Abstract
Background
The signal output of ethylene receptor family members is mediated by unknown mechanisms to activate the Raf-like protein CONSTITUTIVE TRIPLE RESPONSE1 (CTR1) in negatively regulating ethylene signaling. The physical interaction between the ethylene receptor histidine kinase (HK) domain and CTR1 N terminus is essential to the CTR1-mediated receptor signal output. To advance our knowledge of the involvement of CTR1-mediated ethylene receptor signaling, we performed a genetic screen for mutations that enhanced the constitutive ethylene response in the weak ctr1-10 allele.
Results
We isolated a loss-of-function allele of ENHANCING ctr1-10 ETHYLENE RESPONSE2 (ECR2) and found that ecr2-1 ctr1-10 and the strong allele ctr1-1 conferred a similar, typical constitutive ethylene response phenotype. Genetic analyses and transformation studies suggested that ECR2 acts downstream of the ethylene receptors and upstream of the transcription factors ETHYLENE INSENSITIVE3 (EIN3) and EIN3-LIKE1 (EIL1), which direct the expression of ethylene response genes. Signal output by the N terminus of the ethylene receptor ETHYLENE RESPONSE1 (ETR1) can be mediated by a pathway independent of CTR1. Expression of the N terminus of the ethylene-insensitive etr1-1 but not the full-length isoform rescued the ecr2-1 ctr1-10 phenotype, which indicates the involvement of ECR2 in CTR1-mediated but not -independent, ethylene receptor signaling. ECR2 was mapped to the centromere region on chromosome 2. With incomplete sequence and annotation information and rare chromosome recombination events in this region, the cloning of ECR2 is challenging and still in progress.
Conclusions
ECR2 is a novel allele involved in the ethylene receptor signaling that is mediated by CTR1. CTR1 activation by ethylene receptors may require ECR2 for suppressing the ethylene response.
Keywords: Arabidopsis, Ethylene signaling, CTR1, ETR1, ECR2
Background
Ethylene is a gaseous hormone regulating many aspects of plant growth and development. The dicotyledonous model plant Arabidopsis has five ethylene receptors that physically act at the endoplasmic reticulum (ER) with the Raf-like protein CONSTITUTIVE TRIPLE-RESPONSE1 (CTR1) to negatively regulate ethylene signaling [1-4]. In the absence of ethylene, the receptor signal output is mediated by an unknown mechanism(s) to activate CTR1, and CTR1 is presumably activated to phosphorylate the downstream signaling component ETHYLENE INSENSITIVE2 (EIN2). Phosphorylated EIN2 stays at the ER and cannot induce the ethylene response. With ethylene binding to ethylene receptors, the receptor signal output is prevented and CTR1 is not activated. Unphosphorylated EIN2 undergoes proteolytic cleavage by an unknown mechanism to produce a C-terminal fragment, which enters the nucleus to induce the ethylene response [5,6]. Targets of the EIN2 C terminus remain to be identified. The transcription factors EIN3 and EIN3-LIKE1 (EIL1) direct the expression of ethylene-responsive genes [7], and EIN2 C-terminus–induced ethylene responses are prevented in the ein3-1 loss-of-function mutant [6]. Conceivably, the EIN2 C terminus could mediate the ethylene signaling to EIN3 and EIL1.
The ethylene receptors are structurally similar to histidine kinases (HKs) of prokaryotic two-component modules, and studies have revealed the domain functions of the ethylene receptors. The N terminus has three or four transmembrane domains (TMs) that bind a copper cofactor for ethylene binding and are required for localization at the ER [8-11]. Following the TMs is the GAF domain for non-covalent receptor interaction to mediate inter-receptor signaling [12-14]. The C-terminal portion is the HK domain, which is believed to function in the receptor signal output via direct interaction with the CTR1 N terminus [2,4]. CTR1 has serine/threonine kinase activity, and the ethylene response is inversely associated with CTR1 kinase activity [4,15]. These studies suggest that the HK domain mediates ethylene receptor signaling to the CTR1 N terminus, thus activating CTR1 to suppress the ethylene response, although the underlying biochemical mechanisms are elusive.
Recent studies suggest that the HK domain can be dispensable to ethylene receptor signal output and that CTR1 is not the only component mediating the signaling. Mutations that delete ETHYLENE RESPONSE1 (ETR1) HK and receiver domains have little effect on the receptor signal output [13,16], and expression of the truncated etr11-349 isoform that lacks the site for interacting with CTR1 rescues the ctr1 loss-of-function mutant phenotype. Conceivably, the ETR1 receptor signal output can be mediated via the N terminus to an alternative pathway independent of CTR1 [3,14]. Of note, ETR1 receptor signaling in kinase-defective ctr1 mutants occurs only when HK-domain–lacking ETR1 isoforms are expressed. A kinase-defective ctr1 isoform may dock at the ETR1 HK domain and actively prevent ETR1 N-terminal signaling to the CTR1-independent alternative pathway [14,17].
The ctr1-10 mutation results from a T-DNA insertion at the 5′-untranslated region (5′-UTR) of CTR1[18]. The 5′-UTR of a mRNA may be highly structured and contain upstream AUGs (uAUGs) and internal ribosome entry sites that can affect the translation. With increased upstream open reading frames (uORFs) and a highly structured nature, a long 5′-UTR can be translationally inhibitory [19-23]. With a higher CTR1 level in ctr1-10 than in the wild type [18], the T-DNA insertion may substantially increase the 5′-UTR length of the CTR1 transcript to affect translation efficiency, thus reducing the CTR1 level and thus activity as compared with the wild type.
The underlying biochemical mechanisms of CTR1 activation by the ethylene receptors remain to be unraveled. Mutations that enhance the constitutive ethylene response in ctr1-10 could have a role involving CTR1 activation, protein stability or ethylene signaling. Here we report that the loss-of-function mutation of ENHANCING ctr1-10 ETHYLENE RESPONSE2 (ECR2) enhanced the ethylene response in ctr1-10 comparable to the strong allele ctr1-1 and the ethylene-treated wild type. Results from extensive genetic and transformation studies suggested that ECR2 acts downstream of the ethylene receptors and upstream of the positive ethylene response regulators EIN3 and EIL1. We discuss possible roles for ECR2 in the negative regulation of the ethylene response.
Results
ctr1-10 is a weak allele
The ctr1-10 mutation results from a T-DNA insertion at the 5′-UTR, and the mutant shows a weak constitutive ethylene response phenotype [18]. The nature of the mutation that causes the weak phenotype remains to be determined.
Given that the T-DNA inserts at the 5′-UTR, we expected that the T-DNA fragment is transcribed as part of the 5′-UTR of ctr1-10 mRNA (Figure 1A). Indeed, RT-PCR with primers pairing the 5′-UTR and T-DNA sequences showed the presence of a chimeric cDNA fragment containing the sequence of the ctr1-10 5′-UTR and the T-DNA (Figure 1B). For the same gene, a longer 5′-UTR can be translationally more inhibitory than a shorter one [19,20,22,23]. With a 4.3-kb T-DNA at the 5′-UTR, we hypothesized that ctr1-10 could produce a lower level of CTR1 than the wild type, for a weak constitutive ethylene response phenotype.
Figure 1.
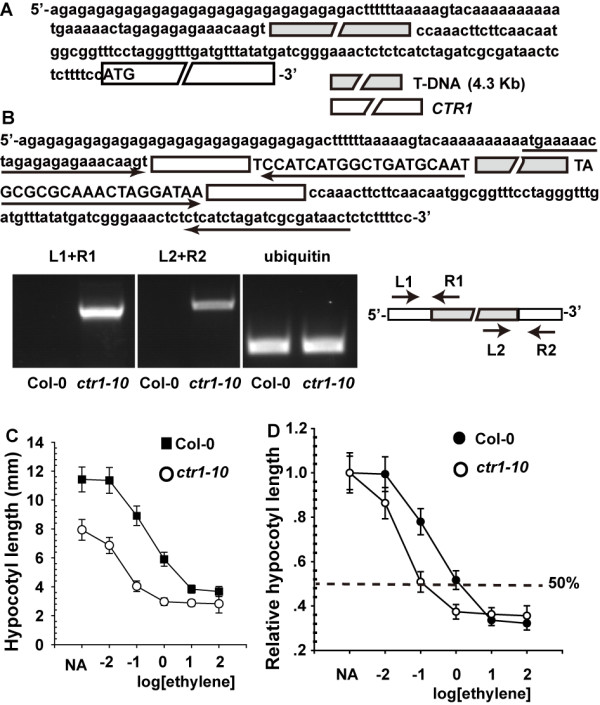
Gene structure and ethylene-dose response assay of the mutant constitutive triple response 1–10 (ctr1-10). (A)ctr1-10 gene structure and cDNA sequence flanking the T-DNA. The T-DNA is in gray and CTR1 coding region white. (B) The T-DNA sequence is transcribed as part of the ctr1-10 5′-untranslated region (5′-UTR). Arrows indicate the primer orientation and corresponding cDNA sequences for RT-PCR. Capital letters are the T-DNA sequence and lowercase letters are the CTR1 cDNA sequence. T-DNA is in gray and the 5′-UTR white. RT-PCR analysis of gene expression. Ethylene dose–response assay for hypocotyl measurement (C) and relative hypocotyl length (D) of etiolated wild-type (Col-0) and ctr1-10 seedlings. The dotted line in (D) indicates 50% growth inhibition. Data are mean ± SD (n ≥ 20).
Ethylene inhibits hypocotyl growth and the ethylene response can be quantified by measurement of hypocotyls. Hypocotyls were shorter for ctr1-10 than wild-type (Col-0) seedlings over a wide range of ethylene concentrations (Figure 1C). When growth for wild-type and ctr1-10 seedlings are normalized to growth with no added ethylene, it becomes apparent that ctr1-10 seedlings are more sensitive to ethylene with a shift from 1 μL L-1 for wild-type to 0.1 μL L-1 for the mutant (Figure 1D). These data suggest increased ethylene sensitivity with the ctr1-10 mutation.
Genetic screen for ctr1-10 enhancer mutations
To isolate components that could be involved in CTR1-mediated ethylene receptor signaling, we performed an enhancer screen for ctr1-10. Alleles that enhanced the ctr1-10 mutant phenotype were designated enhancing ctr1-10 ethylene response (ecr). Here we describe the isolation of ecr2-1 ctr1-10.
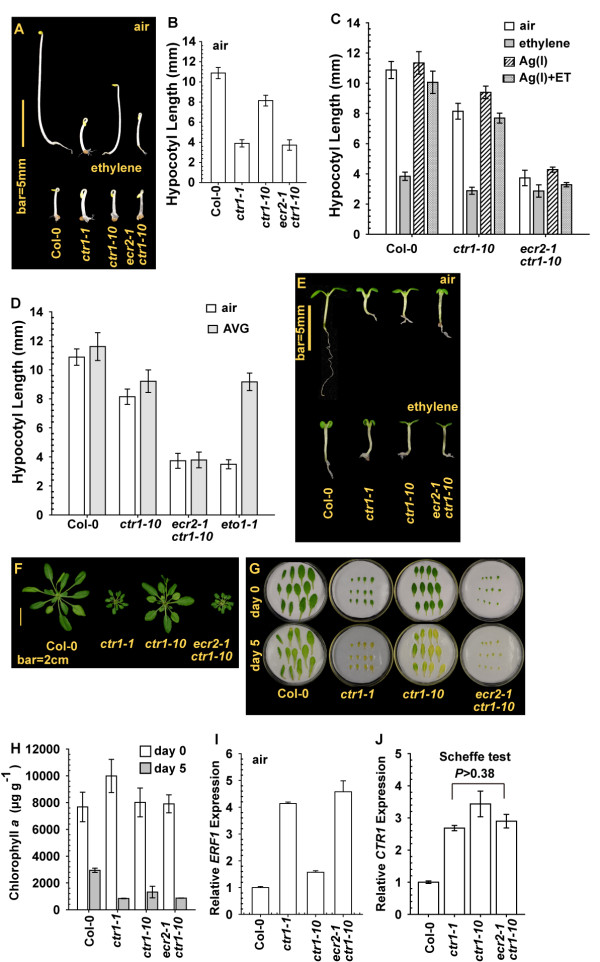
Without ethylene treatment, etiolated seedlings of ecr2-1 ctr1-10 and the strong allele ctr1-1 were identical in phenotype and hypocotyl length (Scheffe test, P = 0.65); ctr1-10 seedlings were longer than the two genotypes and shorter than the wild type (Col-0) (Scheffe test, P < 10-57; Figure 2A and 2B). As expected, ethylene treatment inhibited the seedling growth in the wild type (Col-0), ctr1-10, and ecr2-1 ctr1-10 (Figure 2C). Of note, ecr2-1 ctr1-10 seedlings already showed a constitutive ethylene response phenotype, with a short hypocotyl, so that the effect of ethylene on the seedling growth inhibition was minor (by 0.80458±0.1776 mm; 99% confidence level).
Figure 2.
Characterization of mutant enhancing ctr1-10 ethylene response2-1 (ecr2-1) ctr1-10. (A) Phenotype of etiolated ecr2-1 ctr1-10 seedlings, with the wild type (Col-0), ctr1-10, and ctr1-1 as the control. Hypocotyl measurement of etiolated seedlings (B), with treatment with silver (C) and aminoethoxyvinylglycine (AVG) (D). Phenotype of light-grown seedlings (E) and rosettes (F). Senescence phenotype (G) and chlorophyll a content (H) of leaves before (day 0) and after (day 5) detachment. Quantitative RT-PCR (qRT-PCR) analysis of ERF1(I) and CTR1(J) mRNA expression. Data are mean±SD for seedling hypocotyl and chlorophyll a content and mean±SE for gene expression.
The silver ion Ag(I) may bind ETR1 and prevent ethylene inhibition of ETR1 receptor signaling [11,24]. Silver treatment largely prevented ethylene-induced inhibition of seedling hypocotyl growth in wild-type (Col-0) and ctr1-10 but not ecr2-1 ctr1-10 seedlings (Figure 2C). Of note, the seedling hypocotyls were longer for silver-treated than non-silver–treated ecr2-1 ctr1-10 seedlings (Student’s t test, P < 0.01), regardless of ethylene treatment; the difference was minor and could be of little biological significance. These results suggest that ETR1 receptor signaling that was mediated in part in ctr1-10 was prevented in ecr2-1 ctr1-10.
Aminoethoxyvinylglycine (AVG) prevents the biosynthesis of the immediate ethylene biosynthesis precursor 1-aminocyclopropane-1-carboxylic (ACC), and the treatment can reduce endogenous ethylene production [25]. AVG (10 μM) treatment did not rescue the reduced growth of ecr2-1 ctr1-10 seedlings, so ecr2-1 did not enhance the production of ethylene to cause reduced growth (Figure 2D). ETHYLENE OVERPRODUCER1 (ETO1) is a negative regulator of the ACC-synthesing enzyme ACC SYNTHASE5 (ACS5) and the eto1-1 allele produces a higher level of ACC, and therefore ethylene, than does the wild type [26,27]. As a control, eto1-1 seedlings showed a short hypocotyl, and growth was rescued with AVG treatment (Figure 2D); AVG treatment in this study was sufficient to prevent endogenous ethylene production.
We examined the growth inhibition phenotype at other developmental stages. Grown under light, ethylene treatment inhibited cotyledon expansion and root elongation in wild-type (Col-0) seedlings. Seedlings of ctr1-1, ctr1-10, and ecr2-1 ctr1-10 showed the growth inhibition phenotype regardless of ethylene treatment (Figure 2E). At the adult stage, rosettes were larger for the wild type (Col-0) than the three mutants, with ctr1-1 and ecr2-1 ctr1-10 being phenotypically similar and both producing a smaller rosette than ctr1-10 (Figure 2F).
We examined alterations in other aspects of the ethylene response. Leaves of the wild type, ctr1-1, ctr1-10, and ecr2-1 ctr1-10 showed the senescence phenotype to various degrees 5 days after detachment (Figure 2G). Detached leaves of the mutants showed a similar chlorophyll a level, with a similar or slightly higher level in ctr1-10 than ctr1-1 and ecr2-1 ctr1-10 (Scheffe test, P = 0.075 and 0.03, respectively) (Figure 2H). ETHYLENE RESPONSE FACTOR1 (ERF1) expression is directed by EIN3 and the expression is associated with degrees of ethylene response [7,28]. ERF1 expression was slightly higher in ctr1-10 than the wild type and highly induced in ctr1-1 and ecr2-1 ctr1-10 without ethylene treatment (Figure 2I). CTR1 levels in ctr1-1, ctr1-10, and ecr2-1 ctr1-10 were identical (Scheffe test, P > 0.38) and higher than in the wild type (Figure 2J). The ecr2-1 allele did not affect CTR1 expression in ctr1-10.
ecr2-1 is a recessive, loss-of-function mutation
To genetically evaluate whether the effect of the ecr2-1 mutation on the ctr1-10 mutant phenotype is associated with single or multiple alleles, we crossed ecr2-1 ctr1-10 with the wild type (Col-0) and ctr1-10. In the F2 (filial) generation of the wild type (Col-0) cross, 466 individuals were scored, and only 31 showed the ecr2-1 ctr1-10 growth inhibition phenotype (Figure 3A; segregation ratio = 1:15). In the F2 generation of the ctr1-10 cross, 528 individuals were scored, and 136 showed the ecr2-1 ctr1-10 growth inhibition phenotype (Figure 3B; segregation ratio = 1:3). Results from both genetic analyses suggested that the ecr2-1 loss-of-function mutation was recessive and intergenic to ctr1-10. The independent segregation of ecr2-1 and ctr1-10 suggested that ECR2 and CTR1 are in distinct linkage groups.
Figure 3.
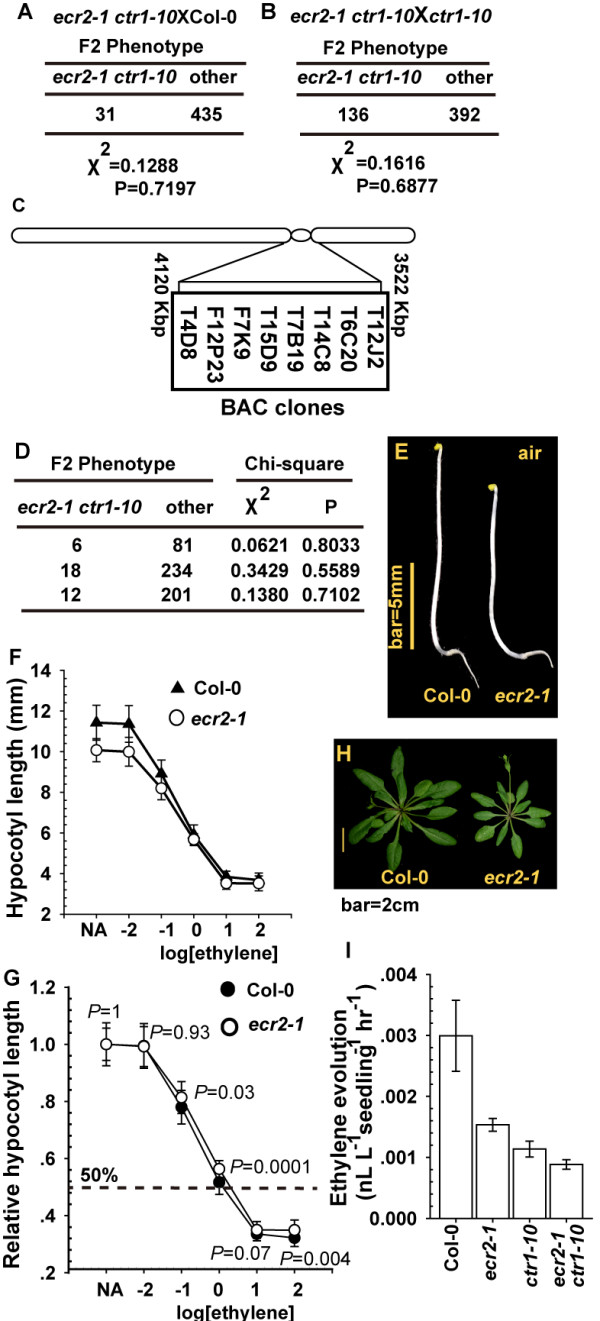
Isolation of ecr2-1. (A) and (B) Genetic analyses and chi-square test of ecr2-1. Numbers are individuals scored in the F2 generation. Other: other phenotype that is not ecr2-1 ctr1-10 growth inhibition phenotype. (C)ECR2 is mapped to a region spanning the centromere at chromosome 2, and the BAC clones are indicated. (D) Results for 3 independent test crosses for ecr2-1 with ctr1-10. Phenotype (E) and ethylene dose–response assay for the hypocotyl measurement (F) and relative hypocotyl length (G) of etiolated wild-type (Col-0) and ecr2-1 seedlings. The dotted line indicates 50% growth inhibition for (G); P values for Student’s t test are indicated. (H) Rosette phenotype of ecr2-1. (I) Ethylene evolution of light-grown seedlings (5 days after germination). Data are mean±SD of 3 independent measurements.
We attempted to clone ECR2 by map-based cloning and mapped ECR2 to a 598-kb region spanning the centromere at chromosome 2 (Figure 3C), which agreed with ECR2 and CTR1 (chromosome 5) being in distinct linkage groups. However, we did not obtain more recombinants to advance the mapping to a narrower region. This region contains sequence gaps (indicated as [gapbp]ExpandNs or a string of Ns in Additional file 1) and fragments with repeated sequences, retrotransposon, transposase, and transposable elements. We sequenced 39 annotated genes (excluding those annotated as retrotransposon, transposase, and transposable elements) but did not identify any mutation (Additional file 1). Among the genes, T-DNA insertion mutants for At2g07981and At2g08986 were each genetically crossed with ctr1-10 to test whether any of these was ECR2, and these mutations did not enhance the ctr1-10 phenotype.
The mutation that affects ECR2 remained to be identified and we attempted to isolate the ecr2-1 single mutant. The F2 seedlings from the genetic crossing of ecr2-1 ctr1-10 and the wild type (Col-0) were crossed with ctr1-10, and the individual that produced the ecr2-1 ctr1-10 mutant phenotype in the F2 generation with a 1:15 segregation ratio was considered the ecr2-1 mutant. We isolated ecr2-1, and results from 3 independent crossings with ctr1-10 showed a 1:15 segregation (Figure 3D). Grown in dark, ecr2-1 seedlings produced a shorter hypocotyl than did the wild type (Col-0) at low ethylene concentrations (Figure 3E and 3F); without ethylene treatment, the height of ecr2-1 seedlings was 1.355±0.62 mm (99% confidence level) shorter than that of wild-type seedlings. When this data was normalized, we found that wild-type and ecr2-1 seedlings had indistinguishable sensitivity to ethylene (Figure 3G). Although the relative hypocotyl length was longer (by 4.6±2.9%; 99% confidence level) for ecr2-1 seedlings than wild-type seedlings with 1 μL L-1 ethylene, the difference was small and could be of little biological significance. At the adult stage, the rosette was smaller for ecr2-1 than the wild type (Figure 3H). We excluded the possibility that the mutant phenotype of ecr2-1 and ecr2-1 ctr1-10 could be due to greater ethylene evolution than the wild type (Col-0), as supported by ethylene evolution (Figure 3I).
These results suggest that the ecr2-1 allele conferred a minor growth inhibition throughout development. As compared with the wild type (Col-0), etiolated ecr2-1 seedlings showed no alteration in ethylene sensitivity.
CTR1-independent but not -dependent ETR1 receptor signaling occurs in ecr2-1 ctr1-10
Silver treatment prevents the inhibition of ETR1 receptor signaling by ethylene [13,24]. Silver treatment reversed the ethylene effect on growth of ctr1-10 but not ecr2-1 ctr1-10 seedlings (Figure 2C), so ETR1 receptor signaling was prevented in ecr2-1 ctr1-10 but partly mediated in ctr1-10. We evaluated whether ETR1 receptor signaling could occur in ecr2-1 ctr1-10.
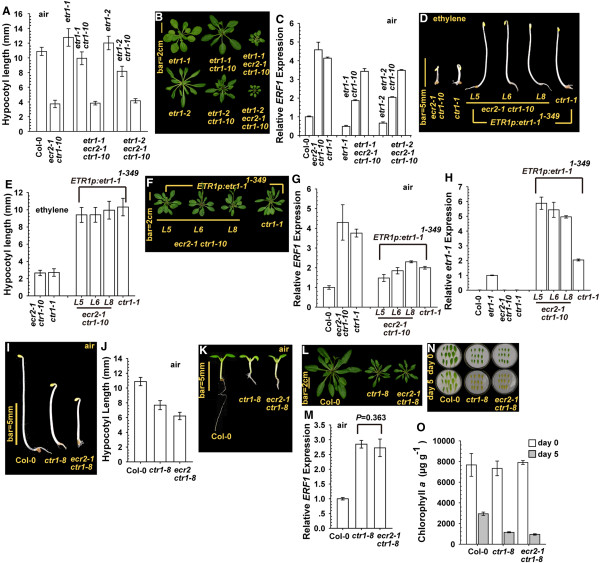
Ethylene-insensitive etr1-1 and etr1-2 mediate receptor signaling by different mechanisms: the former does not require REVERSION-TO-ETHYLENE SENSITIVITY1 (RTE1), whereas the latter does [29,30]. Because of their dominant nature, we used etr1-1 and etr1-2 mutants to evaluate ETR1 receptor signaling. In the absence of ethylene treatment, etiolated etr1-1 and etr1-2 seedlings produced a long hypocotyl, and the ctr1-10 mutation moderately reduced the seedling hypocotyl elongation in each allele; as expected, etr1-1 ecr2-1 ctr1-10 and etr1-2 ecr2-1 ctr1-10 seedlings were short, with a hypocotyl length similar to ecr2-1 ctr1-10 seedlings (Figure 4A). Consistently, at the adult stage, the rosette was larger for etr1-1 ctr1-10 and etr1-2 ctr1-10 than etr1-1 ecr2-1 ctr1-10 and etr1-2 ecr2-1 ctr1-10 plants, respectively (Figure 4B). We scored the ethylene response in these genotypes by measuring ERF1 expression: the ERF1 level was greater in etr1-1 ctr1-10 and etr1-2 ctr1-10 than the wild type and lower than in etr1-1 ecr2-1 ctr1-10 and etr1-2 ecr2-1 ctr1-10 (Figure 4C). ETR1 receptor signaling may be prevented by ecr2-1 ctr1-10 but little affected by ctr1-10.
Figure 4.
ECR2 is involved in the CTR1-dependent but not -independent ethylene receptor signaling. Etiolated seedling hypocotyl measurement (A), rosette phenotype (B), and ERF1 expression (C) of ecr2-1 ctr1-10 with and without the ethylene-insensitive etr1-1 or etr1-2 allele. Etiolated seedling phenotype (D), hypocotyl measurement (E), rosette phenotype (F) and ERF1 mRNA expression (G) in ecr2-1 ctr1-10 lines expressing ETR1p:etr1-11-349. (H)etr1-1 mRNA levels in etr1-1 as the control and etr1-11-349 levels in ecr2-1 ctr1-10 ETR1p:etr1-11-349 lines, respectively. Etiolated seedling phenotype (I) and hypocotyl measurement (J), light-grown seedlings (K), rosette phenotype (L), and ERF1 expression (M) in ecr2-1 ctr1-8. Senescence phenotype (N) and chlorophyll a content (O) in ecr2-1 ctr1-8 before (day 0) and after (day 5) the test. Data are mean ± SD for the seedling hypocotyl measurement and chlorophyll a content, and mean ± SE for gene expression.
ETR1 receptor signaling can be mediated via the N-terminal portion independent of CTR1 [14], so we evaluated whether ECR2 could act in the CTR1-independent pathway. With ethylene treatment, ctr1-1 seedlings expressing the ETR1p:etr1-1 1-349 transgene that encodes the etr1-1 N terminus (residues 1–349) were ethylene insensitive and produced a long hypocotyl [14] (Figure 4D and 4E). Interestingly, the phenotype of ethylene growth inhibition in ecr2-1 ctr1-10 was rescued by the transgene, and ETR1p:etr1-1 1-349 ecr2-1 ctr1-10 seedlings did not show the phenotype of ethylene-inhibited growth (Figure 4D and 4E). Consistently, ETR1p:etr1-1 1-349 expression greatly rescued rosette growth in ecr2-1 ctr1-10 and ctr1-1 (Figure 4F). The expression of the transgene attenuated ERF1 levels in ecr2-1 ctr1-10 and ctr1-1 (Figure 4G). Expression of ETR1p:etr1-1 1-349 in the transformation lines was confirmed by qRT-PCR with etr1-1–specific primers, and etr1-1 expression was detectable only in genotypes that carried the allele or transgene (Figure 4H).
These data suggest that the ecr2-1 allele prevented the receptor signaling with the full-length etr1-1 and etr1-2 but not etr1-1 N terminus in ctr1-10. Therefore, ECR2 may be involved in CTR1-dependent but not -independent receptor signaling in ctr1-10. Given that the signaling of full-length ETR1 but not ETR1 N terminus is prevented by kinase-defective ctr1 isoforms [14,17], ECR2 could be involved in part in CTR1 kinase activity or protein stability.
ctr1-8 is not associated with the ethylene receptors, and the mutant shows a weak constitutive ethylene response. We previously hypothesized that mediation of ethylene receptor signaling in ctr1-8 was independent of CTR1 [4,17]. The possibility that ctr1-8 could mediate in part the receptor signaling, for a weak mutant phenotype, should be considered. Given that ECR2 is involved in CTR1-dependent ethylene receptor signaling, we expected that ecr2-1 would have little effect on ctr1-8 if ctr1-8 did not mediate the ethylene receptor signaling.
We crossed ecr2-1 ctr1-10 with ctr1-8, and the resulting F2 plants were genotyped for the CTR1 allele. All F2 plants that showed the typical constitutive ethylene response phenotype were ctr1-10 or ctr1-8 ctr1-10, so they all carried the homozygous ecr2-1 allele and ecr2-1 did not enhance the ctr1-8 mutant phenotype. The mutant ecr2-1 ctr1-8 ctr1-10 was selfed, and the resulting progenies with the homozygous ctr1-8 allele were ecr2-1 ctr1-8.
The length of etiolated ecr2-1 ctr1-8 seedlings was only 1.4±0.43 mm (99% confidence level) shorter than that of ctr1-8 seedlings (Figure 4I and 4J), which is in line with the result showing that the ecr2-1 allele conferred minor seedling growth inhibition (Figure 3E and 3F) but not increased ethylene sensitivity (Figure 3G). Light-grown seedlings of ctr1-8 and ecr2-1 ctr1-8 were phenotypically similar, with a shorter root and smaller cotyledons than the wild type (Col-0) (Figure 4K). Consistently, the rosette was larger for the wild type than ctr1-8 and ecr2 ctr1-8 plants (Figure 4L). Therefore, the ecr2-1 allele may have little effect on the ethylene response phenotype in ctr1-8. We quantified the ethylene response for the two mutants. ERF1 expression was identical in ctr1-8 and ecr2-1 ctr1-8 (Student’s t test, P = 0.363) and higher than in the wild type (Figure 4M). Both ctr1-8 and ecr2-1 ctr1-8 had the same senescence phenotype, and their chlorophyll a content was identical before (Student’s t test; P = 0.056) and after (Student’s t test; P =0.059) the test (Figure 4N and 4O).
The ecr2-1 allele had little effect on ctr1-8 ethylene response, which suggests that ethylene receptor signaling in ctr1-8 was predominantly mediated by a pathway independent of CTR1.
Genetic analysis of effect of ein3-1 and eil1-1 on the ethylene response in ecr2-1 ctr1-10
The present data suggested the involvement of ECR2 in ethylene receptor signaling mediated by CTR1. ETHYLENE INSENSITIVE2 (EIN2) acts downstream of CTR1, and ein2 loss-of-function mutation may rescue the ecr2-1 ctr1-10 mutant phenotype. Genetic analyses for the effects of ein2 on ecr2-1 ctr1-10 were not successful because EIN2 is tightly linked with CTR1 and ECR2 remains to be cloned.
EIN3 and EIN3-LIKE1 (EIL1) are the transcription factors that direct the expression of ethylene-responsive genes, and EIN3 loss-of-function mutations confer ethylene insensitivity [7,31]. We used genetic analyses to evaluate whether ECR2 acts upstream of EIN3/EIL1. To obtain ein3-1 ecr2-1 ctr1-10 and eil1-1 ecr2-1 ctr1-10 mutants, ein3-1 and eil1-1 were each genetically crossed with ecr2-1 ctr1-10, and the resulting F2 individuals carrying ein3-1 ctr1-10 and eil1-1 ctr1-10 were identified by genotyping. All F2 generations with the ein3-1 ctr1-10 and eil1-1 ctr1-10 genotypes showed relatively normal growth. The F2 individuals were each genetically crossed with ecr2-1 ctr1-10 to test the presence of the ecr2-1 allele, and the individuals that were ein3-1 ecr2-1 ctr1-10 and eil1-1 ecr2-1 ctr1-10 would give a 1:3 segregation (long seedlings: short seedlings) in the resulting F2 generation. Verified by chi-square test for 2 independent test crossings, we isolated ein3-1 ecr2-1 ctr1-10 and eil1-1 ecr2-1 ctr1-10 (Figure 5A and 5B).
Figure 5.
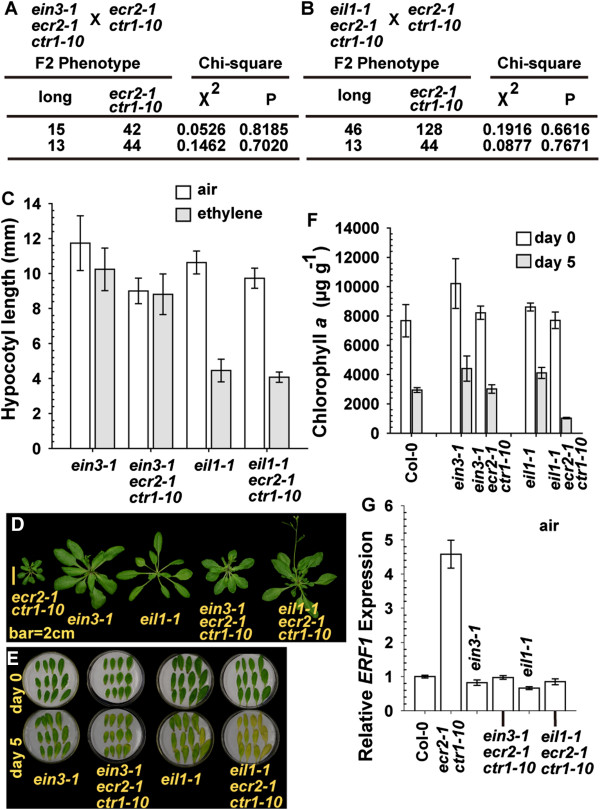
Genetic analysis of the effect of ethylene insensitive3-1 (ein3-1) and ein3-like 1 (eil1-1) on ecr2-1 ctr1-10. Segregation ratio and chi-square analysis of 2 independent genetic crossings of ecr2-1 ctr1-10 with ein3-1 ecr2-1 ctr1-10(A) and eil1-1 ecr2-1 ctr1-10(B). Etiolated seedling hypocotyl measurement (C), rosette phenotype (D), senescence test for detached leaves (E), chlorophyll a content for the senescence test (F), and ERF1 expression (G) in ein3-1 ecr2-1 ctr1-10 and eil1-1 ecr2-1 ctr1-10. Data are mean ± SD for seedling hypocotyl and chlorophyll a measurement, and mean ± SE for ERF1 levels.
Without ethylene treatment, ein3-1 ecr2-1 ctr1-10 and eil1-1 ecr2-1 ctr1-10 seedlings produced a long seedling hypocotyl (Figure 5C) as compared with ecr2-1 ctr1-10 seedlings (Figure 2B). Of note, ein3-1 but not eil1-1 seedlings were ethylene-insensitive, and ethylene treatment inhibited eil1-1 but not ein3-1 seedling growth. Consistently, the ein3-1 allele prevented ethylene-inhibited growth of seedling hypocotyls in ecr2-1 ctr1-10 whereas eil1-1 did not (Figure 5C). At the adult stage, ein3-1 and eil1-1 alleles each rescued ecr2-1 ctr1-10 rosette growth to a great extent, and rosettes were larger for ein3-1 ecr2-1 ctr1-10 and eil1-1 ecr2-1 ctr1-10 than ecr2-1 ctr1-10 plants (Figure 5D). The ethylene response in each triple mutant was quantified by leaf senescence and ERF1 expression. The leaf senescence phenotype was weaker in ein3-1, eil1-1 and ein3-1 ecr2-1 ctr1-10 than ecr2-1 ctr1-10 and eil1-1 ecr2-1 ctr1-10 plants (Figures 2G and 5E), which was consistent with chlorophyll a content (Figures 2H and 5F). Without ethylene treatment, the ERF1 level in ecr2-1 ctr1-10 was highly reduced with the respective addition of ein3-1 and eil1-1 alleles, and ein3-1, eil1-1, ein3-1 ecr2-1 ctr1-10 and eil1-1 ecr2-1 ctr1-10 showed identical ERF1 expression (Scheffe test, P = 0.06-0.995; Figure 5G).
The present data support that ECR2 acts in ethylene signal transduction upstream of the transcription factors EIN3 and EIL1. The eil1-1 allele did not prevent the ethylene-inhibited seedling growth or leaf senescence in ecr2-1 ctr1-10, which indicates that functions of EIN3 and EIL1 differ, with a larger role for EIN3 than EIL1 in ethylene responses.
EBF1 and EBF2 overexpression rescues ecr2-1 ctr1-10 phenotype
EIN3-BINDING F-BOX1 (EBF1) and EBF2 are F-box proteins involved in the ubiquitination of EIN3 and EIL1 to mediate their degradation by the 26S proteosome [32,33]. Excess EBF1 and EBF2 reduces EIN3 and EIL1 levels and suppresses the ethylene response [34]. The argument that ECR2 acts in the ethylene signal transduction pathway upstream of the transcription factors EIN3 and EIL1 can be tested by examining whether overexpression of EBF1 and EBF2 rescues the ecr2-1 ctr1-10 phenotype.
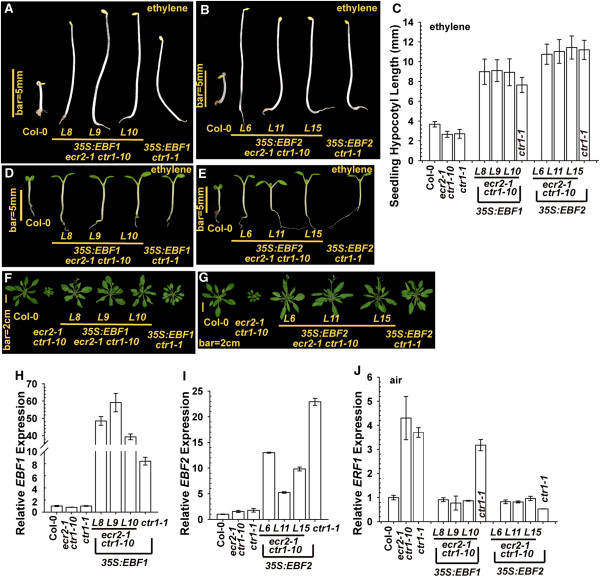
EBF1 and EBF2 were each expressed under the regulation of the constitutive CAULIFLOWER MOSAIC VIRUS 35S promoter in ecr2-1 ctr1-10 and ctr1-1. As expected, with ethylene treatment, hypocotyls were longer for etiolated seedlings of 35S:EBF1 ecr2-1 ctr1-10 and 35S:EBF2 ecr2-1 ctr1-10, than the wild type (Col-0), as well as longer for the strong allele ctr1-1 expressing the transgene than ctr1-1 (Figure 6A, 6B and 6C). Consistently, ethylene treatment inhibited the cotyledon expansion in the wild type (Col-0) but not the transformation lines in light-grown seedlings (Figure 6D and 6E). At the adult stage, the expression of each transgene largely rescued the growth inhibition phenotype in ecr2-1 ctr1-10 and ctr1-1. Rosettes were slightly smaller for 35S:EBF1 ecr2-1 ctr1-10 lines than the wild type, but those for 35S:EBF2 ecr2-1 ctr1-10 lines and the wild type were similar in size (Figure 6F and 6G). Confirmation of the expression of the transgenes by qRT-PCR revealed lower EBF1 level in 35S:EBF1 ctr1-1 than in 35S:EBF1 ecr2 ctr1-10 lines (Figure 6H). The lower EBF1 expression could be associated with the stronger phenotype of rosette growth inhibition in 35S:EBF1 ctr1-1 than 35S:EBF1 ecr2-1 ctr1-10. In contrast, EBF2 level was higher in 35S:EBF2 ctr1-1 than 35S:EBF2 ecr2-1 ctr1-10 lines (Figure 6I).
Figure 6.
Overexpression of EIN3-BINDING F-BOX1 (EBF1) and EBF2 rescues ecr2-1 ctr1-10 mutant phenotype and confers ethylene insensitivity. Phenotype of etiolated, ethylene-treated ecr2-1 ctr1-10 seedlings expressing 35S:EBF1(A) and 35S:EBF2(B) and their seedling hypocotyl measurement (C). Phenotype of light-grown seedlings (with ethylene treatment) (D) and (E) and rosettes (F) and (G) for ecr2-1 ctr1-10 expressing 35S:EBF1 and 35S:EBF2, respectively. qRT-PCR analysis of mRNA levels of EBF1(H), EBF2(I), and ERF1(J) in the transformation lines. Data are mean ± SD for hypocotyl measurement and mean ± SE for gene expression.
We also examined other aspects of the ethylene response affected by the transgenes. ERF1 expression was greater in ecr2-1 ctr1-10 and ctr1-1 than the wild type; the expression of each transgene largely reduced ERF1 levels (Figure 6J). Of note, ERF1 expression was greater in 35S:EBF1 ctr1-1 than 35S:EBF1 ecr2-1 ctr1-10 lines, likely because of its lower EBF1 expression (Figure 6H).
Consistent with the suppression of ecr2-1 ctr1-10 by ein3-1 and eil1-1, the respective elevation in EBF1 and EBF2 levels in ecr2-1 ctr1-10 rescued the inhibition in mutant growth and reduced ERF1 levels, which supports that ECR2 acts upstream of the transcriptions factors EIN3 and EIL1 in the ethylene signal transduction pathway.
Discussion
Arabidopsis ethylene signaling is negatively regulated by ethylene receptors and the Raf-like protein CTR1 [1-3,35]. The biochemical nature of receptor signaling is unknown, as are the underlying mechanisms by which CTR1 is activated [3,14,36]. A genetic screen for enhancers facilitates the isolation of genes or mutations that play a role in the same biological process as the known mutation. Given that the ctr1-10 mutation does not disrupt the CTR1 ORF and that ctr1-10 is a weak allele, an enhancer screen for ctr1-10 could lead to isolation of components involved in CTR1-mediated ethylene receptor signaling.
In this study, we reported the isolation of ecr2-1 as an enhancer mutation for ctr1-10. With a relative weak constitutive ethylene response phenotype for ctr1-10 and a strong phenotype for ecr2-1 ctr1-10, the mutations ecr2-1 and ctr1-10 synergistically facilitated ethylene signaling. The assumption that CTR1 level in ctr1-10 could be reduced needs to be verified biochemically. Nevertheless, extensive studies suggest that for a common transcript, a long 5′-UTR can be translationally inhibitory as compared with a short one [22,37,38]. Conceivably, the T-DNA insertion that increases the 5′-UTR length may add structures of higher order and uORFs to inhibit the translation of CTR1, thus reducing CTR1 level, so that ctr1-10 shows a weak constitutive ethylene response phenotype. We considered 3 scenarios explaining the synergistic effect of ecr2-1 and ctr1-10: 1) ECR2 could be required in part for CTR1 activity or the protein stability, 2) ECR2 and CTR1 act in different pathways, or 3) ECR2 could be a signaling molecule acting with CTR1 to suppress the ethylene signaling. In the first scenario, CTR1 level, and possibly activity, in ctr1-10 is presumably reduced to a level requiring ECR2 to suppress the ethylene signaling.
The nature of ctr1-10 and ctr1-8 mutations is distinct: CTR1 level could be reduced in ctr1-10 and the ctr1-8 protein is not associated with the ethylene receptors [4,18]. The docking of kinase-defective ctr1 isoforms at the HK domain of ETR1 prevents receptor signaling to an alternative CTR1-independent pathway. In ctr1-8, the ethylene receptors are free from association with ctr1-8; thus, the receptor signaling is mediated to the alternative pathway and the mutant shows a weak phenotype [14,17]. Conceivably, in ecr2-1 ctr1-10, the activity of CTR1 docking at the ethylene receptors could be greatly reduced, and ethylene receptor signaling that is mediated by CTR1 as well as the alternative pathway was prevented. In contrast, the ctr1-8 protein does not dock at the ethylene receptors and the receptor signaling can be mediated by the alternative, CTR1-independent pathway; thus, ecr2-1 had little effect on ctr1-8 phenotype. In line with these results, our data showing that ecr2-1 prevented etr1-1 and etr1-2 but not etr1-11-349 receptor signaling in ctr1-10 also support that ECR2 is involved in the ethylene receptor signaling that is dependent on CTR1.
An alternative explanation for the distinct effects of ecr2-1 on ctr1-10 and ctr1-8 could arise from a role of ECR2 in CTR1 stability. In this scenario, very small amount of the ctr1-8 protein that could associate with the ethylene receptors could have a higher turnover rate in the absence of ECR2; meanwhile, ctr1-8 reserved in the soluble fraction could be continuously recruited to the receptors and mediate the receptor signaling. Contrarily, in ctr1-10, CTR1 level is highly reduced without ECR2 and the receptor signaling is prevented. However, this scenario is not consistent with the argument that the ethylene receptor signaling can be mediated to an alternative pathway that is independent of CTR1. In other words, if ECR2 were involved in CTR1 stability, the ethylene receptor signaling that was mediated independent of CTR1 still occurred and the ecr2-1 allele would have little effect on the degree of ctr1-10 ethylene responses.
Conclusions
CTR1 is presumably activated by the ethylene receptors, and the mechanism is unknown [1]. Little is known about the presence of any other components that are involved in the ethylene receptor signaling to CTR1. The present data suggest that ECR2 is a component involved in part in CTR1-mediated ethylene receptor signaling, which indicates a regulatory mechanism for the receptor signaling. We favor the argument that CTR1 activity but not level was affected by ecr2-1 in ctr1-10. With limited sequence information for the 598-kb region that contains ECR2, the cloning of ECR2 is currently hampered. Complementation of ecr2-1 by a large DNA fragment that contains ECR2 is ongoing. However, with repeated sequences, transposons, retrotransposons, and sequence gaps in this region, cloning of a large DNA fragment is still highly challenging. Nevertheless, the cloning of ECR2 will advance our knowledge of ethylene signaling that involves CTR1 activation.
Methods
Plant materials and growth conditions
ctr1-10 and the PCR primers for genotyping were previously described [18]. For mutagenesis, following ethyl methanesulfonate treatment, ctr1-10 seeds (M0) were washed with continuous flowing water and grown on soil as 380 pools (20–25 M1 plants in a pool). Seedlings at the M2 generation showing the constitutive ethylene response phenotype were candidates carrying the enhancer mutation and were characterized for other aspects of the ethylene response. We screened 162 pools, and ecr2-1 ctr1-10 was isolated. For the growth of etiolated seedlings, seeds were stratified at 4°C for 72 hr and then moved to 22°C for germination (80 hr) in the dark. Seedling hypocotyl length was measured by use of Video tesT (Moscow) [29]. For measuring growth of seedlings and rosettes under light, stratified seeds were germinated and grown under 16-hr light/8-hr dark at 25°C; seedlings were phenotyped 7 days after germination and rosettes 4 weeks after germination. The isolation of ecr2-1 and the genetic crossing of ecr2-1 with etr1-1, etr1-2, ctr1-8, ein3-1, and eil1-1 are described in the Results section and the segregation was verified by chi-square test (α=0.01). For ethylene treatment, the ethylene concentration was 20 μL L-1, or otherwise as indicated.
Leaf senescence test
Detached leaves from plants were incubated in a Petri dish with wet filter paper in the dark for 5 days. Leaf senescence was quantified by measuring chlorophyll a content [39].
Quantitative RT-PCR analysis
qRT-PCR of mRNA expression involved use of StepOne Plus (ABI). Each measurement was repeated 3 times with 3 independent biological materials (n = 3 × 3). The primer sequences were ERF1-F (5′-TTTCTCGATGAGAGGGTC-3′) and ERF1-R (5′- AAGCTCCTCAAGGTACTG-3′) for ERF1, etr1-1-NF (5′- GCTTTTATCGTTCTTTA-3′) and etr1-1-NR (5′-GCTTTATTTTTCAAGAAA-3′) for etr1-1 and etr1-1 1-349 , EBF1-F (5′-GGAGATTGATGTTCCTTCCAAGA-3′) and EBF1-R (5′-CAATAGACCGAAGACCAAGATC-3′) for EBF1, EBF2-F (5′-CTTCAGATTTAGTGGTGATGAAG-3′) and EBF2-R (5′-CAAGCACTCCTCTCTTGTCCA-3′) for EBF2, and UBI-F (5′-ATGGAAAATCCCACCTACTAAATT-3′) and UBI-R (5′-TTGAACAACTCGTAGCAACTCATC-3′) for ubiquitin (the calibrator).
Transgenes
The ETR1p:etr1-1 1-349 transgene was previously described [14]. The EBF1 cDNA fragment was released from a cDNA clone with the restriction enzymes BamHI and SmaI and cloned to the binary vector pCAMBIA1301 with the constitutive 35S promoter. An EBF2 genomic fragment was cloned by PCR with the primer sequences EBF2-F-BamHI (5′-TCGGATCCAAATGTCTGGAATCTTCAGATTTAG-3′) and EBF2-R-BamHI (5′-GCGGATCCTTAGTAGAGTATATCG-3′). The genomic EBF2 clone was confirmed by sequencing and cloned to pCAMBIA1301. The transgenes were each transformed to ecr2-1 ctr1-10 by Agrobacterium, with the floral-dip method [40,41], and phenotypes were scored in T3 and higher generations.
Ethylene measurement
Ethylene evolution in light-grown seedlings was measured by use of the Ethylene Detector (ETD-300 by Sensor Sense) with the “stop-and-flow” measurement. In brief, 25–30 seedlings were grown in a vented vial for 5 days after germination. The vial was closed for 3.5 hr to let ethylene accumulate, and the ethylene amount was measured. We measured 3 independent biological samples for each genotype, and ethylene evolution is represented as mean (nL L-1 seedling-1 hr-1)±SD.
Statistical analysis
Data are expressed as mean±SD or mean±SE for gene expression. Student’s t test (α = 0.01) was used for comparing 2 groups and Scheffe test (α = 0.01) for multiple groups. Chi-square test (α = 0.01) was used for testing segregation ratio.
Competing interest
The authors declare that they have no competing interests.
Authors’ contributions
AX conducted the experiments. WZ isolated the allele, and C-KW supervised the work and wrote the paper. All authors read and approved the final manuscript.
Supplementary Material
BAC clones and sequences encompassing ECR2.
Contributor Information
Aibei Xu, Email: aibeixu@163.com.
Wei Zhang, Email: wzhang05@sibs.ac.cn.
Chi-Kuang Wen, Email: qgwen@sibs.ac.cn.
Acknowledgements
This work was supported by the National Natural Sciences Foundation of China (31123006, 31070249, and 31200217) and the Chinese Academy of Sciences (KSCX2-EW-J-12 and 2012KIP303).
References
- Ju C, Chang C. Advances in ethylene signalling: protein complexes at the endoplasmic reticulum membrane. AoB Plants. 2012;2012:pls031. doi: 10.1093/aobpla/pls031. doi:10.1093/aobpla/pls031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark KL, Larsen PB, Wang X, Chang C. Association of the Arabidopsis CTR1 Raf-like kinase with the ETR1 and ERS ethylene receptors. Proc Natl Acad Sci U S A. 1998;95(9):5401–5406. doi: 10.1073/pnas.95.9.5401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shakeel SN, Wang X, Binder BM, Schaller GE. Mechanisms of signal transduction by ethylene: overlapping and non-overlapping signalling roles in a receptor family. AoB Plants. 2013;5:plt010. doi: 10.1093/aobpla/plt010. doi:10.1093/aobpla/plt010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y, Li H, Hutchison CE, Laskey J, Kieber JJ. Biochemical and functional analysis of CTR1, a protein kinase that negatively regulates ethylene signaling in Arabidopsis. Plant J. 2003;33(2):221–233. doi: 10.1046/j.1365-313X.2003.01620.x. [DOI] [PubMed] [Google Scholar]
- Ju C, Yoon GM, Shemansky JM, Lin DY, Ying ZI, Chang J, Garrett WM, Kessenbrock M, Groth G, Tucker ML. et al. CTR1 phosphorylates the central regulator EIN2 to control ethylene hormone signaling from the ER membrane to the nucleus in Arabidopsis. Proc Natl Acad Sci. 2012;109(47):19486–19491. doi: 10.1073/pnas.1214848109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiao H, Shen Z, Huang S-sC, Schmitz RJ, Urich MA, Briggs SP, Ecker JR. Processing and subcellular trafficking of ER-tethered EIN2 control response to ethylene gas. Science. 2012;338(6105):390–393. doi: 10.1126/science.1225974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solano R, Stepanova A, Chao Q, Ecker J. Nuclear events in ethylene signaling: a transcriptional cascade mediated by ETHYLENE-INSENSITIVE3 and ETHYLENE-RESPONSE-FACTOR1. Genes Dev. 1998;12:3703–3714. doi: 10.1101/gad.12.23.3703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y-F, Gao Z, Kerris RJ III, Wang W, Binder BM, Schaller GE. Ethylene receptors function as components of high-molecular-mass protein complexes in Arabidopsis. PLoS ONE. 2010;5(1):e8640. doi: 10.1371/journal.pone.0008640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gamble RL, Coonfield ML, Schaller GE. Histidine kinase activity of the ETR1 ethylene receptor from Arabidopsis. Proc Natl Acad Sci U S A. 1998;95(13):7825–7829. doi: 10.1073/pnas.95.13.7825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen YF, Randlett MD, Findell JL, Schaller GE. Localization of the ethylene receptor ETR1 to the endoplasmic reticulum of Arabidopsis. J Biol Chem. 2002;277(22):19861–19866. doi: 10.1074/jbc.M201286200. [DOI] [PubMed] [Google Scholar]
- Rodriguez FI, Esch JJ, Hall AE, Binder BM, Schaller GE, Bleecker AB. A copper cofactor for the ethylene receptor ETR1 from Arabidopsis. Science. 1999;283(5404):996–998. doi: 10.1126/science.283.5404.996. [DOI] [PubMed] [Google Scholar]
- Gao Z, Wen C-K, Binder BM, Chen Y-F, Chang J, Chiang Y-H, Kerris RJ III, Chang C, Schaller GE. Heteromeric interactions among ethylene receptors mediate signaling in Arabidopsis. J Biol Chem. 2008;283(35):23801–23810. doi: 10.1074/jbc.M800641200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie F, Liu Q, Wen C-K. Receptor signal output mediated by the ETR1 N terminus is primarily subfamily I receptor dependent. Plant Physiol. 2006;142(2):492–508. doi: 10.1104/pp.106.082628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu L, Xie F, Yu J, Wen C-K. Arabidopsis RTE1 is essential to ethylene receptor ETR1 amino-terminal signaling independent of CTR1. Plant Physiol. 2012;159(3):1263–1276. doi: 10.1104/pp.112.193979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeda Y, Men S, Fischer U, Stepanova AN, Alonso JM, Ljung K, Grebe M. Local auxin biosynthesis modulates gradient-directed planar polarity in Arabidopsis. Nat Cell Biol. 2009;11(6):731–738. doi: 10.1038/ncb1879. [DOI] [PubMed] [Google Scholar]
- Gamble RL, Qu X, Schaller GE. Mutational analysis of the ethylene receptor ETR1. Role of the histidine kinase domain in dominant ethylene insensitivity. Plant Physiol. 2002;128(4):1428–1438. doi: 10.1104/pp.010777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie F, Qiu L, Wen C-K. Possible modulation of Arabidopsis ETR1 N-terminal signaling by CTR1. Plant Signal Behav. 2012;7(10):1243–1245. doi: 10.4161/psb.21545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J, Wen C-K. Arabidopsis aux1rcr1 mutation alters AUXIN RESISTANT1 targeting and prevents expression of the auxin reporter DR5:GUS in the root apex. J Exp Bot. 2013;64(4):921–933. doi: 10.1093/jxb/ers371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sobczak K, Krzyzosiak WJ. Structural determinants of BRCA1 translational regulation. J Biol Chem. 2002;277(19):17349–17358. doi: 10.1074/jbc.M109162200. [DOI] [PubMed] [Google Scholar]
- Arrick BA, Lee AL, Grendell RL, Derynck R. Inhibition of translation of transforming growth factor-beta 3 mRNA by its 5′ untranslated region. Mol Cell Biol. 1991;11(9):4306–4313. doi: 10.1128/mcb.11.9.4306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arrick BA, Grendell RL, Griffin LA. Enhanced translational efficiency of a novel transforming growth factor beta 3 mRNA in human breast cancer cells. Mol Cell Biol. 1994;14(1):619–628. doi: 10.1128/mcb.14.1.619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatterjee S, Pal JK. Role of 5′- and 3′-untranslated regions of mRNAs in human diseases. Biol Cell. 2009;101(5):251–262. doi: 10.1042/BC20080104. [DOI] [PubMed] [Google Scholar]
- Medenbach J, Seiler M, Hentze Matthias W. Translational control via protein-regulated upstream open reading frames. Cell. 2011;145(6):902–913. doi: 10.1016/j.cell.2011.05.005. [DOI] [PubMed] [Google Scholar]
- McDaniel BK, Binder BM. Ethylene receptor 1 (ETR1) is sufficient and has the predominant role in mediating inhibition of ethylene responses by silver in Arabidopsis thaliana. J Biol Chem. 2012;287(31):26094–26103. doi: 10.1074/jbc.M112.383034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W, Wen C-K. Preparation of ethylene gas and comparison of ethylene responses induced by ethylene, ACC, and ethephon. Plant Physiol Biochem. 2010;48(1):45–53. doi: 10.1016/j.plaphy.2009.10.002. [DOI] [PubMed] [Google Scholar]
- Wang KLC, Yoshida H, Lurin C, Ecker JR. Regulation of ethylene gas biosynthesis by the Arabidopsis ETO1 protein. Nature. 2004;428(6986):945–950. doi: 10.1038/nature02516. [DOI] [PubMed] [Google Scholar]
- Yoshida H, Nagata M, Saito K, Wang K, Ecker J. Arabidopsis ETO1 specifically interacts with and negatively regulates type 2 1-aminocyclopropane-1-carboxylate synthases. BMC Plant Biol. 2005;5(1):14. doi: 10.1186/1471-2229-5-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Q, Xu C, Wen C-K. Genetic and transformation studies reveal negative regulation of ERS1 ethylene receptor signaling in Arabidopsis. BMC Plant Biol. 2010;10(1):60. doi: 10.1186/1471-2229-10-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X, Liu Q, Xie F, Wen C-K. RTE1 is a Golgi-associated and ETR1-dependent negative regulator of ethylene responses. Plant Physiol. 2007;145(1):75–86. doi: 10.1104/pp.107.104299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivarola M, McClellan CA, Resnick JS, Chang C. ETR1-specific mutations distinguish ETR1 from other Arabidopsis ethylene receptors as revealed by genetic interaction with RTE1. Plant Physiol. 2009;150(2):547–551. doi: 10.1104/pp.109.138461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chao Q, Rothenberg M, Solano R, Roman G, Terzaghi W, Ecker J. Activation of the ethylene gas response pathway in Arabidopsis by the nuclear protein ETHYLENE-INSENSITIVE3 and related proteins. Cell. 1997;89:1133–1144. doi: 10.1016/S0092-8674(00)80300-1. [DOI] [PubMed] [Google Scholar]
- Potuschak T, Lechner E, Parmentier Y, Yanagisawa S, Grava S, Koncz C, Genschik P. EIN3-dependent regulation of plant ethylene hormone signaling by two Arabidopsis F box proteins: EBF1 and EBF2. Cell. 2003;115:679–689. doi: 10.1016/S0092-8674(03)00968-1. [DOI] [PubMed] [Google Scholar]
- Guo H, Ecker JR. Plant responses to ethylene gas are mediated by SCF(EBF1/EBF2)-dependent proteolysis of EIN3 transcription factor. Cell. 2003;115(6):667–677. doi: 10.1016/S0092-8674(03)00969-3. [DOI] [PubMed] [Google Scholar]
- An F, Zhao Q, Ji Y, Li W, Jiang Z, Yu X, Zhang C, Han Y, He W, Liu Y. et al. Ethylene-induced stabilization of ETHYLENE INSENSITIVE3 and EIN3-LIKE1 is mediated by proteasomal degradation of EIN3 binding F-box 1 and 2 that requires EIN2 in Arabidopsis. Plant Cell. 2010;22(7):2384–2401. doi: 10.1105/tpc.110.076588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hua J, Meyerowitz EM. Ethylene responses are negatively regulated by a receptor gene family in Arabidopsis thaliana. Cell. 1998;94(2):261–271. doi: 10.1016/S0092-8674(00)81425-7. [DOI] [PubMed] [Google Scholar]
- Wang W, Hall AE, O’Malley R, Bleecker AB. Canonical histidine kinase activity of the transmitter domain of the ETR1 ethylene receptor from Arabidopsis is not required for signal transmission. Proc Natl Acad Sci U S A. 2003;100(1):352–357. doi: 10.1073/pnas.0237085100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bugaut A, Balasubramanian S. 5′-UTR RNA G-quadruplexes: translation regulation and targeting. Nucleic Acids Res. 2012;40(11):4727–4741. doi: 10.1093/nar/gks068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray NK, Wickens M. Control of translation initiation in animals. Annu Rev Cell Dev Biol. 1998;14(1):399–458. doi: 10.1146/annurev.cellbio.14.1.399. [DOI] [PubMed] [Google Scholar]
- Zhang W, Hu W, Wen C-K. Ethylene preparation and its application to physiological experiments. Plant Signal Behav. 2010;5(4):453–457. doi: 10.4161/psb.5.4.10875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough SJ, Bent AF. Floral dip: a simplified method forAgrobacterium-mediated transformation ofArabidopsis thaliana. Plant J. 1998;16(6):735–743. doi: 10.1046/j.1365-313x.1998.00343.x. [DOI] [PubMed] [Google Scholar]
- Bent A. In: Agrobacterium Protocols, vol. 343. Wang K, editor. Totowa, New Jersey: Humana Press; 2006. Arabidopsis thaliana Floral Dip Transformation Method; pp. 87–104. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
BAC clones and sequences encompassing ECR2.