Abstract
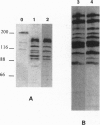
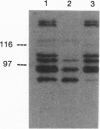
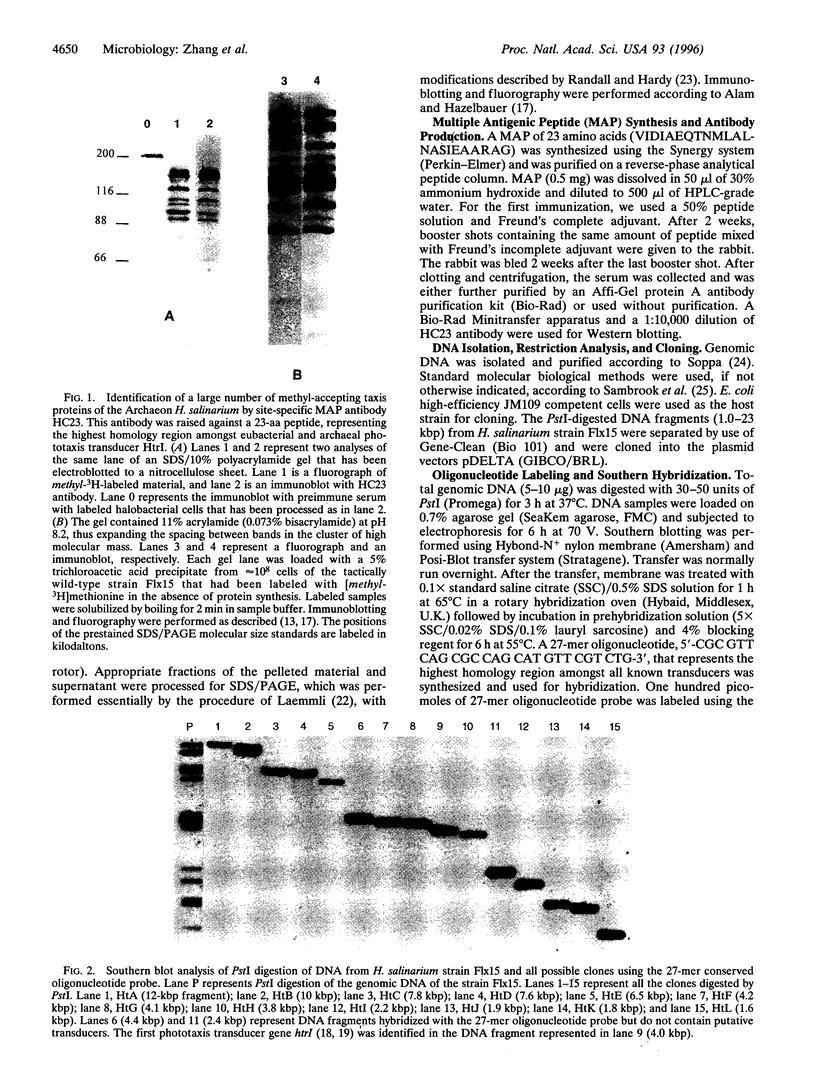
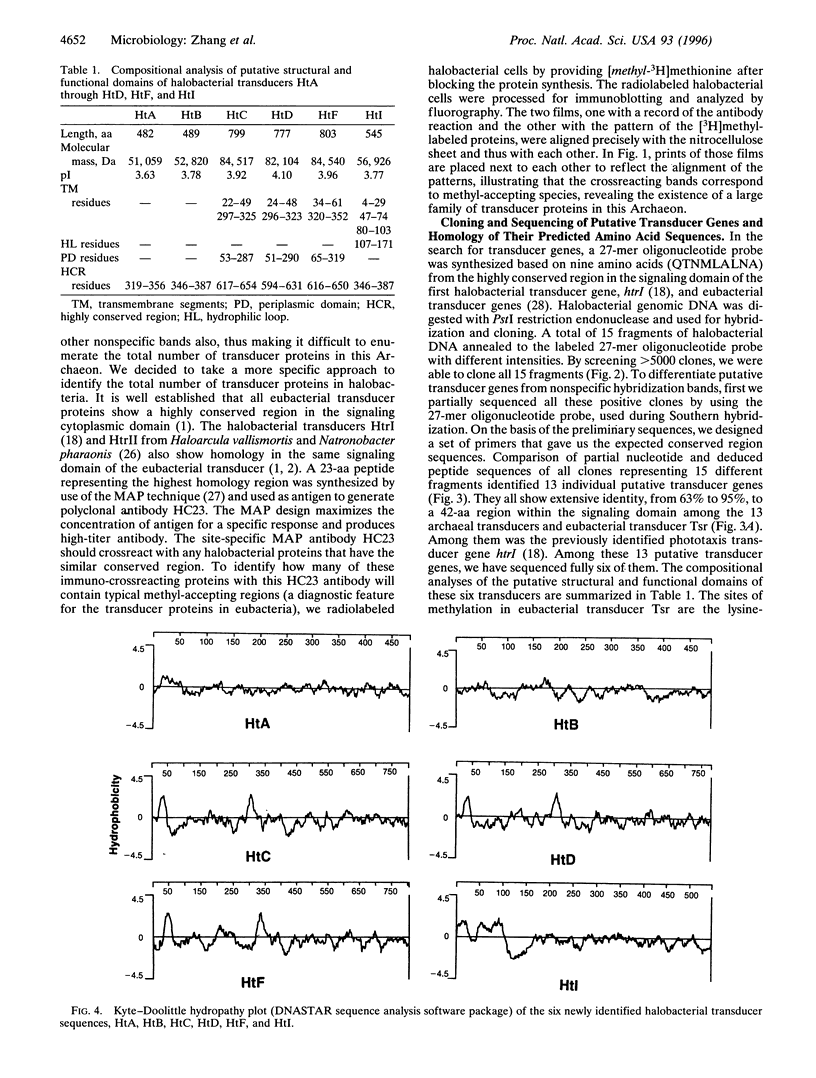
Eubacterial transducers are transmembrane, methyl-accepting proteins central to chemotaxis systems and share common structural features. We identified a large family of transducer proteins in the Archaeon Halobacterium salinarium using a site-specific multiple antigenic peptide antibody raised against 23 amino acids, representing the highest homology region of eubacterial transducers. This immunological observation was confirmed by isolating 13 methyl-accepting taxis genes using a 27-mer oligonucleotide probe, corresponding to conserved regions between the eubacterial and first halobacterial phototaxis transducer gene htrI. On the basis of the comparison of the predicted structural domains of these transducers, we propose that at least three distinct subfamilies of transducers exist in the Archaeon H. salinarium: (i) a eubacterial chemotaxis transducer type with two hydrophobic membrane-spanning segments connecting sizable domains in the periplasm and cytoplasm; (ii) a cytoplasmic domain and two or more hydrophobic transmembrane segments without periplasmic domains; and (iii) a cytoplasmic domain without hydrophobic transmembrane segments. We fractionated the halobacterial cell lysate into soluble and membrane fractions and localized different halobacterial methyl-accepting taxis proteins in both fractions.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Alam M., Hazelbauer G. L. Structural features of methyl-accepting taxis proteins conserved between archaebacteria and eubacteria revealed by antigenic cross-reaction. J Bacteriol. 1991 Sep;173(18):5837–5842. doi: 10.1128/jb.173.18.5837-5842.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alam M., Lebert M., Oesterhelt D., Hazelbauer G. L. Methyl-accepting taxis proteins in Halobacterium halobium. EMBO J. 1989 Feb;8(2):631–639. doi: 10.1002/j.1460-2075.1989.tb03418.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bibikov S. I., Grishanin R. N., Kaulen A. D., Marwan W., Oesterhelt D., Skulachev V. P. Bacteriorhodopsin is involved in halobacterial photoreception. Proc Natl Acad Sci U S A. 1993 Oct 15;90(20):9446–9450. doi: 10.1073/pnas.90.20.9446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blair D. F. How bacteria sense and swim. Annu Rev Microbiol. 1995;49:489–522. doi: 10.1146/annurev.mi.49.100195.002421. [DOI] [PubMed] [Google Scholar]
- Bourret R. B., Borkovich K. A., Simon M. I. Signal transduction pathways involving protein phosphorylation in prokaryotes. Annu Rev Biochem. 1991;60:401–441. doi: 10.1146/annurev.bi.60.070191.002153. [DOI] [PubMed] [Google Scholar]
- Boyd A., Simon M. I. Multiple electrophoretic forms of methyl-accepting chemotaxis proteins generated by stimulus-elicited methylation in Escherichia coli. J Bacteriol. 1980 Aug;143(2):809–815. doi: 10.1128/jb.143.2.809-815.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chelsky D., Dahlquist F. W. Structural studies of methyl-accepting chemotaxis proteins of Escherichia coli: evidence for multiple methylation sites. Proc Natl Acad Sci U S A. 1980 May;77(5):2434–2438. doi: 10.1073/pnas.77.5.2434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke S. Protein carboxyl methyltransferases: two distinct classes of enzymes. Annu Rev Biochem. 1985;54:479–506. doi: 10.1146/annurev.bi.54.070185.002403. [DOI] [PubMed] [Google Scholar]
- Deckers H. M., Voordouw G. Identification of a large family of genes for putative chemoreceptor proteins in an ordered library of the Desulfovibrio vulgaris Hildenborough genome. J Bacteriol. 1994 Jan;176(2):351–358. doi: 10.1128/jb.176.2.351-358.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engström P., Hazelbauer G. L. Multiple methylation of methyl-accepting chemotaxis proteins during adaptation of E. coli to chemical stimuli. Cell. 1980 May;20(1):165–171. doi: 10.1016/0092-8674(80)90244-5. [DOI] [PubMed] [Google Scholar]
- Ferrando-May E., Krah M., Marwan W., Oesterhelt D. The methyl-accepting transducer protein HtrI is functionally associated with the photoreceptor sensory rhodopsin I in the archaeon Halobacterium salinarium. EMBO J. 1993 Aug;12(8):2999–3005. doi: 10.1002/j.1460-2075.1993.tb05968.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hildebrand E., Dencher N. Two photosystems controlling behavioural responses of Halobacterium halobium. Nature. 1975 Sep 4;257(5521):46–48. doi: 10.1038/257046a0. [DOI] [PubMed] [Google Scholar]
- Iwabe N., Kuma K., Hasegawa M., Osawa S., Miyata T. Evolutionary relationship of archaebacteria, eubacteria, and eukaryotes inferred from phylogenetic trees of duplicated genes. Proc Natl Acad Sci U S A. 1989 Dec;86(23):9355–9359. doi: 10.1073/pnas.86.23.9355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krah M., Marwan W., Verméglio A., Oesterhelt D. Phototaxis of Halobacterium salinarium requires a signalling complex of sensory rhodopsin I and its methyl-accepting transducer HtrI. EMBO J. 1994 May 1;13(9):2150–2155. doi: 10.1002/j.1460-2075.1994.tb06491.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krikos A., Mutoh N., Boyd A., Simon M. I. Sensory transducers of E. coli are composed of discrete structural and functional domains. Cell. 1983 Jun;33(2):615–622. doi: 10.1016/0092-8674(83)90442-7. [DOI] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Lindbeck J. C., Goulbourne E. A., Jr, Johnson M. S., Taylor B. L. Aerotaxis in Halobacterium salinarium is methylation-dependent. Microbiology. 1995 Nov;141(Pt 11):2945–2953. doi: 10.1099/13500872-141-11-2945. [DOI] [PubMed] [Google Scholar]
- Marwan W., Oesterhelt D. Signal formation in the halobacterial photophobic response mediated by a fourth retinal protein (P480). J Mol Biol. 1987 May 20;195(2):333–342. doi: 10.1016/0022-2836(87)90654-1. [DOI] [PubMed] [Google Scholar]
- McBride M. J., Weinberg R. A., Zusman D. R. "Frizzy" aggregation genes of the gliding bacterium Myxococcus xanthus show sequence similarities to the chemotaxis genes of enteric bacteria. Proc Natl Acad Sci U S A. 1989 Jan;86(2):424–428. doi: 10.1073/pnas.86.2.424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCleary W. R., McBride M. J., Zusman D. R. Developmental sensory transduction in Myxococcus xanthus involves methylation and demethylation of FrzCD. J Bacteriol. 1990 Sep;172(9):4877–4887. doi: 10.1128/jb.172.9.4877-4887.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olson K. D., Spudich J. L. Removal of the transducer protein from sensory rhodopsin I exposes sites of proton release and uptake during the receptor photocycle. Biophys J. 1993 Dec;65(6):2578–2585. doi: 10.1016/S0006-3495(93)81295-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Posnett D. N., Tam J. P. Multiple antigenic peptide method for producing antipeptide site-specific antibodies. Methods Enzymol. 1989;178:739–746. doi: 10.1016/0076-6879(89)78048-4. [DOI] [PubMed] [Google Scholar]
- Randall L. L., Hardy S. J. Synthesis of exported proteins by membrane-bound polysomes from Escherichia coli. Eur J Biochem. 1977 May 2;75(1):43–53. doi: 10.1111/j.1432-1033.1977.tb11502.x. [DOI] [PubMed] [Google Scholar]
- Rose I. A., Hanson K. R., Wilkinson K. D., Wimmer M. J. A suggestion for naming faces of ring compounds. Proc Natl Acad Sci U S A. 1980 May;77(5):2439–2441. doi: 10.1073/pnas.77.5.2439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schimz A., Hildebrand E. Chemosensory responses of Halobacterium halobium. J Bacteriol. 1979 Dec;140(3):749–753. doi: 10.1128/jb.140.3.749-753.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seidel R., Scharf B., Gautel M., Kleine K., Oesterhelt D., Engelhard M. The primary structure of sensory rhodopsin II: a member of an additional retinal protein subgroup is coexpressed with its transducer, the halobacterial transducer of rhodopsin II. Proc Natl Acad Sci U S A. 1995 Mar 28;92(7):3036–3040. doi: 10.1073/pnas.92.7.3036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spudich E. N., Hasselbacher C. A., Spudich J. L. Methyl-accepting protein associated with bacterial sensory rhodopsin I. J Bacteriol. 1988 Sep;170(9):4280–4285. doi: 10.1128/jb.170.9.4280-4285.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spudich E. N., Spudich J. L. Control of transmembrane ion fluxes to select halorhodopsin-deficient and other energy-transduction mutants of Halobacterium halobium. Proc Natl Acad Sci U S A. 1982 Jul;79(14):4308–4312. doi: 10.1073/pnas.79.14.4308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spudich E. N., Spudich J. L. The photochemical reactions of sensory rhodopsin I are altered by its transducer. J Biol Chem. 1993 Aug 5;268(22):16095–16097. [PubMed] [Google Scholar]
- Spudich E. N., Sundberg S. A., Manor D., Spudich J. L. Properties of a second sensory receptor protein in Halobacterium halobium phototaxis. Proteins. 1986 Nov;1(3):239–246. doi: 10.1002/prot.340010306. [DOI] [PubMed] [Google Scholar]
- Spudich E. N., Takahashi T., Spudich J. L. Sensory rhodopsins I and II modulate a methylation/demethylation system in Halobacterium halobium phototaxis. Proc Natl Acad Sci U S A. 1989 Oct;86(20):7746–7750. doi: 10.1073/pnas.86.20.7746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spudich J. L. Color sensing in the Archaea: a eukaryotic-like receptor coupled to a prokaryotic transducer. J Bacteriol. 1993 Dec;175(24):7755–7761. doi: 10.1128/jb.175.24.7755-7761.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoeckenius W., Wolff E. K., Hess B. A rapid population method for action spectra applied to Halobacterium halobium. J Bacteriol. 1988 Jun;170(6):2790–2795. doi: 10.1128/jb.170.6.2790-2795.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sundberg S. A., Alam M., Lebert M., Spudich J. L., Oesterhelt D., Hazelbauer G. L. Characterization of Halobacterium halobium mutants defective in taxis. J Bacteriol. 1990 May;172(5):2328–2335. doi: 10.1128/jb.172.5.2328-2335.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinberg R. A., Zusman D. R. Evidence that the Myxococcus xanthus frz genes are developmentally regulated. J Bacteriol. 1989 Nov;171(11):6174–6186. doi: 10.1128/jb.171.11.6174-6186.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolff E. K., Bogomolni R. A., Scherrer P., Hess B., Stoeckenius W. Color discrimination in halobacteria: spectroscopic characterization of a second sensory receptor covering the blue-green region of the spectrum. Proc Natl Acad Sci U S A. 1986 Oct;83(19):7272–7276. doi: 10.1073/pnas.83.19.7272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan B., Cline S. W., Doolittle W. F., Spudich J. L. Transformation of a bop-hop-sop-I-sop-II-Halobacterium halobium mutant to bop+: effects of bacteriorhodopsin photoactivation on cellular proton fluxes and swimming behavior. Photochem Photobiol. 1992 Oct;56(4):553–561. doi: 10.1111/j.1751-1097.1992.tb02200.x. [DOI] [PubMed] [Google Scholar]
- Yao V. J., Spudich J. L. Primary structure of an archaebacterial transducer, a methyl-accepting protein associated with sensory rhodopsin I. Proc Natl Acad Sci U S A. 1992 Dec 15;89(24):11915–11919. doi: 10.1073/pnas.89.24.11915. [DOI] [PMC free article] [PubMed] [Google Scholar]