Abstract
FtsZ, an essential protein for bacterial cell division, is a highly promising therapeutic target, especially for the discovery and development of new-generation anti-TB agents. Following up the identification of two lead 2,5,6-trisubstituted benzimidazoles, 1 and 2, targeting Mtb-FtsZ in our previous study, an extensive SAR study for optimization of these lead compounds was performed through systematic modification of the 5 and 6 positions. This study has successfully led to the discovery of a highly potent advanced lead 5f (MIC 0.06 µg/mL) and several other compounds with comparable potencies. These advanced lead compounds possess a dimethylamino group at the 6 position. The functional groups at the 5 position exhibit substantial effects on the antibacterial activity as well. In vitro experiments such as the FtsZ polymerization inhibitory assay and TEM analysis of Mtb-FtsZ treated with 5f and others indicate that Mtb-FtsZ is the molecular target for their antibacterial activity.
Keywords: Mycobacterium tuberculosis, Antibacterial, Benzimidazoles, FtsZ, Structure-activity relationship
Introduction
Tuberculosis (TB) was responsible for 1.4 million deaths in 2011, which was second only to HIV/AIDS related fatalities among all infectious diseases.1, 2 TB is caused by Mycobacterium tuberculosis (Mtb) and is highly contagious when airborne.3 It has been estimated that 90% of humans who are exposed to or infected with the pathogen have latent-TB, which has a 10% chance of progressing to an active-TB diseased state during their lifetime.4, 5 Recent statistics from the World Health Organization (WHO) indicate an estimated 8.7 million new TB cases globally in 2011.1, 2 Among these cases, 13% were HIV-positive individuals with 430,000 deaths, which demonstrates the ability of the bacteria to target immunocompromised patients.1, 2 It has been reported that MDR-TB is the causative agent of 3.7% of the new cases and 20% of the previously reported cases worldwide.1, 2 MDR-TB resistant to at least one of the three injectable “second-line” antibiotics and fluoroquinolones is classified as extensively drug resistant-TB (XDR-TB). About 9% of MDR-TB cases have been classified as XDR-TB by WHO and 84 countries have reported at least one case of XDR-TB.1, 2
The widespread resistance to existing therapeutics has become a key hurdle for effective treatment of the disease. Therefore, in order to counterattack the drug resistance, there is a dire need for the identification of novel therapeutic targets. In this context, FtsZ, an essential bacterial cytokinesis protein, is a highly promising therapeutic target.6–9 In the presence of GTP, FtsZ polymerizes bi-directionally at the center of the cell on the inner membrane to form a highly dynamic helical structure known as the “Z-ring”.10–14 The recruitment of several other cell division proteins leads to Z-ring contraction, resulting in septum formation and eventually cell division.10–14 Accordingly, the inhibition of proper FtsZ assembly would block the septum formation and then cell division, which should lead to bacterial growth inhibition and cell death.6, 8, 9, 15–17
Recently, FtsZ inhibitors have been actively investigated for pathogen specific as well as broadspectrum antibacterial drug discovery against a variety of pathogens.6, 18 Several classes of compounds have been identified as promising leads for antibacterial drug development, including OTBA,19 2-alkoxycarbonylaminopyridines,20 2-carbamoylpteridine,21 taxanes,15 benzimidazoles,22 GTP analogs,23, 24 benzo[c]phenathridines,25, 26 isoquinolines,27–29 PC190723,30, 31 Zantrins,7 and chrysophaentins32. However, against Mtb FtsZ, only taxanes15 and benzimidazoles22 have emerged as promising leads in the last several years after the pioneering work by the Southern Research Institute group.20, 21, 27, 33
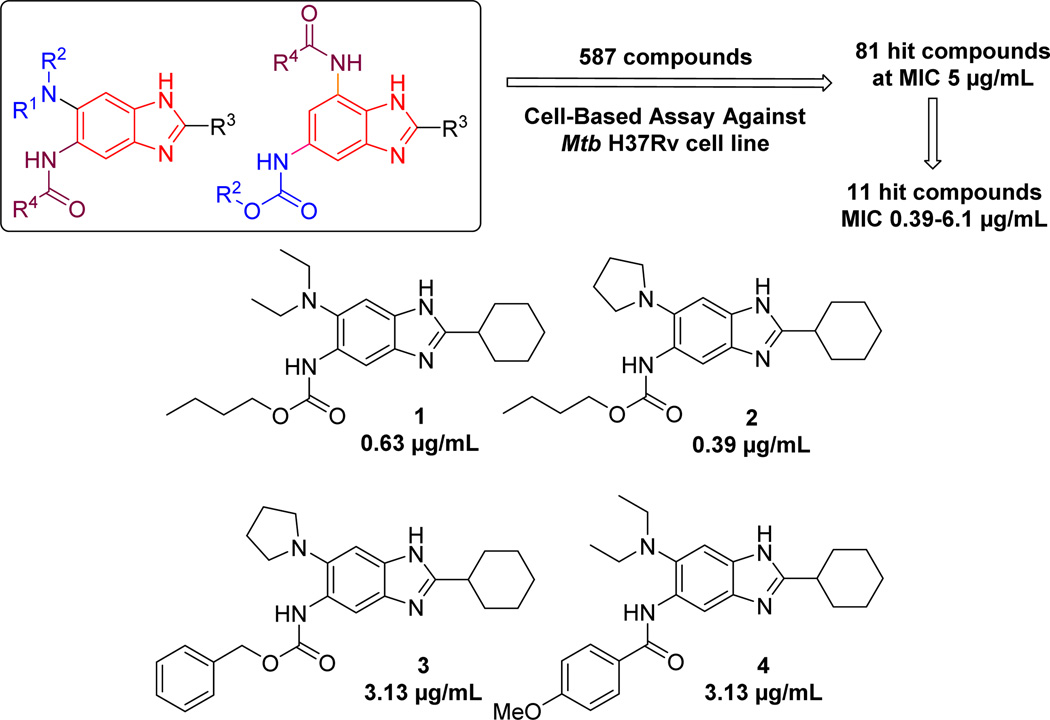
We have previously reported the design and synthesis of libraries of 2,5,6- and 2,5,7-trisubstituted benzimidazoles, which gave a good number of hit compounds through screening against Mtb H37Rv strain at 5 µg/mL concentration.22 Moreover, selected lead compounds from these hits were found to exhibit excellent MIC values in the range of 0.39–6.1 µg/mL against drug sensitive (H37Rv) as well as drug resistant strains (Figure 1).22 These lead compounds inhibited Mtb FtsZ assembly in a dose dependent manner, while enhancing the GTPase activity.22
Figure 1. Early Lead Compounds from Hit Benzimidazoles22.
Subsequently, benzimidazoles 1 and 2 were found to be bactericidal and also active against non-replicating Mtb grown under low oxygen conditions.34 The latter result is particularly exciting since it indicates that these compounds have potential to be effective against latent TB infection. Furthermore, 1 exhibited good efficacy in vivo in the standard acute infection model using immune incompetent GKO mice.35 These promising results will be published elsewhere shortly.
Since these two lead compounds have shown very promising antibacterial activities in vitro and in vivo, we set out for the SAR study and optimization of these leads through systematic structural modifications at the 5 and 6 positions, keeping the cyclohexyl group at the 2 position intact as shown in Figure 2. We describe here our SAR study and successful optimization of the early lead compounds, which has led to the identification of a highly potent lead compound 5f (MIC 0.06 µg/mL) and others with comparable potencies.
Figure 2. Optimization of 2,5,6-Trisubstituted Benzimidazoles.
Chemical Synthesis
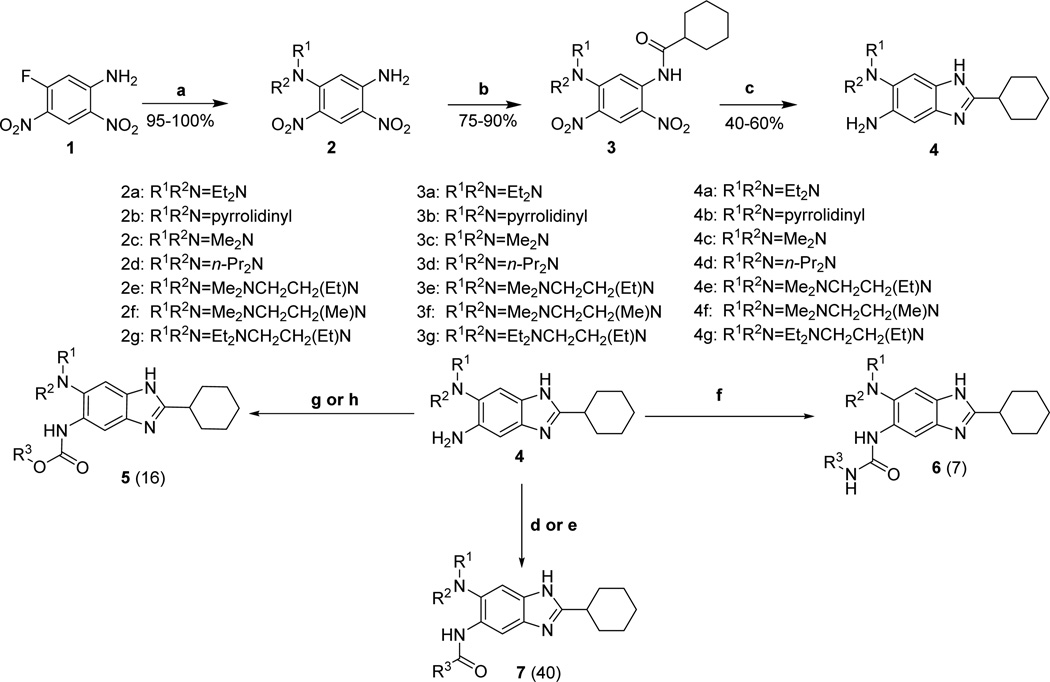
Synthesis of compounds for the optimization library of 2,5,6-trisubstituted benzimidazoles (63 compounds in total) is outlined in Scheme 1, which in most part follows the protocol previously published by our laboratory.22 The aromatic nucleophilic substitution of commercially available 2,4-dinitro-5-fluoroaniline with various amines afforded 5-dialkylaminodinitroanilines 2a–g in 94–98 % yields. The acylation of 2a–g with cyclohexanecarbonyl chloride gave the corresponding N-acylanilines 3a–g in 75–95% yields. One-pot reduction and the subsequent cyclization in the presence of stannous chloride dihydrate and 4 M hydrochloric acid gave 5-aminobenzimidazoles 4a–g in 65–79% yields. The derivatization of 4a–g was carried out using five different methods depending on the functional group to be introduced, and analytically pure compounds 5, 6 and 7 were obtained in 42–97% yields after chromatographic purification. Compounds 5, 6 and 7 bear a carbamate, urea and amide group, respectively, at the 5 position.
Scheme 1. Synthesis of 2,5,6-Trisubstituted Benzimidazoles a22.
a Reagents and conditions: (a) R1R2NH, DIPEA, THF, 2 h, room temperature (RT)/ 12 h, 50 °C; (b) cyclohexanecarbonyl chloride, pyridine, reflux, overnight; (c) SnCl2·2H2O, 4 M HCl, EtOH, reflux,4 h; (d) R3COCl, Et3N, 0 °C-RT, overnight; (e) R3COOH, EDC.HCl, DMAP, CH2Cl2, reflux, overnight; (f) R3NCO, Et3N, reflux, overnight; (g) (i) 1,1’-carbonyldiimidazole, CH2Cl2, reflux, 4 h; (ii) R3OH, CH2Cl2, reflux, overnight; (h) R3OSu ester, CH2Cl2, 0 °C-RT, overnight.
Evaluation of antibacterial activities and SAR study
The 2,5,6-trisubstituted benzimidazoles, 5, 6 and 7 (63 compounds) were evaluated for their activity against Mtb H37Rv using the “Microplate Alamar Blue Assay (MABA)”.36 The antibacterial activities of the compounds are indicated by MIC values.
Since the early lead compounds, 1 (MIC 0.63 µg/mL) and 2 (MIC 0.39 µg/mL), possess a carbamate group at the 5 position, we examined the effect of different carbamate groups at the 5-position as well as different dialkylamino groups at the 6 position on the potency against Mtb (H37Rv). Results are summarized in Table 1.
Table 1.
Antibacterial Activity of Benzimidazoles 5 Against Mtb H37Rv Strain (MIC, µg/mL) and Their Cytotoxicity (IC50, µg/mL)
| Compound | R1R2N | R30 |
MtbH37Rv MIC |
Vero Cytotoxicity |
|---|---|---|---|---|
| 1 |  |
0.63 | >100 | |
| 5a | 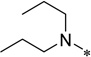 |
6.25 | >100 | |
| 5b | 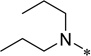 |
 |
>100 | >100 |
| 5c | 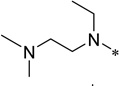 |
100 | >100 | |
| 5d | 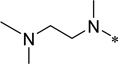 |
>100 | >100 | |
| 5e | 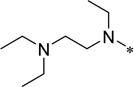 |
50.0 | >50 | |
| 5f | 0.06 | >100 | ||
| 5g | 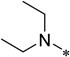 |
>50 | >100 | |
| 5h | 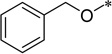 |
0.16 | >100 | |
| 5i | 3.13 | >100 | ||
| 5j | 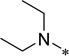 |
1.56 | >100 | |
| 5k | 1.56 | >100 | ||
| 51 | 0.31 | >100 | ||
| 5m | 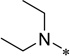 |
12.5 | >100 | |
| 5n | 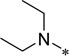 |
1.56 | >100 | |
| 5o | 0.31 | >100 | ||
| 5p | 0.63 | >100 |
As Table 1 shows, the substituents at both 5 and 6 positions have substantial effects on antibacterial activity, but it is very clear that bulky dialkylamino groups at the 6 position are detrimental to the potency of compounds (5b, 5c, 5d, 5e). On the contrary, a smaller dialkyl group, i.e., dimethylamino group, at the 6 position exerts a remarkable enhancement in potency (5f, 5h, 5l, 5p), leading to the discovery of a highly active lead compound, 5f (MIC 0.06 µg/mL), which is 11 times more potent than 1. The nature of carbamate groups at 5 position exhibits moderate effects on the potency. An analog of 1, 5g bearing a branched alkyl carbamate group at the 5-position showed substantially lower potency with an MIC value >50 µg/mL, but another analog 5h with a benzyl carbamate group showed enhanced potency. The introduction of ethyleneglycol moiety into the carbamate group should increase solubility in aqueous media. 2-(Methoxy)ethyl and 2-(ethoxy)ethyl carbamate groups are well tolerated and 5l bearing 5-dimethylamino moiety possesses high potency. However, the use of the MeO(CH2CH2O)2-carbamate group resulted in a substantial decrease in potency (5m). A polar and lipophilic 2,2,2-trifluoroethyl carbamate group is well tolerated (5n, 5o, 5p).
Next, we examined the effect of urea moieties at the 5 position on the antibacterial activity. As Table 2 shows, the introduction of a urea group to the 5 position in place of a carbamate group is detrimental to the potency (6a–f). Nevertheless, compound 6g with a 5-dimethylamino group retains good potency.
Table 2.
Antibacterial Activity of Benzimidazoles 6 Against Mtb H37Rv Strain (MIC, µg/mL) and Their Cytotoxicity (IC50, µg/mL)
| Compound | R1R2N | R3NH |
Mtb H37Rv MIC |
Vera Cytotoxicity |
|---|---|---|---|---|
| 6a | 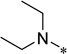 |
>100 | >100 | |
| 6b |  |
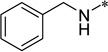 |
>100 | >100 |
| 6c | 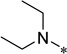 |
>100 | >100 | |
| 6d | 25.0 | >100 | ||
| 6e | 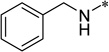 |
25.0 | >100 | |
| 6f | 100 | >100 | ||
| 6g | 6.25 | >100 |
As exemplified in Figure 1, we identified several lead compounds showing good antibacterial activities (MIC 3.1 µg/mL), which bear a benzamide group at the 5 position. In a manner similar to compound 1 and 2, 2-cyclohexyl-6-N,N-diethylamino-5-(4-methoxybenzamido)-1H–benzo[d]imidazole (4) is equally active against drug-sensitive (H37Rv) and drug-resistant clinical isolates of Mtb.22 Since the amide linkage may have better metabolic stability in vivo, we synthesized 40 compounds for the SAR study and evaluated their activity against Mtb H37Rv. Results are summarized in Table 3. Compounds 7a-1∼15 bear a 6-diethylamino group, 7b-1∼7 bear a 6-pyrrolidino group, 7c-1∼17 a 6-dimethylamino group and 7d a 6-dipropylamino group.
Table 3.
Antibacterial Activity of Benzimidazoles 7 Against Mtb H37Rv Strain (MIC, µg/mL) and Their Cytotoxicity (IC50, µg/mL)
| Compound | R1R2N | R3 | Mtb H37Rv | Vero Cytotoxicity |
Compound | R1R2N | R3 |
Mtb H37Rv MIC |
Vero Cytotoxicity |
|---|---|---|---|---|---|---|---|---|---|
| 7a-1 | >100 | >100 | 7b-6 | 6.25 | >100 | ||||
| 7a-2 | >100 | >100 | 7b-7 | 12.5 | >100 | ||||
| 7a-3 | >100 | >100 | 7c-1 | 1.56 | >100 | ||||
| 7a-4 | 12.5 | >100 | 7c-2 | 0.31 | >100 | ||||
| 7a-5 | 1.56 | >100 | 7c-3 | 0.31 | >100 | ||||
| 7a-6 | >100 | n.d* | 7c-4 | 0.16 | >100 | ||||
| 7a-7 | 12.5 | n.d* | 7c-5 | 0.31 | >100 | ||||
| 7a-8 | 10.0 | >100 | 7c-6 | 0.16 | >100 | ||||
| 7a-9 | 100 | >100 | 7c-7 | 0.63 | >100 | ||||
| 7a-10 | >100 | >100 | 7c-8 | 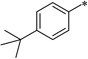 |
1.56 | >100 | |||
| 7a-11 | >100 | >100 | 7c-9 | 5.0 | >100 | ||||
| 7a-12 | >100 | >100 | 7c-10 | 6.25 | >100 | ||||
| 7a-13 | 25.0 | >100 | 7c-11 | 0.31 | >100 | ||||
| 7a-14 | 12.5 | n.d* | 7c-12 | 3.13 | >100 | ||||
| 7a-15 | 25.0 | >100 | 7c-13 | 0.63 | >100 | ||||
| 7b-1 | 6.25 | >100 | 7c-14 | 0.31 | >100 | ||||
| 7b-2 | 6.25 | >100 | 7c-15 | 1.25 | >100 | ||||
| 7b-3 | 3.13 | >100 | 7c-16 | 6.25 | <50 | ||||
| 7b-4 | 1.56 | >100 | 7c-17 | >100 | >100 | ||||
| 7b-5 | 12.5 | >100 | 7d | >100 | >100 |
nd. = not determined
As Table 3 shows, the bulkiness of the 6-dialkylamino group exerts a dramatic influence on the antibacterial activity of these compounds, and it is very clear that the 6-dimethylamino group is by far the best substituent in this library of 2,5,6-trisubstituted benzimidazoles. As anticipated, 5-benzamido-6-dipropylamino-benzimidazole (7d) does not show appreciable activity.
In the 6-diethylamino series of compounds (7a-1∼15), the potency is very sensitive to the substitution pattern of the 5-benzamide moiety. Thus, the compounds with 4-difluoromethoxy-, 4-trifluoromethoxy-, 4-ethoxy-, 4-propyl-, 4-bromo-, 4-fluoro-, 2,4-difluoro- and 3-bromobenzamide moieties at the 5 position do not show appreciable antibacterial activity (MIC >100 µg/mL). 4-Methyl-, 4-ethyl-, 4-dimethylaminobenzamide moieties as well as pyrazin-2-ylcarbonylamino and pentanamide moieties are tolerated. 4-Ethylbenzamide analog of compound 4, 7a-5, shows better potency than the parent compound (MIC 1.56 µg/mL). 4-Dimethylamino- and 3-vinylbenzamide moieties as well as ethoxyacetamide moiety show only modest activities.
In the 6-pyrrolidinylamino series of compounds (7b-1∼7), the potency of the compounds is much less sensitive to the substitution pattern of the 5-amido moiety. Nevertheless, 3-phenylpropanamide and ethoxyacetamide moieties decrease potency. Two compounds bearing 4-methylbenzamide and 4-bromobenzamide moieties, 7b-3 (MIC 3.13 µg/mL) and 7b-4 (MIC 1.56 µg/mL) possess better activity than compound 4.
In the 6-dimethylamino series of compounds (7c-1∼17), all compounds examined possess good to excellent antibacterial activities, wherein 9 compounds have MIC less than 1.0 µg/mL. The results clearly indicate that the introduction of the dimethylamino group to the 6 position is a breakthrough in this SAR study. Also, the 4-substituents on the 5-benzamide moiety are found to be beneficial to increase the potency. The most potent compounds in this series, at present, are 7c-4 and 7c-6 (MIC 0.16 µg/mL). Thus, 7c-4 and 7c-6 have emerged as advanced lead compounds together with 5f for further preclinical drug development. Table 4 lists 9 most potent compounds found based on this SAR study.
Table 4.
Nine Most Potent 6-Dimethylaminobenzimidazoles (Mtb H37Rv, µg/mL)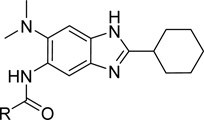
| Compound | R | MIC |
|---|---|---|
| 5f | 0.06 | |
| 5h |  |
0.16 |
| 7c-6 | 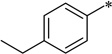 |
0.16 |
| 7c-4 | 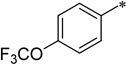 |
0.16 |
| 7c-5 |  |
0.31 |
| 7c-14 | 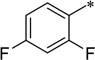 |
0.31 |
| 7c-11 | 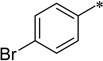 |
0.31 |
| 51 | 0.31 | |
| 5p | 0.63 |
These advanced lead compounds are also highly active against clinical isolates of Mtb, possessing different resistance profiles. Table 5 shows the antibacterial activity of two representative advanced lead compounds, 5f and 7c-6, against four Mtb strains; laboratory strain H37Rv (drug sensitive), clinical strain NHN382 (isoniazide-resistant, Kat G S315t mutation)37, clinical strain TN587 (isoniazide-resistant, Kat G 3315T mutation)37 and clinical strain W210 (drug-sensitive).37 Both 5f and 7c-6 possess substantially higher potency than compound 1 for all strains and more importantly there is no difference in their potencies against drug-resistant Mtb strains. The unbiased ability of these compounds to kill drug-resistant strains of Mtb demonstrates the significance of FtsZ as the drug target to develop new generation anti-TB drugs.
Table 5.
Antibacterial Activity (MIC µg/mL) of 5f and 7c-6 Against Clinical Isolates of Mtb
| Compound |
Mtb strains |
|||
|---|---|---|---|---|
| H37Rv | NHN382 | TN587 | W210 | |
| INH | 0.013 | 0.25 | 0.25 | 0.01 |
| SB-P3G2 | 0.63 | 1.25 | 1.25 | 1.25 |
| 5f | 0.06 | 0.06 | 0.13 | 0.06 |
| 7c-6 | 0.16 | 0.16 | 0.16 | 0.16 |
Mtb-FtsZ Polymerization Inhibitory Assay
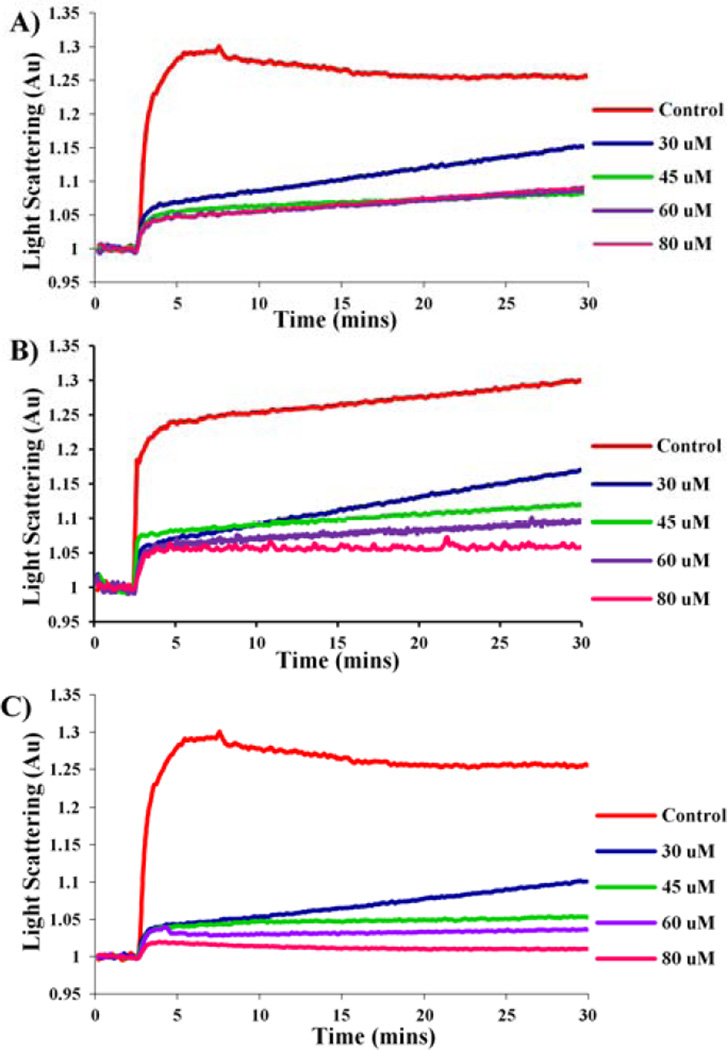
To confirm that this series of compounds actually target Mtb FtsZ, the most potent compound 5f was selected for the light scattering assay.22, 38 In addition, compound 5p and 7c-6 with MIC of 0.63 µg/mL and 0.16 µg/mL, respectively, were also tested. As Figure 3 shows, 5f, 5p and 7c-6 inhibit the assembly of Mtb FtsZ in a dose dependent manner. Since the extent of light scattering correlates to the extent of FtsZ polymerization/aggregation, reduction in light scattering in presence of the compounds demonstrates their inhibitory effect on Mtb FtsZ assembly.
Figure 3. Dose Dependent Inhibition of Mtb FtsZ Polymerization by 5f (A), 5p (B) and 7c-6 (C).
Transmission Electron Microscopy (TEM) Analysis
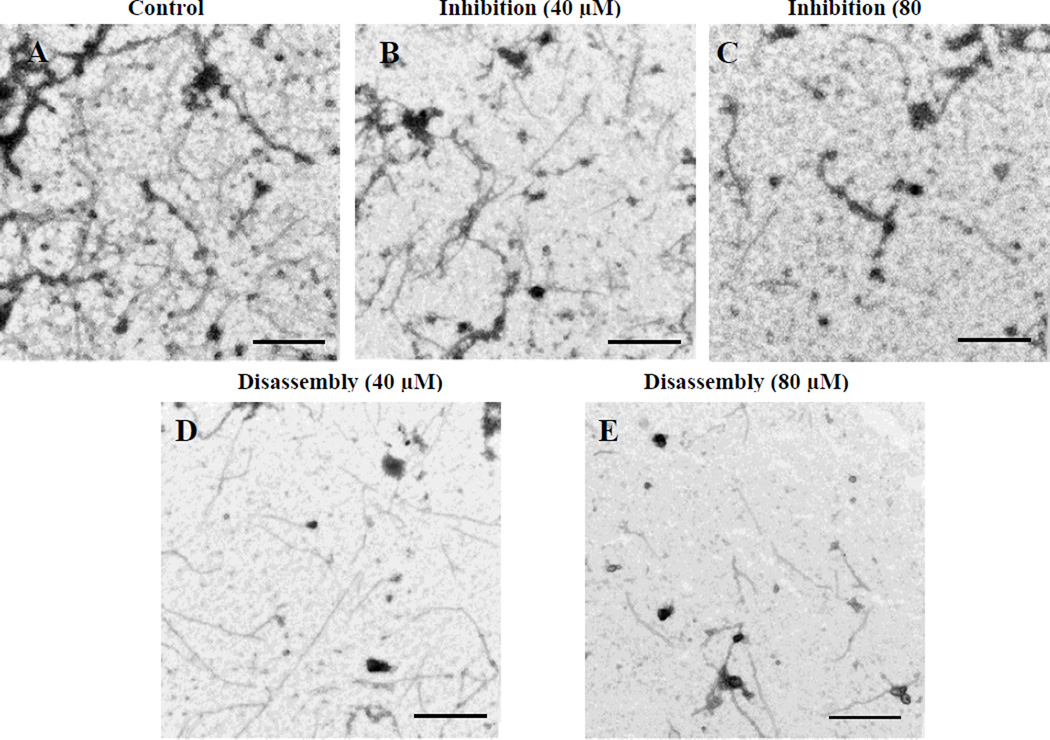
TEM imaging of Mtb FtsZ is also useful to validate the molecular target of lead compounds. 22 Mtb FtsZ (5 µM) was incubated with 40 µM or 80 µM of 5f followed by addition of GTP (25 µM) to initiate polymerization. The TEM images of Mtb FtsZ treated with 40 µM of 5f clearly show reduction in the density and population of FtsZ polymers, protofilaments and aggregates when compared to the untreated protein polymers (Figure 4A, B). The effect is more apparent at 80 µM treatment (Figure 4C). In addition to inhibiting the assembly of Mtb FtsZ in the presence of GTP, 5f was also capable of disrupting the formed FtsZ polymers and aggregates as shown by images D and E. Mtb FtsZ (5 µM) was polymerized by adding GTP (25 µM) and then treated with 40 µM or 80 µM of 5f. The polymerized protein was depolymerized/disassembled after the addition of 5f. These results clearly indicate that 5f interacts with Mtb FtsZ and deters its normal GTP initiated polymerization.
Figure 4. TEM Images of FtsZ.
FtsZ (5 µM) was polymerized by GTP (25 µM) in the absence (A) and presence of 5f at 40 µM (B) and 80 µM (C). Images D and E display the effect of the compound on preformed polymers, wherein FtsZ polymers formed after addition of GTP were treated with 40 µM (D) or 80 µM (E) of 5f. Images are at 49,000x magnification (scale bar 500 nm).
Conclusions
The extensive SAR study on 2-cyclohexyl-5,6-disubstituted benzimidazoles for their antibacterial activities against Mtb H37Rv strain was performed, building upon the identification of promising early lead compounds, 1 and 2, in our previous study. The purpose of this SAR study was to optimize the nitrogen substituents at the 5 and 6 positions of these two lead compounds through systematic modifications. It has been found that the nature of the 6-amino group exerts remarkable effects on the antibacterial activity. For example, long or bulky 6-alkylamino groups are detrimental to the activity. On the contrary, a small dimethylamino group at this position dramatically increases the potency. This breakthrough finding in this SAR study has led to the discovery of 5f with exceptional potency (MIC 0.06 µg/mL), which bears a butoxycarbonylamino group at the 5 position and a dimethylamino group at the 6 position. Also, 12other compounds were found to possess comparable potencies (MIC <0.63 µg/mL), including 7c-6 (MIC 0.16 µg/mL), bearing a 4-ethylbenzamido group at the 5 position and a dimethylamino group at the 6 position. These advanced lead compounds do not show appreciable cytotoxicity against Vero cells (IC50 >100 µg/mL). The advanced leads, 5f and 7c-6, exhibit the same potencies against drug-resistant Mtb clinical isolates, as anticipated. Other important findings in this SAR study include the fact that urea groups at the 5 position are detrimental to the antibacterial activity of this series of benzimidazoles, while carbamate and amide groups are well tolerated although alkylamides and heteroaromatic amides are not favorable. The light scattering assays for FtsZ polymerization inhibition by selected advanced lead compounds, 5f, 5p and 7c-6, show effective dose-dependent inhibition, which validates that Mtb FtsZ is the molecular target of these compounds. The TEM analysis also supports the mechanism of action for these compounds. Furthermore, the TEM analysis has revealed that the pre-formed FtsZ polymers and their aggregates are effectively disassembled by 5f. These observations strongly suggest that the advanced lead benzimidazoles have a novel mechanism of action on the inhibition of Mtb FtsZ assembly and Z-ring formation. Extensive preclinical evaluations for the pharmacological properties of these advanced lead compounds as well as in vivo efficacy evaluations are actively underway, and the results will be reported in due course.
Experimental Section
Methods
1H and 13C NMR spectra were measured on a Brucker or Varian 300, 400 or 500 MHz NMR spectrometer. Melting points were measured on a Thomas Hoover Capillary melting point apparatus and are uncorrected. TLC was performed on silica coated aluminum sheets (thickness 200 µm) or alumina coated (thickness 200 µm) aluminum sheets supplied by Sorbent Technologies and column chromatography was carried out on Siliaflash® P60 silica gel, 40–63 µm (230–400 mesh) supplied by Silicycle. Aluminum oxide, activated, neutral, Brockmann Grade I, 58 Å, was supplied by Alfa Aesar. High-resolution mass spectra were obtained from the Mass Spectrometry Laboratory, University of Illinois at Urbana-Champaign, Urbana, IL. Purity of synthesized compounds was determined by a Shimadzu LC-2010A HT series HPLC assembly or Agilent 1100 series HPLC assembly. Four analytical conditions were used and noted as a part of the characterization data for synthesized compounds. HPLC (1): Adsorbosphere Silica 5 μ, 250 × 4.6 mm column, isopropanol and hexanes, flow rate of 1 mL/min, t = 0–40 min, gradient of 5–50% isopropanol. HPLC (2): Adsorbosphere Silica 5 μ, 250 × 4.6 mm column, isopropanol and hexanes, flow rate of 1.0 mL/min, t = 0–20 min, gradient of 5–50% isopropanol. HPLC (3): Kinetex PFP, 2.6 µm, 4.6 mm × 100 mm column, methanol and water, flow rate of 0.3 mL/min, t = 0–30 min, gradient of 40–95% MeOH. HPLC (4): Kinetex PFP, 2.6 µm, 4.6 mm × 100 mm column, solvent A of water/acetonitrile 95:5 (25 mM ammonium acetate, pH 6.5), solvent B of water/acetonitrile, 5:95 (25mM ammonium acetate, pH 6.5), temperature of 25 °C, flow rate of 1.0 mL/min, t = 0–15 min, gradient of 20–95% solvent B. Measurements were made at 254 nm and 303 nm.
Light scattering assays were performed using a PTI-QM4 spectrofluorimeter. FEI Tecnai12 BioTwinG transmission electron microscope with an AMT XR-60 CCD digital camera system was used to acquire transmission electron microscopy images.
Materials
BL21(DE3) cells, His-Bind protein purification resin and the buffers were purchased from Novagen. Bradford kit for protein concentration determination was purchased from Sigma. Buffer salts (reagent grade or better), solvents (HPLC grade or better), and all the other chemicals were purchased from Fisher Scientific Co. (Pittsburgh, PA). The chemicals were purchased from Aldrich Co., Synquest Inc. and Sigma and purified before use by standard methods. Tetrahydrofuran (THF) was freshly distilled from sodium metal and benzophenone. Dichloromethane was also distilled immediately prior to use under nitrogen from calcium hydride. Compounds, 2a, 2b, 2e-2g, 3a, 3b, 3e-3g, 4a, 4b, and 4e–g were prepared using the procedures previously published by us.22
2,4-Dinitro-5-N,N-di-n-propylaminoaniline (2d)
To a solution of 5-fluoro-2,4-dinitroaniline (1.01 g, 4.97 mmol) and diisopropylethylamine (DIPEA) (1.04 mL, 5.96 mmol) in THF (50 mL) was added di- n-propylamine (818 µL, 5.96 mmol). The reaction mixture was stirred at 50 °C overnight. After completion of the reaction, solvent was evaporated and water was added to give a yellow precipitate. The solution was extracted with dichloromethane (30 mL × 3). The organic layer was collected, dried over anhydrous magnesium sulfate and evaporated to give 2d as a yellow solid (1.25 g, 89% yield); mp 68–70 °C; 1H NMR (400 MHz, CDCl3) δ 0.86 (t, 6 H, J = 7.4 Hz), 1.60 (m, 4 H), 3.12 (t, 4 H, J = 7.28 Hz), 6.09 (s, 1 H), 6.40 (br. s, NH), 8.75 (s, 1 H); 13C NMR (100 MHz, CDCl3) δ 11.28, 20.45, 53.51, 102.9, 123.4, 128.3, 131.7, 147.6, 150.1; MS (ESI) m/z 283.1 (M+1)+.
Same procedure was used for the synthesis of 2c.
5-N,N-Dimethylamino-2,4-dinitroaniline (2c)
Yellow solid (98% yield); mp 164–165 °C; 1H NMR (300 MHz, CDCl3) δ 2.94 (br. s, 6 H), 6.01 (br. s, 1 H), 6.41 (br. s, 2H), 8.8 (br. s, 1 H); 13C NMR (101 MHz, CDCl3) δ 42.5 100.7, 123.6, 128.8, 130.5, 147.9, 150.7; HRMS (ESI) m/z calcd for C8H10N4O4 H+: 227.0775, Found: 227.0778 (Δ = 1.4 ppm).
1-Cyclohexylamido-2,4-dinitro-5-N,N-di-n-propylaminobenzene (3d)
A solution of 2d (1.25 g, 4.40 mmol) and cyclohexanecarbonyl chloride (0.721 mL, 5.34 mmol) in pyridine (10 mL) was refluxed overnight. After completion of the reaction, pyridine was removed in vacuo to give a brown solid. This crude mixture was washed with methanol and collected by filteration to afford 3d as a yellow solid which was used directly in the following step. MS (ESI) m/z 393.2 (M+1)+.
Same procedure was used for the synthesis of 3c.
1-Cyclohexylamido-5-N,N-dimethylamino-2,4-dinitrobenzene (3c)
Yellow solid (90% yield); mp 148–149 °C; 1H NMR (300 MHz, CDCl3) δ 1.21–1.43 (m, 3 H), 1.67–1.79 (m, 1 H), 1.80–1.94 (m, 2 H), 1.97–2.09 (m, 2 H), 2.32–2.45 (m, 1 H), 3.06 (s, 6 H), 8.60 (s, 1 H), 8.85 (s, 1 H), 11.00 (s, 1 H); 13C NMR (101 MHz, CDCl3) δ 25.5, 25.6, 29.4, 42.5, 47.3, 106.1, 125.0, 127.4, 131.3, 138.7, 150.1, 175.9; HRMS (ESI) m/z calcd for C15H20N4O5H+: 337.1507, Found: 337.1511 (Δ = 1.3 ppm).
5-Amino-2-cyclohexyl-6-N,N-di-n-propylamino-1H-benzo[d]imidazole (4d)
To a solution of 3d from previous step (1.75 g, 4.45 mmol) in ethanol (50 mL) was added solid stannous chloride dihydrate (7.03 g, 31.1 mmol). Concentrated hydrochloric acid (15 mL) was added to the reaction mixture such that the final concentration of HCl is 4 M in the reaction flask. The reaction mixture was refluxed for 4 h. Upon completion, the reaction mixture was basified with 30% sodium hydroxide solution. Excess stannous chloride formed a soluble salt in excess base. The desired product was extracted with dichloromethane and purified by flash chromatography on silica gel using ethyl acetate as eluent to afford 4d as a pale green solid (721 mg, 55% yield in two steps); mp 76–78 °C; 1H NMR (400 MHz, CDCl3) δ 0.78 (t, 6 H, J = 7.4 Hz), 1.20–1.23 (m, 3 H), 1.35–1.40 (m, 4 H), 1.59–1.63 (m, 5 H), 2.01 (d, 2 H, J = 7.28 Hz), 2.77 (m, 5 H), 6.80 (s, 1 H), 7.25 (s, 1 H); 13C NMR (100 MHz, CDCl3) δ 11.60, 20.45, 25.64, 25.91, 31.84, 38.43, 57.04, 98.19, 110.0, 132.9, 134.3, 135.0, 140.0, 158.0; HRMS (ESI) m/z calcd for C19H30N4H+: 315.2543, Found: 315.2549 (Δ = 1.9 ppm).
Same procedure was used for the synthesis of 4c.
5-Amino-2-cyclohexyl-6-N,N-dimethylamino-1H-benzo[d]imidazole (4c)
Light brown solid (69% yield); mp 135–137 °C; 1H NMR (300 MHz, CDCl3) δ 1.14–1.46 (m, 4 H), 1.54–1.86 (m, 5 H), 1.99–2.17 (m, 2 H), 2.64 (s, 6 H), 2.73–2.87 (m, 1 H), 6.81 (s, 5 H), 7.26 (s, 1 H); 13C NMR (101 MHz, CDCl3) δ 26.0, 26.2, 32.2, 38.8, 44.8, 98.9, 106.7, 133.1, 134.5, 138.0, 138.3, 158.2; HRMS (ESI) m/z calcd for C15H22N4H+: 259.1917, Found: 259.1914 (Δ = –1.2 ppm).
2-Cyclohexyl-5-(3-propylurea)-6-pyrrolidinyl-1H-benzo[d]imidazole (6f)
To a solution of 4b (100 mg, 0.35 mmol) in dichloromethane (DCM) (3 mL), was added n-propylisocyanate (32.9 mg, 0.39 mmol) and triethylamine (0.39 mmol). The reaction was refluxed for 14 h and then diluted with ethyl acetate, transferred to a separatory funnel and washed with water, sat. sodium bicarbonate solution and brine. The organic layer was dried over magnesium sulfate and concentrated using rotary evaporator. The crude product was purified by flash chromatography (column was packed with DCM, 2% methanol was used steadily increasing to 5% methanol) to obtain the product as a white solid (80% yield); mp 231–232 °C; 1H NMR (400 MHz, Methanol-d4) δ 0.95 (t, J = 7.2 Hz, 3 H), 1.26–1.69 (m, 8 H), 1.75–1.78 (m, 1 H), 1.84–1.88 (m, 2 H), 1.92–2.11 (m, 6 H), 2.83 (m, 1 H), 3.04 (s, 4 H), 3.16 (t, J = 7.03 Hz, 2 H), 7.24 (s, 1 H), 7.92 (s, 1 H); 13C NMR (126 MHz, Methanol-d4) δ 11.9, 24.6, 25.5, 27.1, 27.4, 33.0, 40.0, 42.9, 53.9, 106.3, 107.1, 130.9, 134.5, 136.5, 139.2, 159.2, 160.5; HRMS (FAB) m/z calcd for C21H31N5OH+: 370.2601, Found: 370.2611 (Δ = 2.6 ppm); HPLC (1): t =17.7 min, purity > 99%.
Same procedure was followed for the synthesis of 6a-6g
5-(4-Butylurea)-2-cyclohexyl-6-N,N-diethylamino-1H-benzo[d]imidazole (6a)
White solid (70% yield); mp 197–198 °C; 1H NMR (500 MHz, Methanol-d4) δ 0.95–1.10 (m, 9 H), 1.36–1.66 (m, 7 H), 1.67–1.79 (m, 2 H), 1.80–1.90 (m, 1 H), 1.90–2.01 (m, 2 H), 2.14 (d, J = 13.73 Hz, 2 H), 2.93 (m, 1 H), 3.06 (quin, J = 7.20 Hz, 4 H), 3.29 (q, J = 7.20 Hz, 2 H), 7.42 (d, J = 7.02 Hz, 1 H), 8.27 (d, J = 7.63 Hz, 2 H); 13C NMR (126 MHz, Methanol-d4) δ 13.0, 13.01 14.3, 21.3, 27.1, 27.4, 33.0, 33.5, 40.0, 40.7, 51.7, 103.2, 111.3, 135.3, 135.9, 136.2, 158.7, 160.7; HRMS (FAB) m/z calcd for C22H35N5OH+: 386.2914, Found: 386.2911 (Δ = –0.9 ppm); HPLC (1): t =12.7 min, purity > 99%.
5-(Benzylurea)-2-cyclohexyl-6-N,N-diethylamino-1H-benzo[d]imidazole (6b)
White solid (94% yield); mp 198–199 °C; 1H NMR (400 MHz, Methanol-d4) δ 0.90 (t, J = 7.15 Hz, 6 H), 1.23–1.52 (m, 4 H), 1.57–1.70 (m, 2 H), 1.77 (d, J = 14.31 Hz, 1 H), 1.86 (d, J = 13.05 Hz, 2 H), 1.98–2.12 (m, 2 H), 2.84–2.87 (m, 1 H), 2.95 (q, J = 7.00 Hz, 4 H), 4.34–4.45 (m, 2 H), 7.19–7.26 (m, 1 H), 7.28–7.39 (m, 5 H), 8.22 (s, 1 H); 13C NMR (101 MHz, Methanol-d4) δ 13.0, 27.1, 27.3, 33.0, 40.0, 44.8, 51.6, 103.4, 111.3, 128.2, 128.4, 129.7, 135.1, 136.0, 136.3, 141.3, 158.5, 160.8; HRMS (FAB) m/z calcd for C25H33N5OH+: 420.2758, Found: 420.2761 (Δ = 0.7 ppm); HPLC (1): t =13.4 min, purity > 99%.
2-Cyclohexyl-6-N,N-diethylamino-5-(3-propylurea)-1H-benzo[d]imidazole (6c)
White solid (87% yield); mp 216–217 °C; 1H NMR (400 MHz, Methanol-d4) δ 0.87–1.01 (m, 9 H), 1.23–1.50 (m, 4 H), 1.51–1.70 (m, 4 H), 1.71–1.79 (m, 1 H), 1.85 (dt, J = 13.18, 3.33 Hz, 2 H), 2.05 (d, J = 13.40 Hz, 2 H), 2.84–2.88 (m, 1 H), 2.96 (q, J = 7.2 Hz, 4 H), 3.16 (t, J = 7.20 Hz, 2 H), 7.33 (s, 1 H), 8.20 (s, 1 H); 13C NMR (101 MHz, Methanol-d4) δ 11.9, 13.0, 24.5, 27.1, 27.4, 33.0, 40.0, 42.8, 51.6, 103.2, 111.3, 135.3, 136.0,136.2, 158.7, 160.7; HRMS (FAB) m/z calcd for C21H33N5OH+: 372.2758, Found: 372.2754 (Δ = − 1.0 ppm); HPLC (1): t =12.6 min, purity > 99%.
5-(4-Butylurea)-2-cyclohexyl-6-pyrrolidinyl-1H-benzo[d]imidazole (6d)
White solid (96% yield); mp 206–207 °C; 1H NMR (400 MHz, Methanol-d4) δ 0.95 (t, J = 7.2 Hz, 3 H), 1.26–1.55 (m, 7 H), 1.56–1.70 (m, 2 H), 1.71–1.82 (m, 1 H), 1.82–1.91 (m, 2 H), 1.91–2.11 (m, 6 H), 2.79–2.87 (m, 1 H), 3.03 (br. s, 4 H), 3.20 (t, J = 7.2 Hz, 2 H), 7.24 (s, 1 H), 7.94 (s, 1 H); 13C NMR (101 MHz, Methanol-d4) δ 14.3, 21.2, 27.12, 25.5, 27.3, 33.0, 33.5, 39.9, 40.8, 53.9, 106.3, 107.1, 131.0, 134.4, 136.4, 139.3, 159.2, 160.4; HRMS (FAB) m/z calcd for C22H33N5OH+: 384.2758, Found: 384.2755 (Δ = −0.8 ppm); HPLC (1): t =14.2 min, purity > 99%.
5-(4-Benzylurea)-2-cyclohexyl-6-pyrrolidinyl-1H-benzo[d]imidazole (6e)
White solid (91% yield); mp 225–226 °C; 1H NMR (500 MHz, Methanol-d4) δ 1.26–1.49 (m, 3 H), 1.56–1.68 (m, 2 H), 1.71–1.79 (m, 1 H), 1.80–1.89 (m, 2 H), 1.92 (br. s, 4 H), 1.98–2.10 (m, 2 H), 2.80–2.85 (m, 1 H), 3.01 (br. s, 4 H), 4.39 (s, 2 H), 7.18–7.26 (m, 2 H), 7.26–7.37 (m, 4 H), 7.96 (s, 1 H); 13C NMR (126 MHz, Methanol-d4) δ 25.5, 27.1, 27.3, 33.0, 39.9, 44.8, 53.8, 106.2, 107.4, 128.2, 128.4, 129.7, 130.7, 134.4, 136.5, 139.4, 141.3, 159.1, 160.5; HRMS (FAB) m/z calcd for C25H31N5OH+: 418.2601, Found: 418.2604 (Δ = 0.6 ppm); HPLC (1): t =14.5 min, purity > 99%.
5-(4-Butylurea)-2-cyclohexyl-6-N,N-dimethylamino-1H-benzo[d]imidazole (6g)
White solid (74% yield); mp 185–186 °C; 1H NMR (400 MHz, Methanol-d4) δ 1.71 (t, J = 7.28 Hz, 3 H), 1.98–2.27 (m, 7 H), 2.30–2.44 (m, 2 H), 2.49 (d, J = 12.30 Hz, 1 H), 2.54–2.65 (m, 2 H), 2.73–2.85 (m, 2 H), 3.38 (s, 6 H), 3.57 (m, 1 H), 3.89 (q, J = 6.53 Hz, 2 H), 4.15 (br. s, 1 H), 7.80 (br. s, 1 H), 8.05 (br. s, 1 H), 8.83 (br. s, 1 H), 8.97 (s, 1 H), 12.54 (br. s, 1 H); 13C NMR (126 MHz, Methanol-d4) δ 14.3, 21.2, 27.0, 27.3, 32.9, 33.5, 39.7, 40.7, 45.8, 104.3, 107.7, 132.3, 134.3, 135.0, 141.4, 158.8, 160.1; HRMS (FAB) m/z calcd for C20H31N5OH+: 358.2601, Found: 358.2609 (Δ = 2.1 ppm); HPLC (1): t =15.3 min, purity > 99%.
5-Butoxycarbonylamino-2-cyclohexyl-6-N,N-dipropylamino-1H-benzo[d]imidazole (5a)
To a solution of 4d (124 mg, 0.39 mmol) in dichloromethane (5 mL) was added a solution of N-butoxycarbonyloxysuccinimide (80 mg, 0.39 mmol) in dichloromethane (10 mL) dropwise at room temperature. The reaction mixture was stirred at room temperature overnight and after completion of the reaction, the reaction mixture was concentrated. The crude was purified via flash chromatography on silica gel (gradient 20–40% EtOAc/hexanes) to give 5a as white solid (72 mg, 33% yield): mp 128–130 °C; 1H NMR (400 MHz, CDCl3) δ 0.83 (t, 6 H, J = 7.2 Hz), 0.94 (t, 3 H, J = 7.5 Hz), 1.33 (m, 9 H), 1.65–1.69 (m, 5 H), 1.80–1.82 (m, 2 H, J = 10 Hz), 2.05–2.07 (m, 2 H, J = 10 Hz), 2.79–2.81 (m, 5 H), 4.17 (t, 2 H, J = 6.8 Hz), 7.47 (s, 1 H), 8.22 (s, 1 H), 8.54 (s, 1 H); 13C NMR (100 MHz, CDCl3) δ 11.70, 13.74, 19.09, 20.61, 25.78, 26.00, 31.04, 31.76, 38.42, 58.59, 64.82, 99.39, 112.6, 131.9, 135.4, 154.1, 158.8, 173.3; HRMS (ESI) m/z calcd for C24H38N4O2 H+: 415.3068, Found: 415.3073 (Δ = 1.3 ppm); HPLC (1): t =6.1 min, purity > 97%.
Same procedure was used for the synthesis of 5b.
5-Benzyloxycarbonylamino-2-cyclohexyl-6-N,N-dipropylamino-1H-benzo[d]imidazole (5b)
White solid (51% yield): mp 138–140 °C; 1H NMR (400 MHz, CDCl3) δ 0.80 (t, 6 H, J = 7.2 Hz), 1.32–1.34 (m, 7 H), 1.59–1.62 (m, 3 H), 1.77–1.79 (m, 2 H, J = 10 Hz), 2.04–2.06 (m, 2 H, J = 10 Hz), 2.77–2.80 (m, 5 H), 5.23 (s, 2 H), 7.37 (m, 5 H), 7.49 (s, 1 H), 8.24 (s, 1 H), 8.67 (s, 1 H), 9.84 (s, 1 H); 13C NMR (100 MHz, CDCl3) δ 11.70, 20.57, 25.77, 25.96, 31.76, 38.42, 58.66, 66.57, 98.87, 113.3, 127.9, 128.1, 128.5, 131.7, 135.3, 136.4, 153.6, 158.9; HRMS (ESI) m/z calcd for C27H36N4O2H+: 449.2911, Found: 449.2909 (Δ = −0.4 ppm); HPLC (1): t =6.5 min, purity > 99%.
5-Butyloxycarbonylamino-2-cyclohexyl-6-N,N-dimethylamino-1H-benzo[d]imidazole (5f)
A solution of 4c (100 mg, 0.39 mmol) and 1,1’-carbonyldiimidazole (69 mg, 0.43 mmol) in dichloromethane (2 mL) was stirred under reflux conditions for 4 hours. After all the starting material had disappeared, n-butanol (71 µL, 0.77 mmol) was added and the reaction was refluxed for additional 12 hours. After the completion of the reaction, the reaction mixture was concentrated. The crude was purified by flash chromatography on silica gel (gradient: 30–70% ethyl acetate /hexanes) to give 5f as a white solid (111 mg, 79% yield); mp 182–183 °C; 1H NMR (300 MHz, CDCl3) δ 0.97 (t, J = 7.28 Hz, 3 H), 1.23–1.51 (m, 6 H), 1.55–1.78 (m, 5 H), 1.79–1.91 (m, 2 H), 2.10 (d, J = 12.91 Hz, 2 H), 2.64 (s, 6 H), 2.76–2.92 (m, 1 H), 4.20 (t, J = 6.73 Hz, 2 H), 7.55 (br. s, 1 H), 8.20 (br. s, 2 H), 9.20 (br. s, 1 H); 13C NMR (101 MHz, CDCl3) δ 14.0, 19.3, 26.0, 26.2, 31.3, 32.0, 38.6, 45.8, 65.2, 100.9, 109.5, 129.8, 132.1, 136.6, 139.1, 154.4, 159.0; HRMS (FAB) m/z calcd for C20H30N4O2H+: 359.2442, Found: 359.2443 (Δ = 0.4 ppm); HPLC (1): t =8.9 min, purity > 99%.
Same procedure was used for the synthesis of 5c-5p.
5-Butyloxycarbonylamino-2-cyclohexyl-6-N,N-dimethyl-N-ethylethylenediamino-1H-benzo[d]imidazole (5c)
White solid (44% yield): mp 141–143 °C; 1H NMR (400 MHz, CDCl3) δ 0.86 (t, 3 H, J = 7.2 Hz), 0.94 (t, 3 H, J = 7.2 Hz), 1.23–1.27 (m, 3 H), 1.41–1.43 (m, 2 H), 1.67–1.75 (m, 6 H), 2.00 (m, 2 H, J = 10 Hz), 2.19 (s, 6 H), 2.74–2.75 (m, 1 H), 2.92–2.93 (m, 4 H), 4.16 (t, 2 H, J = 7.2 Hz), 7.47 (s, 1 H), 8.24 (s, 1 H), 9.88 (s, 1 H); 13C NMR (100 MHz, CDCl3) δ 13.08, 13.73, 19.12, 25.77, 26.01, 31.12, 31.70, 38.48, 45.03, 50.99, 55.03, 57.04, 64.51, 100.2, 113.8, 131.8, 132.9, 135.0, 138.8, 154.9, 159.2; HRMS (ESI) m/z calcd for C24H39N5O2H+: 430.3176, Found: 430.3179 (Δ = 0.7 ppm); HPLC (3): t =23.8 min, purity > 98%.
5-Butyloxycarbonylamino-2-cyclohexyl-6-N,N,N-trimethylethylenediamino-1H-benzo[d]imidazole (5d)
White solid (45% yield); mp 116–118 °C; 1H NMR (400 MHz, CDCl3) δ 0.95 (t, 3 H, J = 7.2 Hz), 1.24–1.45 (m, 6 H), 1.60–1.68 (m, 5 H), 1.79–1.80 (m, 2 H), 2.03–2.04 (m, 2 H), 2.24 (s, 6 H), 2.31 (t, 2 H, J = 7.2 Hz), 2.68 (s, 3 H), 2.84–2.85 (m, 3 H), 4.17 (t, 2 H, J = 6.8 Hz), 7.49 (s, 1 h), 8.26 (s, 1 H), 9.80 (s, 1 H); 13C NMR (100 MHz, CDCl3) δ 13.77, 19.15, 25.81, 26.01, 31.16, 31.77, 38.45, 43.65, 45.41, 57.02, 57.23, 64.55, 100.0, 112.3, 131.2, 137.8, 154.8, 158.8; HRMS (ESI) m/z calcd for C23H37N5O2H+: 416.3020, Found: 416.3031 (Δ = 2.6 ppm); HPLC (1): t =6.5, purity > 98%.
5-Butyloxycarbonylamino-2-cyclohexyl-6-N,N,N-triethylethylenediamino-1H-benzo[d]imidazole (5e)
White solid (60% yield); mp 150–151 °C; 1H NMR (400 MHz, CDCl3) δ 0.95 (t, 3 H, J = 7.2 Hz), 0.97–1.00 (m, 9 H), 1.24–1.48 (m, 6 H), 1.57–1.70 (m, 6 H), 1.78–1.79 (m, 2 H), 2.02–2.05 (m, 2 H), 2.37 (t, 2 H, J = 7.2 Hz), 2.50–2.55 (m, 4 H, J = 7.2 Hz), 2.76–2.79 (m, 1 H), 2.93–2.95 (m, 4 H), 4.17 (t, 2 H, J = 6.9 Hz), 7.52 (s, 1 H), 8.23 (s, 1 H), 9.84 (s, 1 H), 10.5 (s, 1 H); 13C NMR (100 MHz, CDCl3) δ 11.10, 12.96, 13.73, 19.12, 25.79, 26.03, 31.30, 31.70, 38.48, 46.81, 50.41, 50.67, 55.34, 64.56, 100.5, 113.6, 131.6, 132.7, 135.4, 138.9, 154.9, 159.1; HRMS (ESI) m/z calcd for C26H43N5O2H+: 458.3489, Found: 458.3489 (Δ = 0.0 ppm); HPLC (1): t =25.5 min, purity > 99%.
2-Cyclohexyl-6-N,N-diethylamino-5-(1-methylbutoxycarbonylamino)-1H-benzo[d]imidazole (5g)
White solid (50% yield): mp 88–90 °C; 1H NMR (400 MHz, CDCl3) δ 0.88–0.92 (m, 9 H), 1.37–1.63 (m, 16 H), 1.79–1.81 (m, 2 H, J = 10 Hz), 2.07–2.10 (m, 2 H, J = 10 Hz), 2.83–2.93 (m, 5 H), 4.90–4.93 (m, 1 H), 7.45 (s, 1 H), 8.22 (s, 1 H), 8.41 (s, 1 H); 13C NMR (100 MHz, CDCl3) δ 12.68, 13.96, 18.72, 20.34, 25.76, 25.97, 29.67, 31.74, 38.35, 50.41, 71.46, 99.98, 112.2, 132.6, 134.5, 153.9, 158.6; HRMS (ESI) m/z calcd for C23H36N4O2H+: 401.2911, Found: 401.2917 (Δ = 1.5 ppm); HPLC (1): t =6.8 min, purity > 97%.
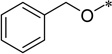
5-Benzyloxycarbonylamino-2-cyclohexyl-6-N,N-dimethylamino-1H-benzo[d]imidazole (5h)
White solid (82% yield); mp 145–146 °C; 1H NMR (300 MHz, CDCl3) δ 1.15–1.40 (m, 3 H), 1.45–1.87 (m, 5 H), 2.03 (d, J = 11.73 Hz, 2 H), 2.60 (s, 6 H), 2.74–2.92 (m, 1 H), 5.23 (s, 2 H), 7.28–7.60 (m, 6 H), 8.14–8.42 (m, 2 H), 10.23 (br. s, 1 H); 13C NMR (126 MHz, CDCl3) δ 21.8, 25.9, 26.1, 31.80, 31.84, 38.3, 38.3, 45.8, 67.1, 100.9, 109.5, 128.5, 128.8, 129.9, 131.6, 136.0 136.4, 139.4, 153.9, 158.9, 175.9; HRMS (FAB) m/z calcd for C23H28N4O2H+: 393.2285, Found: 393.2290 (Δ = 1.3 ppm); HPLC (1): t =9.0 min, purity > 99%.
2-Cyclohexyl-5-methoxyethoxycarbonylamino-6-pyrrolidinyl-1H-benzo[d]imidazole (5i)
White solid (55% yield): mp 75–77 °C; 1H NMR (400 MHz, CDCl3) δ 1.25–1.43 (m, 3 H), 1.57–1.93 (m, 6 H), 1.88 (br. s, 4 H), 2.00–2.02 (m, 2 H, J = 10 Hz), 2.76–2.77 (m, 1 H), 2.89 (br. s, 4 H), 3.39 (s, 3 H), 3.64 (t, 2 H, J = 6.5 Hz), 4.32 (t, 2 H, J = 6.5 Hz), 7.37 (s, 1 H), 8.09 (br. s, 1 H), 8.16 (br. s, 1 H); 13C NMR (100 MHz, CDCl3) δ 24.22, 25.66, 25.88, 31.70, 38.37, 53.55, 58.87, 63.89, 70.81, 101.6, 108.7,129.4, 132.3, 135.7, 136.9, 153.8, 159.1; HRMS (ESI) m/z calcd for C21H30N4O3H+: 387.2390, Found: 387.2400 (Δ = 2.6 ppm); HPLC (1): t =14.2 min, purity > 97%.
2-Cyclohexyl-5-ethoxyethoxycarbonylamino-6-N,N-diethylamino-1H-benzo[d]imidazole (5j)
White solid (42% yield); mp 158–160 °C; 1H NMR (400 MHz, CDCl3) δ 0.84 (t, 6 H, J = 7.2 Hz), 1.19–1.22 (m, 5 H), 1.57–1.60 (m, 3 H), 1.74–1.77 (m, 2 H), 2.02–2.05 (m, 2 H, J = 10 Hz), 2.78–2.88 (m, 5 H), 3.52–3.57 (m, 2 H, J = 7 Hz), 3.68–3.69 (m, 2 H), 4.33 (t, 2 H, J = 6.5 Hz), 7.40 (s, 1 H), 8.23 (br. s, 1 H), 8.58 (s, 1 H); 13C NMR (100 MHz, CDCl3) δ 12.49, 15.03, 25.72, 25.94, 31.74, 38.44, 50.34, 64.10, 66.61, 68.73, 99.27, 112.9, 131.9, 134.2, 138.0, 153.7, 159.1; HRMS (ESI) m/z calcd for C22H34N4O3H+: 403.2703, Found: 403.2711 (Δ = 1.9 ppm); HPLC (1): t =9.1 min, purity > 97%.
2-Cyclohexyl-5-ethoxyethoxycarbonylamino-6-pyrrolidinyl-1H-benzo[d]imidazole (5k)
White solid (56% yield); mp 72–73 °C; 1H NMR (400 MHz, CDCl3) δ 1.19–1.22 (m, 6 H), 1.54–1.57 (m, 3 H), 1.72–1.75 (m, 2 H), 1.89 (br. s, 4 H), 1.99–2.02 (m, 2 H, J = 10 Hz), 2.76–2.78 (m, 1 H), 2.90 (br. s, 4 H), 3.51–3.56 (m, 2 H, J = 7.2 Hz), 3.68 (t, 2 H, J = 6.5 Hz), 4.32 (t, 2 H, J = 6.5 Hz), 7.38 (s, 1 H), 8.08 (bs, 1 H), 8.16 (bs, 1 H); 13C NMR (100 MHz, CDCl3) δ 15.01, 24.25, 25.69, 25.91, 31.72, 38.38, 53.57, 64.14, 66.59, 68.73, 101.5, 108.8, 129.5, 132.1, 135.7, 136.8, 153.8, 159.0; HRMS (ESI) m/z calcd for C22H32N4O3H+: 401.2547, Found: 401.2550 (Δ = 0.7 ppm); HPLC (1): t =11.8 min, purity > 99%.
2-Cyclohexyl-5-ethoxyethoxycarbonylamino-6-N,N-dimethylamino-1H-benzo[d]imidazole (5l)
White solid (86% yield); mp 158–159 °C; 1H NMR (500 MHz, CDCl3) δ 1.20–1.44 (m, 7 H), 1.62 (qd, J = 12.31, 3.05 Hz, 2 H), 1.72 (d, J = 12.82 Hz, 1 H), 1.83 (d, J = 13.12 Hz, 2 H), 2.09 (d, J = 12.51 Hz, 2 H), 2.61 (s, 6 H), 2.77–2.93 (m, 1 H), 3.58 (q, J = 5.00 Hz, 2 H), 3.71 (t, J = 5.00 Hz, 2 H), 4.36 (t, J = 5.00 Hz, 2 H), 7.51 (br. s, 1 H), 8.07–8.48 (m, 2 H), 9.40 (br. s, 1 H); 13C NMR (126 MHz, CDCl3) δ 15.3, 26.0, 26.2, 29.9, 32.0, 38.6, 45.9, 64.4, 66.9, 69.0, 99.5, 111.1, 129.7, 139.0, 154.0, 159.0; HRMS (FAB) m/z calcd for C20H30N4O3H+: 375.2391, Found: 375.2410 (Δ = 5.1 ppm); HPLC (1): t =12.7 min, purity > 99%.
2-Cyclohexyl-6-N, N-diethylamino-5-(2-methoxyethoxyethoxycarbonylamino)-1H-benzo[d]imidazole (5m)
White solid (52% yield); mp 109–110 °C; 1HNMR (400 MHz, CDCl3) δ 0.87 (t, 6 H, J = 7.2 Hz), 1.25–1.32 (m, 3 H), 1.58–1.63 (m, 3 H), 1.70–1.78 (m, 2 H), 2.05–2.08 (m, 2 H, J = 10 Hz), 2.87–2.88 (m, 5 H), 3.37 (s, 3 H), 3.57 (bs, 2 H), 3.67 (t, 2 H, J = 7 Hz), 3.76 (t, 2 H, J = 7 Hz), 4.34 (t, 2 H, J = 6.5 Hz), 7.42 (s, 1 H), 8.22 (bs, 1 H), 8.56 (s, 1 H); 13C NMR (100 MHz, CDCl3) δ 12.53, 25.75, 25.95, 31.74, 38.42, 50.34, 58.98, 63.92, 69.58, 70.43, 71.84, 99.81, 112.5, 132.0, 134.2, 153.6, 159.0; HRMS (ESI) m/z calcd for C23H36N4O4H+: 433.2809, Found: 433.2815 (Δ = 1.4 ppm); HPLC (1): t =16.9 min, purity > 97%.
2-Cyclohexyl-6-N,N-diethylamino-5-trifluoroethoxycarbonylamino-1H-benzo[d]imidazole (5n)
White solid (42% yield); mp 203–204 °C; 1H NMR (400 MHz, CDCl3) δ 0.90 (t, 6 H, J = 6.9 Hz), 1.25–1.37 (m, 4 H), 1.61–1.65 (m, 4 H), 1.81–1.84 (m, 2 H), 2.09–2.10 (m, 2 H), 2.85–2.94 (m, 5H), 4.53–4.57 (m, 2 H, J = 8.5 Hz), 7.51 (s, 1 H), 8.13 (s, 1 H), 8.78 (s, 1 H), 9.76 (s, 1 H); 13C NMR (100 MHz, CDCl3) δ 12.69, 25.81, 25.99, 31.83, 38.49, 50.59, 98.99, 113.5, 119.0, 121.7, 124.5, 127.3, 131.1, 131.4, 139.0, 151.5, 159.2; HRMS (ESI) m/z calcd for C20H27F3N4O2H+: 413.2159, Found: 413.2175 (Δ = 3.8 ppm); HPLC (1): t =7.3 min, purity > 99%.
2-Cyclohexyl-5-trifluoroethoxycarbonylamino-6-prrolidinyl-1H-benzo[d]imidazole (5o)
White solid (69% yield); mp 123–125 °C; 1H NMR (400 MHz, CDCl3) δ 1.16–1.31 (m, 3 H), 1.59–1.62 (m, 3 H), 1.75–1.78 (m, 2 H), 1.92 (bs, 4 H), 2.03–2.07 (m, 2H), 2.79–2.82 (m, 1 H), 2.92–2.93 (m, 4 H), 4.52–4.59 (q, 2 H, J = 7.4 Hz), 7.41 (s, 1 H), 8.12 (s, 1 H), 8.21 (s, 1 H); 13C NMR (100 MHz, CDCl3) δ 24.30, 25.70, 25.92, 31.82, 38.48, 53.65, 102.0, 108.8, 118.9, 121.7, 124.4, 127.2, 128.7, 135.9, 151.7, 159.4; HRMS (ESI) m/z calcd for C20H25F3N4O2H+: 411.2002, Found: 411.2007 (Δ = 1.2 ppm); HPLC (1): t =7.1 min, purity > 97%.
2-Cyclohexyl-5-trifluoroethoxycarbonylamino-6-N,N-dimethylamino-1H-benzo[d]imidazole (5p)
White solid (70% yield); mp 190–191°C;1H NMR (400 MHz, CDCl3) δ 1.05–1.43 (m, 4 H), 1.50–1.91 (m, 5 H), 1.96–2.21 (m, 2 H), 2.61 (s, 6 H), 2.83–2.89 (m, 1 H), 4.56 (q, J = 8.53 Hz, 2 H), 7.45 (s, 1 H), 8.19 (s, 1 H), 8.41 (s, 1 H); 13C NMR (101 MHz, CDCl3) δ 14.3, 22.8, 25.9, 26.2, 32.1, 38.8, 45.8, 60.4, 60.7, 61.1, 61.5, 101.8, 109.7, 119.2, 121.9, 124.7, 127.5, 128.6, 139.1, 151.8, 159.8; HRMS (FAB) m/z calcd for C18H23F3N4O2H+: 385.1846, Found: 385.1851 (Δ = 1.3 ppm); HPLC (1): t =9.1 min, purity > 99%.
5-Benzamido-2-cyclohexyl-6-N,N-dimethylamino-1H-benzo[d]imidazole (7c-1)
To a solution of 4c (70 mg, 0.27 mmoles) in 2 mL of dichloromethane was added 1-ethyl-3-(3-dimethylaminopropyl) carbodiimide (57.5 mg, 0.3 mmol) (EDC) and benzoic acid (36.4 mg, 0.30 mmol). 4-Dimethylaminopyridine (DMAP) (36.4 mg, 0.30 mmol) was added to the above reaction mixture and the reaction was refluxed overnight. After completion of the reaction as per TLC, the reaction mixture was diluted with dichloromethane, transferred to a separatory funnel and the organic layer was washed with water, sodium bicarbonate and finally brine. The organic layer was dried over magnesium sulfate and concentrated using rotary evaporator. The crude product was purified by flash chromatography (column was packed with hexane, 30% ethyl acetate was used with gradual increase to 50% ethyl acetate) to obtain the product as a white solid (83 mg, 85% yield). mp 158–159°C; 1H NMR (300 MHz, CDCl3)δ 1.07–1.19 (m, 3 H), 1.54–1.71 (m, 5 H), 1.97–2.03 (m, 2 H, J = 12.9 Hz), 2.74 (s, 6 H), 2.85 (m, 1 H),7.58 (m, 3 H), 7.67 (s, 1 H), 7.95 (d , 1H, J = 6.6 Hz), 8.91 (s, 1 H), 9.88 (s, 1 H); 13C NMR (126 MHz, CDCl3) δ 25.9, 26.1, 29.9, 32.0, 38.6, 46.2, 101.8, 110.9, 127.2, 129.0, 129.2, 131.4, 132.0, 135.8, 139.4, 139.9, 160.1, 165.7; HRMS (FAB) m/z calcd for C22H26N4OH+: 363.2179, Found: 363.2190 (Δ = 2.9ppm); HPLC (1): t =11.0 min, purity > 99%.
Same procedure was followed for the synthesis of 7d, 7a-1∼3, 7a-5, 7a-7∼10, 7a-15, 7b-1, 7b-4, 7b-7, 7c-3∼6, 7c-11∼12, and 7c-16.
5-Benzamido-2-cyclohexyl-6-N,N-di-n-propylamino-1H-benzo[d]imidazole (7d)
White solid (52% yield): mp 212–213 °C; 1H NMR (400 MHz, CDCl3) δ 0.78 (t, 6 H, J = 7.2 Hz), 1.05–1.08 (m, 3 H), 1.36–1.39 (m, 4 H), 1.54–1.63 (m, 5 H), 1.90–1.95 (m, 2 H, J = 10 Hz), 2.65–2.67 (m, 1 H), 2.88–2.89 (m, 4 H, J = 7.5 Hz), 7.56–7.60 (m, 3 H), 7.99 (d, 2 H, J = 6.9 Hz), 9.00 (s, 1 H), 10.40 (s, 1 H); 13C NMR (100 MHz, CDCl3) δ 11.72, 20.69, 20.81, 25.57, 25.87, 31.71, 38.40, 59.10, 100.8, 113.3, 126.8, 128.9, 131.2, 131.6, 131.7, 135.4, 135.8, 139.7, 160.1, 165.2; HRMS (ESI) m/z calcd for C26H34N4OH+: 419.2805, Found: 419.2813 (Δ = 1.9 ppm); HPLC (1): t =6.3 min, purity > 99%.
2-Cyclohexyl-6-N,N-diethylamino-5-(4-difluoromethoxybenzamido)-1H-benzo[d]imidazole (7a-1)
White solid (66% yield); mp 194–196 °C; 1H NMR (400 MHz, CDCl3) δ 0.95 (t, 6 H, J = 7.2 Hz), 1.07–1.10 (m, 3 H), 1.54–1.64 (m, 5 H), 1.96–1.98 (m, 2 H, J = 10 Hz), 2.69–2.70 (m, 1 H), 2.99 (q, 4 H, J = s Hz), 6.61 (t, 1 H (CHF2), J = 73 Hz), 7.28 (d, 2 H, J = 8.5 Hz), 7.57 (s, 1 H), 8.00 (d, 2 H, J = 8.6 Hz), 8.92 (s, 1 H), 10.40 (s, 1 H); 13C NMR (100 MHz, CDCl3) δ 13.07, 25.55, 25.88, 31.70, 38.48, 50.79, 100.8, 112.7, 113.2, 115.3, 117.9, 119.5, 128.7, 131.5, 131.8, 132.3, 139.7, 153.5, 160.0, 163.8; HRMS (ESI) m/z calcd for C25H30F2N4O2H+: 457.2409, Found: 457.2411 (Δ = 0.4 ppm); HPLC (1): t =8.9 min, purity > 99%.
2-Cyclohexyl-6-N,N-diethylamino-5-(4-trifluoromethoxybenzamido)-1H-benzo[d]imidazole (7a-2)
White solid (71% yield); mp 207–208 °C; 1H NMR (400 MHz, CDCl3) δ 0.96 (t, 6 H, J = 7.2 Hz), 1.09–1.11 (m, 3 H), 1.55–1.66 (m, 5 H), 1.96–1.98 (m, 2 H, J = 10 Hz), 2.68–2.70 (m, 1 H), 3.00 (q, 4 H, J = 7.2 Hz), 7.25 (d, 2 H, J = 8.5 Hz), 7.58 (s, 1 H), 8.02 (d, 2 H, J = 8.6 Hz), 8.92 (s, 1 H), 10.43 (s, 1 H); 13C NMR (100 MHz, CDCl3) δ 13.10, 25.59, 25.88, 31.72, 38.49, 50.84, 100.8, 113.5, 119.0, 121.0, 121.6, 128.7, 131.5, 133.8, 135.0, 151.6, 159.9, 163.5; HRMS (ESI) m/z calcd for C25H29F3N4O2H+: 475.2315, Found: 475.2323 (Δ = 1.7 ppm); HPLC (1): t =7.5 min, purity > 99%.
2-Cyclohexyl-5-(4-ethoxybenzamido)-6-N,N-diethylamino-1H-benzo[d]imidazole (7a-3)
White solid (41% yield); mp 191–193 °C; 1H NMR (400 MHz, CDCl3) δ 0.96 (t, 6 H, J = 7.2 Hz), 1.07–1.10 (m, 3 H), 1.45 (t, 6 H, J = 7.2 Hz), 1.55–1.66 (m, 5 H), 1.94–1.97 (m, 2 H, J = 10 Hz), 2.68–2.70 (m, 1 H), 3.00 (q, 4 H, J = 7.2 Hz), 4.13 (q, 2 H, J = 7 Hz) 7.03 (d, 2 H, J = 8.8 Hz), 7.55 (s, 1 H), 7.94 (d, 2 H, J = 8.8 Hz), 8.95 (s, 1 H), 10.32 (s, 1 H); 13C NMR (100 MHz, CDCl3) δ 13.10, 14.68, 25.60, 25.93, 31.71, 38.43, 50.77, 63.74, 100.7, 113.2, 114.6, 127.4, 128.7, 131.8, 134.9, 139.3, 159.8, 161.8, 164.7; HRMS (ESI) m/z calcd for C26H34N4O2H+: 435.2754, Found: 435.2754 (Δ = 0.0 ppm); HPLC (1): t =8.6 min, purity > 99%.
2-Cyclohexyl-6-N,N-diethylamino-5-(4-ethylbenzamido)-1H-benzo[d]imidazole (7a-5)
White solid (75% yield); mp 175–177 °C; 1H NMR (400 MHz, CDCl3) δ 0.94–1.03 (m, 9 H), 1.26 (t, 3 H, J = 7.6 Hz), 1.46–1.56 (m, 5 H), 1.91–1.94 (m, 2 H, J = 10 Hz), 2.68–2.75 (m, 3 H), 2.99 (q, 4 H, J = 7.2 Hz), 7.36 (d, 2 H, J = 8.5 Hz), 7.56 (s, 1 H), 7.93 (d, 2 H, J = 8.6 Hz), 9.03 (s, 1 H), 10.41 (s, 1 H); 13C NMR (100 MHz, CDCl3) δ 13.02, 15.26, 25.50, 25.82, 28.76, 31.68, 38.37, 50.73, 100.9, 113.1, 126.9, 128.4, 131.6, 131.9, 132.8, 134.8, 139.6, 148.3, 160.0, 165.1; HRMS (ESI) m/z calcd for C26H34N4OH+: 419.2805, Found: 419.2809 (Δ = 0.9 ppm); HPLC (1): t =7.2 min, purity > 99%.
2-Cyclohexyl-6-N,N-diethylamino-5-(4-N,N-dimethylbenzamido)-1H-benzo[d]imidazole (7a-7)
White solid (65% yield); mp 189–191 °C; 1H NMR (400 MHz, CDCl3) δ 0.96 (t, 6 H, J = 7.6 Hz), 1.08–1.11 (m, 3 H), 1.53–1.64 (m, 5 H), 1.90–1.94 (m, 2 H, J = 10 Hz), 2.69 (m, 1 H), 2.99 (q, 4 H, J = 7.2 Hz), 3.06 (s, 6 H), 6.79 (d, 2 H, J = 8.5 Hz), 7.54 (s, 1 H), 7.92 (d, 2 H, J = 8.6 Hz), 8.98 (s, 1 H), 10.26 (s, 1 H); 13C NMR (100 MHz, CDCl3) δ 13.08, 25.58, 25.91, 31.70, 38.37, 40.09, 50.71, 100.6, 111.35, 113.0, 122.0, 128.5, 132.0, 133.0, 134.7, 139.2, 152.5, 159.7, 165.2; HRMS (ESI) m/z calcd for C26H35N5OH+: 434.2914, Found: 434.2914 (Δ = 0.0 ppm); HPLC (1): t =4.6 min, purity > 99%.
2-Cyclohexyl-6-N,N-diethylamino-5-(2-pyrazinamido)-1H-benzo[d]imidazole (7a-8)
White solid (64% yield); mp > 230°C; 1H NMR (400 MHz, CDCl3) δ 0.96 (t, 6 H, J = 7.6 Hz), 1.14–1.16 (m, 3 H), 1.53–1.64 (m, 5 H), 2.01–2.02 (m, 2 H), 2.76–2.8 (m, 1 H), 2.99 (q, 4H, J = 7.2 Hz), 7.55 (s, 1 H), 8.65 (d, 2 H, J = 8.6 Hz), 8.77 (d, 2 H, J = 8.6 Hz), 8.92 (s, 1 H), 9.53 (s, 1 H), 10.91 (s, 1 H); 13C NMR (100 MHz, CDCl3) δ 13.05, 25.66, 25.94, 31.82, 38.64, 50.54, 101.1, 113.6, 131.1, 136.3, 139.9, 142.9, 144.4, 145.6, 147.0, 160.1, 160.6; HRMS (ESI) m/z calcd for C22H28N6OH+: 393.2397, Found: 393.2412 (Δ = 3.8 ppm); HPLC (1): t =10.9 min, purity > 99%.
5-(4-Bromobenzamido)-2-cyclohexyl-6-N,N-diethylamino-1H-benzo[d]imidazole (7a-9)
White solid (61% yield); mp >230 °C; Mixture of rotamers, 1H NMR (400 MHz, CDCl3) δ 0.89 (t, 6 H, J = 7.2 Hz), 1.27–1.40 (m, 3 H), 1.57–1.67 (m, 3 H), 1.76–1.78 (m, 2 H), 1.96–1.98 (m, 2 H, J = 10 Hz), 2.80–2.81 (m, 1 H), 2.95–3.01 (m, 4 H, J = 7 Hz), 7.52 (m, 1 H), 7.81–7.82 (m, 4 H), 8.51 (s, 1 H), 10.1 (m, 2 H), 12.0 (m, 1 H); 13C NMR (100 MHz, CDCl3) δ 12.82, 25.47, 25.54, 31.22, 37.68, 49.92, 100.1, 112.9, 125.4, 128.6, 130.9, 131.5, 132.0, 133.9, 134.4, 139.4, 159.2, 162.4; HRMS (ESI) m/z calcd for C24H29BrN4OH+: 469.1597, Found: 469.1604 (Δ= 1.5 ppm); HPLC (1): t =7.6 min, purity > 99%.
5-(3-Bromobenzamido)-2-cyclohexyl-6-N,N-diethylamino-1H-benzo[d]imidazole (7a-10)
White solid (71% yield); mp 218–220 °C; 1H NMR (400 MHz, CDCl3) δ 0.95 (t, 6 H, J = 7.2 Hz), 1.09–1.10 (m, 3 H), 1.57–1.66 (m, 5 H), 1.94–1.96 (m, 2 H, J = 10 Hz), 2.69–2.70 (m, 1 H), 2.99–3.00 (m, 4 H), 7.41 (t, 1 H, J = 7.8 Hz), 7.58 (s, 1 H), 7.69 (d, 1 H, J = 7.7 Hz), 7.84 (d, 1 H, J = 7.7 Hz), 8.14 (s, 1 H), 8.93 (s, 1 H), 10.42 (s, 1 H), 11.39 (s, 1 H); 13C NMR (100 MHz, CDCl3) δ 13.07, 25.57, 25.84, 31.69, 38.43, 50.69, 100.8, 113.3, 123.2, 124.9, 130.4, 130.5, 131.2, 131.8, 134.6, 135.0, 137.4, 139.8, 160.1, 163.4; HRMS (ESI) m/z calcd for C24H29N4OH+: 469.1597, Found: 469.1604 (Δ = 1.5 ppm); HPLC (1): t =8.0 min, purity > 99%.
2-Cyclohexyl-6-N,N-diethylamino-5-(2-propoxyacetamido)-1H-benzo[d]imidazole (7a-15)
White solid (64% yield); mp 148–150 °C; 1H NMR (400 MHz, CDCl3) δ 0.94 (t, 6 H, J = 7.2 Hz), 1.01 (t, 3 H, J = 7.5 Hz), 1.23–1.46 (m, 3 H), 1.57–1.72 (m, 5 H), 1.85–1.88 (m, 2 H, J = 10 Hz), 2.12–2.15 (m, 2 H, J = 10 Hz), 2.83–2.85 (m, 1 H), 2.94 (q, 4 H, J = 7.2 Hz), 3.59 (t, 2 H, J = 6.5 Hz), 4.15 (s, 2 H), 7.55 (s, 1 H), 8.77 (s, 1 H), 10.44 (s, 1 H); NMR (100 MHz, CDCl3) δ 10.40, 12.75, 22.98, 25.87, 26.11, 31.74, 38.66, 50.42, 70.98, 73.46, 100.7, 113.4, 131.0, 131.6, 135.2, 139.6, 159.6, 167.7; HRMS (ESI) m/z calcd for C22H34N4O2H+: 387.2754, Found: 387.2769 (Δ = 3.9 ppm); HPLC (1): t =8.4 min, purity > 99%.
5-Benzamido-2-cyclohexyl-6-pyrrolinyl-1H-benzo[d]imidazole (7b-1)
White solid (76% yield); mp 197–198 °C; 1H NMR (500 MHz, CDCl3) δ 1.04–1.35 (m, 4 H), 1.51–1.68 (m, 3 H), 1.74 (dt, J = 13.20, 3.32 Hz, 2 H), 1.92–2.11 (m, 6 H), 2.75 (m, 1 H), 3.07 (br. s, 4 H), 7.46–7.71 (m, 4 H), 7.95 (d, J = 8.80 Hz, 2 H), 8.86 (s, 1 H), 9.91 (br. s, 1 H), 10.45 (br. s, 1 H); 13C NMR (126 MHz, CDCl3) δ 24.6, 25.9, 26.2, 32.0, 38.7, 54.2, 101.8, 111.1, 127.1, 129.2, 129.9, 131.9, 135.7, 136.0, 159.8, 165.3; HRMS (FAB) m/z calcd for C24H28N4OH+: 389.2336, Found: 389.2349 (Δ = 3.4 ppm); HPLC (1): t =8.1 min, purity > 99%.
5-(4-Bromobenzamido)-2-cyclohexyl-6-pyrrolidinyl-1H-benzo[d]imidazole (7b-4)
White solid (76% yield); mp 215–216 °C; 1H NMR (500 MHz, DMSO-d6) δ 1.19–1.45 (m, 3 H), 1.58 (qd, J = 12.21, 3.05 Hz, 2 H), 1.64–1.74 (m, 1 H), 1.79 (dt, J = 12.89, 3.17 Hz, 2 H), 1.88 (br. s, 4 H), 2.00 (d, J = 10.68 Hz, 2 H), 2.72–2.86 (m, 1 H), 3.06 (br. s, 4 H), 7.16 (br. s, 1 H), 7.68–7.86 (m, 3 H), 7.91 (d, J = 8.24 Hz, 2 H), 9.82 (s, 4 H), 11.90 (br. s, 1 H); 13C NMR (101 MHz, DMSO-d6 at 55 °C) δ 24.1, 25.2, 25.4, 31.0, 37.5, 51.9, 124.9, 125.8, 128.9, 131.4, 134.0, 138.3, 158.6, 163.4; HRMS (FAB) m/z calcd for C24H27BrN4OH+: 467.1441, Found: 467.1443 (Δ = 0.4 ppm); HPLC (2): t =9.6 min, purity > 98%.
2-Cyclohexyl-5-(2-propoxyacetamido)-6-(pyrrolidin-1-yl)-1H-benzo[d]imidazole (7b-7)
White solid (78% yield); mp 179–181 °C; 1H NMR (300 MHz, CDCl3) δ 1.01 (t, 3 H, J = 7.5 Hz), 1.35–1.38 (m, 3 H), 1.70–1.72 (m, 5 H), 1.80–1.82 (m, 2 H), 1.94 (br. s, 4 H), 2.09–2.12 (m, 2 H), 2.81–2.87 (m, 1 H), 2.99 (br. s, 4 H), 3.56 (t, 2 H, J = 6.5 Hz), 4.12 (s, 2 H), 7.54 (s, 1 H), 8.69 (s, 1 H), 9.93 (s, 1 H); 13C NMR (100 MHz, CDCl3) δ 10.48, 22.96, 24.27, 25.84, 26.07, 31.76, 38.63, 53.46, 70.90, 73.46, 101.5, 110.2, 128.6, 130.8, 135.8, 139.7, 159.5, 167.6; HRMS (ESI) m/z calcd for C22H32N4O2H+: 385.2598, Found: 385.2594 (Δ = −1.0 ppm); HPLC (1): t =9.7 min, purity > 99%.
2-Cyclohexyl-5-(4-difluoromethoxybenzamido)-6-N,N-dimethylamino-1H-benzo[d]imidazole (7c-3)
White solid (65% yield); mp 183–185 °C; 1H NMR (400 MHz, CDCl3) δ 1.02–1.33 (m, 5 H), 1.51–1.79 (m, 6 H), 2.00 (d, J = 13.55 Hz, 2 H), 2.72 (s, 7 H), 6.61 (d, J = 73.53 Hz, 1 H), 7.25–7.31 (m, 2 H), 7.59 (s, 1 H), 7.97 (d, J = 9.03 Hz, 2 H), 8.82 (s, 1 H), 9.87 (s, 1 H); 13C NMR (126 MHz, CDCl3) δ 14.3, 25.9, 26.1, 31.8, 32.0, 34.9, 38.7, 46.1, 101.8, 110.9, 113.5, 115.6, 117.7, 119.7, 128.9, 129.1, 132.7, 139.5, 153.8, 160.1, 164.4; HRMS (FAB) m/z calcd for C23H26F2N4O2H+: 429.2097, Found: 429.2100 (Δ = 0.8 ppm); HPLC (1): t =12.8 min, purity > 99%.
2-Cyclohexyl-5-(4-trifluoromethoxybenzamido)-6-N,N-dimethylamino-1H-benzo[d]imidazole (7c-4)
White solid (70% yield); mp 210–211 °C; 1H NMR (400 MHz, CDCl3) δ 1.08–1.31 (m, 3 H), 1.53–1.70 (m, 3 H), 1.70–1.81 (m, 2 H), 1.99–2.09 (m, 2 H), 2.69–2.83 (m, 7 H), 7.35–7.42 (m, 2 H), 7.60 (s, 1 H), 7.97–8.04 (m, 2 H), 8.81 (s, 1 H), 9.90 (s, 1 H); 13C NMR (101 MHz, CDCl3) δ 25.68, 25.93, 31.78, 38.47, 45.95, 119.08, 121.00, 121.01, 128.85, 128.91, 133.95, 139.43, 151.64, 159.68, 163.91; HRMS (FAB) m/z calcd for C23H25F3N4O2H+: 447.2002, Found: 447.2007 (Δ = 1.0 ppm); HPLC (1): t =10.3 min, purity > 99%.
2-Cyclohexyl-6-N,N-dimethylamino-5-(4-methylbenzamido)-1H-benzo[d]imidazole (7c-5)
White solid (76% yield); mp 179–180 °C; 1H NMR (500 MHz, CDCl3) δ 1.05–1.27 (m, 3 H), 1.50–1.77 (m, 5 H), 1.99 (d, J = 12.51 Hz, 2 H), 2.46 (s, 3 H), 2.66–2.78 (m, 7 H), 7.35 (d, J = 8.24 Hz, 2 H), 7.61 (s, 1 H), 7.87 (d, J = 8.24 Hz, 2 H), 8.89 (s, 1 H), 9.90 (s, 1 H), 10.65 (br. s, 1 H); 13C NMR (126 MHz, CDCl3) δ 21.7, 25.9, 26.2, 32.0, 38.6, 46.1, 101.7, 110.9, 127.2, 129.2, 129.8, 131.3, 133.0, 139.4, 139.8, 142.5, 159.9, 165.7; HRMS (FAB) m/z calcd for C23H28N4OH+: 377.2336, Found: 377.2349 (Δ = 3.5 ppm); HPLC (1): t =10.6 min, purity > 99%.
2-Cyclohexyl-5-(4-ethylbenzamido)-6-N,N-dimethylamino-1H-benzo[d]imidazole (7c-6)
White solid (72% yield); mp 177–178 °C; 1H NMR (400 MHz, CDCl3) δ 1.03–1.23 (m, 3 H), 1.30 (t, J = 7.65 Hz, 3 H), 1.50–1.63 (m, 3 H), 1.68 (d, J = 7.28 Hz, 2 H), 1.97 (d, J = 11.80 Hz, 2 H), 2.60–2.87 (m, 9 H), 7.38 (d, J = 8.03 Hz, 2 H), 7.61 (br. s, 1 H), 7.90 (d, J = 8.03 Hz, 2 H), 8.93 (d, J = 5.02 Hz, 1 H), 9.93 (br. s, 3 H); 13C NMR (101 MHz, CDCl3) δ 15.6, 25.9, 26.1, 29.1, 32.0, 38.6, 46.1, 101.8, 110.8, 127.3, 128.7, 129.1, 129.2, 131.5, 133.3, 139.3, 139.8, 148.7, 160.0, 160.1, 165.8; HRMS (FAB) m/z calcd for C24H30N4OH+: 391.2492, Found: 391.2500 (Δ = 1.9 ppm); HPLC (1): t =9.9 min, purity > 99%.
5-(4-Bromobenzamido)-2-cyclohexyl-6-N,N-dimethylamino-1H-benzo[d]imidazole (7c-11)
Yellow solid (78% yield); mp 207–208 °C; 1H NMR (500 MHz, Methanol-d4) δ 0.80–1.02 (m, 1 H), 1.16–2.21 (m, 13 H), 2.54–3.00 (m, 7 H), 7.44 (s, 1 H), 7.60–7.98 (m, 4 H), 8.46 (s, 1 H); 13C NMR (126 MHz, Methanol-d4) δ 27.1, 27.3, 31.9, 33.0, 40.0, 46.1, 65.6, 106.9, 108.2, 127.6, 129.9, 130.0, 133.4, 135.5, 142.3, 161.6, 166.2; HRMS (FAB) m/z calcd for C22H25BrN4OH+: 441.1285, Found: 441.1287 (Δ = 0.6 ppm); HPLC (1): t =9.9 min, purity > 99%.
5-(3-Bromobenzamido)-2-cyclohexyl-6-N,N-dimethylamino-1H-benzo[d]imidazole (7c-12)
Yellow solid (59% yield); mp 148–149 °C; 1H NMR (500 MHz, Methanol-d4) δ 1.28–1.38 (m, 1 H), 1.40–1.52 (m, 2 H), 1.57–1.93 (m, 5 H), 2.06 (d, J = 11.60 Hz, 2 H), 2.65–2.80 (m, 6 H), 2.80–2.93 (m, 1 H), 7.38–7.49 (m, 2 H), 7.66–7.76 (m, 1 H), 7.80–7.91 (m, 1 H), 8.02–8.13 (m, 1 H), 8.43 (s, 1 H); 13C NMR (126 MHz, Methanol-d4) δ 27.10, 27.31, 32.9, 39.95, 46.06, 106.96, 108.03, 124.08, 126.62, 129.75, 131.56, 131.91, 135.21, 135.95, 137.36, 138.51, 138.53 , 142.33, 142.36, 161.53, 161.55, 165.48, 165.54; HRMS (FAB) m/z calcd for C22H25BrN4OH+: 441.1285, Found: 441.1288 (Δ = 0.8 ppm); HPLC (1): t =10.5 min, purity > 99%.
2-Cyclohexyl-6-N,N-dimethylamino-5-(2-propoxyacetamido)-1H-benzo[d]imidazole (7c-16)
White solid (61% yield); mp 146–148 °C; 1H NMR (400 MHz, CDCl3) δ 1.03 (t, 3 H, J = 7.2 Hz), 1.23–1.33 (m, 3 H), 1.60–1.72 (m, 5 H), 1.81–1.84 (m, 2 H, J = 10 Hz), 2.09–2.12 (m, 2 H, J = 10 Hz), 2.82 (s, 6 H), 2.83–2.85 (m, 1 H), 3.58 (t, 2 H, J = 6.5 Hz), 4.14 (s, 2 H), 7.53 (s, 1 H), 8.71 (s, 1 H), 10.07 (s, 1 H); 13C NMR (100 MHz, CDCl3) δ 10.42, 23.01, 25.89, 26.13, 31.83, 38.71, 45.68, 70.95, 73.53, 100.7, 113.4, 128.1, 131.6, 135.2, 139.2, 159.8, 167.9; HRMS (ESI) m/z calcd for C20H30N4O2H+: 359.2441, Found: 359.2450(Δ = 2.5 ppm); HPLC (1): t =10.2 min, purity > 99%.
2-Cyclohexyl-5-(4-methoxybenzamido)-6-N,N-dimethylamino-1H-benzo[d]imidazole (7c-2)
To a solution of 4c (100 mg, 0.39 mmol) in dichloromethane (2 mL) was added triethylamine (60 µL, 0.43 mmol). To this, 4-methoxybenzoyl chloride (53.3 µL, 0.39 mmol) was added dropwise at 0 °C and reacted at room temperature. After completion of the reaction, the reaction mixture was diluted with ethyl acetate and transferred to a separatory funnel. The organic layer was washed with saturated sodium bicarbonate solution, followed by water and finally brine. The organic layer was dried over magnesium sulfate. The crude product was purified by flash chromatography on silica gel (gradient: 30–70% ethyl acetate /hexanes) to obtain the product which was then treated with activated charcoal to yield the product as a white solid (130 mg, 86%); mp 201-202 °C; 1H NMR (300 MHz, CDCl3) δ 1.06–1.15 (m, 3 H), 1.54–1.68 (m, 5 H), 1.94–1.98 (m, 2 H, J = 12.9 Hz), 2.67–2.71 (m, 7 H), 3.90 (s, 3 H), 7.04 (d, 1 H, J = 9 Hz), 7.60 (s, 1H), 7.93 (d, 2 H, J = 9 Hz), 8.86 (s, 1 H), 9.82 (s, 1 H); 13C NMR (101 MHz, CDCl3) δ 25.8, 26.1, 32.0, 38.7, 46.1, 55.7, 101.9, 110.6, 114.3, 127.9, 129.0, 129.0, 131.6, 139.3, 160.1, 162.6, 165.2; HRMS (FAB) m/z calcd for C23H28N4O2H+: 393.2285, Found: 393.2285 (Δ = 0.0 ppm); HPLC (1): t =13.2 min, purity > 99%.
Same Procedure was used for the synthesis of 7a-4, 7a-6, 7a-11∼14, 7b-2, 7b-3, 7b-5, 7b-6, 7c-2, 7c-7∼10, 7c-13∼15, and 7c-17.
2-Cyclohexyl-6-N,N-diethylamino-5-(4-methylbenzamido)-1H-benzo[d]imidazole (7a-4)
White solid (65% yield); mp 183–184 °C;1H NMR (300 MHz, CDCl3) δ 0.97 (t, 6 H, J = 6 Hz), 1.20–1.30 (m, 3 H), 1.58–1.78 (m, 5 H), 2.06–2.08 (m, 2 H), 2.45 (s, 3 H), 2.76–2.77 (m, 1 H), 3.03 (q, 4 H, J = 7.2 Hz), 7.34 (d, 2 H, J = 8.4 Hz) 7.59 (s, 1 H), 7.87 (d, 2 H, J = 8.1 Hz), 8.91 (s, 1 H), 10.31 (s, 1 H); 13C NMR (100 MHz, CDCl3) δ 13.09, 21.48, 25.63, 25.92, 31.72, 38.40, 50.75, 100.9, 113.1, 126.8, 129.6, 131.8, 132.7, 135.0, 139.4, 142.1, 159.8, 165.1; HRMS (FAB) m/z calcd for C25H32N4OH+: 405.2649, Found: 405.2654 (Δ = 1.2 ppm); HPLC (4): t =10.1 min, purity > 99%.
2-Cyclohexyl-6-N,N-diethylamino-5-(4-propylbenzamido)-1H-benzo[d]imidazole (7a-6)
White solid (56% yield); mp 169–171 °C; 1H NMR (400 MHz, CDCl3) δ 0.98–1.10 (m, 12 H), 1.56–1.69 (m, 7 H), 1.93–1.95 (m, 2 H, J = 10 Hz), 2.67–2.71 (m, 3 H), 3.01 (q, 4 H, J = 7.2 Hz), 7.36 (d, 2 H, J = 8.5 Hz), 7.57 (s, 1 H), 7.92 (d, 2 H, J = 8.6 Hz), 9.00 (s, 1 H), 10.40 (s, 1 H); 13C NMR (100 MHz, CDCl3) δ 13.08, 13.73, 24.29, 25.59, 25.89, 31.72, 37.90, 38.40, 50.78, 100.8, 113.2, 126.9, 129.0, 131.8, 132.8, 134.9, 146.9, 159.9, 165.1; HRMS (ESI) m/z calcd for C27H36N4OH+: 433.2962, Found: 433.2961 (Δ = − 0.2 ppm); HPLC (1): t =7.5 min, purity > 99%..
2-Cyclohexyl-6-N,N-dimethylamino-5-(4-propylbenzamido)-1H-benzo[d]imidazole (7c-7)
White solid (75% yield); mp 190–191°C; 1H NMR (400 MHz, CDCl3) δ 0.98 (t, J = 7.40 Hz, 3 H), 1.03–1.36 (m, 4 H), 1.48–1.77 (m, 8 H), 1.94 (d, J = 13.30 Hz, 2 H), 2.62–2.80 (m, 9 H), 7.36 (d, J = 8.28 Hz, 2 H), 7.61 (s, 1 H), 7.91 (d, J = 8.03 Hz, 2 H), 8.94 (s, 1 H), 9.95 (s, 1 H), 11.15 (br. s, 1 H); 13C NMR (101 MHz, CDCl3) δ 14.0, 24.5, 25.8, 26.1, 32.0, 38.1, 38.6, 46.1, 101.8, 110.8, 127.2, 129.0, 129.3, 131.5, 133.2, 139.3, 139.8, 147.2, 160.2, 165.8; HRMS (FAB) m/z calcd for C25H32N4OH+: 405.2649, Found: 405.2651 (Δ = 0.5 ppm); HPLC (1): t =8.0 min, purity > 99%.
2-Cyclohexyl-6-N,N-diethylamino-5-(4-fluorobenzamido)-1H-benzo[d]imidazole (7a-11)
White solid (71% yield); mp 213–214 °C; 1H NMR (400 MHz, Methanol-d4) δ 0.98 (t, 6 H, J = 7.2 Hz), 1.09–1.46 (m, 3 H), 1.66–170 (m, 3 H), 1.86–1.89 (m, 2 H), 2.08–2.12 (m, 2 H, J = 10 Hz), 2.89–2.92 (m, 1 H), 3.04–3.10 (m, 4 H, J = 7.16 Hz), 7.31 (t, 1 H, J = 7.8 Hz), 7.49 (s, 1 H), 7.97–7.99 (m, 1 H), 8.66 (s, 1 H); 13C NMR (100 MHz, Methanol-d4) δ13.55, 27.11, 27.33, 32.98, 40.03, 51.89, 117.0, 117.2, 130.5, 130.6, 132.8, 133.3, 137.3, 161.7, 165.2, 165.4, 167.7; HRMS (ESI) m/z calcd for C24H29FN4OH+: 409.2398, Found: 409.2401 (Δ = 0.7 ppm); HPLC (1): t =8.3 min, purity > 99%.
2-Cyclohexyl-6-N,N-diethylamino-5-(2,4-difluorobenzamido)-1H-benzo[d]imidazole (7a-12)
White solid (79% yield); mp 204–206 °C; 1H NMR (400 MHz, Methanol-d4) δ 0.94 (t, 6 H, J = 7.2 Hz), 1.32–1.47 (m, 3 H), 1.64–1.78 (m, 5 H), 1.84–1.86 (m, 2 H, J = 10 Hz), 2.05–2.09 (m, 2 H, J = 10 Hz), 2.77–2.79 (m, 1 H), 2.84–2.89 (m, 4 H), 7.16–7.18 (m, 2 H), 7.46 (s, 1 H), 8.13–8.19 (m, 1 H), 8.72 (s, 1 H); 13C NMR (100 MHz, Methanol-d4) δ 13.15, 27.11, 27.33, 32.97, 40.02, 51.70, 105.0, 105.7, 111.5, 113.7, 119.8, 134.7, 135.7, 137.3, 160.9, 161.4, 161.7, 163.5, 165.4, 167.9; HRMS (ESI) m/z calcd for C24H28F2N4OH+: 427.2304, Found: 427.2309 (Δ = 1.1 ppm); HPLC (1): t =7.1 min, purity > 99%.
5-(3-Allylbenzamido)-2-cyclohexyl-6-N,N-diethylamino-1H-benzo[d]imidazole (7a-13)
White solid (69% yield); mp 89–90 °C; 1H NMR (400 MHz, CDCl3) δ 0.96 (t, 6 H, J = 7.2 Hz), 1.09–1.10 (m, 3 H), 1.55–1.58 (m, 5 H), 1.91–1.95 (m, 2 H, J = 10 Hz), 2.67 (m, 1 H), 3.00 (q, 4 H, J = 7.2 Hz), 5.36 (d, 1 H, J = 11.1 Hz), 5.85 (d, 1 H, J = 17.5 Hz), 6.75–6.80 (m, 1 H), 7.50 (t, 1 H, J = 7.7 Hz), 7.55 (s, 1 H), 7.61 (d, 1 H, J = 7.7 Hz), 8.82 (d, 1 H, J = 7.8 Hz), 8.04 (s, 1 H); 13C NMR (100 MHz, CDCl3) δ 13.06, 25.49, 25.78, 31.64, 38.39, 50.66, 101.2, 112.9, 115.3, 124.9, 125.7, 129.0, 129.3, 131.4, 135.0, 135.7, 138.3, 160.0, 164.9; HRMS (ESI) m/z calcd for C26H32N4OH+: 417.2649, Found: 417.2648 (Δ = − 0.2 ppm); HPLC (1): t =7.3 min, purity > 99%.
2-Cyclohexyl-6-N,N-diethylamino-5-(hexanamido)-1H-benzo[d]imidazole (7a-14)
White solid; 56% yield; mp 183–184 °C; 1H NMR (400 MHz, CDCl3) δ 0.91–1.01 (m, 9 H), 1.23–1.39 (m, 7 H), 1.63–1.83 (m, 7 H), 2.10–2.13 (m, 2 H), 2.47 (t, 2 H, J = 7.2 Hz), 2.86–2.88 (m, 1 H), 2.94 (q, 4 H, J = 7.2 Hz), 7.50 (s, 1 H), 8.74 (s, 1 H), 9.46 (s, 1 H); 13C NMR (100 MHz, CDCl3) δ 12.89, 13.81, 22.39, 25.88, 26.11, 31.43, 31.55, 38.59, 38.72, 50.60, 101.0, 112.6, 131.6, 132.0, 134.4, 139.0, 159.7, 171.1; HRMS (ESI) m/z calcd for C23H36N4O: 385.2962, Found:385.2967 (Δ = 1.3 ppm); HPLC (2): t =6.6 min, purity > 98%.
2-Cyclohexyl-5-(4-methoxybenzamido)-6-pyrrolydinyl-1H-benzo[d]imidazole (7b-2)
White solid (56% yield); mp 195–196 °C; 1H NMR (400 MHz, CDCl3) δ 1.08–1.35 (m, 5 H), 1.52–1.69 (m, 3 H), 1.70–1.80 (m, 2 H), 1.96–2.10 (m, 6 H), 2.76 (m, 1 H), 3.06 (br. s, 4 H), 3.90 (s, 3 H), 7.03 (d, J = 8.8 Hz, 2 H), 7.57 (br. s, 1 H), 7.91 (d, J = 8.8 Hz, 2 H), 8.80 (s, 1 H), 9.77 (br. s, 1 H); 13C NMR (101 MHz, CDCl3) δ 24.6, 25.9, 26.2, 32.0, 38.7, 54.1, 55.7, 100.2, 114.4, 128.0, 128.9, 130.1, 136.0, 159.7, 162.6, 164.9; HRMS (FAB) m/z calcd for C25H30N4O2H+: 419.2442, Found: 419.2450 (Δ = 2.0 ppm); HPLC (2): t =12.8 min, purity > 98%.
2-Cyclohexyl-5-(4-methylbenzamido)-6-pyrrolodinyl-1H-benzo[d]imidazole (7b-3)
White solid (40% yield); mp 179–180 °C;1H NMR (500 MHz, CDCl3) δ 1.00–1.37 (m, 4 H), 1.49–1.65 (m, 3 H), 1.71 (dt, J = 13.05, 3.40 Hz, 2 H), 1.92–2.09 (m, 6 H), 2.45 (s, 3 H), 2.73 (m, 1 H), 3.05 (br. s, 4 H), 7.34 (d, J = 8.24 Hz, 2 H), 7.58 (br. s, 1 H), 7.85 (d, J = 8.20 Hz, 2 H), 8.86 (s, 1 H), 9.85 (br. s, 1 H), 10.70 (br. s, 1 H); 13C NMR (126 MHz, CDCl3) δ 21.7, 24.6, 25.9, 26.2, 32.0, 38.7, 54.1, 101.9, 110.9, 127.1, 129.8, 129.9, 130.1, 131.3, 132.9, 136.0, 139.9, 142.4, 159.9, 165.4; HRMS (FAB) m/z calcd for C25H30N4OH+: 403.2492, Found: 403.2496 (Δ = 0.9 ppm); HPLC (2): t =9.1 min, purity > 98%.
2-Cyclohexyl-5-(3-phenylpropanamido)-6-pyrrolidinyl-1H-benzo[d]imidazole (7b-5)
White solid (97% yield); mp 146–147 °C; 1H NMR (500 MHz, CDCl3) δ 1.16–1.40 (m, 4 H), 1.57–1.73 (m, 3 H), 1.80 (dt, J = 13.05, 3.40 Hz, 2 H), 1.84–1.94 (m, 4 H), 2.09 (d, J = 12.90 Hz, 2 H), 2.76 (t, J = 7.40 Hz, 2 H), 2.80–2.92 (m, 5 H), 3.11 (t, J = 7.40 Hz, 2 H), 7.15–7.34 (m, 5 H), 7.47 (s, 1 H), 8.58 (s, 1 H), 8.75 (br. s, 1 H); 13C NMR (126 MHz, CDCl3) δ 24.5, 26.0, 26.2, 29.9, 32.0, 32.2, 38.7, 40.5, 53.7, 103.2, 109.3, 126.6, 128.5, 128.8, 129.4, 131.6, 136.0, 138.5, 140.6, 159.6, 170.4; HRMS (FAB) m/z calcd for C26H32N4OH+: 417.2649, Found: 417.2654 (Δ = 1.2 ppm); HPLC (2): t =11.3 min, purity > 98%.
2-Cyclohexyl-5-hexanamido-6-pyrrolidinyl-1H-benzo[d]imidazole (7b-6)
White solid; 61% yield; mp 127–129 °C; 1H NMR (400 MHz, CDCl3) δ 0.91 (t, 3 H, J = 7.2 Hz), 1.20–1.43 (m, 7 H), 1.63–1.85 (m, 7 H), 1.97 (br. s, 4 H), 2.05–2.07 (m, 2 H), 2.47 (t, 2 H, J = 7.2 Hz), 2.83–2.85 (m, 1 H), 2.99 (br. s, 4 H), 7.52 (s, 1 H), 8.59 (s, 1 H), 8.91 (s, 1 H); 13C NMR (100 MHz, CDCl3) δ 13.86, 22.43, 24.33, 25.84, 26.09, 31.43, 31.78, 38.57, 53.72, 102.3, 109.7, 129.4, 131.4, 135.5, 138.5, 159.4, 171.2; HRMS (ESI) m/z calcd for C23H34N4O: 383.2805, Found: 383.2811 (Δ = 1.5 ppm); HPLC (2): t =8.8 min, purity > 98%.
5-(4-tert-Butylbenzamido)-2-cyclohexyl-6-N,N-dimethylamino-1H-benzo[d]imidazole (7c-8)
White solid (74% yield); mp 222–223 °C; 1H NMR (500 MHz, CDCl3) δ 0.96–1.20 (m, 4 H), 1.39 (s, 9 H), 1.49–1.72 (m, 5 H), 1.96 (d, J = 12.21 Hz, 2 H), 2.74 (s, 7 H), 7.58 (d, J = 8.24 Hz, 3 H), 7.93 (d, J = 8.54 Hz, 2 H), 8.97 (s, 1 H), 9.98 (s, 1 H), 11.00 (s, 1 H); 13C NMR (126 MHz, CDCl3) δ 25.8, 26.1, 31.4, 32.0, 35.2, 38.6, 46.2, 101.7, 110.9, 126.2, 127.1, 129.2, 133.0, 139.3, 139.8, 155.5, 160.1, 165.8; HRMS (FAB) m/z calcd for C26H34N4OH+: 419.2805, Found: 419.2809 (Δ = 0.9 ppm); HPLC (1): t =9.0 min, purity > 99%.
2-Cyclohexyl-6-N,N-dimethylamino-5-(thiophene-2-carboxamido)-1H-benzo[d]imidazole (7c-9)
White solid (75% yield); mp 139–140 °C; 1H NMR (500 MHz, CDCl3) δ 1.00–1.08 (m, 1 H), 1.11–1.24 (m, 2 H), 1.51–1.61 (m, 3 H), 1.63–1.69 (m, 2 H), 1.97 (d, J = 11.90 Hz, 2 H), 2.12 (s, 1 H), 2.69 (s, 6 H), 2.77–2.84 (m, 1 H), 7.16 (dd, J = 4.88, 3.66 Hz, 1 H), 7.56 (d, J = 1.00 Hz, 2 H), 7.67 (dd, J = 3.66, 0.61 Hz, 1 H), 8.72 (s, 1 H), 9.75 (s, 1 H), 10.20 (br. s, 1 H); 13C NMR (126 MHz, CDCl3) δ 22.2, 25.7, 26.1, 31.8, 38.5, 45.9, 102.6, 109.7, 128.2, 128.4, 128.7, 130.8, 131.7, 137.9, 139.5, 140.1, 160.1, 160.1, 175.3; HRMS (FAB) m/z calcd for C20H23N4OSH+: 369.1744, Found: 369.1742 (Δ = −0.4 ppm); HPLC (1): t =11.8 min, purity > 98%.
2-Cyclohexyl-5-(furan-2-carboxamido)-6-N,N-dimethylamino-1H-benzo[d]imidazole (7c-10)
White solid (71% yield); mp 214–215 °C; 1H NMR (400 MHz, CDCl3) δ 1.10–1.37 (m, 3 H), 1.55–1.84 (m, 5 H), 2.06 (d, J = 14.81 Hz, 2 H), 2.67–2.88 (m, 7 H), 6.56–6.66 (m, J = 3.60, 1.90 Hz, 1 H), 7.22–7.32 (m, 1 H), 7.52–7.68 (m, 2 H), 8.79 (s, 1 H), 9.98 (s, 1 H), 10.59 (s, 1 H); 13C NMR (101 MHz, CDCl3) δ 25.9, 26.2, 32.0, 38.7, 46.0, 101.8, 110.8, 112.6, 114.9, 128.6, 131.2, 139.6, 139.9, 144.7, 148.8, 156.4, 159.9; HRMS (FAB) m/z calcd for C20H24N4O2H+: 353.1972, Found: 353.1982 (Δ = 2.8 ppm); HPLC (1): t =11.6 min, purity > 99%.
2-Cyclohexyl-5-(4-fluorobenzamido)-6-N,N-dimethylamino-1H-benzo[d]imidazole (7c-13)
White solid (70% yield); mp 196–197 °C; 1H NMR (500 MHz, Methanol-d4) δ 2.00–2.26 (m, 3 H), 2.34–2.45 (m, 2 H), 2.46–2.55 (m, 1 H), 2.60 (dt, J = 12.97, 3.28 Hz, 2 H), 2.81 (d, J = 13.12 Hz, 2 H), 3.48 (s, 6 H), 3.58–3.67 (m, 1 H), 8.20 (t, J = 8.85 Hz, 2 H), 8.82 (dd, J = 8.85, 5.49 Hz, 2 H), 9.07 (br. s, 1 H), 10.50 (br. s, 1 H), 12.80 (s, 1 H); 13C NMR (126 MHz, Methanol-d4) δ 35.1, 35.2, 40.9, 47.3, 54.9, 111.6, 112.1, 119.5, 121.1, 125.3, 125.5, 136.7, 137.5, 139.2, 139.3, 140.3, 141.1, 141.1, 149.3, 150.7, 168.8, 172.7, 174.7; HRMS (FAB) m/z calcd for C22H25FN4OH+: 381.2085, Found: 381.2088 (Δ = 0.7 ppm); HPLC (1): t =11.4 min, purity > 99%.
2-Cyclohexyl-5-(2,4-difluorobenzamido)-6-N,N-dimethylamino-1H-benzo[d]imidazole (7c-14)
White solid (70% yield); mp 212–213 °C; 1H NMR (400 MHz, METHANOL-d4) δ 1.29–1.55 (m, 3 H), 1.58–1.94 (m, 5 H), 2.07 (d, J = 12.05 Hz, 2 H), 2.72 (s, 6 H), 2.81–2.95 (m, 1 H), 7.10–7.24 (m, 2 H), 7.45 (s, 1 H), 8.06–8.21 (m, 1 H), 8.63 (s, 1 H); 13C NMR (101 MHz, Methanol-d4) δ 27.12, 27.34, 30.81, 32.98, 40.00 46.16 , 105.42, 105.69, 105.71, 105.97, 106.17, 108.39, 113.54, 113.57, 113.76, 113.78, 119.94, 119.97, 120.05, 120.09, 130.57, 134.62, 134.65, 134.72, 134.76, 141.75, 161.04, 161.16, 161.54, 161.90, 161.94, 163.53, 163.65, 165.26, 165.39, 167.79; HRMS (FAB) m/z calcd for C22H24F2N4OH+: 399.1991, Found: 399.1988 (Δ = −0.7 ppm); HPLC (1): t =8.9 min, purity > 99%.
2-Cyclohexyl-5-(hexanamido)-6-N,N-dimethylamino-1H-benzo[d]imidazole (7c-15)
White solid (63% yield); mp 114–117 °C; 1H NMR (400 MHz, CDCl3) δ 0.90 (t, 3 H, J = 7.2 Hz), 1.30–1.39 (m, 7 H), 1.63–1.83 (m, 7 H), 2.06–2.10 (m, 2 H), 2.46 (t, 2 H, J = 7.2 Hz), 2.65 (s, 6 H), 2.82–2.84 (m, 1 H), 7.51 (s, 1 H), 8.62 (s, 1 H), 8.97 (s, 1 H); 13C NMR (100 MHz, CDCl3) δ 13.89, 22.41, 25.80, 26.07, 31.43, 31.77, 38.56, 45.69, 101.8, 110.1, 128.6, 138.7, 159.6, 171.4; HRMS (ESI) m/z calcd for C21H32N4OH+: 357.2649, Found: 357.2650 (Δ = 0.2 ppm); HPLC (2): t =8.9 min, purity > 99%.
2-Cyclohexyl-5-[2-(4-methoxyphenyl)acetamido]-6-N,N-dimethylamino-1H-benzo[d]imidazole (7c-17)
White solid (88% yield); mp 167–168 °C; 1H NMR (400 MHz, CDCl3) δ 1.17–1.42 (m, 3 H), 1.55–1.77 (m, 3 H), 1.83 (d, J = 12.80 Hz, 2 H), 2.12 (d, J = 12.05 Hz, 2 H), 2.34 (s, 6 H), 2.79–3.00 (m, 1 H), 3.78 (s, 2 H), 3.83 (s, 3 H), 6.95 (d, J = 8.28 Hz, 6 H), 7.28 (s, 2 H), 7.44 (s, 1 H), 8.63 (s, 1 H), 9.00 (s, 1 H); 13C NMR (101 MHz, CDCl3) δ 26.0, 26.2, 31.9, 38.7, 44.8, 45.5, 55.6, 101.8, 109.9, 114.0, 114.7, 126.7, 129.3, 130.5, 131.0, 139.5, 159.4, 159.5, 169.9; HRMS (FAB) m/z calcd for C24H30N4O2H+: 407.2442, Found: 407.2246 (Δ = 1.1 ppm); HPLC (1): t =13.4 min, purity > 99%.
Bacterial Strains and Growth.22, 37
H37Rv, the drug sensitive laboratory strain of Mtb, and clinical Mtb strains W210, NHN382, and TN587 exhibiting different resistant profiles to isoniazid were used. For evaluation of drug sensitivity all strains were grown in Difco™ 7H9 Middlebrook liquid media (BD Biosciences, 271310) supplemented with 10% Middlebrook OADC Enrichment (VWR, 9000-614), 0.05% Tween (G-Biosciences, 786-519), and 0.2% Glycerol at 37 °C.
Antibacterial Activity.22, 36
M. tuberculosis H37Rv strain and clinical isolates were used in the antibacterial studies, which were grown at 37 °C in Difco™ 7H9 Middlebrook liquid media supplemented with 10% Middlebrook OADC Enrichment, 0.05% Tween, and 0.2% Glycerol. The minimum inhibitory concentration (MIC) of the compounds was determined by the microplate Alamar Blue assay (MABA) in triplicates. The compounds were prepared in DMSO and were serially diluted 2-fold in 96-well microtiter plates. Bacteria were added to the appropriate wells in the plate and the plates were incubated for 6 days at 37 °C. AlamarBlue was added to the plates, and the plates were incubated for an additional 24 h at 37 °C. The MIC was the lowest concentration (µg/mL) of compound that inhibited bacterial growth and prevented a color change.
Cytotoxicity Assay
The cytotoxicity of the compounds was tested against a cell line that originated from the epithelial cells from the kidneys of Cercopithecus aethiops (Vero). Vero cells were grown without CO2 in L15 media supplemented with antibiotics and heat inactivated calf serum. Serial 2-fold dilutions of the drugs were prepared in the 96-well microplates. The Vero cells, in media containing 2X Alamar Blue, were added to the wells to a final concentration of 1.3 × 104 cells per well. The plates were incubated for 3 days at 37 °C. The IC50 was calculated according to manufacturer directions.
Mtb FtsZ Protein Preparation.22
E.coli expression plasmid encoding the ftsz gene (pET 15b vector) was transformed into 100 µL of BL21(DE3) cells. The transformed cells were plated onto LB plates, containing 100 µg/mL ampicilin. The antibiotic concentration was kept the same for the following steps. The plates were incubated overnight at 37 °C. The colonies were picked and grown in 10 mL of LB media at 37 °C at 250 rpm shake rate. The inoculum was transferred to 1 L of LB media in a 4 L flask and grown to an OD of 0.6 at A600. Then, 1 mM IPTG was added to induce protein expression overnight at 25 °C at 250 rpm shake rate. Next day the cells were harvested at 4,225×g for 15 min and re-suspended in approximately 20–30 mL binding buffer (500 mM NaCl, 20 mM sodium phosphate, pH 7.8). The re-suspended cells were lysed using cell disruptor. The lysate was centrifuged in an ultracentrifuge at 126,603×g, 4 °C for 90 min. The supernatant was filtered and loaded onto Ni2+-NTA column washed with 50 mL of binding buffer and eluted using a gradient of binding buffer with 30–500 mM imidazole. The eluted protein was first dialyzed against the polymerization buffer (50 mM MES, 5mM MgCl2, 50 mM KCl, pH 6.5) and then polymerization buffer containing 10% v/v glycerol. Alternatively, the eluted protein was dialyzed against buffer containing 50 mM Tris, 5mM MgCl2, 50 mM KCl, pH 7.2 followed by buffer containing 10% v/v glycerol. The protein after dialysis was concentrated and stored at −80 °C for further use. Since the number of aromatic residues in Mtb FtsZ protein are low (Tyr: 1, Trp: 0), it is not reliable to follow concentration of protein by scanning at A280. The concentration of protein was therefore ascertained using the Bradford kit from Sigma.
Polymerization Assay.22, 38
FtsZ (15 µM) was equilibrated in polymerization buffer (50 mM MES, 100 mM KCl, 5 mM MgCl2, pH 6.5) at 25 °C. Polymerization was measured by light scattering using a PTI-QM4 spectrofluorimeter set at 400nm excitation/emission, 2 nm slit width, 860 V). Polymerization was initiated with 50 µM GTP and monitored for up to 30 minutes. Compound stock solutions were prepared in DMSO and incubated with Mtb FtsZ prior to initiation of polymerization with GTP. The percentage of DMSO was maintained at 2% for all experiments.
Transmission Electron Microscopy (TEM) Analysis.22
Stock solution of compound 5f was prepared in ethanol. Mtb FtsZ (5 µM) was incubated with 40 or 80 µM of compound 5f in the polymerization buffer (50 mM MES, 5mM MgCl2, 100 mM KCl, pH 6.5) for 15 min on ice. To each solution was added GTP to the final concentration of 25 µM. The resulting solution was incubated at 37 °C for 30 min. The incubated solution was diluted 2 times with the polymerization buffer and immediately transferred to carbon coated 300 mesh formvar copper grid and negatively stained with 1% uranyl acetate. To visualize the effect of compound 5f on preformed polymers, Mtb FtsZ (5 µM) was allowed to polymerize at 37 °C in the presence of GTP (25 µM) for 30 mins. Compound 5f (40 µM or 80 µM) was added to the polymerized protein and incubated for additional 5 min. The resulting solution was diluted 2 times with the polymerization buffer and immediately transferred to carbon coated 300 mesh formvar copper grid and negatively stained with 1% uranyl acetate. The samples were viewed with a FEI Tecnai12 BioTwinG transmission electron microscope at 80 kV. Digital images were acquired with an AMT XR-60 CCD digital camera system.
Supplementary Material
Acknowledgment
This research is supported by grants the National Institutes of Health (Grant AI078251 to I.O. and AI082164 to R.A.S.). The authors gratefully acknowledge the technical support of Susan Van Horn for TEM operation at the Microscopy Imaging Center at Stony Brook University, NY.
Footnotes
Supporting Information Available: Spectral data of new benzimidazole intermediates are available free of charge via the Internet at http://pubs.acs.org.
References
- 1.World Health Organization, Tuberculosis: Data and Country Profiles. Available at http://www.who.int/tb/publications/global_report/gtbr12_executivesummary.pdf.
- 2.Gandhi NR, Moll A, Sturm AW, Pawinski R, Govender T, Lalloo U, Zeller K, Andrews J, Friedland G. Extensively drug-resistant tuberculosis as a cause of death in patients co-infected with tuberculosis and HIV in a rural area of South Africa. Lancet. 2006;368:1575–1580. doi: 10.1016/S0140-6736(06)69573-1. [DOI] [PubMed] [Google Scholar]
- 3.Bloom BR, Murray CJ. Tuberculosis: commentary on a reemergent killer. Science (New York, N.Y.) 1992;257:1055–1064. doi: 10.1126/science.257.5073.1055. [DOI] [PubMed] [Google Scholar]
- 4.Tuberculosis: Basic TB facts. Center for Disease Control and Prevention (CDC) http://www.cdc.gov/tb/topic/basics/default.html.
- 5.Raviglione MC. Issues facing TB control (7). Multiple drug-resistant tuberculosis. Scott Med J. 2000;45:52–55. doi: 10.1177/00369330000450S124. discussion 56. [DOI] [PubMed] [Google Scholar]
- 6.Kumar K, Awasthi D, Berger WT, Tonge PJ, Slayden RA, Ojima I. Discovery of anti-TB agents that target the cell-division protein FtsZ. Future Med. Chem. 2010;2:1305–1323. doi: 10.4155/fmc.10.220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Margalit DN, Romberg L, Mets RB, Hebert AM, Mitchison TJ, Kirschner MW, RayChaudhuri D. Targeting cell division: Small-molecule inhibitors of FtsZ GTPase perturb cytokinetic ring assembly and induce bacterial lethality. [Erratum to document cited in CA141:271048] Proc. Natl. Acad. Sci. U. S. A. 2004;101:13969. doi: 10.1073/pnas.0404439101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vollmer W. The prokaryotic cytoskeleton: A putative target for inhibitors and antibiotics? Appl. Microbiol. Biotechnol. 2006;73:37–47. doi: 10.1007/s00253-006-0586-0. [DOI] [PubMed] [Google Scholar]
- 9.Huang Q, Tonge Peter J, Slayden Richard A, Kirikae T, Ojima I. FtsZ: a novel target for tuberculosis drug discovery. Curr. Top. Med. Chem. 2007;7:527–543. doi: 10.2174/156802607780059790. [DOI] [PubMed] [Google Scholar]
- 10.Ben-Yehuda S, Losick R. Asymmetric cell division in B. subtilis involves a spiral-like intermediate of the cytokinetic protein FtsZ. Cell (Cambridge, MA, United States) 2002;109:257–266. doi: 10.1016/s0092-8674(02)00698-0. [DOI] [PubMed] [Google Scholar]
- 11.Goehring NW, Beckwith J. Diverse paths to midcell: assembly of the bacterial cell division machinery. Curr. Biol. 2005;15:R514–R526. doi: 10.1016/j.cub.2005.06.038. [DOI] [PubMed] [Google Scholar]
- 12.Leung AK, White LE, Ross LJ, Reynolds RC, DeVito JA, Borhani DW. Structure of Mycobacterium tuberculosis FtsZ reveals unexpected, G protein-like conformational switches. J. Mol. Biol. 2004;342:953–970. doi: 10.1016/j.jmb.2004.07.061. [DOI] [PubMed] [Google Scholar]
- 13.Moller-Jensen J, Loewe J. Increasing complexity of the bacterial cytoskeleton. Curr. Opin. Cell Biol. 2005;17:75–81. doi: 10.1016/j.ceb.2004.11.002. [DOI] [PubMed] [Google Scholar]
- 14.Thanedar S, Margolin W. FtsZ Exhibits Rapid Movement and Oscillation Waves in Helix-like Patterns in Escherichia coli. Curr. Biol. 2004;14:1167–1173. doi: 10.1016/j.cub.2004.06.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Huang Q, Kirikae F, Kirikae T, Pepe A, Amin A, Respicio L, Slayden RA, Tonge PJ, Ojima I. Targeting FtsZ for Antituberculosis Drug Discovery: Noncytotoxic Taxanes as Novel Antituberculosis Agents. J. Med. Chem. 2006;49:463–466. doi: 10.1021/jm050920y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Respicio L, Nair PA, Huang Q, Burcu AB, Tracz S, Truglio JJ, Kisker C, Raleigh DP, Ojima I, Knudson DL, Tonge PJ, Slayden RA. Characterizing Septum Inhibition in Mycobacterium tuberculosis for Novel Drug Discovery. Tuberculosis. 2008;88:420–429. doi: 10.1016/j.tube.2008.03.001. [DOI] [PubMed] [Google Scholar]
- 17.Slayden RA, Knudson DL, Belisle JT. Identification of cell cycle regulators in Mycobacterium tuberculosis by inhibition of septum formation and global transcriptional analysis. Microbiology. 2006;152:1789–1797. doi: 10.1099/mic.0.28762-0. [DOI] [PubMed] [Google Scholar]
- 18.Awasthi D, Kumar K, Ojima I. Therapeutic potential of FtsZ inhibition: a patent perspective. Expert Opin. Ther. Pat. 2011;21:657–679. doi: 10.1517/13543776.2011.568483. [DOI] [PubMed] [Google Scholar]
- 19.Beuria TK, Singh P, Surolia A, Panda D. Promoting assembly and bundling of FtsZ as a strategy to inhibit bacterial cell division: A new approach for developing novel antibacterial drugs. Biochem. J. 2009;423:61–69. doi: 10.1042/BJ20090817. [DOI] [PubMed] [Google Scholar]
- 20.White EL, Suling WJ, Ross LJ, Seitz LE, Reynolds RC. 2-Alkoxycarbonylaminopyridines: inhibitors of Mycobacterium tuberculosis FtsZ. J. Antimicrob. Chemother. 2002;50:111–114. doi: 10.1093/jac/dkf075. [DOI] [PubMed] [Google Scholar]
- 21.Reynolds RC, Srivastava S, Ross LJ, Suling WJ, White EL. A new 2-carbamoyl pteridine that inhibits mycobacterial FtsZ. Bioorg. Med. Chem. Lett. 2004;14:3161–3164. doi: 10.1016/j.bmcl.2004.04.012. [DOI] [PubMed] [Google Scholar]
- 22.Kumar K, Awasthi D, Lee S-Y, Zanardi I, Ruzsicska B, Knudson S, Tonge PJ, Slayden RA, Ojima I. Novel Trisubstituted Benzimidazoles, Targeting Mtb FtsZ, as a New Class of Antitubercular Agents. J. Med. Chem. 2011;54:374–381. doi: 10.1021/jm1012006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Laeppchen T, Hartog AF, Pinas VA, Koomen G-J, Den Blaauwen T. GTP Analogue Inhibits Polymerization and GTPase Activity of the Bacterial Protein FtsZ without Affecting Its Eukaryotic Homologue Tubulin. Biochemistry. 2005;44:7879–7884. doi: 10.1021/bi047297o. [DOI] [PubMed] [Google Scholar]
- 24.Paradis-Bleau C, Beaumont M, Sanschagrin F, Voyer N, Levesque RC. Parallel solid synthesis of inhibitors of the essential cell division FtsZ enzyme as a new potential class of antibacterials. Bioorg. Med. Chem. 2007;15:1330–1340. doi: 10.1016/j.bmc.2006.11.015. [DOI] [PubMed] [Google Scholar]
- 25.Parhi A, Kelley C, Kaul M, Pilch DS, LaVoie EJ. Antibacterial activity of substituted 5-methylbenzo[c]phenanthridinium derivatives. Bioorg. Med. Chem. Lett. 2012;22:7080–7083. doi: 10.1016/j.bmcl.2012.09.097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Beuria TK, Santra MK, Panda D. Sanguinarine Blocks Cytokinesis in Bacteria by Inhibiting FtsZ Assembly and Bundling. Biochemistry. 2005;44:16584–16593. doi: 10.1021/bi050767+. [DOI] [PubMed] [Google Scholar]
- 27.Mathew B, Ross L, Reynolds RC. A novel quinoline derivative that inhibits mycobacterial FtsZ. Tuberculosis (Oxford, U. K.) 2013;93:398–400. doi: 10.1016/j.tube.2013.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kelley C, Zhang Y, Parhi A, Kaul M, Pilch DS, LaVoie EJ. 3-Phenyl substituted 6,7-dimethoxyisoquinoline derivatives as FtsZ-targeting antibacterial agents. Bioorg. Med. Chem. 2012;20:7012–7029. doi: 10.1016/j.bmc.2012.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Parhi A, Lu S, Kelley C, Kaul M, Pilch DS, LaVoie EJ. Antibacterial activity of substituted dibenzo[a,g]quinolizin-7-ium derivatives. Bioorg. Med. Chem. Lett. 2012;22:6962–6966. doi: 10.1016/j.bmcl.2012.08.123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Haydon DJ, Stokes NR, Ure R, Galbraith G, Bennett JM, Brown DR, Baker PJ, Barynin VV, Rice DW, Sedelnikova SE, Heal JR, Sheridan JM, Aiwale ST, Chauhan PK, Srivastava A, Taneja A, Collins I, Errington J, Czaplewski LG. An Inhibitor of FtsZ with Potent and Selective Anti-Staphylococcal Activity. Science (Washington, DC, U. S.) 2008;321:1673–1675. doi: 10.1126/science.1159961. [DOI] [PubMed] [Google Scholar]
- 31.Haydon DJ, Bennett JM, Brown D, Collins I, Galbraith G, Lancett P, Macdonald R, Stokes NR, Chauhan PK, Sutariya JK, Nayal N, Srivastava A, Beanland J, Hall R, Henstock V, Noula C, Rockley C, Czaplewski L. Creating an antibacterial with in vivo efficacy: synthesis and characterization of potent inhibitors of the bacterial cell division protein FtsZ with improved pharmaceutical properties. J. Med. Chem. 2010;53:3927–3936. doi: 10.1021/jm9016366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Plaza A, Keffer JL, Bifulco G, Lloyd JR, Bewley CA. Chrysophaentins A–H, antibacterial bisdiarylbutene macrocycles that inhibit the bacterial cell division protein FtsZ. J. Am. Chem. Soc. 2010;132:9069–9077. doi: 10.1021/ja102100h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mathew B, Srivastava S, Ross LJ, Suling WJ, White EL, Woolhiser LK, Lenaerts AJ, Reynolds RC. Novel pyridopyrazine and pyrimidothiazine derivatives as FtsZ inhibitors. Bioorg. Med. Chem. 2011;19:7120–7128. doi: 10.1016/j.bmc.2011.09.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Awasthi D, Park B, Kumar K, Melief E, Knudson S, Slayden RA, Iwao Ojima I. 243rd ACS National Meeting and Exposition. San Diego, CA: Mar 25–29, 2012. Synthesis and biological evaluation of novel antitubercular trisubstituted benzimidazoles and C-seco taxoids targeting FtsZ; p. 388. MEDI. [Google Scholar]
- 35.Kumar K, Awasthi D, Park B, Melief E, Cummings J, Knudson S, Slayden RA, Ojima I. 244th ACS National Meeting & Exposition. Philadelphia, PA: Aug 19–23, 2012. Synthesis, Optimization and Biological Evaluation of Novel Benzimidazoles as Efficacious Antitubercular and Broad–Spectrum Antimicrobial Agents, Targeting FtsZ; p. 18447. MEDI. [Google Scholar]
- 36.Collins L, Franzblau SG. Microplate Alamar blue assay versus BACTEC 460 system for highthroughput screening of compounds against Mycobacterium tuberculosis and Mycobacterium avium. Antimicrob. Agents Chemother. 1997;41:1004–1009. doi: 10.1128/aac.41.5.1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Boyne ME, Sullivan TJ, amEnde CW, Lu H, Gruppo V, Heaslip D, Amin AG, Chatterjee D, Lenaerts A, Tonge PJ, Slayden RA. Targeting Fatty Acid Biosynthesis for the Development of Novel Chemotherapeutics against Mycobacterium tuberculosis: Evaluation of ARing-Modified Diphenyl Ethers as High-Affinity InhA Inhibitors. Antimicrob. Agents Chemother. 2007;51:3562–3567. doi: 10.1128/AAC.00383-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.White EL, Ross LJ, Reynolds RC, Seitz LE, Moore GD, Borhani DW. Slow Polymerization of Mycobacterium tuberculosis FtsZ. J. Bacteriol. 2000;182:4028–4034. doi: 10.1128/jb.182.14.4028-4034.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.