Abstract
Aberrant activation of fibroblast growth factor receptors (FGFRs) contributes to breast cancer growth, progression and therapeutic resistance. Due to the complex nature of the FGF/FGFR axis, and the numerous effects of FGFR activation on tumor cells and the surrounding microenvironment, the specific mechanisms through which aberrant FGFR activity contributes to breast cancer are not completely understood. We show here that FGFR activation induces accumulation of hyaluronan (HA) within the extracellular matrix (ECM) and that blocking HA synthesis decreases proliferation, migration and therapeutic resistance. Furthermore, FGFR-mediated HA accumulation requires activation of the signal transducer and activator of transcription 3 (STAT3) pathway, which regulates expression of hyaluronan synthase 2 (HAS2) and subsequent HA synthesis. Using a novel in vivo model of FGFR-dependent tumor growth, we demonstrate that STAT3 inhibition decreases both FGFR-driven tumor growth and HA levels within the tumor. Finally, our results suggest that combinatorial therapies inhibiting both FGFR activity and HA synthesis is more effective than targeting either pathway alone and may be a relevant therapeutic approach for breast cancers associated with high levels of FGFR activity. In conclusion, these studies indicate a novel targetable mechanism through which FGFR activation in breast cancer cells induces a pro-tumorigenic microenvironment.
Introduction
Recent genomic profiling studies have demonstrated that a number of potentially targetable pathways are aberrantly regulated in breast cancer, including the fibroblast growth factor receptor (FGFR) pathway (1). Members of the FGFR family, comprised of four FGFR genes, are transmembrane receptor tyrosine kinases that are activated by FGFs (2). Aberrant FGFR activity in breast cancers can occur through a variety of potential mechanisms, including amplification of receptor genes, increased protein expression of both ligands and receptors, single nucleotide polymorphisms (SNPs), gene rearrangements and mutations in FGFRs, all of which have been identified in human breast cancer cell lines and patient samples (3, 4). Experimental studies have demonstrated that FGFR activation contributes to breast cancer growth and progression (5–11). Furthermore, a number of clinical trials have been initiated to investigate the safety and efficacy of small molecule FGFR inhibitors in breast and other cancers (3, 5).
To study FGFR1 activation, we use an inducible FGFR1 (iFGFR1) construct containing a dimerization domain that is activated with the synthetic homodimerizer B/B, resulting in sustained activation of FGFR1-induced signaling pathways (11). Using this inducible model, our studies have focused on the mechanisms through which FGFR1 activation in epithelial and tumor cells contributes to tumor initiation and growth (12–16). Specifically, we have shown that aberrant FGFR1 activation in mammary epithelial cells leads to alterations in the stroma, including the generation of a localized inflammatory response and alterations in the ECM (11, 16). In the studies described here, we demonstrate that activation of FGFR signaling pathways leads to structural modifications of the ECM component hyaluronan (HA). HA is a glycosaminoglycan that interacts with cancer cells through various receptors including CD44 and receptor for hyaluronan-mediated motility (RHAMM) to promote proliferation and migration. Furthermore, aberrant HA synthesis has been linked to breast cancer growth and progression (17–21). We demonstrate here that FGFR activation leads to increased synthesis of HA, which contributes to proliferation, migration and resistance to chemotherapy. Thus these studies link aberrant activation of growth factor receptor signaling pathways in tumor cells to pro-tumorigenic modifications in the surrounding stroma.
Because HA is often associated with an inflammatory environment (22), further studies examined the contribution of FGFR induced inflammatory pathways to HA synthesis. We demonstrate that activation of FGFR leads to increased production of proinflammatory cytokines, including members of the IL-6 family, which activate the signal transducer and activator of transcription 3 (STAT3) pathway. STAT3 is a proinflammatory transcription factor that contributes to breast cancer cell proliferation, migration, invasion and chemotherapeutic resistance (23–27). In these studies, we demonstrate that FGFR-induced STAT3 activation contributes to HA synthesis and is important for FGFR-driven mammary tumor growth. These studies are the first to identify HA as a downstream target of FGFR activation and suggest that the addition of microenvironment-targeted therapies may enhance the efficacy of FGFR-specific therapies in cancers associated with high levels of FGFR activity.
Materials and Methods
Cell Culture
Generation of HC-11 cells stably expressing the iFGFR1 construct (HC-11/R1 cells) was described previously (28), and cells were obtained from Dr. Jeff Rosen (Baylor College of Medicine, Houston, TX) and maintained as described (28). Hs578T, MCF-7 and MDA-MB-453 cells were obtained from the American Type Culture Collection (ATCC) and maintained as suggested. For experiments, Hs578T cells were grown on plates coated with 1.2% polyHEMA [poly(2-hydroxyethylmethacrylate)] (Sigma). All cell lines were used for fewer than 6 months after resuscitation. HC-11 cells and HC-11/R1 cells are not maintained for longer than 20 passages and are tested for mycoplasma, β-casein expression and iFGFR1 expression regularly.
Immunoblot analysis
Serum starved cells were treated with B/B (Clontech) or bFGF (Invitrogen). For blocking and inhibitor studies, gp130 blocking antibody (R&D Systems), doxorubicin (Boynton Pharmacy, UMN), Stattic (Sigma) and/or 4-MU (Sigma) were used. ON-TARGETplus SMARTpool STAT3 and non-targeting (NT) siRNA (Thermo Scientific) were used according to manufacturer’s instructions. Cells were lysed in RIPA buffer and equal amounts of protein were analyzed by SDS-PAGE. Immunoblot analysis was performed with the following antibodies: pSTAT3Ser727 (9134), pSTAT3Tyr705 (9131), STAT3 (9132), cleaved caspase-3 (9661), and β-tubulin (2146) (Cell Signaling).
ELISA
Serum starved HC-11/R1 cells were treated with B/B or ethanol as solvent control. Hs578T cells were treated with or without bFGF. At 2,6, and 24 hours, conditioned media was collected and ELISAs for mouse or human LIF, IL-6, and IL-11 were performed according to the manufacturer’s protocol (R&D Systems). ON-TARGETplus SMARTpool-siRNA Has2 or NT siRNA (Thermo Scientific) was transfected into HC-11/R1 cells as described previously (28). Cells were pretreated with 4-MU for 1 hour prior to addition of B/B. To analyze HA synthesis, conditioned media samples were collected and tumor samples were lysed in RIPA and analyzed using the Hyaluronan ELISA (R&D systems) according to the manufacturer’s protocols.
Quantitative reverse transcription-PCR
Cells were treated as described above and qRT-PCR was done as described previously and normalized to cyclophilin B levels (29). The following mouse primers were used: cyclophilin B 5’-TGCAGGCAAAGACACCAATG-3’ and 5’-GTGCTCTCCACCTCCCGTA-3’, Lif 5'-GCCTCCCTGACCAATATCACC-3' and 5'-GACGGCAAAGCACATTGCTG-3', Il-6 5’-TAGTCCTTCCTACCCCAATTTCC-3’ and 5’-TTGGTCCTTAGCCACTCCTTC-3’, Has2 5’-TGTGAGAGGTTTGTATGTGTCCT-3’ and 5’-ACCGTACAGTGGAAATGAGAAGT-3’.
TUNEL assays
Serum starved cells were treated with B/B or ethanol, doxorubicin or saline, and Stattic or DMSO for 20 hours. Cells were fixed and stained using the DeadEnd Fluorometric TUNEL System (Promega) according to the manufacturer’s protocol. Five representative pictures were taken of each treatment, and cell counting was performed in a blinded manner.
Mice
3–4 week old Balb/c female mice were purchased from Harlan Laboratories. 250,000 HC-11/R1 cells in 50% Matrigel (BD Biosciences) were injected into the fourth inguinal mammary fat pads. Mice were given twice weekly intraperitoneal (i.p.) injections of B/B to activate iFGFR1. When tumors reached at least 100 mm3, mice were given Stattic or solvent control (DMSO) by oral gavage five days a week for three weeks. At least three mice were in each treatment group. For the studies with transgenic mice, MMTV-iFGFR1 mice were treated with B/B and mammary glands were isolated as described previously (16). All animal care and procedures were approved by the Institutional Animal Care and Use Committee of the University of Minnesota and were in accordance with the procedures detailed in the Guide for Care and Use of Laboratory Animals.
Clinical cohort and TMA construction
Specimens and associated clinical data were obtained from the UMN BioNet core facility (www.bionet.umn.edu) after approval from the UMN Institutional Review Board (IRB). Archival formalin-fixed paraffin-embedded (FFPE) tissues from breast cancer patients treated at the University of Minnesota were collected. Areas of invasive carcinoma were verified by a pathologist. TMA blocks consisting of quadruplicate 1 mm core carcinoma samples were constructed using a manual tissue arrayer (MTA-1, Beecher Inc). Clinical characteristics were abstracted from pathology reports. Coded specimens and data were provided for this study. Patient identifiers were not available to the authors, but rather were held within the BioNet office per the BioNet IRB approval.
Immunohistochemistry
Mammary glands from mice were fixed, sectioned and stained using sodium citrate antigen retrieval as described previously (14, 16). Antibodies used were pStat3Tyr705 (1:200; Cell Signaling, 9145), Has2 (1:50; Santa Cruz, sc-365263), and pFRS2 (1:40; R&D, AF5126). As a control, sections were stained with the biotinylated anti-rabbit only. For HABP staining, sections were blocked with 3% BSA and then incubated overnight with 2μg/ml biotinylated HABP. Visualization was performed as described above. As a control, tissues were incubated with hyaluronidase before addition of HABP. HABP-positive stromal thickness of at least 50 ducts from three mice per timepoint was quantified using Leica LAS software.
Three-dimensional culture
Primary mammary epithelial cells were isolated from MMTV-iFGFR1 transgenic mice and plated in Matrigel as described previously (30). 10,000 Hs578T cells were plated per well in Matrigel. 4-MU or DMSO (solvent) was added to the cultures for 8 days. At least 50 acini were measured for each condition using Leica LAS software.
Proliferation Assay
For HC-11/R1 cells, Thiazolyl Blue Tetrazolium Bromide (Sigma, M2128) was used. Serum starved HC-11/R1 cells were treated with B/B, 4-MU or Stattic. MTT reagent was added to the wells and after 2 hours proliferation was determined according to the manufacturer protocol. For Hs578T cells, the CellTiter 96 Aqueous Proliferation Assay (Promega) was used according to manufacturer protocol. Briefly, 1×105 cells/ml were resuspended in serum free media in 96 well plates coated with 1.2% polyHEMA. The next day cells were treated with bFGF and either Stattic or 4-MU. The absorbance was read at 490 nm following 24 and 48 hours of treatment.
Migration Assay
Confluent HC-11/R1 cells were serum starved and wound healing assays were performed as described previously (28) in the presence of B/B and/or 4-MU and/or Stattic. Area of wound closure was quantified following 18 hours using Leica LAS software.
Statistical analysis
Experiments were performed at least three separate times. Statistical analysis was performed using the unpaired student’s t-test to compare two means. In all figures, error bars represent the standard error of the mean. For the human samples, the association between pFRS2 and pSTAT3 was evaluated using proportional odds logistic regression with pSTAT3 as the outcome and pFRS2 as a covariate in a univariate regression model and the association between pFRS2 and pSTAT3 was summarized by the odds ratio.
Results
FGFR activation induces synthesis of hyaluronan
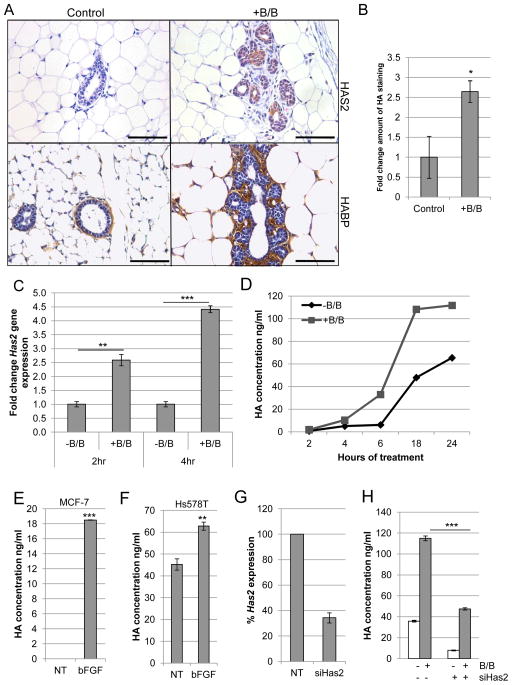
We have previously used the MMTV-iFGFR1 transgenic mouse model to identify mechanisms through which FGFR1 activation in mammary epithelial cells contributes to pro-tumorigenic changes within the stroma (12, 14–16, 28, 30). It has been previously noted that activation of iFGFR1 in mammary epithelial cells results in alterations in the surrounding ECM (11, 16). To gain insights into these changes, previously published microarray studies (16) were examined to identify ECM-related genes that are regulated following iFGFR1 activation. Interestingly, Has2, which stimulates synthesis of the ECM component HA, was significantly induced following 8 hours of B/B treatment. To verify these findings, mammary glands from B/B-treated MMTV-iFGFR1 mice were analyzed for HAS2 expression and HA accumulation using immunohistochemical analysis. Following 48 hours of iFGFR1 activation, increased expression of HAS2 was detectable within the aberrant budding epithelial structures (Figure 1A). Furthermore, increased accumulation of HA in the surrounding stroma was determined by measuring the thickness of HA-positive stroma (Figure 1A,B).
Figure 1. FGFR activation leads to increased production of HA.
A) MMTV-iFGFR1 transgenic mice were treated with 1mg/kg B/B or solvent for 48 hours. Mammary gland sections were stained with HAS2-specific antibody or HA binding protein (HABP). Magnification bars represent 50μm. B) Quantification of HA-positive stromal thickness in mammary glands from solvent or B/B treated mice. C) HC-11/R1 cells were treated with solvent (-B/B) or 30nM B/B and Has2 gene expression was analyzed by qRT-PCR. D) HC-11/R1 cells were treated as described in (C) and HA expression in conditioned media was determined by ELISA. E,F) MCF-7 (E) and Hs578T (F) cells were treated with or without 50ng/ml bFGF for 18 hours and HA in the conditioned media was determined by ELISA. G) HC-11/R1 cells were treated with Has2 siRNA or a non-targeting (NT) control. Expression of Has2 was measured by qRT-PCR. H) Amount of HA was determined by ELISA. *p<0.05, **p<0.01, ***p<0.001.
To verify that activation of iFGFR1 leads to increased expression of Has2 in epithelial cells, HC-11 cells that stably express iFGFR1 (HC-11/R1) (28) were treated with B/B to activate iFGFR1. Has2 expression increased following iFGFR1 activation as shown by qRT-PCR analysis (Figure 1C). Furthermore, increased levels of HA were detected in the media of B/B-treated HC-11/R1 cells using an ELISA-based assay (Figure 1D). Analysis of FGF-responsive human breast cancer cell lines, including the estrogen receptor positive (ER+) cell line MCF-7 and the triple negative cell line Hs578T, demonstrated that basic FGF (bFGF) treatment also led to increased levels of HA in the media (Figure 1E,F). To confirm that HAS2 is the primary hyaluronan synthase that contributes to HA synthesis, Has2 knock-down was performed using siRNA (Figure 1G) and resulted in a significant decrease in HA synthesis (Figure 1H). Together, these studies demonstrate that FGFR activation induces expression of HAS2, which leads to increased synthesis of HA in vitro and in vivo.
Inhibition of HA synthesis leads to decreased proliferation, migration, and chemoresistance
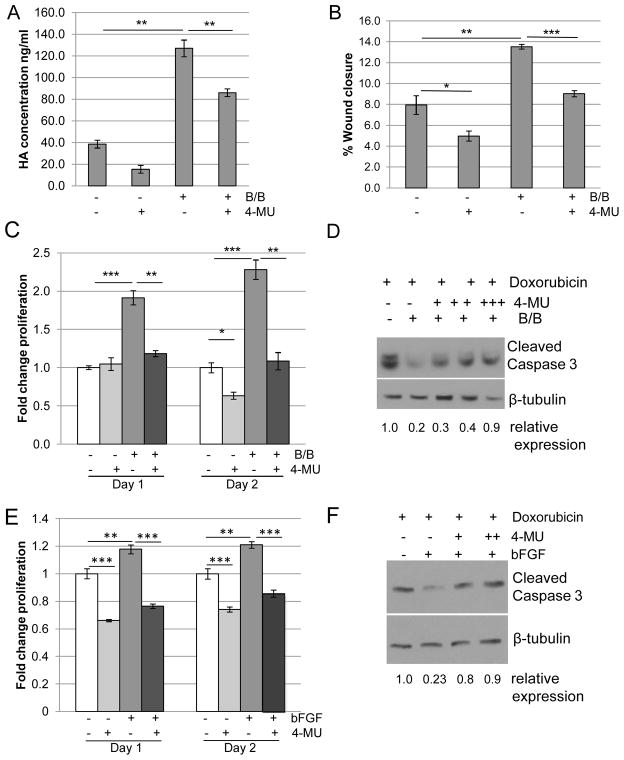
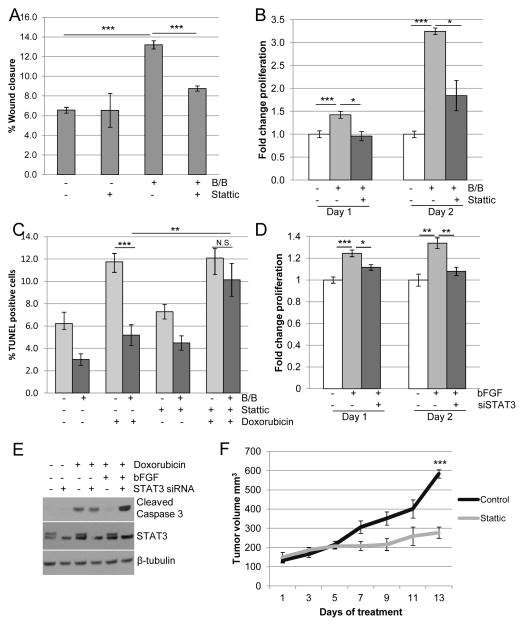
Because HA has been linked to tumor growth and progression (17–19), further studies were performed to examine the contributions of HA to proliferation and migration. For these studies, cells were treated with 4-methylumbelliferone (4-MU), which inhibits HA synthases including HAS2 (31–33). As shown in Figure 2A, treatment of HC-11/R1 cells with 4-MU effectively inhibited both basal and iFGFR1-induced HA synthesis. Furthermore, treatment of HC-11/R1 cells with 4-MU inhibited iFGFR1-induced migration (Figure 2B) and proliferation (Figure 2C). In addition, the contribution of HA to iFGFR1-induced survival in response to chemotherapy was examined by analyzing apoptosis following exposure of cells to doxorubicin. Activation of iFGFR1 significantly decreased doxorubicin-induced apoptosis in the HC-11/R1 cells, which was partially reversed by 4-MU, demonstrating that HA contributes to iFGFR1-induced chemoresistance. Similarly, treatment of Hs578T cells with bFGF and 4-MU inhibited bFGF-induced proliferation (Figure 2E) and restored apoptosis in response to doxorubicin treatment (Figure 2F). Interestingly, we found that treatment of both cell lines with 4-MU had effects on cell behavior independently of FGFR activation, possibly due to the decreased levels in basal HA synthesis (Figure 2A). These results suggest that there are likely to be additional mechanisms that regulate HA synthesis in the absence of FGFR activation, which may contribute to the 4-MU induced inhibition of FGFR-induced phenotypes.
Figure 2. Blocking HA synthesis leads to decreased migration, proliferation, and chemoresistance.
A) HC-11/R1 cells were treated with 250 μM of the HAS2 inhibitor 4-MU or solvent, followed by the addition of 30nM B/B or solvent. HA was detected in conditioned media by ELISA. B,C) HC-11/R1 cells were treated with B/B, 250μM 4-MU and/or solvent. The change in wound closure was determined at 18 hours (B) and proliferation was measured by an MTT assay (C) at day 1 or 2. D) HC-11/R1 cells were treated with B/B, 4-MU (62.5μM (+), 125μM (++), 250μM 9 (+++)), and 2μM doxorubicin for 24 hours. Levels of cleaved caspase-3 and the loading control β-tubulin were examined by immunoblot analysis. E) Hs578T cells were treated with 50ng/ml bFGF and 25μM 4-MU or solvent. Proliferation was measured at either day 1 or 2 relative to solvent-only treated samples. F) Hs578T cells were treated with bFGF, 4-MU (125μM (+) and 250μM (++)), and doxorubicin and levels of cleaved caspase-3 and β-tubulin were examined. *P<0.05; **p<0.01; and ***p<0.001.
FGFR activation leads to increased phosphorylation of STAT3
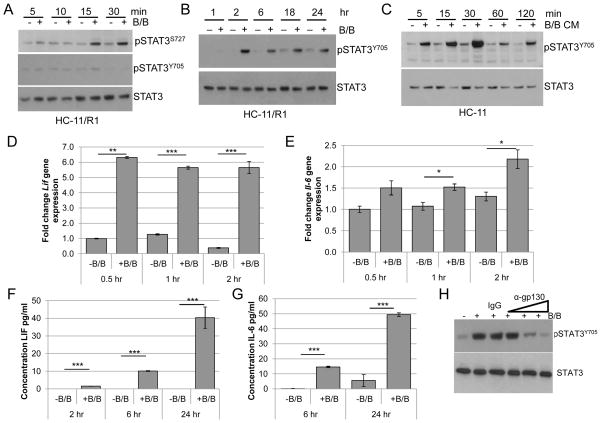
Next, we examined the mechanisms involved in mediating FGFR-induced HA synthesis. Because HA is involved in inflammation (22, 34), our initial studies focused on examining inflammatory mediators such as STAT3, which regulates inflammation-related genes including Has2 (35). To examine STAT3 activation in the iFGFR1 model, HC-11/R1 cells were treated with B/B and phosphorylation of STAT3Ser727 and STAT3Tyr705 was assessed by immunoblot analysis. Similar to what has been shown previously (36), STAT3Ser727 was phosphorylated within 15 minutes of iFGFR1 activation, although pSTAT3Tyr705 was not detected at these timepoints (Fig. 3A). However, analysis of later timepoints demonstrated that pSTAT3Tyr705 was detectable following 2 hours of iFGFR1 activation and remained elevated throughout the 24-hour time course (Figure 3B).
Figure 3. Activation of iFGFR1 leads to increased pSTAT3Tyr705 in a gp130-dependent manner.
HC-11/R1 cells were stimulated with solvent (-B/B) or 30nM B/B, followed by protein and RNA extraction or collection of conditioned media. A, B) pSTAT3S727, pSTAT3Y705, and STAT3 (loading control) were examined using immunoblot analysis. C) Conditioned media (CM) samples from treated HC-11/R1 cells were collected and added to HC-11 cells. pSTAT3Y705 and STAT3 was examined using immunoblot analysis. D,E) qRT-PCR analysis was performed to assess Lif or Il-6 gene expression. F,G) LIF and IL-6 expression in conditioned media was assessed by ELISA. H) HC-11/R1 were treated as above with the addition of IgG control or gp130 blocking antibody (0.05, 0.5, 5 μg/ml) for 6 hours. Expression of pSTAT3Y705 and STAT3 was examined by immunoblot analysis. *p<0.05, **p<0.01, ***p<0.001.
The timing of STAT3Tyr705 phosphorylation led to the hypothesis that FGFR induces STAT3Tyr705 phosphorylation indirectly through inducing production of soluble factors that activate STAT3. To address this possibility, HC-11/R1 cells were treated with B/B for 18 hours and conditioned media were used to stimulate parental HC-11 cells. pSTAT3Tyr705 was elevated in the HC-11 cells as early as 5 minutes after treatment with conditioned media (Figure 3C), consistent with the hypothesis that soluble factors produced by the cells following iFGFR1 activation contribute to STAT3 activation.
Further studies were performed to identify soluble factors that induce STAT3Tyr705 phosphorylation in our system. Because STAT3 is a well-established downstream target of IL-6 family cytokines (37), the ability of iFGFR1 to induce expression of genes in the IL-6 family was assessed. Initial screening of HC-11/R1 cells demonstrated that iFGFR1 activation led to increased gene expression of Il-6, Lif and Il-11 (Figure 3D,E and data not shown). ELISA analysis of conditioned media confirmed that soluble LIF was detectable within 2 hours of iFGFR1 activation (Figure 3F) and soluble IL-6 was detectable within 6 hours (Figure 3G). However, increased IL-11 was not found in the media at any timepoint (data not shown), suggesting that IL-6 and LIF are the primary IL-6 family cytokines produced by HC-11/R1 cells in response to iFGFR1 activation. Initial studies using cytokine-specific blocking antibodies demonstrated that blocking a single cytokine was unable to completely abolish STAT3 phosphorylation (data not shown), possibly due to ligand redundancy. IL-6 family cytokines utilize the common receptor subunit gp130 to transmit their signals (38). As shown in Figure 3H, treatment of HC-11/R1 cell with a gp130-blocking antibody prior to activation of iFGFR1 led to a dose-dependent reduction of STAT3Tyr705 phosphorylation. These results demonstrate that activation of FGFR1 leads to production of IL-6 family cytokines, which act through gp130 to induce phosphorylation of STAT3Tyr705.
FGFR activation induces expression of IL-6 family cytokines and STAT3 activation in human breast cancer cells
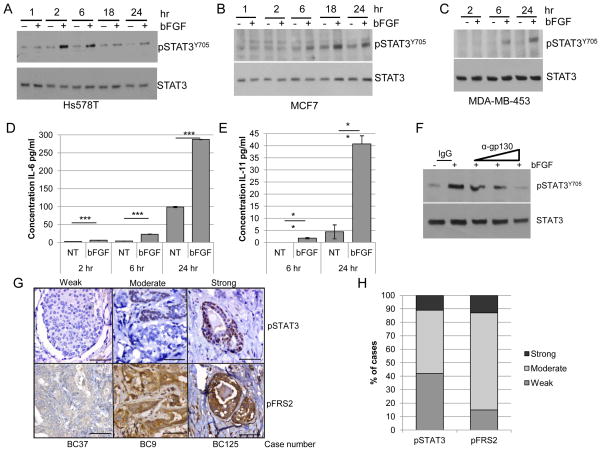
Further studies focused on validating FGFR-induced STAT3Tyr705 phosphorylation in other FGF-responsive cell lines including MCF-7, MDA-MB-453 and Hs578T (12, 39, 40). Following treatment of cells with bFGF, pSTAT3Tyr705 was observed at later time points, including 2 and 6 hours post-treatment, in all cell types (Figure 4A–C), similar to what was observed with the HC-11/R1 cells. Furthermore, bFGF treatment of Hs578T cells led to increased production of IL-6 family cytokines, including IL-6 (Figure 4D) and IL-11 (Figure 4E), although LIF was not induced in these cells (data not shown). Finally, treatment of cells with a gp130-blocking antibody led to decreased pSTAT3Tyr705 in a dose dependent manner (Figure 4F). These results verify our findings from the HC-11/R1 model that activation of endogenous FGFR induces expression of IL-6 family cytokines, which contribute to phosphorylation of pSTAT3Tyr705
Figure 4. FGFR activation leads to pSTAT3Tyr705 in human breast cancer cells.
Hs578T (A), MCF7 (B), and MDA-MB-453 (C) were treated with or without 50ng/ml bFGF and expression of pSTAT3Y705 and STAT3 was examined by immunoblot analysis. D,E) Hs578T cells were treated as described above for the indicated times, and conditioned media samples were collected to examine protein expression of IL-6 (D) or IL-11 (E) by ELISA. F) Hs578T cells were treated as above with the addition of IgG control or gp130 blocking antibody (0.1, 1, 10 μg/ml) for 6 hours. G,H) A human breast cancer tissue microarray was stained for pSTAT3Tyr705 and pFRS2 using IHC. G) Representative images of weak, moderate, or strong staining intensity are shown. Magnification bars represent 50μm. H) Percentage of cases based on staining intensity. **p<0.01 and ***p<0.001.
To determine whether the FGFR and STAT3 signaling pathways are correlated in human breast cancers, a tissue microarray (TMA) was generated (Supplemental Table 1) and stained with antibodies that recognize pSTAT3Tyr705 and phosphorylated fibroblast growth factor receptor substrate 2 (pFRS2), which is indicative of activated FGFR (41). As control, tissues were stained with secondary antibody only and positive staining was not detected (Supplemental Figure 1A). Both sets of samples were scored for weak, moderate and strong staining (Figure 4G). Similar to previously published results (24, 38, 42), pSTAT3Tyr705 was observed at moderate to strong levels in approximately 60% of breast cancers (Figure 4H). pFRS2 staining was found to be detectable at some level in all samples, with approximately 85% of tumors expressing moderate to high levels of staining (Figure 4H), suggesting that FGFR activity is present in a large percentage of breast cancer samples. Analysis of all samples revealed a significant association between pFRS2 and pSTAT3 (odds ratio=4.8, 95% CI: 1.91, 12.05, p<0.001), demonstrating a correlation between FGFR activation and STAT3Tyr705 phosphorylation in a proportion of breast cancers (Supplemental Table 1).
STAT3 contributes to FGFR-induced migration, proliferation and resistance to chemotherapy
To assess the functional contributions of STAT3 activation to FGFR-induced tumorigenic phenotypes, we used the pharmacological inhibitor Stattic (43). Inhibition of STAT3 in HC-11/R1 cells led to decreased iFGFR1-induced migration (Figure 5A) and proliferation (Figure 5B). Because STAT3 has also been linked to resistance to chemotherapy (44), the contribution of STAT3 activation to FGFR-induced chemoresistance was examined. Stattic restored the sensitivity of HC-11/R1 cells to doxorubicin, suggesting that STAT3 contributes to iFGFR1-induced chemoresistance (Figure 5C).
Figure 5. STAT3 promotes FGFR-induced migration, proliferation, and chemoresistance.
A) HC-11/R1 cells were treated with solvent or 30nM B/B in the presence of 1μM Stattic or DMSO, and wound closure was measured after 18 hours. B) HC-11/R1 cells were treated with B/B, 2μM Stattic or solvent for 1 or 2 days. Proliferation rate was calculated by MTT assay and given relative to the solvent treated samples. C) Apoptosis was determined by TUNEL assay for HC-11/R1 cells treated with B/B, 4μM Stattic, or 2μM doxorubicin for 24 hours. D) Hs578T cells were treated with NT or STAT3 siRNA for 24 hours, followed by 1 or 2 days treatment with 50ng/ml bFGF, and proliferation was calculated relative to solvent treated samples. E) Hs578T cells were treated as described above, and 2μM doxorubicin was added to the indicated groups for 24 hours. Expression levels of cleaved caspase-3 and β-tubulin were examined by immunoblotting. F) HC-11/R1 cells were injected into the fat pads of Balb/c mice. Mice were given twice weekly injections of 1mg/kg B/B. Once tumors reached a size of 100 mm3, mice received either DMSO or 20mg/kg Stattic and tumor growth was assessed. *p<0.05, **p<0.01, ***p<0.001.
To verify the contribution of STAT3 to proliferation and chemotherapeutic resistance following endogenous FGFR activation, these processes were assessed in Hs578T cells. Consistent with the results from the HC-11/R1 cells, treatment of Hs578T cells with bFGF in the presence of Stattic decreased bFGF-induced proliferation and reversed resistance to doxorubicin-induced apoptosis (Supplemental Figure 2). To demonstrate that these effects are due to loss of STAT3 activity and not due to off-target effects of Stattic, STAT3 expression was decreased using siRNA. As shown in Figure 5E, STAT3 siRNA decreased expression of STAT3α, while leaving the STAT3β splice variant intact. Loss of STAT3α expression correlated with decreased bFGF-induced proliferation of Hs578T cells (Figure 5D) and restoration of chemosensitivity as shown by an increase in cleaved caspase-3 (Figure 5E).
A novel orthotopic mammary tumor model was used to evaluate the contribution of STAT3 activation to FGFR-induced tumor growth in vivo. HC-11/R1 cells were injected into fat pads of Balb/c mice and the mice were administered B/B to induce tumor growth. Average time to tumor growth in this model is 5.5 weeks, compared with parental HC-11 cells, which do not form palpable tumors within this timeframe (Supplemental Figure 3). To determine the effects of STAT3 inhibition on iFGFR1-induced tumor growth, mice bearing 100 mm3 tumors were treated with either Stattic or solvent control. Treatment of mice with Stattic led to tumor stabilization, resulting in a significant (p<0.001) reduction in the size of end stage tumors (Figure 5F). Together, these results demonstrate that STAT3 is an important mediator of FGFR-induced proliferation, migration, therapeutic resistance and tumor growth.
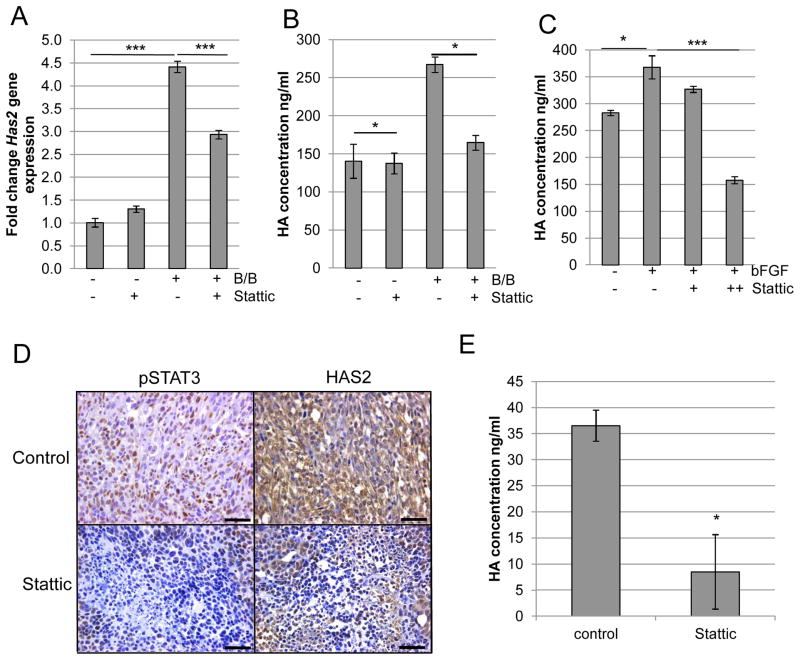
FGFR activation induces HA accumulation in a STAT3-dependent manner
Because HAS2 was previously identified as a STAT3 target gene (35), we determined the contribution of STAT3 to iFGFR1-mediated Has2 expression and HA synthesis. As shown in Figure 6A, treatment of HC-11/R1 cells with Stattic led to decreased Has2 gene expression within 2 hours of B/B treatment. Furthermore, inhibition of STAT3 also led to decreased HA synthesis following iFGFR1 activation (Figure 6B). To verify the link between FGFR, STAT3 and HA in human breast cancer cells, Hs578T cells were treated with bFGF in the presence or absence of Stattic and HA synthesis was analyzed. Although Hs578T cells already express high basal levels of HA (45), inhibition of STAT3 activity led to a decrease in bFGF-induced HA synthesis (Figure 6C).
Figure 6. STAT3 regulates expression of HAS2 and production of HA.
A) HC-11/R1 cells were treated with 30nM B/B, 4μM Stattic or solvent for 2 hours. RNA was collected to examine expression of Has2 by qRT-PCR. B) HC-11/R1 cells were treated as in (A) for 18 hours. Conditioned media was collected to examine expression of HA by ELISA. C) Hs578T cells were treated with 50ng/ml bFGF and Stattic (2μM (+) or 4μM (++)) or solvent for 18 hours, and levels of HA in the conditioned media were determined by ELISA. D) Serial tumor sections from mice treated with solvent or 20mg/kg Stattic were stained for pSTAT3Tyr705 and HAS2. E) Amount of HA in tumors from mice in D) was assessed by ELISA. *p<0.05, **p<0.01, ***p<0.001.
To confirm these findings in vivo, immunohistochemical analysis was performed on HC-11/R1-derived tumors (Figure 5F). Both pSTAT3Tyr705 and HAS2 staining were observed in the tumors from solvent-treated mice (Figure 6D). Analysis of serial sections of tumors from Stattic-treated mice revealed decreased pSTAT3Tyr705 and HAS2 staining in the same areas within the tumor. Furthermore, analysis of tumor-associated HA by ELISA revealed decreased HA levels in tumors from mice treated with Stattic (Figure 6E), consistent with the observed decrease in HAS2 expression.
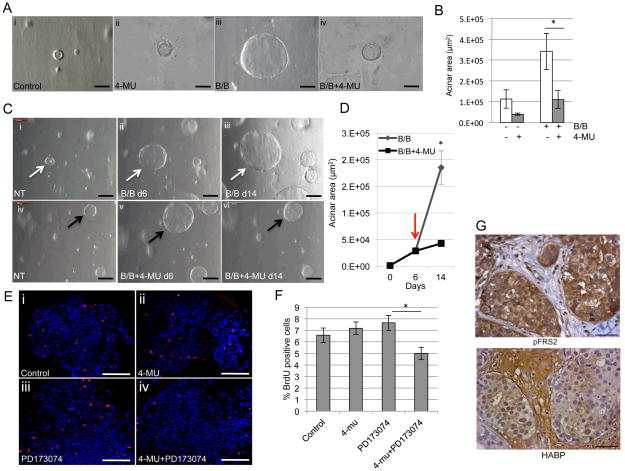
Inhibition of HA synthesis decreases acinar growth in three-dimensional culture
Further studies were performed to determine the effects of inhibiting HA synthesis using three-dimensional (3D) culture models, which provide a more relevant environment than cells in traditional two-dimensional culture (46). We have previously demonstrated that primary mammary epithelial cells isolated from MMTV-iFGFR1 transgenic mice form large acinar structures in 3D culture upon iFGFR1 activation (30). To assess effects HA inhibition on acinar growth, 4-MU was added to the 3D culture at the same time as activating iFGFR1, which led to inhibition of iFGFR1-induced acinar growth (Figure 7A,B). Furthermore, treatment of established structures with 4-MU led to inhibition of further acinar growth (Figure 7C,D). These studies demonstrate that blocking HA synthesis leads to inhibition of iFGFR1-dependent growth of both developing and established acinar structures.
Figure 7. Inhibition of HA synthesis leads to decreased FGFR-induced growth in 3D culture.
A) Primary mammary epithelial cells were isolated from MMTV-iFGFR1 transgenic mice and plated in 3D culture. Cells were treated with 30nM B/B,10μM 4-MU or solvent. Light microscopy images were obtained after 10 days in culture. B) Quantification of acinar area. C) Structures were treated with B/B to activate iFGFR1 for 6 days, followed by treatment with solvent or 4-MU for 8 days. Images were obtained from the same structures. D) Quantification of acinar area. Red arrow indicates addition of 4-MU. E) Hs578T cells were plated in Matrigel. The cells were treated with solvent (Control), 10μM 4-MU, 1μM PD173074 or both for 6 days. The cultures were stained with phospho-histone H3 and analyzed by confocal microscopy. F) Quantification of phospho-histone H3 positive cells. *p<0.05. G) Representative images of pFRS2 and HABP-stained sections of human breast cancers. Magnification bars represent 50μm.
Further studies were performed using the Hs578T cells, which exhibit high levels of both FGFR activation (40) and high levels of HA synthesis (45). The cells were plated in 3D culture and structures were treated with either PD173074 to inhibit FGFR activation, 4-MU to inhibit HA synthesis or both for 6 days, and proliferation was assessed using phospho-histone H3 staining. Surprisingly, treatment of cells with either PD173074 or 4-MU alone did not affect proliferation (Figure 7E,F). However, when treated with both inhibitors together, proliferation was significantly decreased (Figure 7E,F). Taken together, the 3D culture studies suggest that HA inhibition may be a relevant approach for targeting both FGFR-driven cancers and cancers that have high levels of FGFR activation, but are not necessarily FGFR-driven. To assess whether high levels of FGFR activation and HA co-exist in human patient samples, the TMA described above was stained for HA. No staining was observed in hyaluronidase treated samples, demonstrating specificity for the staining (Supplemental Figure 1B). HA was found in all samples and was therefore present in all breast cancer samples exhibiting high levels of pFRS2 (Figure 7G). Therefore, combination therapies targeting both FGFR activity and HA synthesis may be considered for patients with high levels of FGFR activation.
Discussion
The results from these studies reveal a novel link between FGFR activation and synthesis of HA by tumor cells, thus linking intracellular signaling to alterations in the microenvironment (Supplemental Figure 4). HA is an important component of the ECM that is normally involved in a number of physiological processes including maintenance of tissue integrity, morphogenesis, wound healing and inflammation (22, 34). HA is a prevalent component of the normal human breast ECM and alterations in HA contribute to breast cancer growth and progression (17–19). Tumor-associated HA alterations are complex and can include increased HA synthesis, changes in HA localization and increased HA fragmentation (47, 48). High levels of HA accumulate in breast cancer, in part due to increased HA synthesis by HAS enzymes including HAS2; these high levels of HA are associated with reduced survival and poor response to therapy (49, 50). HA is typically produced in a high molecular weight form and can be cleaved into lower molecular weight fragments by hyaluronidases or reactive oxygen species under specific conditions, such as during inflammation and within the tumor microenvironment (51–53). While high molecular weight HA inhibits tumor formation (54), low molecular weight forms have been shown to stimulate cancer cell migration and invasion possibly through differential interactions with its receptors, such as CD44 and RHAMM (47, 48, 55). While our current studies have not specifically assessed HA fragmentation, they demonstrate that FGFR can increase the levels of HA within the tumor microenvironment creating the necessary substrate for HA fragment production.
Because changes in HA are often associated with inflammation, further studies focused on the contributions of inflammatory pathways to FGFR-mediated changes in HA, specifically the transcription factor STAT3. In our current studies, we found differences in the kinetics of the two STAT3 phosphorylation sites, Ser727 and Tyr705, following FGFR activation. Phosphorylation at Ser727 has been previously identified as a rapid site of phosphorylation following FGFR activation (36). However, the observation of cytokine mediated indirect phosphorylation at Tyr705 at later time points is novel and provides a potential feed forward mechanism through which FGFR activation induces inflammation in breast cancer. Although initially thought to be secondary to phosphorylation of Tyr705 and involved in maximal STAT3 activation, recent studies have suggested that pSTAT3Ser727 may have different functions than pSTAT3Tyr705 (56). Interestingly, pSTAT3Ser727 has been identified in early stage tumors in melanoma, whereas pSTAT3Tyr705 has been associated with later stage cancer (56). Further studies are required to elucidate whether these two phosphorylation sites have different roles in mediating FGFR-driven tumor growth.
Further analysis of the mechanisms driving STAT3 phosphorylation demonstrated that FGFR activation induces various IL-6 family members, which contributed to phosphorylation of STAT3Tyr705. While these studies focused on actions of IL-6-mediated signaling in the tumor cells, it is likely that the IL-6 family members also act on neighboring epithelial cells (Supplemental Figure 4) and cells within the tumor reactive stroma, including infiltrating inflammatory cells, to promote tumor growth and progression. Further studies are in progress to determine the relative contributions of IL-6 family cytokine-induced signaling in tumor cells compared with other cell types in the microenvironment.
These studies raise the possibility that patients with high levels of FGFR activity and tumor-associated HA may be candidates for combinatorial therapies targeting both of these molecules. Consistent with this, our 3D culture studies suggest that while inhibition of FGFR in FGF-responsive cells alone does not affect cell proliferation, combinatorial treatment inhibiting both FGFR and HA synthesis leads to decreased proliferation. Whether this is due to cooperative effects of decreased signaling of both pathways or due to removal of HA surrounding the tumor cell and thus providing access of the drug to the cells, which has been demonstrated in other models (57), remains to be determined. Regardless of the mechanism, these findings provide an example of the potential therapeutic impact of designing therapeutic strategies that target both tumor cell-specific oncogenic pathways and the pro-tumorigenic microenvironment.
In conclusion, these studies define a novel pathway involving FGFR, STAT3 and HA synthesis that contribute to tumor growth. Because all of the components of these pathways are known to contribute to other tumor types as well as inflammation-related diseases, this pathway likely represents a general mechanism that can be applied more broadly than just breast cancer. Finally, these results provide important insights into the potential need for targeting growth factor driven signaling pathways within the tumor cells in combination with inhibiting HA/tumor cell interaction to more effectively treat breast cancer in patients.
Supplementary Material
Acknowledgments
The authors thank Sarah Bowell, Kimberly Casey, Colleen Forster, Anthony Rizzardi, and Xingchu Shen of UMN BioNet for assistance with TMA construction and tissue sectioning. The authors also thank Dr. Jeff Rosen for providing the HC-11/R1 cells and the MMTV–iFGFR1 transgenic mice and Dr. Mariya Farooqui for providing comments on the article. BioNet is supported by NIH grants P30 CA77598, P50 CA101955, and KL2 RR033182, and by the University of Minnesota Academic Health Center. This study was also funded by NIH/NCI R01CA132827 and American Cancer Society RSG-09-192-01-LIB (K.L. Schwertfeger). We would like to acknowledge the use of the confocal microscope at the Masonic Cancer Center made available through an NCRR Shared Instrumentation Grant (#1 S10 RR16851).
Footnotes
Conflict of interest statement: The authors disclose no potential conflicts of interest.
References
- 1.Greenman C, Stephens P, Smith R, et al. Patterns of somatic mutation in human cancer genomes. Nature. 2007;446:153–8. doi: 10.1038/nature05610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Schwertfeger KL. Fibroblast growth factors in development and cancer: insights from the mammary and prostate glands. Curr Drug Targets. 2009;10:632–44. doi: 10.2174/138945009788680419. [DOI] [PubMed] [Google Scholar]
- 3.Dieci MV, Arnedos M, Andre F, Soria JC. Fibroblast growth factor receptor inhibitors as a cancer treatment: from a biologic rationale to medical perspectives. Cancer Discov. 2013;3:264–79. doi: 10.1158/2159-8290.CD-12-0362. [DOI] [PubMed] [Google Scholar]
- 4.Wu YM, Su F, Kalyana-Sundaram S, et al. Identification of targetable FGFR gene fusions in diverse cancers. Cancer Discov. 2013;3:636–47. doi: 10.1158/2159-8290.CD-13-0050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Andre F, Bachelot T, Campone M, et al. Targeting FGFR with Dovitinib (TKI258): Preclinical and Clinical Data in Breast Cancer. Clin Cancer Res. 2013;19:3693–702. doi: 10.1158/1078-0432.CCR-13-0190. [DOI] [PubMed] [Google Scholar]
- 6.Chioni AM, Grose R. FGFR1 cleavage and nuclear translocation regulates breast cancer cell behavior. J Cell Biol. 2012;197:801–17. doi: 10.1083/jcb.201108077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dey JH, Bianchi F, Voshol J, Bonenfant D, Oakeley EJ, Hynes NE. Targeting fibroblast growth factor receptors blocks PI3K/AKT signaling, induces apoptosis, and impairs mammary tumor outgrowth and metastasis. Cancer Res. 2010;70:4151–62. doi: 10.1158/0008-5472.CAN-09-4479. [DOI] [PubMed] [Google Scholar]
- 8.Hynes NE, Dey JH. Potential for targeting the fibroblast growth factor receptors in breast cancer. Cancer Res. 2010;70:5199–202. doi: 10.1158/0008-5472.CAN-10-0918. [DOI] [PubMed] [Google Scholar]
- 9.Issa A, Gill JW, Heideman MR, et al. Combinatorial targeting of FGF and ErbB receptors blocks growth and metastatic spread of breast cancer models. Breast Cancer Res. 2013;15:R8. doi: 10.1186/bcr3379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tarkkonen KM, Nilsson EM, Kahkonen TE, et al. Differential roles of fibroblast growth factor receptors (FGFR) 1, 2 and 3 in the regulation of S115 breast cancer cell growth. PLoS One. 2012;7:e49970. doi: 10.1371/journal.pone.0049970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Welm BE, Freeman KW, Chen M, Contreras A, Spencer DM, Rosen JM. Inducible dimerization of FGFR1: development of a mouse model to analyze progressive transformation of the mammary gland. J Cell Biol. 2002;157:703–14. doi: 10.1083/jcb.200107119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bade LK, Goldberg JE, Dehut HA, Hall MK, Schwertfeger KL. Mammary tumorigenesis induced by fibroblast growth factor receptor 1 requires activation of the epidermal growth factor receptor. J Cell Sci. 2011;124:3106–17. doi: 10.1242/jcs.082651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pond AC, Herschkowitz JI, Schwertfeger KL, et al. Fibroblast growth factor receptor signaling dramatically accelerates tumorigenesis and enhances oncoprotein translation in the mouse mammary tumor virus-Wnt-1 mouse model of breast cancer. Cancer Res. 2010;70:4868–79. doi: 10.1158/0008-5472.CAN-09-4404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Reed JR, Leon RP, Hall MK, Schwertfeger KL. Interleukin-1beta and fibroblast growth factor receptor 1 cooperate to induce cyclooxygenase-2 during early mammary tumourigenesis. Breast Cancer Res. 2009;11:R21. doi: 10.1186/bcr2246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Reed JR, Stone MD, Beadnell TC, Ryu Y, Griffin TJ, Schwertfeger KL. Fibroblast growth factor receptor 1 activation in mammary tumor cells promotes macrophage recruitment in a CX3CL1-dependent manner. PLoS One. 2012;7:e45877. doi: 10.1371/journal.pone.0045877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schwertfeger KL, Xian W, Kaplan AM, Burnett SH, Cohen DA, Rosen JM. A critical role for the inflammatory response in a mouse model of preneoplastic progression. Cancer Res. 2006;66:5676–85. doi: 10.1158/0008-5472.CAN-05-3781. [DOI] [PubMed] [Google Scholar]
- 17.Auvinen P, Tammi R, Parkkinen J, et al. Hyaluronan in peritumoral stroma and malignant cells associates with breast cancer spreading and predicts survival. Am J Pathol. 2000;156:529–36. doi: 10.1016/S0002-9440(10)64757-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gotte M, Yip GW. Heparanase, hyaluronan, and CD44 in cancers: a breast carcinoma perspective. Cancer Res. 2006;66:10233–7. doi: 10.1158/0008-5472.CAN-06-1464. [DOI] [PubMed] [Google Scholar]
- 19.Urakawa H, Nishida Y, Wasa J, et al. Inhibition of hyaluronan synthesis in breast cancer cells by 4-methylumbelliferone suppresses tumorigenicity in vitro and metastatic lesions of bone in vivo. Int J Cancer. 2012;130:454–66. doi: 10.1002/ijc.26014. [DOI] [PubMed] [Google Scholar]
- 20.Solis MA, Chen YH, Wong TY, Bittencourt VZ, Lin YC, Huang LL. Hyaluronan regulates cell behavior: a potential niche matrix for stem cells. Biochem Res Int. 2012;2012:346972. doi: 10.1155/2012/346972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Toole BP. Hyaluronan: from extracellular glue to pericellular cue. Nat Rev Cancer. 2004;4:528–39. doi: 10.1038/nrc1391. [DOI] [PubMed] [Google Scholar]
- 22.Itano N. Simple primary structure, complex turnover regulation and multiple roles of hyaluronan. J Biochem. 2008;144:131–7. doi: 10.1093/jb/mvn046. [DOI] [PubMed] [Google Scholar]
- 23.Wagner KU, Schmidt JW. The two faces of Janus kinases and their respective STATs in mammary gland development and cancer. J Carcinog. 2011;10:32. doi: 10.4103/1477-3163.90677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Diaz N, Minton S, Cox C, et al. Activation of stat3 in primary tumors from high-risk breast cancer patients is associated with elevated levels of activated SRC and survivin expression. Clin Cancer Res. 2006;12:20–8. doi: 10.1158/1078-0432.CCR-04-1749. [DOI] [PubMed] [Google Scholar]
- 25.Hsieh FC, Cheng G, Lin J. Evaluation of potential Stat3-regulated genes in human breast cancer. Biochem Biophys Res Commun. 2005;335:292–9. doi: 10.1016/j.bbrc.2005.07.075. [DOI] [PubMed] [Google Scholar]
- 26.Lieblein JC, Ball S, Hutzen B, et al. STAT3 can be activated through paracrine signaling in breast epithelial cells. BMC Cancer. 2008;8:302. doi: 10.1186/1471-2407-8-302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Leslie K, Lang C, Devgan G, et al. Cyclin D1 is transcriptionally regulated by and required for transformation by activated signal transducer and activator of transcription 3. Cancer Res. 2006;66:2544–52. doi: 10.1158/0008-5472.CAN-05-2203. [DOI] [PubMed] [Google Scholar]
- 28.Xian W, Schwertfeger KL, Vargo-Gogola T, Rosen JM. Pleiotropic effects of FGFR1 on cell proliferation, survival, and migration in a 3D mammary epithelial cell model. J Cell Biol. 2005;171:663–73. doi: 10.1083/jcb.200505098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bohrer LR, Schwertfeger KL. Macrophages promote fibroblast growth factor receptor-driven tumor cell migration and invasion in a CXCR2-dependent manner. Mol Cancer Res. 2012;10:1294–305. doi: 10.1158/1541-7786.MCR-12-0275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Xian W, Schwertfeger KL, Rosen JM. Distinct roles of fibroblast growth factor receptor 1 and 2 in regulating cell survival and epithelial-mesenchymal transition. Mol Endocrinol. 2007;21:987–1000. doi: 10.1210/me.2006-0518. [DOI] [PubMed] [Google Scholar]
- 31.Nakamura T, Funahashi M, Takagaki K, et al. Effect of 4-methylumbelliferone on cell-free synthesis of hyaluronic acid. Biochem Mol Biol Int. 1997;43:263–8. doi: 10.1080/15216549700204041. [DOI] [PubMed] [Google Scholar]
- 32.Kakizaki I, Kojima K, Takagaki K, et al. A novel mechanism for the inhibition of hyaluronan biosynthesis by 4-methylumbelliferone. J Biol Chem. 2004;279:33281–9. doi: 10.1074/jbc.M405918200. [DOI] [PubMed] [Google Scholar]
- 33.Kultti A, Pasonen-Seppanen S, Jauhiainen M, et al. 4-Methylumbelliferone inhibits hyaluronan synthesis by depletion of cellular UDP-glucuronic acid and downregulation of hyaluronan synthase 2 and 3. Exp Cell Res. 2009;315:1914–23. doi: 10.1016/j.yexcr.2009.03.002. [DOI] [PubMed] [Google Scholar]
- 34.Wang A, de la Motte C, Lauer M, Hascall V. Hyaluronan matrices in pathobiological processes. FEBS J. 2011;278:1412–8. doi: 10.1111/j.1742-4658.2011.08069.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Saavalainen K, Pasonen-Seppanen S, Dunlop TW, Tammi R, Tammi MI, Carlberg C. The human hyaluronan synthase 2 gene is a primary retinoic acid and epidermal growth factor responding gene. J Biol Chem. 2005;280:14636–44. doi: 10.1074/jbc.M500206200. [DOI] [PubMed] [Google Scholar]
- 36.Dudka AA, Sweet SM, Heath JK. Signal transducers and activators of transcription-3 binding to the fibroblast growth factor receptor is activated by receptor amplification. Cancer Res. 2010;70:3391–401. doi: 10.1158/0008-5472.CAN-09-3033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yu H, Pardoll D, Jove R. STATs in cancer inflammation and immunity: a leading role for STAT3. Nat Rev Cancer. 2009;9:798–809. doi: 10.1038/nrc2734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Berishaj M, Gao SP, Ahmed S, et al. Stat3 is tyrosine-phosphorylated through the interleukin-6/glycoprotein 130/Janus kinase pathway in breast cancer. Breast Cancer Res. 2007;9:R32. doi: 10.1186/bcr1680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liu JF, Crepin M, Liu JM, Barritault D, Ledoux D. FGF-2 and TPA induce matrix metalloproteinase-9 secretion in MCF-7 cells through PKC activation of the Ras/ERK pathway. Biochem Biophys Res Commun. 2002;293:1174–82. doi: 10.1016/S0006-291X(02)00350-9. [DOI] [PubMed] [Google Scholar]
- 40.Sharpe R, Pearson A, Herrera-Abreu MT, et al. FGFR signaling promotes the growth of triple-negative and basal-like breast cancer cell lines both in vitro and in vivo. Clin Cancer Res. 2011;17:5275–86. doi: 10.1158/1078-0432.CCR-10-2727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Turner N, Grose R. Fibroblast growth factor signalling: from development to cancer. Nat Rev Cancer. 2010;10:116–29. doi: 10.1038/nrc2780. [DOI] [PubMed] [Google Scholar]
- 42.Dolled-Filhart M, Camp RL, Kowalski DP, Smith BL, Rimm DL. Tissue microarray analysis of signal transducers and activators of transcription 3 (Stat3) and phospho-Stat3 (Tyr705) in node-negative breast cancer shows nuclear localization is associated with a better prognosis. Clin Cancer Res. 2003;9:594–600. [PubMed] [Google Scholar]
- 43.Schust J, Sperl B, Hollis A, Mayer TU, Berg T. Stattic: a small-molecule inhibitor of STAT3 activation and dimerization. Chem Biol. 2006;13:1235–42. doi: 10.1016/j.chembiol.2006.09.018. [DOI] [PubMed] [Google Scholar]
- 44.Dave B, Landis MD, Tweardy DJ, et al. Selective small molecule Stat3 inhibitor reduces breast cancer tumor-initiating cells and improves recurrence free survival in a human-xenograft model. PLoS One. 2012;7:e30207. doi: 10.1371/journal.pone.0030207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Li Y, Li L, Brown TJ, Heldin P. Silencing of hyaluronan synthase 2 suppresses the malignant phenotype of invasive breast cancer cells. Int J Cancer. 2007;120:2557–67. doi: 10.1002/ijc.22550. [DOI] [PubMed] [Google Scholar]
- 46.Debnath J, Brugge JS. Modelling glandular epithelial cancers in three-dimensional cultures. Nat Rev Cancer. 2005;5:675–88. doi: 10.1038/nrc1695. [DOI] [PubMed] [Google Scholar]
- 47.Veiseh M, Turley EA. Hyaluronan metabolism in remodeling extracellular matrix: probes for imaging and therapy of breast cancer. Integr Biol (Camb) 2011;3:304–15. doi: 10.1039/c0ib00096e. [DOI] [PubMed] [Google Scholar]
- 48.Sironen RK, Tammi M, Tammi R, Auvinen PK, Anttila M, Kosma VM. Hyaluronan in human malignancies. Exp Cell Res. 2011;317:383–91. doi: 10.1016/j.yexcr.2010.11.017. [DOI] [PubMed] [Google Scholar]
- 49.Auvinen P, Tammi R, Kosma VM, et al. Increased hyaluronan content and stromal cell CD44 associate with HER2 positivity and poor prognosis in human breast cancer. Int J Cancer. 2013;132:531–9. doi: 10.1002/ijc.27707. [DOI] [PubMed] [Google Scholar]
- 50.Karihtala P, Soini Y, Auvinen P, Tammi R, Tammi M, Kosma VM. Hyaluronan in breast cancer: correlations with nitric oxide synthases and tyrosine nitrosylation. J Histochem Cytochem. 2007;55:1191–8. doi: 10.1369/jhc.7A7270.2007. [DOI] [PubMed] [Google Scholar]
- 51.Heldin P, Karousou E, Bernert B, Porsch H, Nishitsuka K, Skandalis SS. Importance of hyaluronan-CD44 interactions in inflammation and tumorigenesis. Connect Tissue Res. 2008;49:215–8. doi: 10.1080/03008200802143323. [DOI] [PubMed] [Google Scholar]
- 52.Monzon ME, Fregien N, Schmid N, et al. Reactive oxygen species and hyaluronidase 2 regulate airway epithelial hyaluronan fragmentation. J Biol Chem. 2010;285:26126–34. doi: 10.1074/jbc.M110.135194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Eberlein M, Scheibner KA, Black KE, et al. Anti-oxidant inhibition of hyaluronan fragment-induced inflammatory gene expression. J Inflamm (Lond) 2008;5:20. doi: 10.1186/1476-9255-5-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tian X, Azpurua J, Hine C, et al. High-molecular-mass hyaluronan mediates the cancer resistance of the naked mole rat. Nature. 2013;499:346–9. doi: 10.1038/nature12234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tolg C, Hamilton SR, Zalinska E, et al. A RHAMM mimetic peptide blocks hyaluronan signaling and reduces inflammation and fibrogenesis in excisional skin wounds. Am J Pathol. 2012;181:1250–70. doi: 10.1016/j.ajpath.2012.06.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sakaguchi M, Oka M, Iwasaki T, Fukami Y, Nishigori C. Role and regulation of STAT3 phosphorylation at Ser727 in melanocytes and melanoma cells. J Invest Dermatol. 2012;132:1877–85. doi: 10.1038/jid.2012.45. [DOI] [PubMed] [Google Scholar]
- 57.Toole BP. Hyaluronan-CD44 Interactions in Cancer: Paradoxes and Possibilities. Clin Cancer Res. 2009;15:7462–8. doi: 10.1158/1078-0432.CCR-09-0479. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.