Summary
Human RecQ4 affects cancer and aging, but it is difficult to study because it is a fusion between a helicase and an essential replication factor. Budding yeast Hrq1 is homologous to the disease-linked helicase domain of RecQ4 and, like hRecQ4, was a robust 3’–5’ helicase. Additionally, Hrq1 had the unusual property of forming heptameric rings. Cells lacking Hrq1 exhibited two particularly dangerous DNA damage phenotypes: hypersensitivity to DNA inter-strand crosslinks (ICLs) and telomere addition to DNA breaks. Both activities are rare: their co-existing in a single protein is unprecedented. Resistance to ICLs required helicase activity, but suppression of telomere addition did not. Hrq1 also affected telomere length by a non-catalytic mechanism, as well as telomerase-independent telomere maintenance. As Hrq1 bound telomeres in vivo, it likely affects them directly. Thus, the tumor suppressing activity of RecQ4 could be due to a role in ICL repair and/or suppressing de novo telomere addition.
Introduction
Helicases are motor proteins that use the energy of nucleotide hydrolysis to separate duplex nucleic acids into their component single strands (Abdelhaleem, 2010). RecQ family helicases are involved in many aspects of DNA replication, recombination, and repair (Bernstein et al., 2010). Humans encode 5 RecQs (hRecQ1, hBLM, hWRN, hRecQ4, and hRecQ5), and mutations in 3 of these enzymes (hBLM, hWRN, and hRecQ4) are linked to cancers and/or premature aging. This paper presents in vitro and in vivo studies of the Saccharomyces cerevisiae Hrq1 helicase, a homolog of hRecQ4.
Mutation of hRecQ4 is linked to three distinct diseases with related and overlapping symptoms and which are all characterized by genome instability, premature aging, and increased cancer risk (Capp et al., 2010; Larizza et al., 2010). However, determining how loss of hRecQ4 promotes human disease is complicated because its N-terminus is homologous to the essential S. cerevisiae Sld2 DNA replication factor (Fig. 1A) (Liu, 2010). As 95% of the known disease-causing alleles of hRecQ4 are found C-terminal to its Sld2-like domain (Larizza et al., 2010), these diseases are likely due to loss of its helicase activities rather than loss of its replication function, which would presumably be lethal. Thus, a simple model to determine the non-replication functions of RecQ4 would be useful.
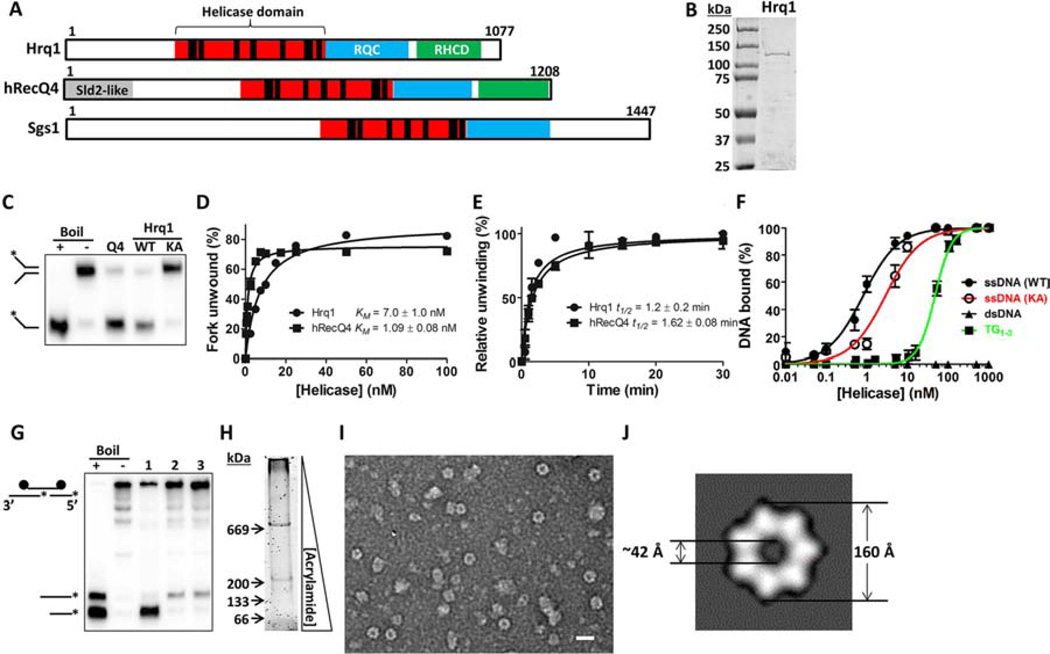
Figure 1. Purified Hrq1 is an active 3’–5’ helicase.
(A) Domain schematics of Hrq1, hRecQ4, and Sgs1. The amino acid length of each is given on the right. The black bars in the helicase domain correspond to conserved ATPase/helicase motifs. RQC: RecQ C-terminal domain. RHCD: RecQ4/Hrq1-conserved domain. Sld2-like: portion of hRecQ4 homologous to S. cerevisiae Sld2. (B) Coomassie-stained gel of purified S. cerevisiae Hrq1. The expected molecular weight is ~130 kDa. (C) hRecQ4 (Q4) and Hrq1 (WT) (both 50 nM) unwind a fork substrate; 100 nM Hrq1-KA (KA) does not. (D) Hrq1 and hRecQ4 unwind the fork with similar apparent KMs ([protein] necessary to unwind 50% of the DNA). (E) The rates of fork unwinding by 50 nM Hrq1 and hRecQ4 were similar (t1/2 = time necessary to unwind 50% of the DNA). (F) Hrq1 and Hrq1-KA bind ssDNA by gel shifts. Hrq1 also preferentially bound ss- vs. dsDNA, as well as telomeric repeat ssDNA (TG1–3). (G) Directionality of Pif1 (lane 1), hRecQ4 (lane 2), and Hrq1 (lane 3) unwinding. The fastest migrating band corresponds to 5’-3’ unwinding; the slower migrating band indicates 3’–5’ activity. (H) Sypro Orange-stained native gradient PAGE gel of Hrq1. (I) TEM image of negative-stained Hrq1; white bar = 200 Å. (J) Two-dimensional reconstruction of the Hrq1 heptamer. The inner and outer diameters of the ring are shown. All gel images are representative of ≥3 independent experiments, plotted data represents the average of ≥3 independent experiments, and error bars correspond to the standard deviation. See also Figures S1 and S5.
Fungi such as S. cerevisiae and Schizosaccharomyces pombe were previously described as encoding only one RecQ helicase (Sgs1 and Rqh1, respectively) that is functionally homologous to hBLM (Mirzaei et al., 2011). However, computational analyses recently identified the product of the S. cerevisiae YDR291W gene as a homolog of hRecQ4 (Lee et al., 2005) and found similar RecQ4 homologs in many fungal and plant genomes, naming these proteins Hrq1 (Barea et al., 2008).
Here, we purified S. cerevisiae Hrq1 and showed that it is a 3’–5’ DNA helicase. Mutation of the S. cerevisiae HRQ1 resulted in strong sensitivity to DNA inter-strand crosslinks (ICLs), a phenotype also reported for hRecQ4-deficient fibroblasts (Jin et al., 2008). In addition, Hrq1, like other RecQ helicases, had multiple telomere functions. HRQ1 suppressed telomere addition (TA) to DSBs, an activity it shares with Pif1, a yeast DNA helicase whose human counterpart is proposed to be a tumor suppressor gene (Chisholm et al., 2012). HRQ1 also suppressed telomere hyper-elongation in pif1 mutant cells. However, unlike Pif1, which acts catalytically at both DSBs and telomeres (Boule et al., 2005; Myung et al., 2001a; Zhou et al., 2000), neither of these telomeric functions required the helicase activity of Hrq1. Like hBLM (Stavropoulos et al., 2002) and Sgs1 (Huang et al., 2001; Johnson et al., 2001), Hrq1 was also important for telomerase-independent telomere maintenance.
Results
Purified Hrq1 displays robust helicase activity
To compare the biochemical functions of Hrq1 and RecQ4, full-length S. cerevisiae Hrq1 and hRecQ4, as well as catalytically inactive Hrq1-KA, were purified from E. coli (Fig. 1B and data not shown). In Hrq1-KA, the invariant lysine (K318) in the Walker A box was mutated to alanine (hereafter called KA alleles). The identity of the purified proteins was verified by western blotting and mass spectrometry (data not shown). Two earlier studies on fungal Hrq1 found that the S. pombe Hrq1 has minimal unwinding activity (Groocock et al., 2012), while S. cerevisiae Hrq1 requires a long (≥70 nt) 3’-tail for activity (Kwon et al., 2012). In contrast, our recombinant S. cerevisiae Hrq1 displayed robust helicase activity, similar to that of hRecQ4 (Suzuki et al., 2009) (Fig. 1C–E, G), on a fork substrate with 25-nt single-stranded DNA (ssDNA) tails. Hrq1-KA had no activity (Fig. 1C and data not shown), but it did bind ssDNA almost as well as wild type (WT) Hrq1 (Fig. 1F). We also tested the ability of WT Hrq1 to bind double-stranded DNA (dsDNA) and a ssDNA substrate comprised of the S. cerevisiae telomeric repeat sequence TG1–3. Hrq1 did not bind dsDNA (Fig. 1F) but did bind TG1–3 with weaker affinity (Kd =48±2 nM) than for a poly(dT) substrate (Kd =800±69 pM) but stronger than that for a poly(dG) or random sequence substrate (apparent Kd =260±60 and 560±20 nM, respectively; Fig. 1F and data not shown).
All tested RecQ family helicases unwind DNA in the 3’–5’direction. Using a universal directionality substrate (Shin and Kelman, 2006), hRecQ4 and Hrq1 produced only the expected 3’–5’ unwinding product, while purified S. cerevisiae Pif1 (a 5’-3’ DNA helicase) yielded only the 5’-3’ unwinding product (Fig. 1G). Thus, like other known RecQs, S. cerevisiae Hrq1 is a 3’–5’ DNA helicase and unwinds DNA with similar efficiency as hRecQ4.
To assess the oligomeric state of Hrq1, we subjected the purified protein to gel filtration and native gradient PAGE analysis. Gel filtration indicated that Hrq1 exists as a high molecular weight oligomer in solution (Fig. S1). In native gradient PAGE, Hrq1 migrated as two prominent bands (Fig. 1H) with apparent molecular weights >669 kDa (67% of the protein) and >200 kDa (33%). Negative staining and transmission electron microscopy (TEM) analysis demonstrated a prominent toroidal organization for Hrq1 (Fig. 1I), and two-dimensional image averaging revealed that the particles are heptameric rings (Fig. 1J).
Deletion of HRQ1 sensitizes cells to ICLs
Mutation of disease-linked helicases often sensitizes cells to DNA damage. For instance, deletion of the BLM homolog SGS1 renders S. cerevisiae sensitive to a wide range of genotoxic agents (see http://www.yeastgenome.org/cgi-bin/locus.fpl?locus=sgs1 for complete list). To compare the effects of loss of HRQ1 to loss of SGS1, we deleted each gene and plated serial dilutions of cultures on rich media ± DNA damaging agents. Catalytic inactivation of a helicase can be more detrimental than deletion of the gene encoding it if the inactive protein binds its sites of action and prevents a compensating activity from accessing those sites (Wu and Brosh, 2010). Therefore, we also tested inactive KA alleles of each gene, which were expressed from their native loci and produced stable protein in vivo (Fig. S2). Finally, because hRecQ4 and hBLM interact in vivo (Singh et al., 2012), we also examined the effects of double HRQ1/SGS1 mutants to determine if they have overlapping functions in DNA repair (Table 1, Fig. 2A,B).
Table 1.
Relative sensitivity to DNA damaging agents.
| DNA damaging agent | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Genotype | None* | Bleo | CPT | cisplatin | 4NQO | HU | MMC | MMS | UV |
| WT | +++++ | +++++ | +++++ | +++++ | +++++ | +++++ | +++++ | +++++ | +++++ |
| hrq1Δ | +++++ | +++++ | +++++ | ++++ | ++++ | +++++ | ++ | ++++ | +++++ |
| hrq1-KA | +++++ | +++++ | +++++ | ++++ | +++ | +++++ | + | ++++ | +++++ |
| sgs1Δ | +++++ | ++++ | +++ | + | + | ++ | +++ | +/− | +++ |
| sgs1-KA | +++++ | ++++ | +++ | + | + | ++ | ++++ | +/− | +++ |
| hrq1Δ sgs1Δ | +++++ | +++++ | +++ | + | +/− | ++ | + | +/− | +++ |
| hrq1Δ sgs1-KA | +++++ | +++++ | +++++ | ++++ | + | Res | ++ | ++++ | +++++ |
| hrq1-KA sgs1Δ | ++++ | +++ | ++ | + | +/− | ++ | +/− | +/− | ++ |
| hrq1-KA sgs1-KA | +++++ | +++++ | +++ | + | +/− | ++ | +/− | +/− | ++++ |
Cells were diluted and plated as in Figure 2A on YEPD with (UV) or without (None) exposure to 100 J/m2 ultraviolet radiation or on YEPD containing 5 µg/mL bleomycin (Bleo), 5 µg/mL camptothecin (CPT), 250 µg/mL cisplatin, 125 ng/mL 4-nitroquinoline-n-oxide (4NQO), 100 mM HU, 100 µg/mL MMC, or 0.03% methyl methanesulfonate (MMS). The plates were incubated for 2 (YEPD, Bleo, CPT, 4NQO, and UV), 3 (cisplatin, MMC, and MMS), or 4 (HU) days at 30°C in the dark, and sensitivity was scored relative to growth of the WT strain on each plate. +++++ indicates no sensitivity to the DNA damaging agent, ++++ indicates no or poor growth of the 10−4 dilution (i.e., 10-fold sensitivity relative to WT), +++ indicates 100-fold sensitivity, etc. +/− indicates little to no growth of the OD660=1 spot. “Res” = resistance to the drug relative to WT.
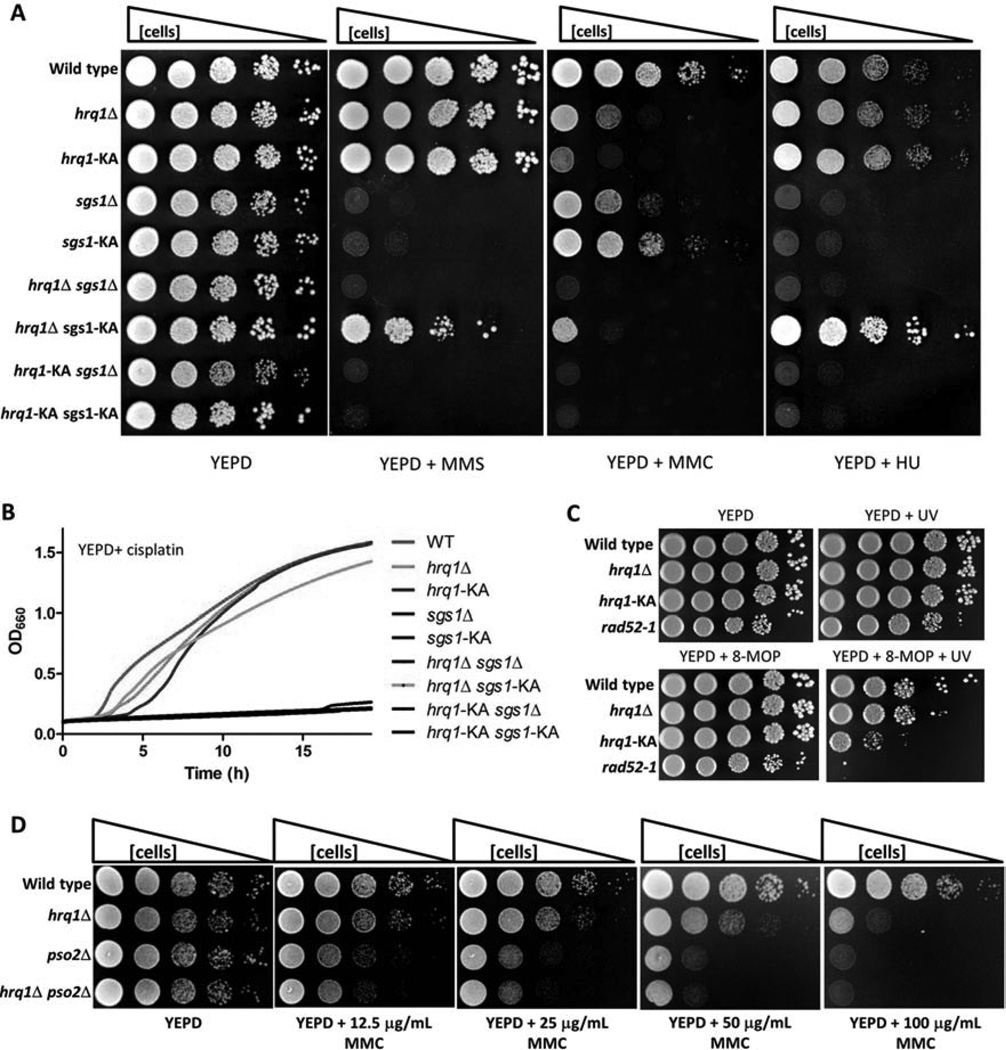
Figure 2. Comparison of hrq1 and sgs1 DNA damage sensitivity.
(A) Growth of the indicated strains on YEPD and YEPD containing 0.03% MMS, 100 µg/mL MMC, or 100 mM HU. Cells of the indicated genotype were grown in liquid culture, diluted to OD660=1, and 10-fold serial dilutions were spotted onto plates, which were then incubated for 2 (YEPD), 3 (MMS and MMC), or 4 (HU) days in the dark. (B) Growth curves of the indicated strains displaying relative sensitivity to cisplatin. Cells were grown overnight in YEPD, diluted to OD660=0.1 in a 96-well plate, and incubated at 30°C in a BioTek EON plate reader with shaking. The OD660 was then measured every 15 min for 24 h. The plotted values are the means of ≥3 independent experiments per strain. (C) 8-MOP+UV sensitivity of the indicated strains. Cells were grown, diluted, and spotted (as in A) on YEPD plates or YEPD plates containing 20 µM 8-MOP and either placed in an opaque container (YEPD and YEPD+8-MOP) or exposed to 365-nm UV for 5 (YEPD+8-MOP+UV) or 15 min (YEPD+UV) and then incubated in the dark for 2 days. The images are representative of results from triplicate experiments. (D) hrq1Δ and pso2Δ are epistatic for MMC sensitivity. Cells were grown, diluted, and spotted (as in A) on YEPD or YEPD+MMC plates and incubated for 2 days. See also Figures S2 and S3.
On rich media, there was no apparent growth difference in hrq1Δ, hrq1-KA, sgs1Δ, or sgs1-KA relative to WT (Fig. 2A), though there were initial lags in the growth of some strains in liquid culture (Fig. S3). We recapitulated the published sgs1Δ sensitivity to 8/8 agents (Table 1, Fig. 2A,B) and showed that sgs1-KA cells had similar patterns of sensitivity. With one exception, mutation of HRQ1 had much milder effects than mutating SGS1; hrq1Δ and hrq1-KA cells had either WT or modestly reduced growth on seven agents to which sgs1 cells showed strong sensitivity (bleomycin, camptothecin (CPT), cisplatin, 4-nitroquinoline-n-oxide (4NQO), hydroxyurea (HU), methyl methanesulfonate (MMS), and UV; see also (Choi et al., 2013)).
However, hrq1Δ and hrq1-KA cells were highly sensitive to MMC, a DNA crosslinker that generates mostly (~80%) inter-strand dG-dG crosslinks (Tomasz, 1995). Thus, Hrq1 acts catalytically during ICL repair. To determine if this sensitivity was specific to dG-dG ICLs, we tested the sensitivity of hrq1 cells to 8-methoxypsoralen (8-MOP), which damages DNA only by inducing ICLs (usually dT-dT) after UV exposure (Averbeck and Averbeck, 1998). Both hrq1Δ and hrq1-KA cells were more sensitive to 8-MOP+UV treatment than WT (Fig. 2C), again indicating that Hrq1 acts catalytically to promote ICL repair. However, hrq1-KA cells were much more sensitive than hrq1Δ (similar to the rad52-1 control) (Henriques and Moustacchi, 1981), suggesting that Hrq1-KA binding to its ICL repair substrate blocks an alternative ICL repair pathway. sgs1Δ cells were not tested on 8-MOP+UV because they are sensitive to UV alone. While sgs1Δ and sgs1-KA cells were only mildly MMC sensitive (Table 1, Fig. 2A), hrq1Δ sgs1Δ cells were dead on MMC. Thus, Hrq1 has a more important role than Sgs1 in suppressing ICL damage, though Sgs1 may have a backup function in cells lacking Hrq1.
Previously, PSO2, which encodes a nuclease (Brendel et al., 2003), was the only S. cerevisiae gene known to suppress ICL damage specifically. We asked if Hrq1 and Pso2 act in the same ICL repair pathway by comparing the MMC sensitivity of WT, hrq1Δ, pso2Δ, and hrq1Δ pso2Δ strains. Although pso2Δ cells were ~10-fold more sensitive than hrq1Δ cells, the hrq1Δ pso2Δ double mutant displayed the same MMC sensitivity as pso2Δ over a range of MMC concentrations (Fig. 2D). Thus, hrq1Δ is epistatic to pso2Δ, suggesting that Hrq1 and Pso2 act in the same ICL repair pathway.
A reverse pattern of sensitivities was seen for cisplatin, which induces mostly (90%) 1,2-intra-strand crosslinks (Table 1, Fig. 2B). More rarely, cisplatin generates dG-dG ICLs (Brabec, 2002). As reported, sgs1Δ cells were highly cisplatin-sensitive (Liao et al., 2007), as were sgs1-KA cells. In contrast, hrq1 and hrq1-KA cells had only modest cisplatin sensitivity (see also (Choi et al., 2013; Dittmar et al., 2013)). Remarkably, deletion of HRQ1 in the sgs1-KA strain strongly suppressed the sgs1-KA cisplatin sensitivity (Fig. 2B). This suppression was not seen for other hrq1 sgs1 combinations, and its mechanism is unknown.
In general, deletion and KA alleles had similar effects on DNA damage sensitivity (Table 1). However, hrq1Δ sgs1-KA cells were more HU resistant than WT cells (Fig. 2A). Although we did not explore the reason for these differences, they may reflect the dual roles of Sgs1 in activating the intra-S phase checkpoint, only one of which is helicase dependent (Frei and Gasser, 2000).
Deleting HRQ1 inhibits de novo TA
In addition to drug sensitivities, helicase mutations often result in high rates of spontaneous DNA damage, such as gross-chromosomal rearrangements (GCRs), which are common in many cancers (Bernstein et al., 2010). The S. cerevisiae GCR assay provides a quantitative measure of such events by selecting for the simultaneous loss of expression of two counter-selectable markers on the left arm of chromosome V (Chr V-L): URA3 and CAN1 (Fig. 3A). Mutation of most yeast genes that are homologs of human tumor suppressor genes (e.g., SGS1) results in increased GCR rates (Myung et al., 2001b).
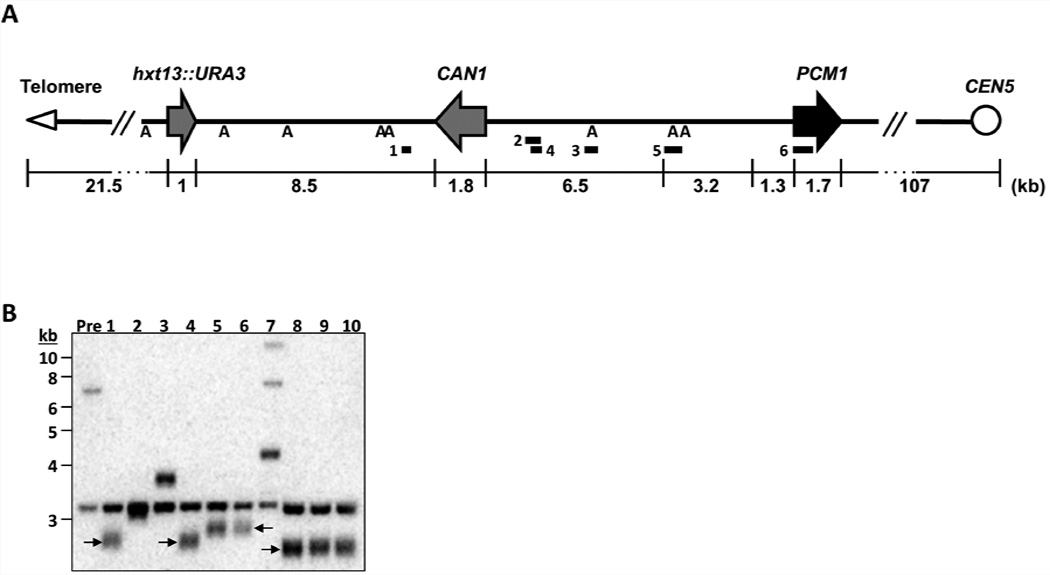
Figure 3. Deletion of HRQ1 affects de novo telomere addition at DSBs.
(A) Schematic of Chr V-L in the GCR strain. The numbered bars indicate the positions of the multiplex PCR products used to determine the relative locations of the GCR events. “A” denotes AlwNI restriction sites. URA3 and CAN1: counter-selectable markers; PCM1: first essential gene to the right of the V-L telomere. (B) Southern blot analysis of representative hrq1Δ GCR clones. The blot was probed with the CIN8 PCR product (primer pair 3 from A). TAs are denoted with arrows.
The sgs1Δ GCR rate was 16-fold higher than in WT cells (p=0.025), similar to the ~20-fold increase reported previously (Myung et al., 2001b). The sgs1-KA strain had the same high GCR rate as sgs1Δ (p=0.020), suggesting that the number of DNA lesions in the two strains was similar (Table 2). In contrast, the GCR rates in both hrq1Δ and hrq1-KA cells were only modestly higher than WT (p=0.043 and 0.048, respectively) (Table 2). The low GCR rate in hrq1 mutants is consistent with our finding that these strains were insensitive to most types of DNA damage except ICLs (Table 1 and Fig. 3), which are rare for cells grown in laboratory conditions (Scharer, 2005). Indeed, pre-growth of hrq1Δ cells in media containing MMC prior to plating resulted in a ~58-fold increase in the GCR rate relative to cells treated with solvent alone (Table 2 and data not shown). The GCR rate of WT cells grown under the same conditions also increased but only 5-fold.
Table 2.
Gross-chromosomal rearrangement rates and analysis of events in independent clones grown in the absence or presence of mitomycin C.
| Genotype | GCR rate*‡ | 95% confidence interval |
Telomere addition (clones analyzed) |
|---|---|---|---|
| WT | 1.0 ± 0.5† | 0.18–2.27 | 0% (10) † |
| WT + MMC | 5 ± 4 | 2.52–11.5 | N.D. |
| sgs1Δ | 16 ± 5† | 5.15–37.4 | 0% (15) † |
| sgs1-KA | 14 ± 3 | 13.1–40.1 | 46±14% (24) |
| hrq1Δ | 4 ± 2 | 3.19–12.6 | 77 ± 2.9% (26) |
| hrq1Δ + MMC | 115 ± 30 | 89.6–188 | N.D. |
| hrq1-KA | 5 ± 3 | 3.48–16.7 | 4.5 ± 3.9% (22) |
| hrq1-KA + MMC | 132 ± 42 | 46.5–246 | N.D. |
| pif1-m2 | 76 ± 8† | 36.4–125 | 93 ± 7.6% (56) † |
Gross-chromosomal rearrangement (GCR) rates are the average of ≥3 independent experiments and are normalized to the WT rate (1.5±0.7×10−10 events/generation). ± denotes standard deviation. + MMC denotes growth in media containing mitomycin C.
p-values were calculated for the GCR rates for all pairwise combinations of strains. All mutant rates are significantly different from WT (p=0.025, sgs1Δ; p=0.020, sgs1-KA; p=0.043, hrq1Δ; p=0.048, hrq1-KA; and p<0.00005, pif1-m2). All rates are significantly different from pif1-m2 (all p≤0.008). The hrq1Δ and hrq1-KA GCR rates are not significantly different from either the sgs1Δ or sgs1-KA rates (all p≥0.078). For telomere additions, the frequency in pif1-m2 cells was significantly different from all other strains (all p<0.030), and the frequencies between hrq1Δ and hrq1-KA were also significantly different (p=0.0002).
Data from (Paeschke et al., 2013), though it was collected at the same time as the other data.
If breaks occur between CAN1 and PCM1 (the most distal essential gene on Chr V-L; Fig. 3A), FOAR CanR cells can theoretically be generated by TA to DSBs. However, in WT cells and virtually all single mutants, GCR events arise from recombination events that delete or move URA3 and CAN1 to new locations (Kolodner et al., 2002). The one published exception is pif1 cells, where most GCR events are due to TA (Myung et al., 2001a). To determine if GCR events were due to TA or recombination, we determined the structure of Chr V-L in multiple independent GCR clones from each mutant (Fig. 3A,B, Table 2, and data not shown).
As anticipated (Myung et al., 2001b), none of the WT (0/10) or sgs1Δ (0/15) GCR clones were due to TA, while 93% of the GCR events in pif1-m2 (an allele that eliminates most of the nuclear Pif1 (Schulz and Zakian, 1994)) cells were TAs (52/56). Remarkably, ~80% (20/26) of the hrq1Δ GCR events were TAs (Table 2 and Fig. 3B). Also surprising, the events leading to the GCRs were different in hrq1-KA (4.5% TA, 1/22) and hrq1Δ (77% TAs) cells, even though the GCR rates were the same in these strains (p=0.264). Thus, unlike Pif1, which requires helicase activity to inhibit telomerase (Myung et al., 2001a; Zhou et al., 2000), Hrq1 inhibited TA non-catalytically. In addition HRQ1 and PIF1 did not act synergistically to suppress TAs, as the two helicases did not have additive effects on TA: the fraction of TAs in hrq1Δ pif1-m2 cells was <50%, lower than in either single mutant. (See discussion for model to explain these data.)
We also determined the structure of Chr V-L in sgs1-KA GCR clones. Although TAs are not detected in sgs1Δ cells (Myung et al., 2001b) (Table 2), ~50% of the GCR events in sgs1-KA cells were TAs (11/24; Table 2). TAs are also detected in sgs1Δ exo1Δ cells, presumably because both pathways for DSB resection are blocked (Lydeard et al., 2010; Marrero and Symington, 2010). As Sgs1-KA binds ssDNA (Cejka and Kowalczykowski, 2010), we suggest that it reduces Exo1 access (Fig. 5A). Thus, both pathways to generate the ssDNA for strand invasion are inhibited, shifting the recombination-TA equilibrium toward TAs.
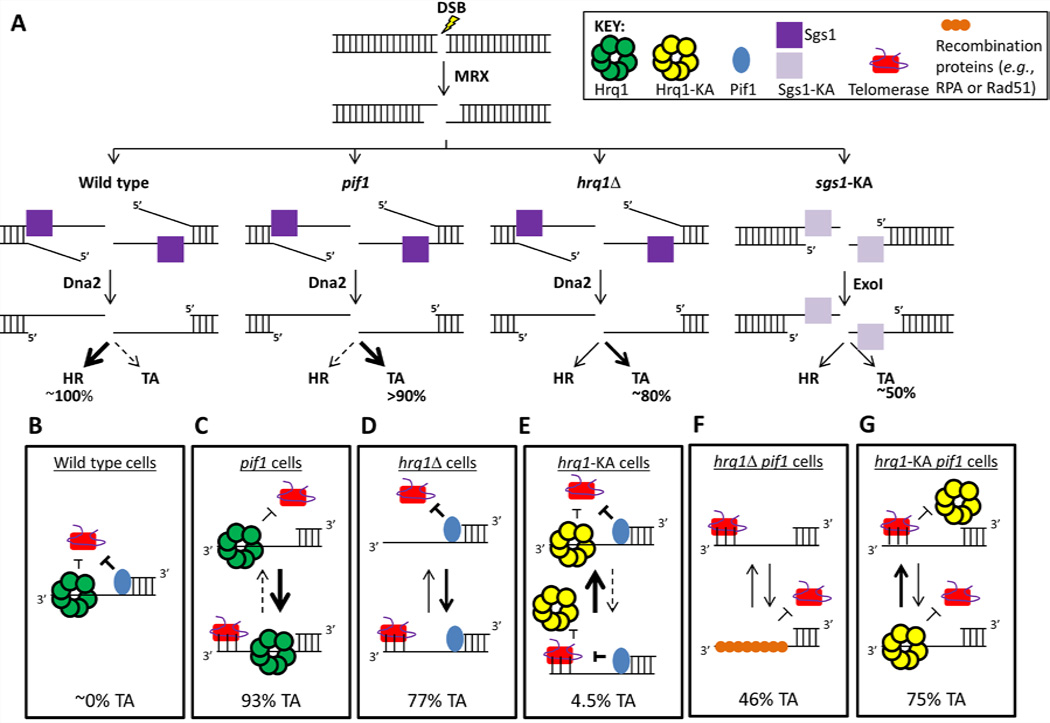
Figure 5. Model for effects of helicases on TA at DSBs.
(A) DSB processing by 5’ end resection in the presence (dark purple) and absence (light purple) of Sgs1 activity (Zhu et al., 2008). A DSB is initially processed by the MRX complex, and then 5’ end resection occurs via the action of Sgs1 and the nuclease Dna2 in WT cells or Exo1 in sgs1Δ (not shown) and sgs1-KA cells. In WT cells, virtually all DSBs are healed by HR rather than TA. Three ways to shift this balance toward TA are to mutate PIF1, delete HRQ1, and express inactive Sgs1. (B–G) Likelihood of TA at a DSB in (B) WT, (C) pif1, (D) hrq1Δ, (E) hrq1-KA, (F) hrq1Δ pif1, and (G) hrq1-KA pif1 cells. See the discussion for a detailed explanation. Note that Pif1 is shown bound to the ss/dsDNA junction because single-molecule analysis indicates that this is its preferred binding position, and it does not translocate from this position toward the 3’ end of the recessed strand (Zhou, Bochman, Zakian, and Ha, in preparation). See also Figures S1 and S5.
Hrq1 limits telomere lengthening in pif1-m2 cells
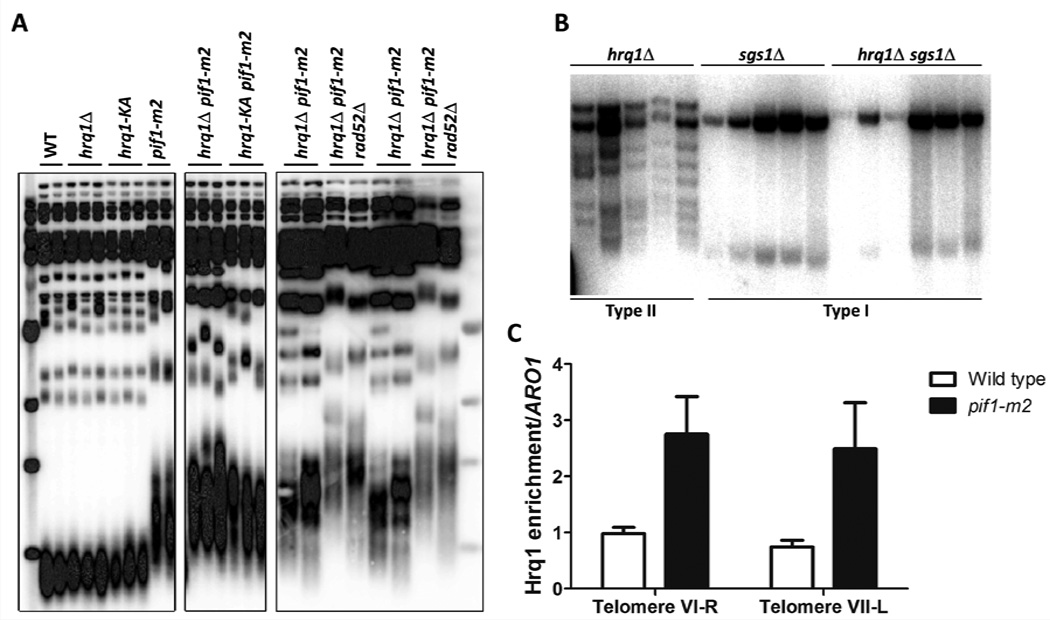
The ability of Hrq1 to suppress TAs to DSBs suggested that like Pif1, it might also inhibit telomerase lengthening of existing telomeres. However, telomere length was indistinguishable from WT in hrq1Δ and hrq1-KA cells (Fig. 4A). In contrast, telomeres in hrq1Δ pif1-m2 cells were even longer than in pif1-m2.
Figure 4. Hrq1 affects telomere maintenance.
(A) Telomere blots of gDNA from the indicated strains. Two leftmost panels are cropped from a gel with intervening lanes removed; see Figure S4. (B) Telomere blot of gDNA from tlc1Δ survivors from the indicated strains after growth in the absence of TLC1 and the indicated helicases. (C) Increased Hrq1-Myc binding to telomeres. Binding was normalized to input DNA and ARO1. The data are the mean of 5 independent experiments, and the error bars correspond to the standard deviation. See also Figure S4.
The hyper-lengthening of telomeres in hrq1Δ pif1-m2 cells could be due to recombination or telomerase. To distinguish between these possibilities, we analyzed telomere length in spore clones from a pif1-m2/WT hrq1Δ/WT rad52Δ/WT diploid (Rad52 is a protein required for virtually all homologous recombination (HR) (Hiom, 1999)). Telomeres were even longer in hrq1Δ pif1-m2 rad52Δ cells than in recombination-proficient cells (Fig. 4A), demonstrating that the hyper-lengthening was not recombination dependent. Telomeres are also longer in rad52 versions of other mutants with long telomeres (e.g., rif1 and rif2), suggesting that HR suppresses elongation in cells that already have long telomeres (Teng et al., 2000; Zhou et al., 2000). Thus, Hrq1 limits telomerase, not recombination, at pif1-m2 telomeres.
Hrq1 did not act catalytically to inhibit telomerase at DSBs (Table 2). To determine if it acts catalytically to affect telomere length in pif1-m2 cells, we analyzed telomeres in hrq1-KA pif1-m2 and hrq1-KA pif1-m2 rad52Δ cells. Telomere length was unaffected in the hrq1-KA pif1-m2 background relative to pif1-m2 (Fig. 4A) (as expected, deletion of RAD52 did increase hrq1-KA pif1-m2 telomere length as seen for pif1-m2 vs. rad52Δ cells (Zhou et al., 2000)). Thus, Hrq1 acts structurally to inhibits lengthening of pif1-m2 telomeres, just as it does during TA at DSBs.
Telomerase-independent telomere maintenance by the type I pathway is Hrq1 dependent
In telomerase-deficient yeast and human cells, survivors arise that maintain telomeres by mechanisms that usually involve HR (i.e., alternative lengthening of telomeres or ALT) (Wellinger and Zakian, 2012). In S. cerevisiae, telomerase-independent survivors come in two types: type I, which have tandem copies of the sub-telomeric Y’ element and very short tracts of telomeric DNA; and type II, which have heterogeneous length telomeres. Because hBLM (Stavropoulos et al., 2002) and Sgs1 (Huang et al., 2001; Johnson et al., 2001) both influence ALT, we asked if Hrq1 does as well. We sporulated an hrq1Δ/WT sgs1Δ/WT tlc1Δ/WT diploid strain (TLC1 encodes telomerase RNA), serially restreaked spore clones until survivors appeared, and examined telomere structure in 20 survivors from each strain. As previously reported, sgs1Δ tlc1Δ spore clones generated only type I survivors. In contrast, all hrq1Δ tlc1Δ survivors had type II telomeres; sgs1Δ was epistatic to hrq1Δ because all hrq1Δ sgs1Δ tlc1Δ survivors were type I (Fig. 4B). Thus, Hrq1 is required to generate type I survivors in the presence of Sgs1.
Hrq1 binds telomeres in vivo
If Hrq1 affects telomeres directly, it will bind telomeres, which might be detectable by chromatin immunoprecipitation (ChIP). In WT cells, Hrq1-13xMyc bound significantly to telomere VI-R (relative to a control site; p<0.001); binding to the VII-L telomere was detectable but not significant (p=0.0567; Fig. 4C). However, in pif1-m2 cells, Hrq1 binding was significant at both (p<0.001; ~2.5-fold higher to pif1-m2 vs. WT telomeres). These results suggest that Hrq1 is telomere associated in WT cells but that Pif1 can displace it from telomeres. This interpretation also explains why telomeres in hrq1Δ cells are of WT length (Fig. 4A).
Discussion
Hrq1 protects against ICLs
Unlike sgs1 cells, hrq1 cells were resistant to most DNA damaging agents (Table 1). This pattern is reminiscent of the damage sensitivities for their human homologs, as hRecQ4-deficient cells are less sensitive than hBLM mutant cells to many DNA damaging agents, including cisplatin (see (Mao et al., 2010) and references therein). However, a different pattern was seen for two ICL-inducing agents: hrq1Δ and hrq1-KA cells were highly sensitive to both MMC and 8-MOP. Thus, Hrq1 acts catalytically to promote ICL repair. Cells expressing Hrq1-KA were much more sensitive to these agents than hrq1Δ cells (Fig. 2A,C). This finding suggests that Hrq1 is the main helicase in ICL repair, and its backup is hindered from accessing the DNA because Hrq1-KA is bound to sites of damage. The even higher MMC-sensitivity of hrq1 sgs1 cells (Fig. 2A) suggests that Sgs1 is the backup for Hrq1 at ICLs. Even the modest sensitivity of hrq1 cells to cisplatin, a predominantly intra-strand crosslinker (Table 1 and Fig. 2B), can be explained by its role in ICL repair, as ~5% of cisplatin lesions are ICLs (Brabec, 2002).
Although our study is the first report of MMC or 8-MOP sensitivity for hrq1 S. cerevisiae, S. pombe hrq1Δ cells are highly sensitive to both cisplatin and MMC (Groocock et al., 2012). However, because 8-MOP was not tested in that study, it is possible that the S. pombe hrq1Δ MMC sensitivity is due to the low level of intra-strand crosslinks generated by MMC. In any case, as S. cerevisiae Sgs1 has quite different in vivo functions from its S. pombe homolog Rqh1 (Ashton and Hickson, 2010; Cromie et al., 2008), it is not surprising that Hrq1 also functions differently in these distantly related yeasts.
ICLs are dangerous because covalent linkage of the two DNA strands prevents both transcription and DNA replication. In addition, ICLs are of medical interest as patients with Fanconi’s anemia (FA), an inherited disease arising from mutation in any of 16 FA genes, are defective in their repair (Clauson et al., 2013). Recently, putative yeast homologs of some of the FA proteins have been identified, but single mutants in these genes are not sensitive to ICL agents (Daee et al., 2012; Daee and Myung, 2012; Ward et al., 2012). Whereas mammals have multiple helicases that suppress ICL damage (e.g., HELQ acts in a pathway parallel to FA to suppress ICL damage (Adelman et al., 2013)), our study is the first to identify a helicase, Hrq1, whose elimination renders S. cerevisiae highly ICL-sensitive. Indeed, HRQ1 and PSO2 are the only S. cerevisiae genes that specifically suppress ICL damage, and genetic data indicate that they act in the same pathway (Fig. 2D). We do not know how Hrq1 acts to facilitate ICL repair, but one possibility is that it functions analogously to HEL308, a human 3’–5’ helicase involved in crosslink repair (Moldovan et al., 2010). hRecQ4 is also implicated in ICL repair (Larizza et al., 2010), and Hrq1 may be its functional homolog in yeast ICL repair.
Hrq1 affects diverse aspects of telomere biology
Our analysis revealed multiple telomere functions for Hrq1. Given that Hrq1 was present by ChIP at telomeres, especially in pif1-m2 cells (Fig. 4C), Hrq1 likely affects telomeres directly. Hrq1 impacts the two major pathways of telomere maintenance: telomerase and recombination. It inhibited telomerase-mediated telomere lengthening in pif1-m2 cells (Fig. 4A) and promoted type I survivor formation in tlc1Δ cells (Fig. 4B). Generation of type II survivors is Sgs1 dependent (Huang et al., 2001; Johnson et al., 2001) (Fig. 4B). Thus, as with crosslink repair, the two S. cerevisiae RecQ helicases have complementary effects on telomerase-independent telomere maintenance.
The most unexpected telomere effect of Hrq1 is its non-catalytic inhibition of TA. Remarkably, 77% of the GCR events in hrq1Δ cells were TAs (Table 2). Until this report, pif1 was the only single mutant in which TAs are easily detected (93% TA, Table 2) (Myung et al., 2001a; Paeschke et al., 2013). In vivo and in vitro, Pif1 uses its ATPase activity to remove telomerase from DNA ends, and thus, TA increases in pif1Δ, pif1-m2, and pif1-KA cells (Boule et al., 2005; Myung et al., 2001a; Zhou et al., 2000). If Hrq1 also removes telomerase, hrq1-KA cells should have high TA rates, and nearly all hrq1Δ pif1-m2 GCR events should be TAs. In contrast, TAs were rare in hrq1-KA (4.5%) and considerably lower in hrq1Δ pif1-m2 (46%) cells than in either single mutant. This structural role of Hrq1 is the key to understanding its mechanism of action at both DSBs and telomeres.
There are two major pathways for DSB repair in yeast: HR and TA. These pathways can be thought of as being in competition with each other, even though TA is rare in WT cells and virtually all tested mutants. Inhibiting TA is critical for genome integrity as it results in aneuploidy for all sequences distal to the TA.
The balance between TA and recombination can be altered by preventing HR, as in sgs1Δ exo1Δ (Lydeard et al., 2010; Marrero and Symington, 2010) or sgs1-KA cells (Table 2) or by eliminating a telomerase inhibitor, as in pif1 cells. Hrq1 is unlikely to affect the HR-TA balance by promoting recombination, as it has not been recovered in the large number of screens for genes that affect recombination. Its binding to telomeres (Fig. 4C) and inhibition of telomerase at pif1-m2 telomeres (Fig. 4A) also argue that it affects telomerase, not recombination, at DSBs.
We propose a working model in which Hrq1 inhibits telomerase by competing with it for ssDNA binding (Fig. 5D,E). According to this model, TA is frequent in hrq1Δ GCR clones because telomerase has better access to its substrate (but not as frequent as in pif1-m2 cells because Pif1 is still there to remove telomerase; Fig. 5D). TA is infrequent in hrq1-KA cells because inactive Hrq1-KA still binds ssDNA (Fig. 1F) and competes with telomerase for ssDNA binding (Fig. 5E). We propose that Hrq1/ Hrq1-KA also compete with recombination proteins (e.g., RPA or Rad51) for binding to ssDNA. This hypothesis would explain why TA was not as frequent in hrq1Δ pif1-m2 cells as in either single mutant because recombination proteins and telomerase then compete with each other for ssDNA in the absence of both Pif1 and Hrq1 (Fig. 5F). Consistent with this model, Hrq1 bound preferentially to ss- vs. dsDNA (Fig. 1F).
The same model can explain how Hrq1 affects telomeres (Fig. 4A). It predicts that Pif1 expels both telomerase and Hrq1 from telomeres, so hrq1Δ telomeres are WT in length. However, telomeres were longer in hrq1Δ pif1-m2 than pif1-m2 cells (Fig. 4A,B and 5A) because when Pif1 is absent, Hrq1 (or Hrq1-KA) limits telomerase by competing with it for binding to telomeric ssDNA. Consistent with this hypothesis, Hrq1 bound single-stranded telomeric DNA in vitro (Fig. 1F).
In summary, our studies show that Hrq1 has two roles that promote genome integrity: 1) it acts catalytically to promote ICL repair. Indeed, the very strong ICL sensitivity of hrq1-KA cells suggests that Hrq1 may be the first line of defense against these dangerous lesions. 2) We also found that Hrq1 affects several aspects of telomere biology, including inhibition of telomerase at DSBs and telomeres. Two of the defects in hrq1 cells, sensitivity to ICLs and frequent TAs, are rare phenotypes; the demonstration of both functions in one protein is unprecedented. If hRecQ4 has either or both activities, it can explain why its mutation results in genomic instability and disease.
Experimental Procedures
Yeast strains, media, and other reagents
All strains (Supplemental Table 1) were created by standard methods and are derivatives of the YPH background (Sikorski and Hieter, 1989). Cells were grown in standard S. cerevisiae media at 30°C unless indicated. 32P-dCTP and 32P-ATP were purchased from PerkinElmer, and unlabeled ATP was from GE Healthcare. DNA damaging agents were from Sigma. All restriction enzymes were from NEB, and all oligonucleotides were from IDT.
Protein purification
Details on HRQ1 and hrq1-KA cloning and expression vector construction, as well as the complete protein purification protocol, can be found in the Supplemental Materials. Briefly, expression plasmids were transformed into Rosetta™ 2(DE3) pLysS cells, and recombinant protein was expressed using the autoinduction method (Studier, 2005). Cells were harvested by centrifugation. The pellets were resuspended in buffer and lysed by adding n-dodecyl β-D-maltoside (DM; Sigma) to a final concentration of 0.05% (w/v) and 1× FastBreak (Promega).
The soluble fraction was clarified by centrifugation and filtering the through a 0.22-µm membrane. This mixture was then loaded onto a Strep-Tactin Sepharose column (IBA), and protein was eluted with 3 column volumes (CVs) of desthiobiotin buffer after extensive washing. Peak fractions were pooled and loaded onto a His60 Ni column (Clontech). After washing, protein was eluted with 6 CVs of imidazole buffer, and peak fractions were pooled and concentrated by ultrafiltration. The protein was then buffer-exchanged into storage buffer using a desalting column.
The hRecQ4 expression plasmid pGEX-RecQ4-His9 was a gift from Patrick Sung, Yale University. hRecQ4 was purified as described (Macris et al., 2006). The protein concentration and purity of the final preparations were determined on SYPRO orange-stained SDS-PAGE gels using known amounts of a standard protein for comparison. In all cases, protein purity was ≥95%.
Helicase assays
The fork substrate was constructed by end labeling oligonucleotide MB733 (Table S2) with T4 PNK and γ32P-ATP and separating the ssDNA from free label using a G-50 micro column (GE Healthcare). Labeled MB733 was then annealed to oligonucleotide MB734 in 1× NEB buffer #2 by boiling and slowly cooling to room temperature. The directionality substrate was similarly constructed by end labeling an equimolar mixture of oligonucleotides MB453 and 454 (Table S2) and removing free label using a G-50 micro column. The labeled oligonucleotides were then annealed to MB452 as above. Both substrates were separated from contaminating ssDNA by gel purification, followed by electroelution into a dialysis membrane.
Helicase reactions were performed in 1× binding buffer (25 mM Na-HEPES (pH 8.0), 5% glycerol, 50 mM NaOAc, 150 µM NaCl, 7.5 mM MgOAc, and 0.01% DM) and contained 0.1 nM radiolabeled substrate, protein as indicated, 5 mM ATP, and 15 nM unlabeled oligonucleotide MB733 (fork) or MB453 and MB454 (directionality). Hrq1 helicase activity was unaffected by omitting these unlabeled ssDNA traps (e.g., Fig. S5). Reactions containing the directionality substrate additionally contained 100 µg/mL Neutravidin (Pierce). The reactions were incubated at 37°C for 30 min and stopped by the addition of 1× stop-load buffer (25% (w/v) Ficoll (type 400), 100 mM EDTA, 0.1% SDS, 0.25% bromophenol blue, and 0.25% xylene cyanol). The samples were then separated on 8% 29:1 acrylamide:bis-acrylamide gels in 1× TBE buffer at 100 V/cm for 30–45 min, dried, and imaged/quantified using a Typhoon 9410 scanner and Image Gauge software.
Native gradient gel electrophoresis
Native gradient gels were poured following the protocol in the Supplemental Experimental procedures. Protein (≥0.5 µg) was loaded into the wells in the absence of loading dye and run at 17 V/cm for 3–6 h in 20 mM Tris-HCl (pH 8.8) and 200 mM glycine. After electrophoresis, gels were incubated in 0.05% SDS for 15 min, rinsed with water, and stained with SYPRO orange overnight prior to analysis as described above for protein concentration.
TEM
Hrq1 was analyzed by TEM essentially as previously described for the Mcm2-7 helicase (Bochman and Schwacha, 2007). Briefly, the protein was diluted to 25 µg/ml in storage buffer, absorbed to glow-discharged copper grids (Ted Pella, Redding, CA), and negatively stained with 2% uranyl acetate. Grids were visualized with a LEO OMEGA 912 electron microscope (Carl Zeiss) at 80 kV and 40,000× magnification. Micrographs were taken with a 7.5-megapixel EMCCD camera (Peltier-cooled Hamamatsu ORCA) and visualized with AMT (ver. 602) software. Two-dimensional image averaging of Hrq1 complexes was performed using EMAN2 (Tang et al., 2007).
GCR assays
GCR assays were performed essentially as described by Putnam and Kolodner (Cold Spring HarbProtoc; 2010; doi:10.1101/pdb.prot5492). Briefly, ≥3 sets of five or more 5-mL cultures of each S. cerevisiae GCR strain (Table 1) were grown to saturation in YEPD medium ±25 µg/mL MMC at 30°C for 36–48 h. Cells (2 mL) from each culture were pelleted, resuspended in sterile water, plated on drop-out medium lacking uracil and arginine (US Biologicals) supplemented with 1g/L 5-FOA and 60 mg/L canavanine sulfate (FOA+Can), and incubated at 30°C for 4 days. GCR rates (per 10−9 mutations/generation) and 95% confidence intervals were calculated using the FALCOR web server and MMS Maximum Likelihood Method (Hall et al., 2009). The rates presented are the means±standard deviations of ≥3 experiments per strain. p values were calculated using Student’s t-test. We define GCR clones as colonies that grew on the FOA+Can plates. Such FOAR CanR clones were selected for post-GCR analyses (multiplex PCR (Supplemental Experimental Procedures) and Southern blotting, below).
Southern blotting
When colonies arose after GCR events, they were restreaked onto FOA+Can plates to verify the FOAR CanR phenotype, grown overnight in YEPD liquid media at 30°C, and genomic DNA (gDNA) was isolated. The gDNA was analyzed by multiplex PCR and Southern blotting as described (Paeschke et al., 2013). Briefly, gDNA from clones retaining the CIN8 PCR product was digested overnight at 37°C with AlwNI, run on 0.7% agarose gels, and blotted on Hybond-XL membranes (GE Life Sciences). For the Southerns, the 400-bp CIN8 PCR product was used a probe (hybridization sites are shown in Fig. 3A), resulting in a 3.2-kb background band in all lanes. A non-GCR event yields a 6.9-kb band, TAs produce fuzzy bands <5.5 kb, and non-TAs result in sharp bands.
For telomere blots, gDNA was isolated from cells as described above. For the experiments examining the effect of recombination on telomere length, heterozygous diploids (Table S1) were sporulated, tetrads were dissected, and 3 spore clones of the desired genotypes were serially restreaked on YEPD for ~100 generations prior to gDNA isolation. For standard telomere length detection, gDNA was digested overnight with PstI and XhoI, whereas for telomere blots from tlc1Δ survivors, the gDNA was digested overnight with XhoI. Digests were separated on 1% agarose gels and transferred as described above. In both types of telomere blots, the probe used was a previously described C1–3A/TG1–3 restriction fragment (Runge and Zakian, 1989).
tlc1Δ survivor analysis
The generation and analysis of telomeric survivors were performed as described previously (Teng and Zakian, 1999). Briefly, a HRQ1/hrq1Δ SGS1/sgs1Δ TLC1/tlc1Δ diploid strain (KP386) was generated, sporulated, and hrq1Δ tlc1Δ and sgs1Δ tlc1Δ spores were identified by plating. Spores where restreaked 4–5 times (~25 generations/streak) on YC plates until survivors formed. For each spore clone, 10 colonies where picked for the next restreak. In total, gDNA was isolated from 20 different spores for each strain background and analyzed as described above.
Chromatin immunoprecipitation (ChIP)
ChIP of asynchronous yeast cells growing in YEPD was performed as described (Azvolinsky et al., 2009) and analyzed using an iCycleriQ Real-Time PCR detection system (Bio-Rad Laboratories). Hrq1 was C-terminally tagged with 13 Myc epitopes, and ChIP was performed using an anti-Myc monoclonal antibody (Clontech #631206). The amount of DNA in the immunoprecipitate was normalized to the amount in input samples. The ChIP experiment was analyzed by quantitative PCR (qPCR) in duplicate or triplicate to obtain an average value for each sample. The ChIP experiment was repeated 3 times at each locus. For each qPCR experiment, the amount of signal in the Hrq1 immunoprecipitate was normalized to input and to the immunoprecipitated signal from ARO1.
Supplementary Material
Highlights.
Hrq1 is a functional homolog of hRecQ4.
Purified Hrq1 is a potent in vitro helicase and forms heptameric rings.
Hrq1 acts catalytically to protect against DNA inter-strand crosslinks.
Hrq1 has many telomere roles, e.g., non-catalytic suppression of telomere addition.
Acknowledgements
M.L.B. dedicates this article to the memory of Megan Davey who passed away during the course of this work. We thank Kathy Friedman, Margaret Platts, Kristina Schmidt, Raymund Wellinger, and Nadine Guenther for sharing their methods, suggesting experiments, for useful discussions, and assistance with experiments. Research in the Zakian laboratory is supported by grants from the National Institutes of Health (GM26938 and GM GM43265) and postdoctoral fellowships from the American Cancer Society (to M.L.B.; PF-10-145-01-DMC), the Deutsche Forschungsgemeinschaft (to K.P.), and the New Jersey Commission on Cancer Research (to K.P.). Research in the Paeschke laboratory is supported by the Emmy-Noether Program of the Deutsche Forschungsgemeinschaft.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
The authors declare no conflicts of interest.
References
- Abdelhaleem M. Helicases: an overview. Methods Mol Biol. 2010;587:1–12. doi: 10.1007/978-1-60327-355-8_1. [DOI] [PubMed] [Google Scholar]
- Adelman CA, Lolo RL, Birkbak NJ, Murina O, Matsuzaki K, Horejsi Z, Parmar K, Borel V, Skehel JM, Stamp G, et al. HELQ promotes RAD51 paralogue-dependent repair to avert germ cell loss and tumorigenesis. Nature. 2013 doi: 10.1038/nature12565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashton TM, Hickson ID. Yeast as a model system to study RecQ helicase function. DNA Repair (Amst) 2010;9:303–314. doi: 10.1016/j.dnarep.2009.12.007. [DOI] [PubMed] [Google Scholar]
- Averbeck D, Averbeck S. DNA photodamage, repair, gene induction and genotoxicity following exposures to 254 nm UV and 8-methoxypsoralen plus UVA in a eukaryotic cell system. Photochemistry and photobiology. 1998;68:289–295. [PubMed] [Google Scholar]
- Azvolinsky A, Giresi P, Lieb J, Zakian V. Highly transcribed RNA polymerase II genes are impediments to replication fork progression in Saccharomyces cerevisiae. Mol Cell. 2009;34:722–734. doi: 10.1016/j.molcel.2009.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barea F, Tessaro S, Bonatto D. In silico analyses of a new group of fungal and plant RecQ4-homologous proteins. Comput Biol Chem. 2008;32:349–358. doi: 10.1016/j.compbiolchem.2008.07.005. [DOI] [PubMed] [Google Scholar]
- Bernstein KA, Gangloff S, Rothstein R. The RecQ DNA helicases in DNA repair. Annu Rev Genet. 2010;44:393–417. doi: 10.1146/annurev-genet-102209-163602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bochman ML, Schwacha A. Differences in the single-stranded DNA binding activities of MCM2-7 and MCM467: MCM2 and MCM5 define a slow ATP-dependent step. J Biol Chem. 2007;282:33795–337804. doi: 10.1074/jbc.M703824200. [DOI] [PubMed] [Google Scholar]
- Boule J, Vega L, Zakian V. The Yeast Pif1p helicase removes telomerase from DNA. Nature. 2005;438:57–61. doi: 10.1038/nature04091. [DOI] [PubMed] [Google Scholar]
- Brabec V. DNA modifications by antitumor platinum and ruthenium compounds: their recognition and repair. Prog Nucleic Acid Res Mol Biol. 2002;71:1–68. doi: 10.1016/s0079-6603(02)71040-4. [DOI] [PubMed] [Google Scholar]
- Brendel M, Bonatto D, Strauss M, Revers LF, Pungartnik C, Saffi J, Henriques JA. Role of PSO genes in repair of DNA damage of Saccharomyces cerevisiae. Mutat Res. 2003;544:179–193. doi: 10.1016/j.mrrev.2003.06.018. [DOI] [PubMed] [Google Scholar]
- Capp C, Wu J, Hsieh TS. RecQ4: the second replicative helicase? Crit Rev Biochem Mol Biol. 2010;45:233–242. doi: 10.3109/10409231003786086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cejka P, Kowalczykowski SC. The full-length Saccharomyces cerevisiae Sgs1 protein is a vigorous DNA helicase that preferentially unwinds holliday junctions. J Biol Chem. 2010;285:8290–8301. doi: 10.1074/jbc.M109.083196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chisholm KM, Aubert SD, Freese KP, Zakian VA, King MC, Welcsh PL. A genomewide screen for suppressors of Alu-mediated rearrangements reveals a role for PIF1. PLoS One. 2012;7:e30748. doi: 10.1371/journal.pone.0030748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi DH, Lee R, Kwon SH, Bae SH. Hrq1 functions independently of Sgs1 to preserve genome integrity in Saccharomyces cerevisiae. J Microbiol. 2013;51:105–112. doi: 10.1007/s12275-013-3048-2. [DOI] [PubMed] [Google Scholar]
- Clauson C, Scharer OD, Niedernhofer L. Advances in understanding the complex mechanisms of DNA interstrand cross-link repair. Cold Spring Harbor perspectives in medicine. 2013;3:a012732. doi: 10.1101/cshperspect.a012732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cromie GA, Hyppa RW, Smith GR. The fission yeast BLM homolog Rqh1 promotes meiotic recombination. Genetics. 2008;179:1157–1167. doi: 10.1534/genetics.108.088955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daee DL, Ferrari E, Longerich S, Zheng XF, Xue X, Branzei D, Sung P, Myung K. Rad5-dependent DNA repair functions of the Saccharomyces cerevisiae FANCM protein homolog Mph1. J Biol Chem. 2012;287:26563–26575. doi: 10.1074/jbc.M112.369918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daee DL, Myung K. Fanconi-like crosslink repair in yeast. Genome integrity. 2012;3:7. doi: 10.1186/2041-9414-3-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dittmar JC, Pierce S, Rothstein R, Reid RJ. Physical and genetic-interaction density reveals functional organization and informs significance cutoffs in genome-wide screens. Proc Natl Acad Sci U S A. 2013;110:7389–7394. doi: 10.1073/pnas.1219582110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frei C, Gasser SM. The yeast Sgs1p helicase acts upstream of Rad53p in the DNA replication checkpoint and colocalizes with Rad53p in S-phase-specific foci. Genes Dev. 2000;14:81–96. [PMC free article] [PubMed] [Google Scholar]
- Groocock LM, Prudden J, Perry JJ, Boddy MN. The RecQ4 orthologue Hrq1 is critical for DNA interstrand cross-link repair and genome stability in fission yeast. Mol Cell Biol. 2012;32:276–287. doi: 10.1128/MCB.06184-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall BM, Ma CX, Liang P, Singh KK. Fluctuation analysis CalculatOR: a web tool for the determination of mutation rate using Luria-Delbruck fluctuation analysis. Bioinformatics. 2009;25:1564–1565. doi: 10.1093/bioinformatics/btp253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henriques JA, Moustacchi E. Interactions between mutations for sensitivity to psoralen photoaddition (pso) and to radiation (rad) in Saccharomyces cerevisiae. J Bacteriol. 1981;148:248–256. doi: 10.1128/jb.148.1.248-256.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiom K. Dna repair: Rad52 - the means to an end. Curr. Biol. 1999;9:R446–R448. doi: 10.1016/s0960-9822(99)80278-4. [DOI] [PubMed] [Google Scholar]
- Huang P, Pryde F, Lester D, Maddison R, Borts R, Hickson I, Louis E. SGS1 is required for telomere elongation in the absence of telomerase. Curr. Biol. 2001;11:125–129. doi: 10.1016/s0960-9822(01)00021-5. [DOI] [PubMed] [Google Scholar]
- Jin W, Liu H, Zhang Y, Otta SK, Plon SE, Wang LL. Sensitivity of RECQL4-deficient fibroblasts from Rothmund-Thomson syndrome patients to genotoxic agents. Hum Genet. 2008;123:643–653. doi: 10.1007/s00439-008-0518-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson F, Marciniak R, McVey M, Stewart S, Hahn W, Guarente L. The Saccharomyces cerevisiae WRN homolog Sgs1p participates in telomere maintenance in cells lacking telomerase. EMBO J. 2001;20:905–913. doi: 10.1093/emboj/20.4.905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolodner RD, Putnam CD, Myung K. Maintenance of genome stability in Saccharomyces cerevisiae. Science. 2002;297:552–557. doi: 10.1126/science.1075277. [DOI] [PubMed] [Google Scholar]
- Kwon SH, Choi DH, Lee R, Bae SH. Saccharomyces cerevisiae Hrq1 requires a long 3'-tailed DNA substrate for helicase activity. Biochem Biophys Res Commun. 2012;427:623–628. doi: 10.1016/j.bbrc.2012.09.109. [DOI] [PubMed] [Google Scholar]
- Larizza L, Roversi G, Volpi L. Rothmund-Thomson syndrome. Orphanet J Rare Dis. 2010;5:2. doi: 10.1186/1750-1172-5-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee W, St Onge RP, Proctor M, Flaherty P, Jordan MI, Arkin AP, Davis RW, Nislow C, Giaever G. Genome-wide requirements for resistance to functionally distinct DNA-damaging agents. PLoS Genet. 2005;1:e24. doi: 10.1371/journal.pgen.0010024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao C, Hu B, Arno MJ, Panaretou B. Genomic screening in vivo reveals the role played by vacuolar H+ ATPase and cytosolic acidification in sensitivity to DNA-damaging agents such as cisplatin. Mol Pharmacol. 2007;71:416–425. doi: 10.1124/mol.106.030494. [DOI] [PubMed] [Google Scholar]
- Liu Y. Rothmund-Thomson syndrome helicase, RECQ4: On the crossroad between DNA replication and repair. DNA Repair (Amst) 2010;9:325–330. doi: 10.1016/j.dnarep.2010.01.006. [DOI] [PubMed] [Google Scholar]
- Lydeard JR, Lipkin-Moore Z, Jain S, Eapen VV, Haber JE. Sgs1 and exo1 redundantly inhibit break-induced replication and de novo telomere addition at broken chromosome ends. PLoS Genet. 2010;6:e1000973. doi: 10.1371/journal.pgen.1000973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macris MA, Krejci L, Bussen W, Shimamoto A, Sung P. Biochemical characterization of the RECQ4 protein, mutated in Rothmund-Thomson syndrome. DNA Repair (Amst) 2006;5:172–180. doi: 10.1016/j.dnarep.2005.09.005. [DOI] [PubMed] [Google Scholar]
- Mao FJ, Sidorova JM, Lauper JM, Emond MJ, Monnat RJ. The human WRN and BLM RecQ helicases differentially regulate cell proliferation and survival after chemotherapeutic DNA damage. Cancer Res. 2010;70:6548–6555. doi: 10.1158/0008-5472.CAN-10-0475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marrero VA, Symington LS. Extensive DNA end processing by exo1 and sgs1 inhibits break-induced replication. PLoS Genet. 2010;6:e1001007. doi: 10.1371/journal.pgen.1001007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirzaei H, Syed S, Kennedy J, Schmidt KH. Sgs1 truncations induce genome rearrangements but suppress detrimental effects of BLM overexpression in Saccharomyces cerevisiae. J Mol Biol. 2011;405:877–891. doi: 10.1016/j.jmb.2010.11.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moldovan GL, Madhavan MV, Mirchandani KD, McCaffrey RM, Vinciguerra P, D'Andrea AD. DNA polymerase POLN participates in cross-link repair and homologous recombination. Mol Cell Biol. 2010;30:1088–1096. doi: 10.1128/MCB.01124-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myung K, Chen C, Kolodner RD. Multiple pathways cooperate in the suppression of genome instability in Saccharomyces cerevisiae. Nature. 2001a;411:1073–1076. doi: 10.1038/35082608. [DOI] [PubMed] [Google Scholar]
- Myung K, Datta A, Chen C, Kolodner RD. SGS1, the Saccharomyces cerevisiae homologue of BLM and WRN, suppresses genome instability and homeologous recombination. Nat Genet. 2001b;27:113–116. doi: 10.1038/83673. [DOI] [PubMed] [Google Scholar]
- Paeschke K, Bochman ML, Garcia PD, Cejka P, Friedman KL, Kowalczykowski SC, Zakian VA. Pif1 family helicases suppress genome instability at G-quadruplex motifs. Nature. 2013 doi: 10.1038/nature12149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Runge KW, Zakian VA. Introduction of extra telomeric DNA sequences into Saccharomyces cerevisiae results in telomere elongation. Mol. Cell. Biol. 1989;9:1488–1497. doi: 10.1128/mcb.9.4.1488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scharer OD. DNA interstrand crosslinks: natural and drug-induced DNA adducts that induce unique cellular responses. Chembiochem. 2005;6:27–32. doi: 10.1002/cbic.200400287. [DOI] [PubMed] [Google Scholar]
- Schulz VP, Zakian VA. The Saccharomyces PIF1 DNA helicase inhibits telomere elongation and de novo telomere formation. Cell. 1994;76:145–155. doi: 10.1016/0092-8674(94)90179-1. [DOI] [PubMed] [Google Scholar]
- Shin JH, Kelman Z. DNA unwinding assay using streptavidin-bound oligonucleotides. BMC Mol Biol. 2006;7:43. doi: 10.1186/1471-2199-7-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sikorski RS, Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh DK, Popuri V, Kulikowicz T, Shevelev I, Ghosh AK, Ramamoorthy M, Rossi ML, Janscak P, Croteau DL, Bohr VA. The human RecQ helicases BLM and RECQL4 cooperate to preserve genome stability. Nucleic Acids Res. 2012;40:6632–6648. doi: 10.1093/nar/gks349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stavropoulos DJ, Bradshaw PS, Li X, Pasic I, Truong K, Ikura M, Ungrin M, Meyn MS. The Bloom syndrome helicase BLM interacts with TRF2 in ALT cells and promotes telomeric DNA synthesis. Hum Mol Genet. 2002;11:3135–3144. doi: 10.1093/hmg/11.25.3135. [DOI] [PubMed] [Google Scholar]
- Studier FW. Protein production by auto-induction in high density shaking cultures. Protein expression and purification. 2005;41:207–234. doi: 10.1016/j.pep.2005.01.016. [DOI] [PubMed] [Google Scholar]
- Suzuki T, Kohno T, Ishimi Y. DNA helicase activity in purified human RECQL4 protein. J Biochem. 2009;146:327–335. doi: 10.1093/jb/mvp074. [DOI] [PubMed] [Google Scholar]
- Tang G, Peng L, Baldwin PR, Mann DS, Jiang W, Rees I, Ludtke SJ. EMAN2: an extensible image processing suite for electron microscopy. J Struct Biol. 2007;157:38–46. doi: 10.1016/j.jsb.2006.05.009. [DOI] [PubMed] [Google Scholar]
- Teng S-C, Chang J, McCowan B, Zakian VA. Telomerase-independent lengthening of yeast telomeres occurs by an abrupt Rad50p-dependent, Rif-inhibited recombinational process. Mol. Cell. 2000;6:947–952. doi: 10.1016/s1097-2765(05)00094-8. [DOI] [PubMed] [Google Scholar]
- Teng S-C, Zakian VA. Telomere-telomere recombination is an efficient bypass pathway for telomere maintenance in Saccharomyces cerevisiae. Mol. Cell. Biol. 1999;19:8083–8093. doi: 10.1128/mcb.19.12.8083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomasz M. Mitomycin C: small, fast and deadly (but very selective) Chem Biol. 1995;2:575–579. doi: 10.1016/1074-5521(95)90120-5. [DOI] [PubMed] [Google Scholar]
- Ward TA, Dudasova Z, Sarkar S, Bhide MR, Vlasakova D, Chovanec M, McHugh PJ. Components of a Fanconi-like pathway control Pso2-independent DNA interstrand crosslink repair in yeast. PLoS Genet. 2012;8:e1002884. doi: 10.1371/journal.pgen.1002884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wellinger RJ, Zakian VA. Everything you ever wanted to know about Saccharomyces cerevisiae telomeres: beginning to end. Genetics. 2012;191:1073–1105. doi: 10.1534/genetics.111.137851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y, Brosh RM., Jr Helicase-inactivating mutations as a basis for dominant negative phenotypes. Cell Cycle. 2010;9:4080–4090. doi: 10.4161/cc.9.20.13667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J-Q, Monson EM, Teng S-C, Schulz VP, Zakian VA. The Pif1p helicase, a catalytic inhibitor of telomerase lengthening of yeast telomeres. Science. 2000;289:771–774. doi: 10.1126/science.289.5480.771. [DOI] [PubMed] [Google Scholar]
- Zhu Z, Chung WH, Shim EY, Lee SE, Ira G. Sgs1 helicase and two nucleases Dna2 and Exo1 resect DNA double-strand break ends. Cell. 2008;134:981–994. doi: 10.1016/j.cell.2008.08.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.