Abstract
Pharmacogenetics is being used to develop personalized therapies specific to individuals from different ethnic or racial groups. Pharmacogenetic studies to date have been primarily performed in trial cohorts consisting of non-Hispanic whites of European descent. A “bottleneck” or collapse of genetic diversity associated with the first human colonization of Europe during the Upper Paleolithic period, followed by the recent mixing of African, European, and Native American ancestries has resulted in different ethnic groups with varying degrees of genetic diversity. Differences in genetic ancestry may introduce genetic variation which has the potential to alter the therapeutic efficacy of commonly used asthma therapies, for example β2-adrenergic receptor agonists (beta agonists). Pharmacogenetic studies of admixed ethnic groups have been limited to small candidate gene association studies of which the best example is the gene coding for the receptor target of beta agonist therapy, ADRB2. Large consortium-based sequencing studies are using next-generation whole-genome sequencing to provide a diverse genome map of different admixed populations which can be used for future pharmacogenetic studies. These studies will include candidate gene studies, genome-wide association studies, and whole-genome admixture-based approaches which account for ancestral genetic structure, complex haplotypes, gene-gene interactions, and rare variants to detect and replicate novel pharmacogenetic loci.
Keywords: asthma, genes, pharmacogenetics, response heterogeneity, single nucleotide polymorphism, admixture mapping, ethnic group
Introduction
It is well known that frequencies and even severity of disease may differ between races. Two simple examples include sickle cell anemia caused by a mutation which arose in Africa while the mutations that cause Cystic Fibrosis primarily arose in individuals of European white descent. A key question is whether an individual’s ethnic or ancestral background determine individual response to a medication? There is robust evidence from clinical trials for different medical conditions which shows that individuals from different ethnic groups experience variable responses to specific therapeutic agents. For instance, there is a class of anti-hypertensive drugs, β1-adrenergic receptor blockers or beta blockers, which may be less effective in a subgroup of African Americans for the management of congestive heart failure.1,2 This reduced efficacy might be related to gene variants in two genes related to the G-protein coupled pathway of the β1-adrenergic receptor, the β1-adrenergic receptor (ADRB1) and G-protein receptor kinase 5 (GRK5) genes. These gene variants are overrepresented in African ancestral populations and result in changes in the amino-acid code: a code change from arginine to a glycine in amino acid position 389 on ADRB1 (Arg389Gly) and Gln41Leu on GRK5.3 The Arg389 allele in ADRB1 and the Leu41 allele in GRK5 have both been associated with a reduction of mortality in human subjects with heart failure and coronary ischemia treated with a beta blocker in different pharmacogenetic studies.3,4 These examples of variable drug responses in cardiovascular disease illustrate the challenge of developing personalized approaches not only through recognizing ethnic or racial subgroups which show variable therapeutic drug responses but also identifying them through pharmacogenetic biomarkers related to a common genetic ancestral origin.
Pharmacogenetics is the study of the role of genetic variability in determining inter-individual (between individual) variability in responses to a pharmacological therapy. Pharmacogenetics represents a gene-by-environment interaction whereby variation in a gene interacts with an exposure to a drug (the “environment”) to alter a measurable phenotype related to drug efficacy or toxicity. The ultimate goal of pharmacogenetics research is the development of personalized medicine through genetic markers (individual genetic profiles) which would accurately predict which individuals with a particular condition would respond to a specific medical therapy, not respond to a therapy, or experience adverse effects.
The Rationale for Pharmacogenetics in the Management of Asthma in Different Ethnic Groups
Asthma is a complex, chronic inflammatory disease of the airways which results from the interaction of multiple genetic and environmental factors. Asthma is a heterogeneous disease with variability in its phenotype expression and variability in inter-individual therapeutic responses to different pharmacologic therapies.5–7 Variability in therapeutic responses may result from the interaction of multiple genes from different biologic pathways and even shared environmental influences.8–10 People with shared ancestry and physical traits are most commonly categorized by ethnic or racial groups; however, this designation also implies common genetic ancestral backgrounds which may potentially impact therapeutic responses.11
Pharmacogenetic studies to date have been primarily performed in trial cohorts consisting of non-Hispanic asthma subjects of European descent; however, a small number of studies have also evaluated study cohorts of recently admixed ethnic groups such as African American or Hispanics. There are inherent challenges related to the genetic study of recently admixed ethnic or racial subgroups from varying ancestries. These challenges have been primarily related to sample size, complex ancestral population structures, and genotype data primarily based on a genetic background from populations of European descent. In this review article, we will summarize the genetic and epidemiologic basis for the variable genetic backgrounds observed between different, recently admixed ethnic groups, outline the rationale for pharmacogenetics research in these ethnic groups, discuss the contribution of pharmacogenetic studies in identifying ethnic group-specific genetic variants for therapeutic responses in asthma, and outline how admixture-based analytical methods and next-generation sequencing will contribute to future pharmacogenetic studies.
Why Genetic Diversity Varies Between Different Ancestries
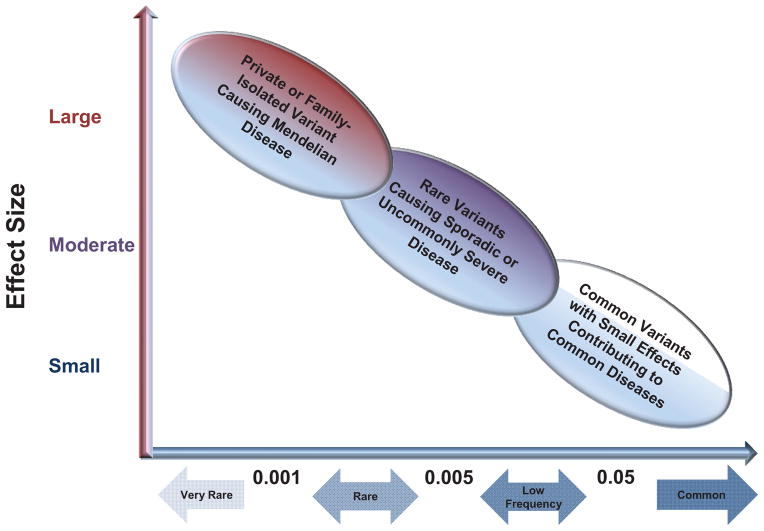
Information about the demographic history of our species can be evaluated through the distribution of gene variants located throughout the human genome. Initially, most publicly available databases contained mostly common gene variants such as single nucleotide polymorphisms (SNP’s) based on the Human Genome Project.12,13 The early ascertainment strategies of the Human Genome Project favored the identification of common variants which were of particular interest based on the “common disease, common allele hypothesis” which states that multiple common gene variation with mild to modest effects influence susceptibility to complex, common phenotypes (figure 1).14–18
Figure 1. Common and Rare Genetic Variants in Human Disease.
Based on the “common disease-common allele” hypothesis, multiple common genetic polymorphisms (alleles or variants) with small to modest effect sizes contribute additively to a common disease. Genome-wide association studies (GWAS) have detected multiple common variants associated with risk for common diseases such as hypertension or asthma. The “common disease-rare allele” hypothesis states that rare genetic variants with a large effect size contribute to risk for common diseases. Rare genetic variants cannot be detected with classic GWAS and have been evaluated with family-based genetic studies, admixture mapping, and DNA sequencing. Adapted from Tsuji S, et al. Hum Mol Genet 2010;19(R1):R65–70.14
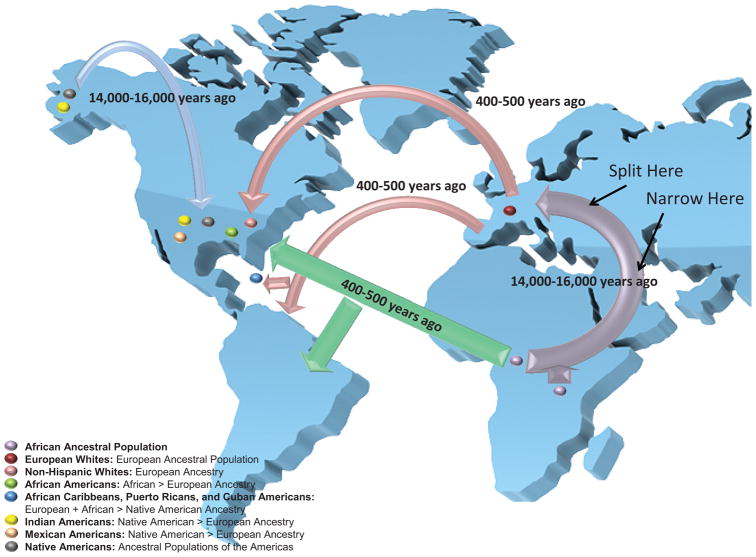
SNP’s are the most common form of gene variation in the human genome and can act as heritable landmarks for genetic diversity and population structure. Based on the diversity of these common variants, different recombination models have estimated that the human population has experienced a “bottleneck” history.18,19 This “bottleneck” history likely began nearly 40,000 years or 1,600 generations ago when the first modern humans migrated from sub-Saharan Africa to colonize Europe.18 This initial colonization of Europe by modern humans was accompanied by a loss or “collapse” of genetic diversity followed by modest growth of the human population during the Upper Paleolithic period (figure 2).19 An important consequence of this “bottleneck” event is that populations from a primarily European ancestry have gene variants which remain co-inherited through linkage equilibrium (often due to being physically close together on a chromosome) as a result of fewer recombination events since the last “collapse.” In contrast, populations with a primarily ancient African ancestry have genomic regions with a greater diversity of gene variation resulting from a greater number of recombination events and shorter regions of linkage disequilibrium.18
Figure 2. Ancestries of Recently Admixed Ethnic Groups in the United States.
The first human colonization of Europe during the Upper Paleolithic period (purple arrow) was accompanied by a “bottleneck” or collapse of genetic diversity in the resulting European White ancestral population. Recent mixing between more genetically diverse, ancient African ancestral populations with European Whites, and Native Americans (blue arrow represents the first human colonization of the Americas) during the European colonization of the Americas (red arrows) and the African slave trade (green arrows) resulted in different, recently admixed ethnic groups with varying degrees of genetic diversity. The flow of genetic diversity is represented by the thickness of the arrows: thicker arrows reflect greater genetic diversity (i.e. resulting from a greater number of recombination events, shorter genomic regions of linkage disequilibrium, and a greater frequency of rare variants).
Over the past 400 generations, the human population has experienced a marked increase in growth by at least three orders of magnitude.20 This accelerated growth has resulted in departures from population genetic equilibrium and a recent growth of rare variation due to recent mutations.21 The phenomenon of increased rare variant load has only been recently appreciated with the advent of next-generation sequencing technologies which have allowed for whole-genome or whole-exome sequencing. Large-scale whole-genome sequencing projects have already provided insight into the load of rare variants in different populations through multi-center studies which include the 1000 Genomes Project Consortium; the National Institutes of Health (NIH) National Heart, Lung, and Blood Institute (NHLBI) GO Exome Sequencing Project (GO ESP); and the NIH NHLBI Consortium on Asthma among African-ancestry Populations in the Americas (CAAPA) Consortium.22,23 For instance, whole-genome sequencing data from 1,097 individual genomes from 14 different populations from the 1,000 Genomes Project Consortium recently identified an enrichment of rare variants defined as an allele frequency less than 0.005 in subjects consistent with the recent explosive expansion of the human population.21,22
The genetic diversity of different ancestral populations has been shown to have implications for the frequency of rare genetic variants. For instance, the 1000 Genomes Project investigators reported that each individual had at least 240 rare coding variants (allele frequency less than 0.05) and hundreds of non-coding variants at conserved motifs such as transcription factor binding sites, thus, the total number of individual rare variants was greater than that of common variants in the total 1,000 Genomes Project study population.22 In addition, these investigators demonstrated that populations from an African ancestry had three times as many rare variants compared to those of European and Asian origin reflecting the ancestral bottlenecks of the descendant, non-African populations (figure 2).18,22 The availability of high-throughput sequencing, the recognition that rare genetic variants are frequent in the human population, and the potential for strong rare variant genetic effects on common diseases has resulted in a recent increase in the study of rare variants in common, complex disease based on the “common disease-rare allele hypothesis.” (figure 1)14,24–27
The analysis of a rapidly expanding catalogue of rare variants remains a formidable challenge because of the low statistical power of conventional association studies and the potential for misclassification of the functional impact of individual variants. Analytical methods for rare variant analysis have evolved from simpler methods which “collapsed” rare variants at the gene-level to more complex methods which assign directionality of effect to individual rare variants and take advantage of predictive functional algorithms to optimize power while minimizing the potential for variant function misclassification.28–30 The role of variable genetic diversity and rare variants will be important to consider in future pharmacogenetic studies of subjects of African ancestry since rare variants appear to be more frequent in these ethnic or racial groups and, thus, may account for inter-ethnic differences in drug responses especially for rare, adverse events.10
Genetic Diversity from Different Ancestries is a Critical Consideration for Genetic Studies in Admixed Populations
Individuals from recently admixed ethnic groups have varying proportions of different ancestries represented within their genomes. The European colonization of the Americas and the African slave trade resulted in mixed genetic ancestries which constitute the complex population structures identified in the genomes of African Americans, African Caribbeans, Puerto Ricans, and Mexican Americans (figure 1). The variable demographic histories of recently admixed populations with a common, ancient African ancestral component has resulted in a population structure with greater genetic diversity compared to non-Hispanic subjects of European White descent. This increased genetic diversity has not been captured by most whole-genome genotyping platforms which were designed for populations from a primarily European ancestry. Thus, efforts are currently underway to develop more comprehensive whole-genome genotyping platforms to capture the genetic diversity of recently admixed populations, for example, through the CAAPA consortium.23
In general, genetic clusters based on genome-wide genotype data have closely reflected self-reported ethnicity; however, subsequent genetic studies of recently admixed populations have demonstrated marked variability in genetic ancestry particularly even between different Hispanic subgroups.31–33 For instance, African Americans, have estimated mean global African ancestry of 80 percent (20 percent European) while Hispanic ethnic subgroups such as Puerto Ricans and Mexican Americans have varying combinations of three different ancestries: African, European, and Amerindian or Native American (figure 2).32,33 These percentages reflect the subgroup overall, but it is extremely important to remember that an individual patient may have a very different level of admixture.
Besides sharing common genetic ancestries, individuals from different ethnic groups also share common languages, cultural behaviors, religious affiliations, and often environmental exposures. These common characteristics may result in gene-by-environment interactions which are implicit to individual ethnic groups and further confound genetic studies by making it difficult to dissect the effect of environment in determining differential drug responses between individuals from these various ethnic subpopulations.
Association Between Genetic Ancestry and Asthma-Related Phenotypes: A Rationale for Pharmacogenetic Studies in Admixed Populations
Some admixed populations show measurable differences in asthma-related phenotypes not attributed to environmental factors. For instance, asthma-related morbidity and mortality is higher among Puerto Ricans and African Americans compared to non-Hispanic Whites and Mexican Americans.34–36 These inter-ethnic differences in disease severity may result from healthcare disparities but also may relate to genetic variants inherited from a specific ancestry associated with disease severity or therapeutic response to different therapies. The impact of global genetic ancestry on asthma and related phenotypes has been evaluated using SNP’s with variable allele frequencies between different ancestral populations. These genetic markers are informative of genetic ancestry and, thus, are known as ancestry informative markers or AIM’s which provide estimates of individual genetic ancestry. The majority of previous admixture-based genetic studies have been related to asthma susceptibility and severity; yet, there are potential applications for determining the role of genetic ancestry on drug response.
Large cohorts of well-characterized asthma subjects have consistently demonstrated the importance of lung function as a fundamental determinant of asthma disease severity.37,38 Admixture-based genetic studies in African Americans from the general population have suggested that genetic variation related to African ancestry might impact both asthma susceptibility and severity based on lung function. The first of these studies was performed by Kumar and colleagues which demonstrated that global estimates of percentage African ancestry was inversely correlated with with baseline lung function in three independent African American populations (figures 3A and 3B).33
Figure 3. 3A and 3B: Relationship of Global African Ancestry with Lung Function in African Americans.
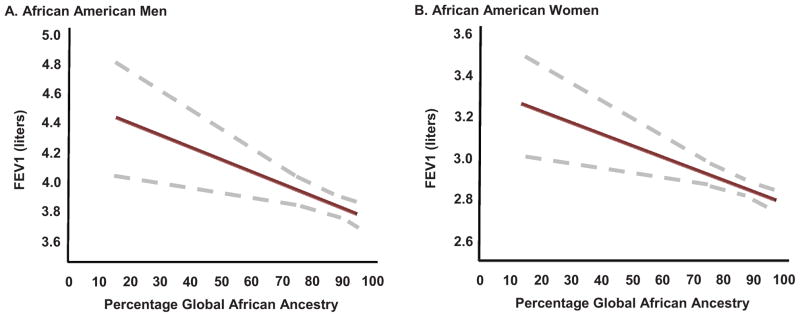
Kumar and colleagues demonstrated an inverse relationship between percentage of global African ancestry and baseline forced expiratory volume in 1 second (FEV1 measured in liters) in self-identified African American men and women shown in Panels A and B, respectively. Global African ancestry was estimated using ancestry informative genetic markers. Reproduced from Kumar R, et al. NEJM 2010;363(4):321–330.33
The observed inverse association between African ancestry and lung function was consistent with the trend in predicted baseline lung function described by Hankinson and coworkers where self-designated African race predicted a lower lung function measure compared to self-designated white individuals of similar age, height, and sex.39 In addition, calculations of baseline lung function based on estimated ancestry using genetic methods revealed that classic predictive equations misclassified asthma severity based on percentage predicted FEV1 in a subset of African American asthma subjects. The potential for misclassification of disease severity in this African American cohort is because each individual asthmatic showed variable proportions of African versus European ancestries (i.e. a continuous variable of percentage ancestry) not captured by classic predictive methods based on self-reported race (i.e. a dichotomous designation of self-reported race).33 In 392 asthma subjects, African ancestry was associated with asthma exacerbations suggesting that there is genetic variation more common in Africans leading to an increased risk for exacerbations; important findings requiring further replication and identification of genetic risk variants.40
Admixture-based genetic studies in the Genetics of Asthma in Latino Americans (GALA) cohort have also evaluated the role for genetic ancestry in determining asthma susceptibility and severity in Puerto Ricans and Mexican Americans.32 Despite being considered from the same Hispanic or Latino ethnic or racial group, Puerto Ricans and Mexican Americans are ethnic subgroups that show remarkably differences in asthma disease severity which might be related to genetic variation from their uniquely different ancestral origins. For instance, Puerto Ricans experience among the highest asthma-related morbidity and mortality when compared to all other ethnic groups in the United States, while Mexican Americans experience the lowest.41 Admixture-based studies from the GALA study group have demonstrated that Puerto Ricans have a higher degree of African and European Ancestry compared to Mexican Americans who have a higher degree of Amerindian or Native American ancestry (figure 2).42,32
An inverse association between African ancestry and baseline lung function was observed in 943 Puerto Rican asthma cases and controls from the GALA cohort, a similar relationship to what had been previously observed in African Americans.43 An inverse relationship between European ancestry and baseline lung function measures was also observed in 181 Mexican American asthmatics from GALA.42 In contrast, Native American ancestry potentially exhibits a beneficial effect on lung function in Mexican Americans and has been associated with a reduced risk of rapid lung function decline and chronic obstructive pulmonary disease in a combined cohort of 305 Hispanics and 1,404 non-Hispanic Whites from New Mexico.44
Differences in genetic ancestry might also be important in the therapeutic efficacy of commonly used asthma therapies. The most relevant is related to a commonly used class of pharmacologic agents: β2-adrenergic receptor agonists (i.e. beta agonists) which are divided into short-acting beta agonists (SABA: fenoterol, isoproterenol, pirbuterol, levalbuterol, and albuterol) used for as-needed, rescue therapy and long-acting beta agonists (LABA: salmeterol and formoterol) used in combination with an inhaled corticosteroid (ICS) for chronic management. Recent surveillance studies and meta-analyses derived from these studies have suggested that LABA increase the risk for life-threatening asthma exacerbations and asthma-related death when LABA therapy was administered without concomitant ICS therapy.45–47 The most cited of these studies is the Salmeterol Multicenter Asthma Research Trial (SMART) in which 26,355 subjects, including 4,685 African Americans, were randomized to either salmeterol or placebo in addition to “usual therapy.”
The SMART trial was terminated early due to an increase in asthma or respiratory-related life-threatening exacerbations and death in the subgroup of African Americans treated with salmeterol. This study was limited by its cursory surveillance design which did not monitor compliance or concomitant medication use (such as regular SABA use) and study participants were not routinely treated with an ICS. In addition, the African American subgroup consisted of asthmatics who were less frequently treated with an ICS and at least twice as likely to have experienced an asthma-related hospitalization or life-threatening exacerbations prior to trial enrollment compared to their non-Hispanic White counterparts.46 Before and after the SMART trial, randomized controlled clinical trials and large meta-analyses have consistently demonstrated that the combination of a LABA and an ICS is safe and efficacious for the management of asthma.48–51 This LABA safety controversy has resulted in two advisory panel meetings by the United States Food and Drug Administration (FDA), subsequent public health advisories, and a boxed warning for all LABA-containing inhalers.52 This controversy is currently being evaluated in an international study of over 40,000 asthmatics mandated by the FDA.52,53
Data from different clinical trials suggest that African Americans respond differently to combination therapies containing a LABA compared to non-Hispanic Whites.8,54 The NIH NHLBI Childhood Asthma Research and Education (CARE) Network Best Add-on Therapy Giving Effective Response (BADGER) trial was a triple cross-over clinical trial consisting of a multi-ethnic cohort of 182 children with uncontrolled asthma who were randomized to a “step-up” from low-dose ICS therapy to different combination therapies. These children were assessed for preferential therapeutic response based on the composite endpoint of lung function decline, exacerbation, and a reduction of days with well-controlled symptoms. In the BADGER trial, African American children did not show a preferential response to a “step-up” from low-dose ICS therapy to a higher-dose ICS compared to a “step-up” to LABA and ICS combination therapy, thus, the addition of a LABA to ICS therapy was no more efficacious than increasing the ICS dose. In contrast, Hispanics and non-Hispanic Whites showed a preferential response to a “step-up” from ICS therapy to LABA and ICS combination therapy compared to the other “step-up” combination therapies.55
In addition, a combined analysis of 795 non-Hispanic White and 233 African American adult asthma subjects from 10 NIH NHLBI Asthma Clinical Research Network Trials showed that African Americans were more likely to experience treatment failures during randomized treatment with LABA therapy compared to non-Hispanic Whites.54 African ancestry may also have a role in differential beta agonist response in other admixed ethnic groups. For instance, Puerto Ricans from the GALA cohort were less likely to show a significant increase in lung function in response to acute bronchodilation with a short-acting beta agonist.56 These findings suggest that African ancestry may introduce genetic variation associated with a greater likelihood for alteration in therapeutic responses to inhaled beta agonists and, thus, provide the rationale for pharmacogenetic studies.
Pharmacogenetic Studies: A Focus on Beta Agonists
Pharmacogenetic studies in multi-ethnic populations have focused on the gene encoding for the β2-adrenergic receptor, ADRB2. ADRB2 is located on chromosome 5q31 and encodes for a seven-transmembrane receptor which activates a Gs-protein coupled receptor pathway consisting of adenylyl cyclase to mediate smooth muscle relaxation.57,58 A common coding SNP, Gly16Arg, has been the most studied polymorphism; however, nearly 49 polymorphisms have also been identified through DNA sequencing in multi-ethnic asthma populations which include polynucleotide insertion-deletions and rare functional variants. The minor allele of this SNP has variable allele frequencies between different ancestral populations but is more frequent in African and Asian populations (table 1).57,58 In vitro, beta agonist stimulation results in enhanced downregulation of receptors expressing the Gly16 variant compared to Arg16.59,60 Additional ADRB2 polymorphisms have been identified in most ethnic groups include additional coding variants (Cys19(BUP)Arg, Gln27Glu, Thr164Ile) with varying allele frequencies resulting in complex haplotypes, some of which are specific to certain ethnic groups.57,58,61
Table 1.
Variant Allele Frequencies of Asthma Pharmacogenetic Loci in Different Ancestral Populations and United States Ethnic Groups.
| Drug Classes | Gene | Previously Associated Pharmacogenetic Locia | Alleles Major/Minor |
Minor Allele Frequency by Racial Groupb | |||||
|---|---|---|---|---|---|---|---|---|---|
| CEU | YRI | ASW | MEX | CHB | JPT | ||||
| Leukotriene Receptor Modifiers: 5-Lipooxygenase Inhibitors (Zileuton) | ALOX5 | rs2029253101 | A/G | 0.33 | 0.23 | 0.29 | 0.52 | 0.56 | 0.56 |
| rs2115819101 | A/G | 0.37 | 0.21 | 0.28 | 0.53 | 0.73 | 0.82 | ||
|
|
|||||||||
| Cysteinyl leukotriene antagonists (Montelukast) | ALOX5 | rs211581999,100 | A/G | 0.37 | 0.21 | 0.28 | 0.53 | 0.73 | 0.82 |
| LTC4S | rs73001299 | A/C | 0.32 | 0.007 | 0.06 | 0.22 | 0.14 | 0.18 | |
| LTA4H | rs26684599 | T/G | 0.17 | 0.007 | 0.06 | 0.22 | 0.44 | 0.36 | |
|
|
|||||||||
| Inhaled Glucocorticoids: (Fluticasone, Budesonide, Flunisolide, Triamcinolone) | STIP1 | rs223664798 | C/T | 0.43 | 0.76 | 0.71 | 0.6 | 0.43 | 0.35 |
| rs101121998 | G/A | 0.2 | 0.08 | NA | NA | 0.08 | 0.14 | ||
| GLCCI1 | rs3797276 | C/T | 0.45 | 0.14 | 0.29 | 0.46 | 0.43 | 0.38 | |
| T Gene | rs312741297 | T/C | 0.35 | 0.36 | 0.3 | 0.15 | 0.13 | 0.11 | |
| rs645604297 | C/A | 0.35 | 0.36 | 0.3 | 0.15 | 0.13 | 0.11 | ||
| rs309926697 | C/T | 0.36 | 0.15 | 0.16 | 0.3 | 0.26 | 0.28 | ||
| rs230508997 | T/C | 0.41 | 0.79 | 0.63 | 0.38 | 0.64 | 0.7 | ||
|
|
|||||||||
| Inhaled β2-Adrenergic Receptor Agonists: Short-Acting Beta Agonists (Albuterol) | ADCY9 | rs2230739 (Met772Ile)96 | T/C | 0.36 | 0.12 | 0.12 | 0.22 | 0.4 | 0.36 |
| ADRB2 | rs1042713 (Gly16Arg)62–64,93–95 | G/A | 0.36 | 0.53 | 0.57 | 0.47 | 0.58 | 0.44 | |
| ARG1 | rs2781659102,103 | A/G | 0.34 | 0.73 | NA | NA | 0.28 | 0.4 | |
| rs278166790 | C/T | 0.31 | 0.79 | 0.72 | 0.36 | 0.32 | 0.37 | ||
| ARG2 | rs714031090 | T/G | 0.11 | 0.24 | 0.21 | 0.12 | 0.25 | 0.26 | |
| rs1048380190 | C/A | 0.16 | 0.42 | 0.36 | 0.29 | 0.4 | 0.42 | ||
| SPATS2L | rs29513777 | C/T | 0.42 | 0.59 | 0.41 | 0.31 | 0.19 | 0.2 | |
|
|
|||||||||
| Long-Acting Beta Agonists (Salmeterol and Formoterol) | ADRB2 | rs1042713 (Gly16Arg)66,67,91,92 | G/A | 0.36 | 0.53 | 0.57 | 0.47 | 0.58 | 0.44 |
Pharmacogenetic loci classified by specific drug class where associations with therapeutic responses were observed.62–64,66,67,76,77,90–103
Minor or less common, variant allele frequencies are based on data from the International HapMap Project Genome Browser release 28 accessed on August 27, 2013 (http://hapmap.ncbi.nlm.nih.gov/cgi-perl/gbrowse/hapmap28_B36/).
Abbreviations from each group are as follows: CEU=Utah residents with ancestry from northern and western Europe; YRI=Individuals from Yoruba in Ibadan, Nigeria; ASW=African Americans from the southwest United States; MEX=Mexican Americans from Los Angeles, CA; CHB=Han Chinese from Beijing, China; JPT=Japanese from Tokyo, Japan.
Pharmacogenetic studies have evaluated the genotypic effects of the Gly16Arg locus on the response to chronic, regular exposure to beta agonist therapies revealing genetic effects which may impact populations with a significant African ancestry. Two early studies were genetic association studies of prior clinical trials which randomized asthma subjects to regular, chronic albuterol exposure. These studies consistently demonstrated that asthma subjects homozygote for the Arg16 allele were more likely to experience adverse effects on peak flow rate during regular SABA treatment compared to Gly16 homozygotes.62,63 These pharmacogenetic findings were confirmed in a prospective, genotype-stratified, cross-over trial performed by NHLBI Asthma Clinical Research Network (ACRN) investigators, the Beta Agonist Response by Genotype (BARGE) trial. In the BARGE trial, 37 Arg16 homozygotes and 41 Gly16 homozygotes were prospectively randomized to regular albuterol or placebo for 16 weeks with ipratropium provided as the rescue inhaler. In this study, Arg16 homozygotes did not experience a change in PEFR and a deterioration of symptom control during albuterol treatment. In contrast, Gly16 homozygotes demonstrated improvements in PEFR and symptom control during albuterol treatment.64
In the BARGE study, 22 percent of Arg16 homozygote subjects were African Americans while 17 percent were Gly16 homozygotes; however, the allele frequency of the Arg16 allele is higher among African Americans (allele frequency=0.53 in an African population, table 1) compared to non-European Whites (allele frequency=0.36, table 1) in the general population.64 This difference in allele frequency is likely because Gly16 is the ancestral allele at the Gly16Arg locus; thus, chromosomes from an ancient African ancestry have had more time to distribute the more recent Arg16 variant compared to chromosomes from a European ancestry.65 This difference in allele frequency at the Gly16Arg locus has the potential to contribute to a disparity in adverse responses between ethnic groups during chronic beta agonist exposure and has been further evaluated in subsequent, prospective, genotype stratified trials for LABA therapy in asthma subjects (table 1).66,67
In contrast to the BARGE trial, the NHLBI ACRN Long-Acting Beta Agonist Response by Genotype (LARGE) trial did not identify differential therapeutic LABA responses related to Gly16Arg genotype in 42 asthmatic Arg16 homozygotes and 45 Gly16 homozygotes randomized to salmeterol or placebo in combination with ICS therapy using a similar cross-over, genotype stratified study design. Despite the fact that both genotypes (Arg16 or Gly16 homozygotes) experienced equal improvements in PEFR during either treatment, Gly16 homozygotes experienced a bronchoprotective effect or an decrease in methacholine bronchial reactivity which was not observed in Arg16 homozygotes.67 In the subgroup of African Americans from the LARGE trial (eight Gly16 homozygotes and nine Arg16 homozygotes), Gly16 homozygotes experienced significantly greater improvements in morning and evening peak flow rates with salmeterol compared to placebo while no such effect was observed among Arg16 homozygotes.68 A second genotype-stratified trial of Gly16Arg genotypes by Bleecker and colleagues randomized 183 Gly16 homozygotes, 182 Gly16 Arg heterozygotes, and 179 Arg16 homozygotes to salmeterol monotherapy or salmeterol with an ICS for 16 weeks without a cross-over design. Similar improvements in PEFR were observed in all subjects and in all ethnic subgroups during LABA therapy with or without an ICS therapy, independent of Gly16Arg genotype.66
Lessons Learned from Pharmacogenetic Studies Related to Admixed Ethnic Populations
There has been conflicting pharmacogenetic evidence for prospective, pharmacogenetic studies of ADRB2 gene variation which highlights the challenges of performing these studies in different ethnic groups. First, admixed ethnic groups represent the minority of subjects enrolled in most pharmacogenetic studies, thus, resulting in sample sizes which may be underpowered to detect genetic associations. Second, there are often environmental factors related to cultural or habitual exposures, for example, access to health care, compliance with medications, and different environmental exposures such as cigarette smoke.
Third, the study of admixed populations is further complicated by the increased genetic diversity at the individual level. This increased genetic diversity complicates genetic association studies due to altered allele frequencies and potentially altered gene-gene interactions between subjects who are self-reporting the same ethnicities. For instance, a SNP in GSNOR (rs11547772) was shown to interact with Gly16Arg in ADRB2 to determine albuterol bronchodilator response in 336 Puerto Rican asthmatic families, a pharmacogenetic interaction not observed among Mexican Americans.69 In addition, 426 SNP’s from 254 plausible candidate genes were evaluated using Bayesian networks to identify predictive gene pathways for acute albuterol bronchodilator response in 308 non-Hispanic White subjects from the Childhood Asthma Management Program (CAMP). This systems biology approach used genetic data to generate a network of 15 SNP’s within 15 genes which predicted SABA response with greater accuracy than each individual locus.70 Systems biology approaches will be particularly important for pharmacogenomic studies in admixed populations where variable allele frequencies alter complex gene-gene interactions and potentially contribute to variable therapeutic responses.71
A final consideration is that there is an increased frequency of rare variants in populations with African ancestry compared to non-Hispanic White populations.22,57 Life-threatening, adverse responses to LABA are uncommon events but represent a strong effect that might be determined by rare genetic variants, particularly among individuals of African descent where rare variants have been most frequently identified. Rare genetic variants have a greater potential for stronger effects on phenotype compared to common variants such as Gly16Arg (figure 1). For instance, the rare Thr164Ile variant in ADRB2 is located in the fourth transmembrane domain and has been associated with receptor ligand binding and decreased receptor coupling to Gs protein in response to different SABA’s.72,73 Similar in vitro effects for this rare variant have also been observed in response to LABA’s resulting in impaired binding of salmeterol to its receptor “exosite.”73 The rare Ile164 allele was also associated with reduced baseline lung function and an increased susceptibility for airflow obstruction in 62,748 subjects from the Copenhagen General Population Study and the Copenhagen City Heart Study which included a subset with self-reported asthma (N=1,300); however, the impact of this rare variant on therapeutic responses to beta agonist remains to be studied in an asthma trial cohort.74 A poly-cytosine (poly-C) repeat variant in the 3′ untranslated region of ADRB2 was not associated with therapeutic responses to LABA and ICS combination therapy in 2,192 asthma subjects; however, nearly half of the poly-C allele lengths had allele frequencies less than five percent consistent with rare variation that remains to be studied.75 Sequencing of ADRB2 has also identified additional potentially functional rare variants at an increased frequency among African American compared to non-Hispanic White asthmatics.22,57 Of these rare functional variants, a novel rare coding variant, Ser220Cys, and a rare promoter 25 base-pair insertion at nucleotide position -376 were only identified in African Americans while a rare coding SNP, Thr164Ile was identified at a great frequency in non-Hispanic Whites.57
Pharmacogenetic Genome-Wide Association Studies and Future Studies of Multi-Ethnic Populations
While there have been no pharmacogenetic genome-wide association studies (GWAS) related to asthma therapies in African American or Hispanic ethnic groups, recent GWAS in non-Hispanic White asthma populations have provided a sound rationale for gene variants with potential pharmacogenetic effects. A GWAS of ICS response in 118 probands from the CAMP and 935 asthmatics from 4 replication cohorts consisting primarily of non-Hispanic Whites revealed a SNP in the promoter region of the glucocorticoid-induced transcript 1 gene (GLCCl1), rs37972, associated with lung function response to ICS.76 More recently, A GWAS of albuterol bronchodilator response in 1,644 non-Hispanic White asthma subjects from 6 clinical trials identified SNP’s adjacent to a gene that determines β2-adrenergic receptor expression, rs295137 and rs295114 in SPATS2L, associated with the acute bronchodilator response to SABA.77
The less common or minor allele of rs37972 in GLCCl1 was associated with a diminished response to ICS and is less common in populations of African descent compared to a European population (table 1).76,78,79 In contrast, the minor allele of rs295137 in SPATS2L was associated with a greater acute SABA bronchodilator response and is more common in an African population compared to populations of European descent (table 1).77–79 Thus, the difference in allele frequencies at these pharmacogenetic loci potentially contributes to a greater frequency of treatment failures or adverse responses to LABA treatment and a relative preference for ICS therapy among African Americans when compared to non-Hispanic Whites.46,54,55
Admixture Mapping as a More powerful Approach to Genome-Wide Studies
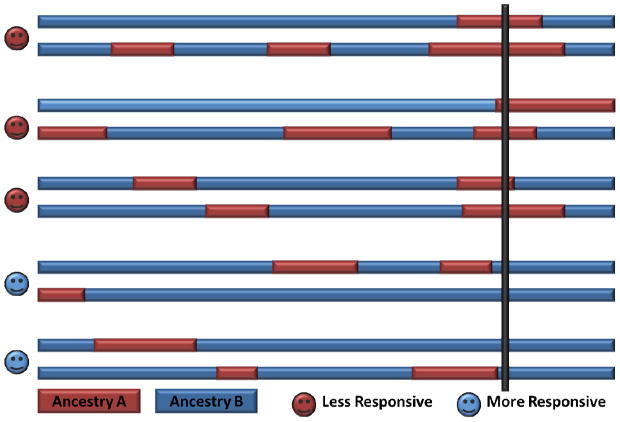
The greatest challenge in performing GWAS in admixed populations is the ability to enroll a sufficient sample size of these populations which have historically been underrepresented in clinical trial cohorts. The recent mixing of ancestries in these populations provides for an alternative method for genome-wide studies which takes advantage of inter-individual differences in genetic ancestry in admixed populations: Mapping by admixture linkage or admixture mapping. Admixture mapping is a genome-wide approach which utilizes the variable allele frequencies of multiple SNP’s between different ancestral populations (traditionally AIM’s, but higher-resolution methods have also used arbitrary SNP’s) to estimate genetic ancestry throughout the genome.80–83
Admixture mapping uses estimates of ancestry at each SNP to test for associations with a phenotype in contrast to GWAS which compares allele frequencies with phenotype (figure 4). Admixture mapping has greater statistical power than GWAS because it requires a smaller number of genetic markers while providing coverage for genomic regions which contain rare variants.80–83 Admixture mapping is best applied in recently admixed populations where a marked racial disparity exists in disease phenotype which is not clearly attributed to environmental factors and has successfully identified novel asthma susceptibility loci in African Americans, Puerto Ricans, and Mexicans. 82,84–89 Admixture mapping estimates ancestry at genetic markers which represent the surrounding genomic region (i.e. region of admixture linkage disequilibrium, figure 4) and, therefore, does not have the resolution of GWAS or whole-genome sequencing. Despite this limitation, admixture mapping is still a powerful method with the potential to identify regions containing pharmacogenetic loci unique to a particular ancestry within admixture mapping association peaks. Thus, the optimal setting for admixture-based approaches in future pharmacogenetic studies will be in combination with whole-genome genotyping or sequencing to potentially identify genetic variation which influence differential responses to asthma therapies observed between different ethnic or racial groups (figure 4).9,10,55
Figure 4. Illustration of Admixture Mapping.
The hypothesis behind mapping by admixture linkage disequilibrium (MALD) or admixture mapping is that chromosomes from an admixed population (shown with red and blue genetic regions from a specific ancestry) contain a susceptibility allele for reduced therapeutic responsiveness which is more frequent in the red (ancestry A) ancestral region versus the blue (ancestry B). A hypothetical MALD for a pharmacogenetic study would identify an increased proportion of ancestry A at a susceptibility locus in individuals who are less likely to respond to a pharmacologic therapy (region intersected by thick black line). Reproduced from Montana G, et al. Am J Hum Genet 2004;75:771–789.83
Next-Generation Sequencing and the Future Path to Personalized Medicine in Admixed Populations
Most pharmacogenetic studies of admixed ethnic groups with asthma have been small candidate gene association studies which have identified “tagging” SNP’s that are potentially co-inherited with another causative variant. Large consortium-based sequencing studies such as the NIH HLBI GO ESP, CAAPA, and the 1,000 Genomes Project are currently using next-generation whole-exome and whole-genome sequencing studies to provide an increasingly diverse genetic map of genomes from different admixed populations.22,23 These large-scale sequencing projects have revealed that admixed ethnic groups demonstrate a remarkable degree of genetic diversity related to an ancient African ancestry. Such genetic diversity has resulted in shorter regions of shared chromosomal segments (i.e. linkage disequilibrium) and a greater frequency of rare variants in ethnic groups with an African ancestry compared to European ancestral populations.22
Future pharmacogenetic approaches will need to enroll a larger number of subjects from underrepresented ethnic groups into clinical trials for GWAS, admixture mapping, and genotype-stratified trials based on comprehensive whole-genome genotyping platforms which account for the genetic diversity of different ancestral backgrounds. Eventually our understanding of the complex genetic factors influencing treatment response will improve and the costs of genotyping, DNA sequencing, and the issues of data storage will progress to the point that we will be able to develop genetic biomarker panels which will predict the most efficacious therapies at the least risk for an individual asthma patient. The path to personalized medicine for all ethnic groups will require that we improve our ability to decipher the expansive volumes of genotype and, now, sequence data using different analytical methods which account for ancestral genetic structure, complex haplotypes, gene-gene interactions, and rare variants in order to detect and replicate novel pharmacogenetic loci.
Abbreviations Used
- ADRB1
β1-adrenergic receptor
- GRK5
G-protein receptor kinase 5
- SNP’s
single nucleotide polymorphisms
- NIH
National Institutes of Health
- NHLBI
National Heart, Lung, and Blood Institute
- GO ESP
GO Exome Sequencing Project
- CAAPA
Consortium on Asthma among African-ancestry Populations in the Americas
- FEV1
forced expiratory volume in one second
- GALA
Genetics of Asthma in Latino Americans
- SABA
short-acting beta agonist
- LABA
long-acting beta agonist
- ICS
inhaled corticosteroid
- SMART
Salmeterol Multicenter Asthma Research Trial
- FDA
Food and Drug Administration
- CARE
Childhood Asthma Research and Education Network
- BADGER
Network Best Add-on Therapy Giving Effective Response Trial
- ADRB2
β2-adrenergic receptor
- ACRN
Asthma Clinical Research Network
- BARGE
Beta Agonist Response by Genotype Trial
- LARGE
Long-acting Beta Agonist Response by Genotype Trial
- GWAS
genome-wide association study
- GLCCl1
glucocorticoid-induced transcript 1 gene
- SPATS2L
spermatogenesis associated, serine-rich 2-like gene
- CEU
Utah residents with ancestry from northern and western Europe
- YRI
Individuals from Yoruba in Ibadan, Nigeria
- ASW
African Americans from the southwest United States
- MEX
Mexican Americans from Los Angeles, CA
- CHB
Han Chinese from Beijing, China
- JPT
Japanese from Tokyo, Japan
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Dries DL, Exner DV, Gersh BJ, Cooper HA, Carson PE, Domanski MJ. Racial differences in the outcome of left ventricular dysfunction. New Engl J Med. 1999;340(8):609–16. doi: 10.1056/NEJM199902253400804. [DOI] [PubMed] [Google Scholar]
- 2.Beta-Blocker Evaluation of Survival Trial I. A trial of the beta-blocker bucindolol in patients with advanced chronic heart failure. New Engl J Med. 2001;344(22):1659–67. doi: 10.1056/NEJM200105313442202. [DOI] [PubMed] [Google Scholar]
- 3.Liggett SB, Cresci S, Kelly RJ, Syed FM, Matkovich SJ, Hahn HS, et al. A GRK5 polymorphism that inhibits beta-adrenergic receptor signaling is protective in heart failure. Nat Med. 2008;14(5):510–7. doi: 10.1038/nm1750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Liggett SB, Mialet-Perez J, Thaneemit-Chen S, Weber SA, Greene SM, Hodne D, et al. A polymorphism within a conserved beta(1)-adrenergic receptor motif alters cardiac function and beta-blocker response in human heart failure. Proc Natl Acad Sci U S A. 2006;103(30):11288–93. doi: 10.1073/pnas.0509937103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Vesell ES. Pharmacogenetic perspectives gained from twin and family studies. Pharmacol Ther. 1989;41(3):535–52. doi: 10.1016/0163-7258(89)90130-7. [DOI] [PubMed] [Google Scholar]
- 6.Kalow W, Tang BK, Endrenyi L. Hypothesis: comparisons of inter- and intra-individual variations can substitute for twin studies in drug research. Pharmacogenetics. 1998;8(4):283–9. doi: 10.1097/00008571-199808000-00001. [DOI] [PubMed] [Google Scholar]
- 7.Drazen JM, Silverman EK, Lee TH. Heterogeneity of therapeutic responses in asthma. Br Med Bull. 2000;56(4):1054–70. doi: 10.1258/0007142001903535. [DOI] [PubMed] [Google Scholar]
- 8.Lemanske RF, Jr, Mauger DT, Sorkness CA, Jackson DJ, Boehmer SJ, Martinez FD, et al. Step-up therapy for children with uncontrolled asthma receiving inhaled corticosteroids. N Engl J Med. 2010;362(11):975–85. doi: 10.1056/NEJMoa1001278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wechsler ME, Castro M, Lehman E, Chinchilli VM, Sutherland ER, Denlinger L, et al. Impact of race on asthma treatment failures in the asthma clinical research network. Am J Respir Crit Care Med. 2011;184(11):1247–53. doi: 10.1164/rccm.201103-0514OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nelson HS, Weiss ST, Bleecker ER, Yancey SW, Dorinsky PM, Group SS. The Salmeterol Multicenter Asthma Research Trial: a comparison of usual pharmacotherapy for asthma or usual pharmacotherapy plus salmeterol. Chest. 2006;129(1):15–26. doi: 10.1378/chest.129.1.15. [DOI] [PubMed] [Google Scholar]
- 11.Sankar P, Cho MK, Mountain J. Race and ethnicity in genetic research. Am J Med Genet A. 2007;143A(9):961–70. doi: 10.1002/ajmg.a.31575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Altshuler D, Pollara VJ, Cowles CR, Van Etten WJ, Baldwin J, Linton L, et al. An SNP map of the human genome generated by reduced representation shotgun sequencing. Nature. 2000;407(6803):513–6. doi: 10.1038/35035083. [DOI] [PubMed] [Google Scholar]
- 13.Irizarry K, Kustanovich V, Li C, Brown N, Nelson S, Wong W, et al. Genome-wide analysis of single-nucleotide polymorphisms in human expressed sequences. Nat Genet. 2000;26(2):233–6. doi: 10.1038/79981. [DOI] [PubMed] [Google Scholar]
- 14.Tsuji S. Genetics of neurodegenerative diseases: insights from high-throughput resequencing. Hum Mol Genet. 2010;19(R1):R65–70. doi: 10.1093/hmg/ddq162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sunyaev SR, Lathe WC, 3rd, Ramensky VE, Bork P. SNP frequencies in human genes an excess of rare alleles and differing modes of selection. Trends Genet. 2000;16(8):335–7. doi: 10.1016/s0168-9525(00)02058-8. [DOI] [PubMed] [Google Scholar]
- 16.Cargill M, Altshuler D, Ireland J, Sklar P, Ardlie K, Patil N, et al. Characterization of single-nucleotide polymorphisms in coding regions of human genes. Nat Genet. 1999;22(3):231–8. doi: 10.1038/10290. [DOI] [PubMed] [Google Scholar]
- 17.Sherry ST, Harpending HC, Batzer MA, Stoneking M. Alu evolution in human populations: using the coalescent to estimate effective population size. Genetics. 1997;147(4):1977–82. doi: 10.1093/genetics/147.4.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Marth G, Schuler G, Yeh R, Davenport R, Agarwala R, Church D, et al. Sequence variations in the public human genome data reflect a bottlenecked population history. Proc Natl Acad Sci U S A. 2003;100(1):376–81. doi: 10.1073/pnas.222673099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Harpending H, Rogers A. Genetic perspectives on human origins and differentiation. Annu Rev Genomics Hum Genet. 2000;1:361–85. doi: 10.1146/annurev.genom.1.1.361. [DOI] [PubMed] [Google Scholar]
- 20.Roberts L. 9 billion? Science. 2011;333(6042):540–3. doi: 10.1126/science.333.6042.540. [DOI] [PubMed] [Google Scholar]
- 21.Keinan A, Clark AG. Recent explosive human population growth has resulted in an excess of rare genetic variants. Science. 2012;336(6082):740–3. doi: 10.1126/science.1217283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Abecasis GR, Auton A, Brooks LD, DePristo MA, Durbin RM, et al. Genomes Project C. An integrated map of genetic variation from 1,092 human genomes. Nature. 2012;491(7422):56–65. doi: 10.1038/nature11632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Exome Variant Server, NHLBI GO Exome Sequencing Project (ESP) Seattle, WA: [cited 2013 May 1]. Available from: (URL: http://evs.gs.washington.edu/EVS/) [Google Scholar]
- 24.Cohen JC, Pertsemlidis A, Fahmi S, Esmail S, Vega GL, Grundy SM, et al. Multiple rare variants in NPC1L1 associated with reduced sterol absorption and plasma low-density lipoprotein levels. Proc Natl Acad Sci U S A. 2006;103(6):1810–5. doi: 10.1073/pnas.0508483103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cohen JC, Kiss RS, Pertsemlidis A, Marcel YL, McPherson R, Hobbs HH. Multiple rare alleles contribute to low plasma levels of HDL cholesterol. Science. 2004;305(5685):869–72. doi: 10.1126/science.1099870. [DOI] [PubMed] [Google Scholar]
- 26.McClellan J, King MC. Genetic heterogeneity in human disease. Cell. 2010;141(2):210–7. doi: 10.1016/j.cell.2010.03.032. [DOI] [PubMed] [Google Scholar]
- 27.Walsh T, King MC. Ten genes for inherited breast cancer. Cancer cell. 2007;11(2):103–5. doi: 10.1016/j.ccr.2007.01.010. [DOI] [PubMed] [Google Scholar]
- 28.Adzhubei IA, Schmidt S, Peshkin L, Ramensky VE, Gerasimova A, Bork P, et al. A method and server for predicting damaging missense mutations. Nat Methods. 2010;7(4):248–9. doi: 10.1038/nmeth0410-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Morris AP, Zeggini E. An evaluation of statistical approaches to rare variant analysis in genetic association studies. Genet Epidemiol. 2010;34(2):188–93. doi: 10.1002/gepi.20450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wu MC, Lee S, Cai T, Li Y, Boehnke M, Lin X. Rare-variant association testing for sequencing data with the sequence kernel association test. Am J Hum Genet. 2011;89(1):82–93. doi: 10.1016/j.ajhg.2011.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tang H, Quertermous T, Rodriguez B, Kardia SL, Zhu X, Brown A, et al. Genetic structure, self-identified race/ethnicity, and confounding in case-control association studies. Am J Hum Genet. 2005;76(2):268–75. doi: 10.1086/427888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Choudhry S, Burchard EG, Borrell LN, Tang H, Gomez I, Naqvi M, et al. Ancestry-environment interactions and asthma risk among Puerto Ricans. Am J Respir Crit Care Med. 2006;174(10):1088–93. doi: 10.1164/rccm.200605-596OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kumar R, Seibold MA, Aldrich MC, Williams LK, Reiner AP, Colangelo L, et al. Genetic ancestry in lung-function predictions. N Engl J Med. 2010;363(4):321–30. doi: 10.1056/NEJMoa0907897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Carter-Pokras OD, Gergen PJ. Reported asthma among Puerto Rican, Mexican-American, and Cuban children, 1982 through 1984. Am J Public Health. 1993;83(4):580–2. doi: 10.2105/ajph.83.4.580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Homa DM, Mannino DM, Lara M. Asthma mortality in U.S. Hispanics of Mexican, Puerto Rican, and Cuban heritage, 1990–1995. Am J Respir Crit Care Med. 2000;161(2 Pt 1):504–9. doi: 10.1164/ajrccm.161.2.9906025. [DOI] [PubMed] [Google Scholar]
- 36.Wenzel SE, Busse WW National Heart L Blood Institute’s Severe Asthma Research P. Severe asthma: lessons from the Severe Asthma Research Program. J Allergy Clin Immunol. 2007;119(1):14–21. doi: 10.1016/j.jaci.2006.10.025. quiz 2–3. [DOI] [PubMed] [Google Scholar]
- 37.Moore WC, Bleecker ER, Curran-Everett D, Erzurum SC, Ameredes BT, Bacharier L, et al. Characterization of the severe asthma phenotype by the National Heart, Lung, and Blood Institute’s Severe Asthma Research Program. J Allergy Clin Immunol. 2007;119(2):405–13. doi: 10.1016/j.jaci.2006.11.639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Moore WC, Meyers DA, Wenzel SE, Teague WG, Li H, Li X, et al. Identification of asthma phenotypes using cluster analysis in the Severe Asthma Research Program. Am J Respir Crit Care Med. 2010;181(4):315–23. doi: 10.1164/rccm.200906-0896OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hankinson JL, Odencrantz JR, Fedan KB. Spirometric reference values from a sample of the general U.S. population. Am J Respir Crit Care Med. 1999;159(1):179–87. doi: 10.1164/ajrccm.159.1.9712108. [DOI] [PubMed] [Google Scholar]
- 40.Rumpel JA, Ahmedani BK, Peterson EL, Wells KE, Yang M, Levin AM, et al. Genetic ancestry and its association with asthma exacerbations among African American subjects with asthma. J Allergy Clin Immunol. 2012;130(6):1302–6. doi: 10.1016/j.jaci.2012.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rose D, Mannino DM, Leaderer BP. Asthma prevalence among US adults, 1998–2000: role of Puerto Rican ethnicity and behavioral and geographic factors. Am J Public Health. 2006;96(5):880–8. doi: 10.2105/AJPH.2004.050039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Salari K, Choudhry S, Tang H, Naqvi M, Lind D, Avila PC, et al. Genetic admixture and asthma-related phenotypes in Mexican American and Puerto Rican asthmatics. Genet Epidemiol. 2005;29(1):76–86. doi: 10.1002/gepi.20079. [DOI] [PubMed] [Google Scholar]
- 43.Brehm JM, Acosta-Perez E, Klei L, Roeder K, Barmada MM, Boutaoui N, et al. African ancestry and lung function in Puerto Rican children. J Allergy Clin Immunol. 2012;129(6):1484–90. e6. doi: 10.1016/j.jaci.2012.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bruse S, Sood A, Petersen H, Liu Y, Leng S, Celedon JC, et al. New Mexican Hispanic smokers have lower odds of chronic obstructive pulmonary disease and less decline in lung function than non-Hispanic whites. Am J Respir Crit Care Med. 2011;184(11):1254–60. doi: 10.1164/rccm.201103-0568OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Castle W, Fuller R, Hall J, Palmer J. Serevent nationwide surveillance study: comparison of salmeterol with salbutamol in asthmatic patients who require regular bronchodilator treatment. BMJ. 1993;306(6884):1034–7. doi: 10.1136/bmj.306.6884.1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nelson HS, Weiss ST, Bleecker ER, Yancey SW, Dorinsky PM. The Salmeterol Multicenter Asthma Research Trial: a comparison of usual pharmacotherapy for asthma or usual pharmacotherapy plus salmeterol. Chest. 2006;129(1):15–26. doi: 10.1378/chest.129.1.15. [DOI] [PubMed] [Google Scholar]
- 47.Salpeter SR, Buckley NS, Ormiston TM, Salpeter EE. Meta-analysis: effect of long-acting beta-agonists on severe asthma exacerbations and asthma-related deaths. Ann Intern Med. 2006;144(12):904–12. doi: 10.7326/0003-4819-144-12-200606200-00126. [DOI] [PubMed] [Google Scholar]
- 48.Peters SP, Prenner BM, Mezzanotte WS, Martin P, O’Brien CD. Long-term safety and asthma control with budesonide/formoterol versus budesonide pressurized metered-dose inhaler in asthma patients. Allergy Asthma Proc. 2008;29(5):499–516. doi: 10.2500/aap.2008.29.3147. [DOI] [PubMed] [Google Scholar]
- 49.Sears MR, Ottosson A, Radner F, Suissa S. Long-acting beta-agonists: a review of formoterol safety data from asthma clinical trials. Eur Respir J. 2009;33(1):21–32. doi: 10.1183/09031936.00145006. [DOI] [PubMed] [Google Scholar]
- 50.Anderson HR, Ayres JG, Sturdy PM, Bland JM, Butland BK, Peckitt C, et al. Bronchodilator treatment and deaths from asthma: case-control study. BMJ. 2005;330(7483):117. doi: 10.1136/bmj.38316.729907.8F. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ni Chroinin M, Greenstone IR, Danish A, Magdolinos H, Masse V, Zhang X, et al. Long-acting beta2-agonists versus placebo in addition to inhaled corticosteroids in children and adults with chronic asthma. Cochrane Database Syst Rev. 2005;(4):CD005535. doi: 10.1002/14651858.CD005535. [DOI] [PubMed] [Google Scholar]
- 52.US Food and Drug Administration. FDA Drug Safety Communication: FDA requires post-market safety trials for Long-Acting Beta-Agonists (LABAs) 2011 [updated 04/15/2011; cited 2012 July 10]. Available from: http://www.fda.gov/Drugs/DrugSafety/ucm251512.htm.
- 53.Chowdhury BA, Seymour SM, Levenson MS. Assessing the safety of adding LABAs to inhaled corticosteroids for treating asthma. N Engl J Med. 2011;364(26):2473–5. doi: 10.1056/NEJMp1104375. [DOI] [PubMed] [Google Scholar]
- 54.Wechsler ME, Castro M, Lehman E, Chinchilli VM, Sutherland ER, Denlinger L, et al. Impact of race on asthma treatment failures in the asthma clinical research network. Am J Respir Crit Care Med. 2011;184(11):1247–53. doi: 10.1164/rccm.201103-0514OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lemanske RF, Jr, Mauger DT, Sorkness CA, Jackson DJ, Boehmer SJ, Martinez FD, et al. Step-up therapy for children with uncontrolled asthma receiving inhaled corticosteroids. N Engl J Med. 2010;362(11):975–85. doi: 10.1056/NEJMoa1001278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Naqvi M, Thyne S, Choudhry S, Tsai HJ, Navarro D, Castro RA, et al. Ethnic-specific differences in bronchodilator responsiveness among African Americans, Puerto Ricans, and Mexicans with asthma. J Asthma. 2007;44(8):639–48. doi: 10.1080/02770900701554441. [DOI] [PubMed] [Google Scholar]
- 57.Hawkins GA, Tantisira K, Meyers DA, Ampleford EJ, Moore WC, Klanderman B, et al. Sequence, haplotype, and association analysis of ADRbeta2 in a multiethnic asthma case-control study. Am J Respir Crit Care Med. 2006;174(10):1101–9. doi: 10.1164/rccm.200509-1405OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Drysdale CM, McGraw DW, Stack CB, Stephens JC, Judson RS, Nandabalan K, et al. Complex promoter and coding region beta 2-adrenergic receptor haplotypes alter receptor expression and predict in vivo responsiveness. Proc Natl Acad Sci U S A. 2000;97(19):10483–8. doi: 10.1073/pnas.97.19.10483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Green SA, Turki J, Bejarano P, Hall IP, Liggett SB. Influence of beta 2-adrenergic receptor genotypes on signal transduction in human airway smooth muscle cells. Am J Respir Cell Mol Biol. 1995;13(1):25–33. doi: 10.1165/ajrcmb.13.1.7598936. [DOI] [PubMed] [Google Scholar]
- 60.Green SA, Turki J, Innis M, Liggett SB. Amino-terminal polymorphisms of the human beta 2-adrenergic receptor impart distinct agonist-promoted regulatory properties. Biochemistry. 1994;33(32):9414–9. doi: 10.1021/bi00198a006. [DOI] [PubMed] [Google Scholar]
- 61.Ortega VE, Hawkins GA, Peters SP, Bleecker ER. Pharmacogenetics of the beta 2-adrenergic receptor gene. Immunol Allergy Clin North Am. 2007;27(4):665–84. vii. doi: 10.1016/j.iac.2007.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Taylor DR, Drazen JM, Herbison GP, Yandava CN, Hancox RJ, Town GI. Asthma exacerbations during long term beta agonist use: influence of beta(2) adrenoceptor polymorphism. Thorax. 2000;55(9):762–7. doi: 10.1136/thorax.55.9.762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Israel E, Drazen JM, Liggett SB, Boushey HA, Cherniack RM, Chinchilli VM, et al. The effect of polymorphisms of the beta(2)-adrenergic receptor on the response to regular use of albuterol in asthma. Am J Respir Crit Care Med. 2000;162(1):75–80. doi: 10.1164/ajrccm.162.1.9907092. [DOI] [PubMed] [Google Scholar]
- 64.Israel E, Chinchilli VM, Ford JG, Boushey HA, Cherniack R, Craig TJ, et al. Use of regularly scheduled albuterol treatment in asthma: genotype-stratified, randomised, placebo-controlled cross-over trial. Lancet. 2004;364(9444):1505–12. doi: 10.1016/S0140-6736(04)17273-5. [DOI] [PubMed] [Google Scholar]
- 65.Meyer LR, Zweig AS, Hinrichs AS, Karolchik D, Kuhn RM, Wong M, et al. The UCSC Genome Browser database: extensions and updates 2013. Nucleic Acids Res. 2013;41(D1):D64–9. doi: 10.1093/nar/gks1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Bleecker ER, Nelson HS, Kraft M, Corren J, Meyers DA, Yancey SW, et al. Beta2-receptor polymorphisms in patients receiving salmeterol with or without fluticasone propionate. Am J Respir Crit Care Med. 2010;181(7):676–87. doi: 10.1164/200809-1511OC. [DOI] [PubMed] [Google Scholar]
- 67.Wechsler ME, Kunselman SJ, Chinchilli VM, Bleecker E, Boushey HA, Calhoun WJ, et al. Effect of beta2-adrenergic receptor polymorphism on response to longacting beta2 agonist in asthma (LARGE trial): a genotype-stratified, randomised, placebo-controlled, crossover trial. Lancet. 2009;374(9703):1754–64. doi: 10.1016/S0140-6736(09)61492-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lee DK, Currie GP, Hall IP, Lima JJ, Lipworth BJ. The arginine-16 beta2-adrenoceptor polymorphism predisposes to bronchoprotective subsensitivity in patients treated with formoterol and salmeterol. Br J Clin Pharmacol. 2004;57(1):68–75. doi: 10.1046/j.1365-2125.2003.01955.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Choudhry S, Que LG, Yang Z, Liu L, Eng C, Kim SO, et al. GSNO reductase and beta2-adrenergic receptor gene-gene interaction: bronchodilator responsiveness to albuterol. Pharmacogenet Genomics. 2010;20(6):351–8. doi: 10.1097/FPC.0b013e328337f992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Himes BE, Wu AC, Duan QL, Klanderman B, Litonjua AA, Tantisira K, et al. Predicting response to short-acting bronchodilator medication using Bayesian networks. Pharmacogenomics. 2009;10(9):1393–412. doi: 10.2217/pgs.09.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Dahlin A, Tantisira KG. Integrative systems biology approaches in asthma pharmacogenomics. Pharmacogenomics. 2012;13(12):1387–404. doi: 10.2217/pgs.12.126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Green SA, Cole G, Jacinto M, Innis M, Liggett SB. A polymorphism of the human beta 2-adrenergic receptor within the fourth transmembrane domain alters ligand binding and functional properties of the receptor. J Biol Chem. 1993;268(31):23116–21. [PubMed] [Google Scholar]
- 73.Green SA, Rathz DA, Schuster AJ, Liggett SB. The Ile164 beta(2)-adrenoceptor polymorphism alters salmeterol exosite binding and conventional agonist coupling to G(s) Eur J Pharmacol. 2001;421(3):141–7. doi: 10.1016/s0014-2999(01)01049-4. [DOI] [PubMed] [Google Scholar]
- 74.Thomsen M, Nordestgaard BG, Sethi AA, Tybjaerg-Hansen A, Dahl M. beta2-adrenergic receptor polymorphisms, asthma and COPD: two large population-based studies. Eur Respir J. 2012;39(3):558–66. doi: 10.1183/09031936.00023511. [DOI] [PubMed] [Google Scholar]
- 75.Ambrose HJ, Lawrance RM, Cresswell CJ, Goldman M, Meyers DA, Bleecker ER. Effect of beta2-adrenergic receptor gene (ADRB2) 3′ untranslated region polymorphisms on inhaled corticosteroid/long-acting beta2-adrenergic agonist response. Respir Res. 2012;13:37. doi: 10.1186/1465-9921-13-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Tantisira KG, Lasky-Su J, Harada M, Murphy A, Litonjua AA, Himes BE, et al. Genomewide association between GLCCI1 and response to glucocorticoid therapy in asthma. N Engl J Med. 2011;365(13):1173–83. doi: 10.1056/NEJMoa0911353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Himes BE, Jiang X, Hu R, Wu AC, Lasky-Su JA, Klanderman BJ, et al. Genome-Wide Association Analysis in Asthma Subjects Identifies SPATS2L as a Novel Bronchodilator Response Gene. PLoS Genet. 2012;8(7):e1002824. doi: 10.1371/journal.pgen.1002824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Altshuler DM, Gibbs RA, Peltonen L, Dermitzakis E, Schaffner SF, Yu F, et al. Integrating common and rare genetic variation in diverse human populations. Nature. 2010;467(7311):52–8. doi: 10.1038/nature09298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Frazer KA, Ballinger DG, Cox DR, Hinds DA, Stuve LL, Gibbs RA, et al. A second generation human haplotype map of over 3.1 million SNPs. Nature. 2007;449(7164):851–61. doi: 10.1038/nature06258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Alexander DH, Novembre J, Lange K. Fast model-based estimation of ancestry in unrelated individuals. Genome Res. 2009;19(9):1655–64. doi: 10.1101/gr.094052.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Tandon A, Patterson N, Reich D. Ancestry informative marker panels for African Americans based on subsets of commercially available SNP arrays. Genet Epidemiol. 2011;35(1):80–3. doi: 10.1002/gepi.20550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Patterson N, Hattangadi N, Lane B, Lohmueller KE, Hafler DA, Oksenberg JR, et al. Methods for high-density admixture mapping of disease genes. Am J Hum Genet. 2004;74(5):979–1000. doi: 10.1086/420871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Montana G, Pritchard JK. Statistical tests for admixture mapping with case-control and cases-only data. Am J Hum Genet. 2004;75(5):771–89. doi: 10.1086/425281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Torgerson DG, Capurso D, Ampleford EJ, Li X, Moore WC, Gignoux CR, et al. Genome-wide ancestry association testing identifies a common European variant on 6q14.1 as a risk factor for asthma in African American subjects. J Allergy Clin Immunol. 2012;130(3):622–9. e9. doi: 10.1016/j.jaci.2012.03.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Zhu X, Tang H, Risch N. Admixture mapping and the role of population structure for localizing disease genes. Adv Genet. 2008;60:547–69. doi: 10.1016/S0065-2660(07)00419-1. [DOI] [PubMed] [Google Scholar]
- 86.Smith MW, Patterson N, Lautenberger JA, Truelove AL, McDonald GJ, Waliszewska A, et al. A high-density admixture map for disease gene discovery in african americans. Am J Hum Genet. 2004;74(5):1001–13. doi: 10.1086/420856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Choudhry S, Taub M, Mei R, Rodriguez-Santana J, Rodriguez-Cintron W, Shriver MD, et al. Genome-wide screen for asthma in Puerto Ricans: evidence for association with 5q23 region. Hum Genet. 2008;123(5):455–68. doi: 10.1007/s00439-008-0495-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Beavitt SJ, Harder KW, Kemp JM, Jones J, Quilici C, Casagranda F, et al. Lyn-deficient mice develop severe, persistent asthma: Lyn is a critical negative regulator of Th2 immunity. J Immunol. 2005;175(3):1867–75. doi: 10.4049/jimmunol.175.3.1867. [DOI] [PubMed] [Google Scholar]
- 89.Torgerson DG, Gignoux CR, Galanter JM, Drake KA, Roth LA, Eng C, et al. Case-control admixture mapping in Latino populations enriches for known asthma-associated genes. J Allergy Clin Immunol. 2012;130(1):76–82. e12. doi: 10.1016/j.jaci.2012.02.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Vonk JM, Postma DS, Maarsingh H, Bruinenberg M, Koppelman GH, Meurs H. Arginase 1 and arginase 2 variations associate with asthma, asthma severity and beta2 agonist and steroid response. Pharmacogenet Genomics. 2010;20(3):179–86. doi: 10.1097/FPC.0b013e328336c7fd. [DOI] [PubMed] [Google Scholar]
- 91.Bleecker ER, Postma DS, Lawrance RM, Meyers DA, Ambrose HJ, Goldman M. Effect of ADRB2 polymorphisms on response to longacting beta2-agonist therapy: a pharmacogenetic analysis of two randomised studies. Lancet. 2007;370(9605):2118–25. doi: 10.1016/S0140-6736(07)61906-0. [DOI] [PubMed] [Google Scholar]
- 92.Bleecker ER, Yancey SW, Baitinger LA, Edwards LD, Klotsman M, Anderson WH, et al. Salmeterol response is not affected by beta2-adrenergic receptor genotype in subjects with persistent asthma. J Allergy Clin Immunol. 2006;118(4):809–16. doi: 10.1016/j.jaci.2006.06.036. [DOI] [PubMed] [Google Scholar]
- 93.Cho SH, Oh SY, Bahn JW, Choi JY, Chang YS, Kim YK, et al. Association between bronchodilating response to short-acting beta-agonist and non-synonymous single-nucleotide polymorphisms of beta-adrenoceptor gene. Clin Exp Allergy. 2005;35(9):1162–7. doi: 10.1111/j.1365-2222.2005.02319.x. [DOI] [PubMed] [Google Scholar]
- 94.Choudhry S, Ung N, Avila PC, Ziv E, Nazario S, Casal J, et al. Pharmacogenetic differences in response to albuterol between Puerto Ricans and Mexicans with asthma. Am J Respir Crit Care Med. 2005;171(6):563–70. doi: 10.1164/rccm.200409-1286OC. [DOI] [PubMed] [Google Scholar]
- 95.Lima JJ, Thomason DB, Mohamed MH, Eberle LV, Self TH, Johnson JA. Impact of genetic polymorphisms of the beta2-adrenergic receptor on albuterol bronchodilator pharmacodynamics. Clin Pharmacol Ther. 1999;65(5):519–25. doi: 10.1016/S0009-9236(99)70071-8. [DOI] [PubMed] [Google Scholar]
- 96.Tantisira KG, Small KM, Litonjua AA, Weiss ST, Liggett SB. Molecular properties and pharmacogenetics of a polymorphism of adenylyl cyclase type 9 in asthma: interaction between beta-agonist and corticosteroid pathways. Hum Mol Genet. 2005;14(12):1671–7. doi: 10.1093/hmg/ddi175. [DOI] [PubMed] [Google Scholar]
- 97.Tantisira KG, Damask A, Szefler SJ, Schuemann B, Markezich A, Su J, et al. Genome-wide association identifies the T gene as a novel asthma pharmacogenetic locus. Am J Respir Crit Care Med. 2012;185(12):1286–91. doi: 10.1164/rccm.201111-2061OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Hawkins GA, Lazarus R, Smith RS, Tantisira KG, Meyers DA, Peters SP, et al. The glucocorticoid receptor heterocomplex gene STIP1 is associated with improved lung function in asthmatic subjects treated with inhaled corticosteroids. J Allergy Clin Immunol. 2009;123(6):1376–83. e7. doi: 10.1016/j.jaci.2009.01.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Lima JJ, Zhang S, Grant A, Shao L, Tantisira KG, Allayee H, et al. Influence of leukotriene pathway polymorphisms on response to montelukast in asthma. Am J Respir Crit Care Med. 2006;173(4):379–85. doi: 10.1164/rccm.200509-1412OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Telleria JJ, Blanco-Quiros A, Varillas D, Armentia A, Fernandez-Carvajal I, Jesus Alonso M, et al. ALOX5 promoter genotype and response to montelukast in moderate persistent asthma. Respir Med. 2008;102(6):857–61. doi: 10.1016/j.rmed.2008.01.011. [DOI] [PubMed] [Google Scholar]
- 101.Tantisira KG, Lima J, Sylvia J, Klanderman B, Weiss ST. 5-lipoxygenase pharmacogenetics in asthma: overlap with Cys-leukotriene receptor antagonist loci. Pharmacogenet Genomics. 2009;19(3):244–7. doi: 10.1097/FPC.0b013e328326e0b1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Litonjua AA, Lasky-Su J, Schneiter K, Tantisira KG, Lazarus R, Klanderman B, et al. ARG1 is a novel bronchodilator response gene: screening and replication in four asthma cohorts. Am J Respir Crit Care Med. 2008;178(7):688–94. doi: 10.1164/rccm.200709-1363OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Duan QL, Gaume BR, Hawkins GA, Himes BE, Bleecker ER, Klanderman B, et al. Regulatory haplotypes in ARG1 are associated with altered bronchodilator response. Am J Respir Crit Care Med. 2011;183(4):449–54. doi: 10.1164/rccm.201005-0758OC. [DOI] [PMC free article] [PubMed] [Google Scholar]