiltration of CCR5(+)CXCR3(+) T lymphocytes in human colorectal carcinoma. Int J Cancer. 2005;116:949–56. doi: 10.1002/ijc.21135. [DOI] [PubMed] [Google Scholar]
- 40.Schimanski CC, Moehler M, Gockel I, Zimmermann T, Lang H, Galle PR, Berger MR. Expression of chemokine receptor CCR5 correlates with the presence of hepatic molecular metastases in K-ras positive human colorectal cancer. J Cancer Res Clin Oncol. 2011;137:1139–45. doi: 10.1007/s00432-011-0980-6. [DOI] [PubMed] [Google Scholar]
- 41.Petrulio C, Kim-Schulze S, Deraffele G, Morozowiecz D, Mitcham J, Schrama D, et al. P239: CCR5 polymorphism as a predictor of clinical resonse to high-dose IL-2 therapy in melanoma and renal cell carcinoma. J Surg Res. 2007;137:327. [Google Scholar]
- 42.Gan Y, Reilkoff R, Peng X, Russell T, Chen Q, Mathai SK, et al. Role of semaphorin 7a signaling in transforming growth factor β1–induced lung fibrosis and scleroderma-related interstitial lung disease. Arth Rheum. 2011;63:2484–2494. doi: 10.1002/art.30386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Vaday GG, Peehl DM, Kadam PA, Lawrence DM. Expression of CCL5 (RANTES) and CCR5 in prostate cancer. Prostate. 2006;66:124–134. doi: 10.1002/pros.20306. [DOI] [PubMed] [Google Scholar]
- 44.Velasco-Velázquez M, Jiao X, De La Fuente M, Pestell TG, Ertel A, Lisanti MP, et al. CCR5 antagonist blocks metastasis of basal breast cancer cells. Can Res. 2012;72:3839–3850. doi: 10.1158/0008-5472.CAN-11-3917. [DOI] [PubMed] [Google Scholar]
- 45.Karnoub AE, Dash AB, Vo AP, Sullivan A, Brooks MW, Bell GW, et al. Mesenchymal stem cells within tumour stroma promote breast cancer metastasis. Nature. 2007;449:557–563. doi: 10.1038/nature06188. [DOI] [PubMed] [Google Scholar]
- 46.Wang SW, Wu HH, Liu SC, Wang PC, Ou WC, Chou WY, et al. CCL5 and CCR5 interaction promotes cell motility in human osteosarcoma. PLoS One. 2012;7:e35101. doi: 10.1371/journal.pone.0035101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Borczuk AC, Papanikolaou N, Toonkel RL, Sole M, Gorenstein LA, Ginsburg ME, et al. Lung adenocarcinoma invasion in TGFbetaRII-deficient cells is mediated by CCL5/RANTES. Oncogene. 2008;27:557–564. doi: 10.1038/sj.onc.1210662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kim HJ, Song SB, Yang Y, Eun YS, Cho BK, Park HJ, et al. Erythroid differentiation regulator 1 (ERDR1) is a proapototic factor in human keratinocytes. Exp Dermatol. 2011;20:920–925. doi: 10.1111/j.1600-0625.2011.01354.x. [DOI] [PubMed] [Google Scholar]
- 49.van Deventer HW, Hoffert T, West M, Mango R, Wu QP, Serody J. Bone marrow stromal cells inhibit apoptosis in chronic lymphocytic leukemia cells by expressing erythroid differentiation regulator 1. Blood. 2011;118 Abstract 1764. [Google Scholar]
- 50.Dorr P, Westby M, Dobbs S, Griffin P, Irvine B, Macartney M, et al. Maraviroc (UK-427,857), a potent, orally bioavailable, and selective small-molecule inhibitor of chemokine receptor CCR5 with broad-spectrum anti-human immunodeficiency virus type 1 activity. Antimicrob Agents Chemother. 2005;49:4721–4732. doi: 10.1128/AAC.49.11.4721-4732.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.