Abstract
Puberty is a major developmental milestone controlled by the interaction of genetic factors and environmental cues of mostly metabolic and circadian nature. An increased pulsatile release of the decapeptide gonadotropin releasing hormone (GnRH) from hypothalamic neurosecretory neurons is required for both the initiation and progression of the pubertal process. This increase is brought about by coordinated changes that occur in neuronal and glial networks associated with GnRH neurons. These changes ultimately result in increased neuronal and glial stimulatory inputs to the GnRH neuronal network and a reduction of transsynaptic inhibitory influences. While some of the major players controlling pubertal GnRH secretion have been identified using gene-centric approaches, much less is known about the system-wide control of the overall process. Because the pubertal activation of GnRH release involves a diversity of cellular phenotypes, and a myriad of intracellular and cell-to-cell signaling molecules, it appears that the overall process is controlled by a highly coordinated and interactive regulatory system involving hundreds, if not thousands, of gene products. In this article we will discuss emerging evidence suggesting that these genes are arranged as functionally connected networks organized, both internally and across sub-networks, in a hierarchical fashion. According to this concept, the core of these networks is composed of transcriptional regulators that, by directing expression of downstream subordinate genes, provide both stability and coordination to the cellular networks involved in initiating the pubertal process. The integrative response of these gene networks to external inputs is postulated to be coordinated by epigenetic mechanisms.
Keywords: Female puberty, hypothalamus, transcriptional regulation, gene networks, systems biology, neuroendocrine control, neurotransmission, glial-neuronal communication
The biological underpinnings of the cellular systems controlling puberty
The basic neuroendocrine mechanisms controlling the initiation of female reproductive capacity are well characterized. An increase in pulsatile release of gonadotropin-releasing hormone (GnRH) is ultimately responsible for setting in motion the endocrine manifestations of puberty. This change is, in turn, determined by modifications in transsynaptic (Kordon and others, 1994; Ojeda and Terasawa, 2002) and glial (Ojeda, Lomniczi, and Sandau, 2010; Ojeda, Lomniczi, and Sandau, 2008b) inputs to the GnRH neuronal network. While the transsynaptic changes involve an increase in excitatory inputs and a reduction in inhibitory influences (Ojeda et al., 2002; Plant and Witchel, 2006; Terasawa and Fernandez, 2001), the glial component is predominantly facilitatory, and exerted by growth factors and small molecules that stimulate GnRH secretion (Lomniczi and Ojeda, 2009; Ojeda and Skinner, 2006).
In the last ten years, significant progress has been made towards identifying neuronal subsets involved in both the excitatory and inhibitory control of GnRH secretion. We now know that the excitatory control of puberty is not only provided by glutamatergic neurons (Ojeda et al., 2006; Plant et al., 2006), but even more conspicuously by neurons that produce kisspeptin [reviewed in (d’Anglemont, X and Colledge, 2010; Oakley, Clifton, and Steiner, 2009)]. Kisspeptins are a family of peptides encoded by the KISS1/Kiss1 gene and that act as powerful stimulators of GnRH release (Kauffman, Clifton, and Steiner, 2007; Oakley et al., 2009; Shahab and others, 2005); in their absence or in the absence of its receptor (known as GPR54 or Kiss1R), puberty fails to occur (de Roux and others, 2003; Lapatto and others, 2007; Seminara and others, 2003; Topaloglu and others, 2012). In primates, kisspeptin neurons are mostly located in the arcuate nucleus (ARC) of the medial basal hypothalamus (Shahab et al., 2005). In rodents, there is a second population of kisspeptin neurons located in the periventricular region of the anteroventral periventricular nucleus (AVPV) (Clarkson and others, 2009; Gottsch and others, 2004). It appears now clear that kisspeptin neurons of the ARC are required for pulsatile GnRH release (Navarro and others, 2011; Wakabayashi and others, 2010), and that at least in rodents AVPV kisspeptin neurons are needed for the pre-ovulatory surge of gonadotropins (Khan and Kauffman, 2011; Smith and others, 2006).
In all mammalian species so far studied the first neuroendocrine manifestation of puberty is a diurnal increase in pulsatile LH release [reviewed in (Ojeda et al., 2006)], which is in all likelihood driven by kisspeptin neurons of the ARC. These cells have been termed KNDy neurons (Lehman, Coolen, and Goodman, 2010; Navarro et al., 2011), because they produce Kisspeptin, Neurokinin B (NKB) and Dynorphin (Navarro et al., 2011; Wakabayashi et al., 2010). It is currently believed that KNDy neurons release NKB, which acts on other KNDy neurons via specific receptors to stimulate kisspeptin release (Navarro et al., 2011; Wakabayashi et al., 2010) [but see (Kinsey-Jones and others, 2012)]. Like KISS1 and its receptor, inactivating mutations of TAC3/Tac2 (which encodes NKB) or TACR3 (the gene encoding the NKB receptor), results in pubertal failure (Topaloglu and others, 2008). NKB and kisspeptin are released periodically, and this oscillatory behavior is thought to be determined by a phase-delayed inhibitory feedback of dynorphin on NKB release (Navarro et al., 2011; Wakabayashi et al., 2010). Dynorphin is an opioid peptide that inhibits GnRH secretion (Kinoshita and others, 1982; Navarro and others, 2009; Schulz and others, 1981). Diagrams describing these interactions have been published (d’Anglemont, X et al., 2010; Lehman et al., 2010; Wakabayashi et al., 2010).
The inhibitory transsynaptic circuitry controlling GnRH release involves at least three different neuronal subsets. GABAergic and opiatergic neurons have been known for many years to be central players [reviewed in (Terasawa et al., 2001)]. More recently, evidence has been provided suggesting that RFamide-related peptide (RFRP), the mammalian ortholog of the peptide gonadotrophin-inhibiting hormone (GnIH) in birds (Ebling and Luckman, 2008), is a physiological inhibitor of GnRH neurons in mammals (Ducret, Anderson, and Herbison, 2009; Gibson and others, 2008; Tsutsui and others, 2010). As such, RFRP-containing neurons may be significant components of the restraining mechanism that maintains GnRH secretion in check during reproductive maturation. RFRP neurons may use one or two peptides (RFRP1 and RFRP3) for transsynaptic communication; these peptides are recognized by a high-affinity receptor termed GPR147 or NPFFR1 (Hinuma and others, 2000; Tsutsui et al., 2010), and a low-affinity receptor termed GPR74 or NPFFR2 (Fukusumi, Fujii, and Hinuma, 2006). Because GPR147 is expressed in GnRH neurons (Ducret et al., 2009; Poling and others, 2012), RFRP-containing neurons can directly repress GnRH neuronal function. In contrast to this simplicity, GABAergic neurons can affect GnRH secretion indirectly, via inhibitory effects exerted on neurons connected to the GnRH neuronal network (Ojeda et al., 2006; Terasawa et al., 2001), or directly using excitatory mechanisms set in motion by the activation of GABAA receptors located on GnRH neurons themselves (DeFazio and others, 2002; Moenter and DeFazio, 2005). Opiatergic neurons appear to exert a pure inhibitory tone to GnRH neurons, but this input is provided by different peptides acting on different receptors (Kordon et al., 1994); as in the case of GABAergic inputs, opiatergic inhibition may be exerted directly on GnRH neurons (Dudas and Merchenthaler, 2006) or indirectly on neurons involved in the stimulatory control of the GnRH neuronal network, such as KNDy neurons of the ARC (Navarro et al., 2009). Additional components of the regulatory system controlling the onset of female puberty include novel molecules required for glutamate release (Choi and others, 2008; Ha and others, 2008) and GnRH neuron excitability (Garcia-Rudaz and others, 2008).
In addition to neurons, the pubertal activation of GnRH secretion involves the participation of glial cells (Ojeda et al., 2010; Ojeda et al., 2008b). Astrocytes and tanycytes (ependymoglial cells lining the ventro-lateral surface of the third ventricle) produce signaling molecules that stimulate GnRH release, and contribute to determining the timing of puberty [reviewed in (Lomniczi et al., 2009; Ojeda et al., 2010)]. Glial cells facilitate GnRH secretion via two complementary mechanisms. One of them involves growth factors of at least four different families [reviewed in (Mahesh, Dhandapani, and Brann, 2006; Prevot, 2002)]. Transforming growth factor-beta (TGFβ) is recognized by cell-membrane receptors endowed with serine-threonine kinase; the epidermal growth factor (EGF) family, basic fibroblast growth factor (bFGF), and insulin-like growth factor 1 (IGF-I) are recognized by receptors with tyrosine kinase activity. Genetic disruption of erbB (erythroblastosis B) receptors (which recognize the members of the EGF family) delays female sexual development due, at least in part, to impaired erbB ligand-induced glial prostaglandin E2 (PGE2) release (Lomniczi et al., 2009). Preventing the proteolytic processing of erbB ligands in astrocytes delays puberty (Lomniczi and others, 2006) and disrupting astrocytic PGE2 production drastically diminishes the electrophysiological activity of GnRH neurons (Clasadonte and others, 2011).
The second mechanism of glia-to-GnRH neuron communication involves plastic rearrangement in cell adhesiveness provided by at least three different cell-cell communications systems: One involves the sialylated form of the neural cell adhesion molecule NCAM (PSA-NCAM) (Parkash and Kaur, 2005; Perera, Lagenaur, and Plant, 1993). Another requires the adhesion molecule Synaptic Cell Adhesion Molecule 1 (SynCAM1) (Sandau and others, 2011a; Sandau and others, 2011b), and a third one is based on the interaction of neuronal contactin with glial Receptor-like Protein Tyrosine Phosphatase-β (RPTPβ) (Parent and others, 2007). All three systems use adhesive proteins containing intracellular signaling domains, suggesting that the interaction of glial cells with GnRH neurons not only involve secreted bioactive molecules (growth factors, prostaglandins, etc.), but also intracellular signaling mechanisms set in motion by adhesive molecules [reviewed in (Lomniczi et al., 2009)]. A more comprehensive coverage of the mechanisms controlling the onset of puberty can be found in earlier reviews (Ojeda et al., 2006; Ojeda and Urbanski, 1994; Tena-Sempere, 2006; Terasawa et al., 2001), and in several chapters of this book (Tena-Sempere, Megan Hagenauer/Terri Lee, Dennis Styne).
The neuroendocrine control of puberty involves many genes of different functions
The demonstration in 2003 by two independent groups that the lack of functional GPR54 receptors results in pubertal failure (de Roux et al., 2003; Seminara et al., 2003) energized the field of puberty to an unprecedented extent, leading to a flurry of publications suggesting a vital role of kisspeptin for a diversity of reproductive functions both at central and peripheral levels. It would appear intuitively evident, however, that the kisspeptin system does not work in isolation. Maintaining reproductive function is such an essential requirement for the preservation of the species that the robustness of the system might be seriously compromised if it did not have built-in safeguard mechanisms to ensure functional continuity in the event of loss of a component. Accordingly, the neuroendocrine regulatory complex within which Kiss1 operates would be expected to follow the same general principles governing other biological networks. Though controlled by a small number of genes, these networks are robust (that is, they have a significant degree of internal redundancy) and are endowed with a high degree of error tolerance (Basso and others, 2005). This general feature appears to be valid for the control of GnRH secretion, because the early removal of kisspeptin neurons appears to be followed by the activation of compensatory mechanisms (Mayer and Boehm, 2011) that reestablish normal reproductive function. There are also compensatory mechanisms that allow reproductive capacity to be sustained in the absence of Tac3R (Yang and others, 2012), indicating that in each case loss of a pivotal element of the network is followed by activation of alternative pathways. A note of caution needs to be introduced here because ablation of kisspeptin neurons did not eliminate all of these neurons, which raises the possibility that only a few kisspeptin neurons are required for normal function.
These compensatory pathways may require some of the many genes implicated in the neuroendocrine control of puberty (Eaves and others, 2004; Gajdos and others, 2008; Krewson and others, 2004; Ojeda and others, 2006; Seminara and Crowley, Jr., 2001). Although monogenic mutations affecting genes such as GNRHR (Bedecarrats and Kaiser, 2007), GPR54 (de Roux et al., 2003; Seminara et al., 2003), KiSS1 (Lapatto et al., 2007; Topaloglu et al., 2012), TAC3 and TACR3 (Topaloglu et al., 2008), result in absence of pubertal development, the combined fraction of individuals affected by these mutations does not surpass 5% of the total population affected by pubertal disorders. The contribution of additional genes is suggested by the results of genome-wide association studies demonstrating that sequence variations in at least 30 genes are associated with an early age at menarche (He and others, 2009; Ong and others, 2009; Perry and others, 2009; Sulem and others, 2009). Prominent among these genes are the post-transcriptional repressor LIN28b (He et al., 2009; Ong et al., 2009; Perry et al., 2009) and several genes encoding proteins associated with body mass index, energy homeostasis and hormone regulation (Elks and others, 2010). Also of interest is the unexpected association of two Zinc-finger (ZNF) genes, ZNF462 (Perry et al., 2009) and ZNF483 (Elks et al., 2010) with the age at menarche. ZNFs constitute a large family of transcriptional repressors (Filion and others, 2006; Shannon and others, 1996; Urrutia, 2003; Vogel and others, 2006), some of which can bind to both DNA and RNA (Burdach and others, 2012) to inhibit gene expression. In addition to these findings, earlier studies from our laboratory identified several transcriptional regulators of puberty, such as the POU-domain gene Oct2 (Ojeda and others, 1999), the homeodomain gene Ttf1/Nkx2.1 (Mastronardi and others, 2006), and a novel gene (Rampazzo and others, 2000), which we termed Eap1 (Enhanced At Puberty1) (Heger and others, 2007).
The above considerations make it clear that there are numerous neuroendocrine genes that are necessary for the onset of puberty, but that there are redundancies in the system. Therefore, it is import to focus on the upper echelon genes regulating the expression or repression of the neuroendocrine axis. Accordingly, this review will focus on the upstream transcriptional regulators of the pubertal process. We will first provide the reader with some basic information about the approaches one may use to identify, interpret and integrate the information derived from high throughput data into a biologically testable model.
General Structure of a Genetic Network
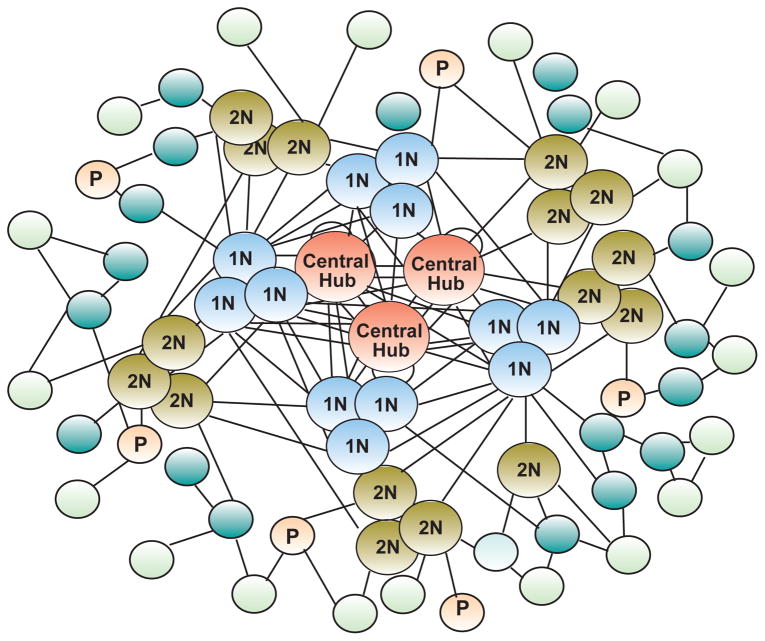
Genetic networks are composed of individual elements that interact with each other (Klipp and others, 2005). Some of these elements act as “portals” receiving and processing information from the environment; others, function mostly within the network. All networks contain central “hubs”, which are strongly interconnected and direct the flow of information throughout the entire network. Subordinate “nodes”, located at various hierarchical positions (and within different intra– and inter-cellular levels), respond to commands emanating from the central hubs, and relayed to them either directly or via first and second “neighbors”. This organization allows for the regulation of a complex system using relatively few genes, since the complexity of the system comes from the connectivity and not so much from the number of genes or hubs. Signal propagation within the network may be both fast, but evanescent, and slow, but stable (Luscombe and others, 2004). Because the network’s nodes are strongly interconnected and this connectivity does not increase (i.e. it is not “scaled up”) as the complexity of the network increases, genetic networks are considered to be scale-free (Basso et al., 2005). In addition, they are hierarchical, because highly connected nodes exhibit preferential interactions among themselves (Klipp et al., 2005). Due to this architectural arrangement, networks contain “upper echelon” hubs that are highly interconnected, and a multitude of peripheral nodes that become increasing less connected as they move away from the major hubs (Fig. 1). This loss of connectivity is not surprising because the size (and complexity) of a network can increase markedly as it expands from its core (see for instance (Basso et al., 2005; Carro and others, 2010; Luscombe et al., 2004). Groups of genes exhibiting a high overlap of connected nodes are considered as “modules” within a network. It is important to also consider that the interactions between transcriptional nodes may vary in response to different stimuli, suggesting that rewiring is a biological feature of network function (Luscombe et al., 2004). An additional feature worth mentioning is that the “edges” of the network (i.e., the connectivity) can consist of multiple types of interactions (co-expression, physical contact, transcriptional control, etc.) all of which complement and overlay one another.
Figure 1.
The general organization of a biological network. The individual elements composing the network interact with each other. Some of these elements (P, portals) receive information from the environment; others, function mostly within the network. The central “hubs” or “nodes” are strongly interconnected and direct the flow of information throughout the entire network. Subordinate nodes respond to commands emanating from the central hubs; this information can be relayed to them either directly or via first (1N) and (2N) second “neighbors”. The architectural arrangement of the network is such that they contain “upper echelon” nodes that are highly interconnected, and a multitude of peripheral nodes that become increasing less connected as they move away from the major hubs. The arrangement of first and second nodes in clusters of three illustrates the concept that the network contains “modules”, i.e. sub-groups of genes with highly related functions. Depending on the data type, interactions between nodes (termed “edges”) may represent co-expression, physical association, transcriptional control, or other types of gene or gene product interactions. The nodes are colored according to their relative position within the network: orange = central nodes; light blue = first neighbors; light brown = second neighbors; light orange = portal nodes; dark blue = subordinate nodes located sub-peripherally; light green = most peripherally located nodes.
Tools for construction and visualization of biological networks
An initial step in the identification of biological networks is the use of high throughput approaches (such as DNA arrays and proteomics). A late step involves perturbation of the system (by either overexpressing or silencing key components of the presumed network) (Davidson and others, 2002). Below, we describe our workflow, which is representative of current methodologies in biological network construction. A main computational framework that can be employed to identify the main modules, which define the high-level architecture of the network, is Weighted Gene Co-expression Network Analysis (WGCNA) (Zhang and Horvath, 2005). This framework allows the investigator to (i) construct gene co-expression networks, (ii) identify network modules and their relationships, (iii) relate the identified modules and module interactions to external information, such as functional enrichment and protein-protein interaction, and (iv) find key drivers in modules of interest with respect to the central hypothesis in order to identify new targets for experimental validation via RNAi silencing/gene overexpression (see for instance (Carro et al., 2010; Iancu and others, 2010; Ponomarev and others, 2012)). Alternative strategies for the construction of biological networks have been employed by other investigators (for instance see (Carro et al., 2010; Ideker and others, 2001; Longabaugh, Davidson, and Bolouri, 2005; Ueda and others, 2005).
To aid our analysis we have developed an extended version of the WGCNA R package (Langfelder and Horvath, 2008) (http://www.genetics.ucla.edu/labs/horvath/CoexpressionNetwork/Rpackages/WGCNA/) (augmented with local connectivity analysis and alternative topological overlap measures). Using this tool, we first compute the Pearson correlation coefficient for all pairs of genes in an expression data set and produce a matrix of pairwise correlations. We then transform the correlation matrix into an adjacency matrix using a power function that computes the connection strength between a pair of probes by raising the correlation value to a power determined by the scale-free topology criterion (Zhang et al., 2005). It may also be possible to employ the Maximal Information-based Nonparametic Exploration (MINE) association metric (Reshef and others, 2011) (http://www.exploredata.net/) as an alternative or supplement to the Pearson correlation. Because MINE can detect nonlinear associations between variables, it can capture biologically relevant relationships between genes that the Pearson correlation may not detect.
Module identification
With the connection strengths determined in an adjacency matrix, the next step is to identify modules in the network. The degree to which a pair of genes share neighbors is measured by the ‘topological overlap’ metric (Ravasz and others, 2002; Zhang et al., 2005), which accounts for the number of genes connected to both genes normalized by the total number of connections. We have extended the topological overlap metric by using graphlet counts (Przulj, 2007), a technique for measuring refined local connectivity. We have developed a publicly available CytoScape plug-in, GraphletCounter (Whelan and Sonmez, 2012) that computes the number of subnetworks (up to 5 nodes) to which a node is connected, producing a connectivity feature vector. We use inner products of the connectivity vectors as an alternative topological metric. Using a topological overlap measure as opposed to thresholding the raw adjacency values reduces the spurious connection strengths and results in a more robust method for estimating putative modules. With the topological overlap matrix, we identify the network modules as branches of the clustering tree resulting from the application of the “dynamic tree cutting algorithm” (Langfelder, Zhang, and Horvath, 2008), which takes advantage of the internal structure of the dendrogram in cutting the branches and identifying modules.
Functional enrichment
Each module’s gene make-up is tested for GO category enrichment (Ashburner and others, 2000) using the GOstats R package (Falcon and Gentleman, 2007) (http://www.bioconductor.org/packages/2.10/bioc/html/GOstats.html). To remove biases due to the nested structure of the GO terms, a graph decorrelation procedure (Alexa, Rahnenfuhrer, and Lengauer, 2006) is employed. The resulting p-values are adjusted for multiple hypotheses testing using False Discovery Rate (FDR) and transformed into q-values (Storey, 2003), as well as by the Bonferroni correction.
Static models of biological networks
A gene network for the control of puberty can be constructed by combining a large variety of functional association data in the meta-network framework GeneMANIA (Warde-Farley and others, 2010), thus allowing the prediction of the function of modules and/or gene sets. GeneMANIA finds interactions and complementary genes that are related to a set of input genes. For the control of puberty, the input sets may be generated using (i) existing gene lists from preliminary data explained in detail in the preceding sections, (ii) genes that display a specified temporal expression pattern in microarray experiments, or (iii) a combination of both. Association data available for forming complementary network edges include protein and genetic interactions, pathways, co-expression, co-localization and protein domain similarity. The basic idea is to find new members of a pathway or complex, find additional genes that may have been missed or augment the network with new genes with a specific function.
GeneMANIA accomplishes this goal by searching for related genes in many large, publicly available biological datasets, including protein-protein, protein-DNA and genetic interactions, pathways, reactions, gene and protein expression data, protein domains and phenotypic screening profiles. Main datasets such as BioGRID, PathwayCommons, Pfam are part of the search as well as datasets from a wealth of datasets from recent papers. The data are available for human, mouse and rat, and the static networks built for the three organisms can be compared, complemented, and novel orthologs identified.
Proteome interactions: transcriptome co-expression and transcription factor (TF) binding sites
To perform this analysis, the gene network co-expression patterns are compared with a manually compiled protein-protein interactions (PPI) database retrieved from the Human Protein Reference Database (HPRD) (Keshava Prasad and others, 2009; Peri and others, 2003). Using Entrez IDs, one can select the network genes that are also present in the list of HPRD gene products. The network genes with PPI interactions can then be selected and used to compute the average topological overlap. Genomic upstream DNA sequences are analyzed for TF binding sites using TRANSFAC (http://www.biobase.de), and the results are incorporated into the network model. We are currently attempting to incorporate epigenetics information to the analysis of predicted transcription factor binding sites by applying a technique we recently developed (Wright and others, 2011) to estimate occupancy of predicted sites based on nearby epigenetic markers.
Dynamic developmental models via BioTapestry
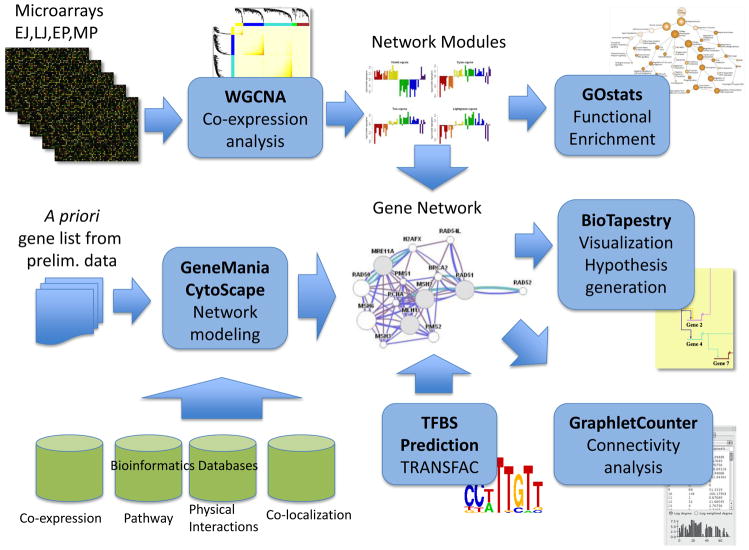
Because network models can be generated to include multiple developmental time point variations in gene expression, a model of gene action over time can be created. The resulting networks can be visualized using the CytoScape software (Shannon and others, 2003) (http://www.cytoscape.org), which allows for interactive exploration of interactions in a network. The CytoScape network can be overlaid with expression data to help identify active pathways in the network. To facilitate this process one can explore the KEGG pathway database for all known pathways containing the highest number of genes from the set of interest. The resulting CytoScape models for each time point can then be saved in a Systems Biology Markup Language (SBML) format (Finney and Hucka, 2003) allowing it to be viewed, analyzed, and refined using software packages, such as the Systems Biology Workbench (SBW) (Sauro and others, 2003) (http://sbw.sourceforge.net). SBML also allows the investigator to share models easily with other research groups. Incorporating temporal and spatial characteristics of gene expressions in the model can be achieved using the BioTapestry software (Longabaugh et al., 2005) (http://www.biotapestry.org). The BioTapestry model can be used to capture the regulation of gene transcription network and animate observed changes in gene expression patterns over time, such as those that occur during pre- and peripubertal development. The different components of a pipeline that can be used for the computational analysis and representation of biological networks are shown in Fig. 2.
Figure 2.
Bioinformatics pipeline and network construction framework. High-throughput experimental data (such as expression) are analyzed both (i) de novo, via Weighted Gene Co-expression Network Analysis (WGCNA), resulting in network modules and their associated enrichment in Gene Ontology categories, such as biological process, molecular function and subcellular localization, and (ii) by building gene networks based on genes identified in preliminary experiments and/or demonstrating a certain temporal expression profile, drawing from a rich battery of datasets via GeneMANIA, a network construction framework that works within CytoScape. Prediction of functional elements such as transcription factor binding sites further elucidates the gene network through the use of motifs in TRANSFAC and the latest ChIP-seq data in ENCODE. The resulting network is visualized and overlaid with expression data in either CytoScape or BioTapestry, allowing the generation of hypotheses for experimental validation (for instance, via RNAi-mediated silencing). The network can also be analyzed for connectivity using GraphletCounter, a publicly available CytoScape plug-in that computes the number of subnetworks (up to 5 nodes) to which a node is connected. EJ = early juvenile period; LJ = late juvenile period; EP = early proestrus, the phase of puberty in the rat when uterine fluid begin to accumulate indicating increased estrogen secretion; MP = midpuberty, the pubertal phase that correspond to the day of the first preovulatory surge of gonadotropins in the rat; A =adulthood; TFBS = transcription factor binding sites.
A gene network controlling puberty
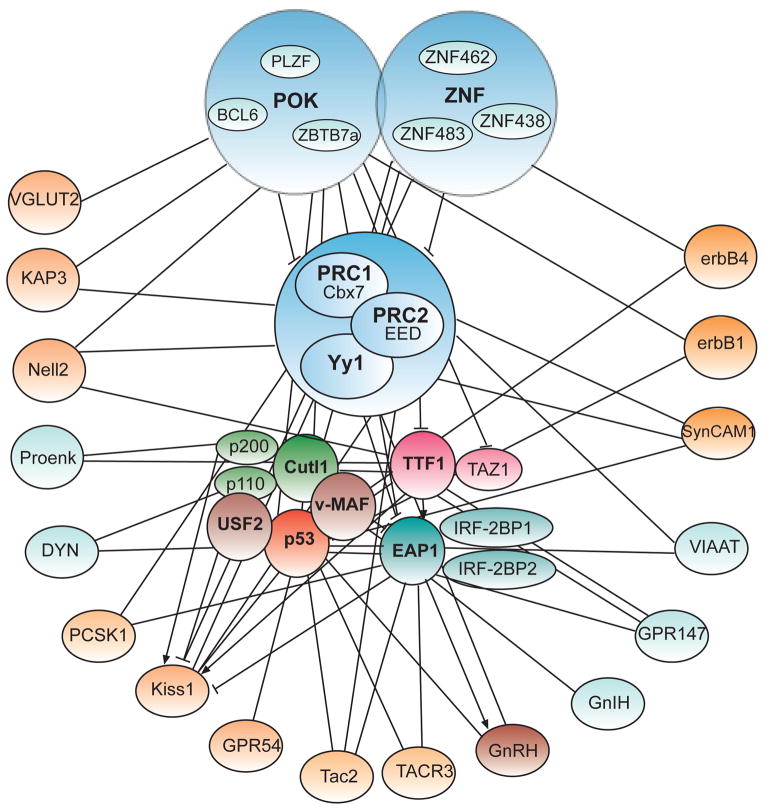
An issue raised by these observations is whether these genes interact with other elements of the neuroendocrine brain to form a biological network with functional significance. Using high-throughput, genomic and proteomic approaches (Choi et al., 2008; Ha et al., 2008; Heger et al., 2007; Lee and others, 2001; Mastronardi et al., 2006; Ojeda et al., 1999; Ojeda et al., 2006; Parent and others, 2008; Roth and others, 2006), in combination with some of the tools described above, we obtained an initial glimpse into the general structure and hierarchical arrangement of one of these networks (Ojeda and others, 2010; Ojeda et al., 2006; Roth and others, 2007; Roth et al., 2006). We used the medial basal hypothalamus of both female rats and female rhesus monkeys for DNA array analysis. The rats were euthanized at three different stages, juvenile (25-days of age), early puberty (30–35 days of age) and on the day of the first proestrus (32–37 days of age). These stages were defined according to established criteria (Ojeda et al., 2006). Accumulation of uterine fluid (an indication of estradiol secretion) begins during early puberty and becomes maximal on the day of the first proestrus. However, vaginal opening does not usually occur until the next day, which is the day of the first ovulation. The monkeys were also euthanized at three developmental phases: juvenile (8.9 month–1.8 year-old), early pubertal (2–3-year old) and midpubertal (3–4 year-old). LH levels are low in juvenile animals and begin to increase during the early pubertal period to become distinctly elevated by midpuberty. Analysis of the array data followed by quantitative PCR verification revealed that expression of several genes earlier identified as involved in tumor suppression/tumor formation increases in the hypothalamus at the time of puberty in both rats and monkeys (Roth et al., 2007). Cis-regulatory analysis of shared binding sites predicted the existence of five central hubs (Cutl1/Cdp/Cux1, Maf, p53, Yy1, and Usf2) controlling the network at the transcriptional level (Roth et al., 2007). The model also predicted the existence of many subordinate genes, including KiSS1 (previously known as suppressor of metastasis) (Kotani and others, 2001; Steeg and others, 2003) and SynCAM1 (previously known as tumor suppressor of lung cancer, TSLC1) (Kuramochi and others, 2001; Watabe and others, 2003) (Fig. 3). While KiSS1 is essential for the occurrence of puberty in mice and humans, we now know that SynCAM1 plays a pivotal role in the developmental control of glia-GnRH neuron adhesive communication (Sandau et al., 2011b), and in the mechanism by which astrocytes facilitate GnRH release and control normal female reproductive function (Ojeda, Lomniczi, and Sandau, 2008a; Sandau et al., 2011a).
Figure 3.
Key features of the TRG network. Two modules composed of different types of transcriptional regulators are postulated to be strongly interconnected and to directly control the activity of peripheral nodes via either trans-activational or repressive mechanisms (← = trans-activation; ⊥ = transcriptional repression). In one of these modules, CUTL1 and p53, function as central hubs; in the other, this role is played by TTF1 and EAP1. Both modules regulate the expression of a host of subordinate genes, including puberty activating genes (e.g., KISS1) and genes involved in the inhibitory control of puberty (e.g. opioid peptides, RFRP3/GnIH). The model predicts that the TRG network is subjected to a two-tiered hierarchy of repressive control provided by two modules. One consisting of POK/ZNF genes, would serve are repressor of repressors. The other repressive module is composed of PcG genes. YY1, which in our initial model was considered to be a central TRG node (Roth et al., 2007), can now be assigned to the PcG complex. According to the current model, POK/ZNF proteins repress PcG genes, and in turn PcG proteins repress downstream puberty activating genes. POK/ZNF proteins may also suppress directly downstream genes. Large blue modules = “upstream” transcriptional repressors consisting of three different families of transcription factors; smaller centrally clustered modules of different colors = secondary central modules composed of genetically unrelated genes; peripheral light blue nodes = genes considered to be inhibitory to the pubertal process; peripheral orange nodes = genes considered to be activators of puberty. Lines ending without arrows or blunt ends denote that the nodes are predicted to be connected by in silico analysis (for instance, the presence of transcription factor binding sites in the downstream target gene), but experimental evidence is lacking.
Interestingly, in silico analysis of the network predicted a connection between these central hubs and Oct2, Ttf1, and Eap1, the three genes implicated by other studies as upstream transcriptional regulators of the pubertal process (Heger et al., 2007; Mastronardi et al., 2006) (Fig. 3). Immunohistochemistry and in situ hybridization analysis demonstrated that these genes, as well as other subordinate nodes of the network, are expressed in neuronal and/or glial subsets involved in the control of GnRH secretion, including GnRH neurons themselves (Heger et al., 2007; Mastronardi et al., 2006; Ojeda et al., 2008b; Roth et al., 2007). A recent assessment of some of these in silico predictions (Mueller and others, 2011) showed that that KISS1 expression is not only controlled by TTF1 and EAP1, but also by CUTL1/CUX1 and YY1, two putative central hubs of the TRG network. While TTF1 and the long form of CUX1 (known as p200) are trans-activational, a short form of CUTL1 (p110) and YY1, and to some extent, EAP1 are repressive (Mueller et al., 2011). Using ChIP assays, we observed that all four of these transcription factors are recruited to the KiSS1 promoter; immunohistofluorescence studies showed that they are present in kisspeptin neurons in vivo (Mueller et al., 2011). We further observed that EAP1 gene expression is also under dual transcriptional regulation imposed by TTF1 (trans-activation) and YY1 and CUX1 (repression), and that EAP1 itself appears to control its own expression via a negative auto-feedback loop mechanism (Mueller and others, 2012). Consistent with the general features of a transcriptional regulatory network, TTF1, YY1 and CUX1 associate with the EAP1 promoter region and are expressed in hypothalamic EAP1 neurons, indicating that they are physiologically involved in the control of EAP1 gene expression. Although we initially considered EAP1 and TTF1 as non-TRGs molecules (Ojeda et al., 2010), it has been recently shown that both are implicated in the biology of cancer (Winslow and others, 2011; Yeung and others, 2011). Based on this information, we now believe that these genes can be considered as part of a distinct module within the TRG network (Fig. 3).
To assess the importance of TTF1 and EAP1 in the control of reproductive function we used loss-of function approaches. Conditional deletion of the Ttf1 gene in mice resulted in delayed puberty and disrupted estrous cyclicity (Mastronardi et al., 2006). Microinjection of lentiviral particles carrying a siRNA against Eap1 mRNA into the preoptic area of juvenile female rats, also delayed puberty and disrupted estrous cyclicity (Heger et al., 2007). Similar injections into the medial basal hypothalamus of rhesus monkeys obliterated menstrual cyclicity (Dissen and others, 2012) indicating that EAP1 is crucial for the ARC to maintain menstrual cyclicity in higher primates. This role is supported by the recent finding that a single nucleotide polymorphism in the 5′-flanking region of the EAP1 gene is associated with reduced EAP1 transcription and oligomenorrhea/amenorrhea in nonhuman primates (Lomniczi and others, 2012a).
The draft model of this TRG network is imperfect in many different ways. Firstly, although based on interactions taking place at different stages of pubertal development, it essentially depicts a static view of these interactions. Secondly, it does not identify the cell type (neurons or glia) where these interactions may be occurring. Lastly, it does not appropriately considered the homeostatic process of post-translational processing that - required for formation of biological active peptides (Artenstein and Opal, 2011) – may be essential for both the short and the long-term activity of the regulatory system.
The potential involvement of additional transcriptional regulatory networks in the control of puberty
Although some central TRG nodes can function as transcriptional repressors (YY1, EAP1, CUX1), there are at least four additional transcriptional and post-transcriptional repressors that may contribute to controlling the timing of puberty. One of them is LIN28b, which encodes an RNA binding protein that inhibits the maturation of let7 miRNAs (Hagan, Piskounova, and Gregory, 2009; Heo and others, 2009; Lehrbach and others, 2009), a family of microRNAs with tumor suppressor activity (Chang and others, 2009). The potential contribution of LIN28b to the regulation of puberty was suggested by the finding that a single nucleotide polymorphism near the LIN28B gene in human chromosome 6(q21) is associated with earlier puberty and shorter stature in girls (Ong et al., 2009; Perry et al., 2009; Sulem et al., 2009).
A broad mechanism of transcriptional repression imposed by members of the ZNF superfamily (Filion et al., 2006; Shannon et al., 1996; Urrutia, 2003; Vogel et al., 2006) may be another regulatory component of the pubertal process. This notion, still in its infancy, derives from high-throughput studies of the agonadal male monkey hypothalamus showing that a reduced expression of several ZNFs accompanies the initiation of the pubertal process in primates (Matagne and others, 2009). Finally, we recently found that members of two additional families of transcriptional repressors are differentially expressed in the peripubertal female rat hypothalamus. One of these families is composed of POZ-ZF proteins, which control a variety of developmental processes via transcriptional repression. Alteration of their function leads to tumorigenesis and developmental disorders (Kelly and Daniel, 2006). We found that expression of three genes belonging to a subset of the POZ-ZF gene family, known as the POK subfamily (Costoya, 2007), increases in the hypothalamus of female rats at puberty (unpublished observations), suggesting that these genes may function as repressors of other downstream repressors of puberty. In contrast to this pattern of expression, expression of two key members of the PcG (Polycomb) complex of gene silencers (Cbx7 and Eed) decreases in the hypothalamus at puberty (Lomniczi, Loche, and Ojeda, 2010), suggesting that – as described below – these genes might prevent the untimely, earlier initiation of puberty by inhibiting the expression of downstream genes whose activation is ultimately required for puberty to occur. The PcG silencing complex is known for its ability to impose gene silencing at various developmental stages (Kohler and Villar, 2008; Schwartz and Pirrotta, 2007; Simon and Kingston, 2009).
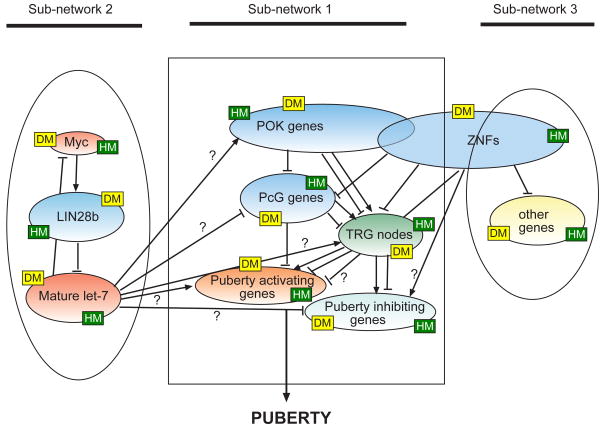
We currently believe that POK and PcG genes may operate within the TRG network itself (Fig. 4), because a) both have tumor suppressor activity (Classen and others, 2009; Kelly et al., 2006; Martinez and others, 2009), b) PcGs are recruited to the promoter of a puberty-activating gene (Kiss1) and restrain Kiss1 expression both in vivo and in vitro (Lomniczi and others, 2012b), and c) POK proteins are, in turn, recruited to PcG gene promoters to repress their activity (unpublished data). We speculate that LIN28b and ZNFs, form associated subnetworks; however, the boundaries of presumed ZNF-driven network may have to be redefined, because some ZNFs display features that makes them structurally similar to the POK family (Costoya, 2007). Because LIN28b delays development via post-transcriptional suppression of microRNA maturation (Hagan et al., 2009; Heo et al., 2009; Lehrbach et al., 2009) instead of transcriptional silencing of target genes, we infer that Lin28b is a central hub of an associated repressive network (Fig. 4). Needless to say, more work is required before all these genes can be accurately assigned to specific modules within the topological architecture of each network (Fig. 4).
Figure 4. General organization of hypothetical transcriptional regulatory networks controlling the onset of female puberty.
This draft model predicts the existence of three functionally connected sub-networks. One of them uses TRGs as central hubs; another uses LIN28b, and a third, less well-defined, is formed by ZNF genes. The latter may overlap considerably with the POK/ZNF module of the TRG network, because several POK genes also contain a kr ppel C2H2 zinc-finger domain core that is characteristic of many ZNFs. Intra and inter sub-networks information is postulated to be dynamically coordinated by epigenetic mechanisms. According to existing information derived from a vast body of literature, including studies performed in our laboratory, these mechanisms involve changes in DNA methylation (DM) and changes in association of modified histones (HM) to gene promoters. The model predicts that the transcriptional inhibition of puberty activating genes is lifted at or before puberty, and replaced by increased trans-activation of gene expression. Simultaneously, puberty-inhibiting genes may be either repressed or experience a reduction of activating inputs. ← = stimulation; ⊥ = Blue denotes transcriptional/post-transcriptional repressors; light red denotes activators; darker green = central nodes with either trans-activating or repressive activity; lighter green = subordinate genes involved in the inhibitory control of puberty, but that are not transcription factors; yellow = other, not yet identified genes.
The contribution of epigenetics
Despite the fact that most, if not all, of the aforementioned genes are subjected to epigenetic regulation (Ball and others, 2009; Guenther and others, 2007; Koch and others, 2007; Miao and Natarajan, 2005; Mikkelsen and others, 2007; Weber and others, 2005; Weber and others, 2007) (see also http://www.epigenome.org/, http://epigenome.usc.edu/ and http://epigenomegateway.wustl.edu/), it is only recently that the potential contribution of epigenetics to the regulation of puberty has been addressed. Although not dealing with puberty itself, two recent reports relevant to the subject need to be mentioned here. One of them demonstrated that the GnRH gene itself is subjected to epigenetic regulation by showing that methylation of the GnRH promoter decreases as GnRH expression increase during in vitro fetal maturation of primate GnRH neurons (Kurian, Keen, and Terasawa, 2010). The other demonstrated that sex differences in Kiss1 expression in the AVPV of mice are related to differences in the degree of Kiss1 promoter methylation between males and females (Semaan and others, 2012). Using a combination of DNA microarrays, genome-wide DNA methylation arrays, ChIP and gene expression analyses we discovered that an epigenetic mechanism of transcriptional repression operating in the hypothalamus plays a significant role in timing female puberty (Lomniczi et al., 2012b). Our results identified the PcG of transcriptional silencers (Kohler et al., 2008; Schwartz et al., 2007; Simon et al., 2009) as a major contributor to this repressive mechanism, and implicated two PcG genes (Cbx7 and Eed) as core components of the PcG complex operating in the ARC of the prepubertal medial basal hypothalamus.
Since kisspeptin and NKB work coordinately within a single cell type to stimulate GnRH secretion, their encoding genes (Kiss1 and Tac2) can be considered as components of a unique class of puberty-activating genes. Using the Kiss1 gene as a prototype of this class, we showed that both the Eed and Cbx7 genes are expressed in kisspeptin neurons of the ARC, and that the CBX7 and EED proteins are associated with the 5′ flanking region of the Kiss1 gene. At the end of juvenile development, coinciding with the pubertal augmentation of pulsatile LH release (Urbanski and Ojeda, 1985), the Eed and Cbx7 promoters become more methylated and the expression of both Eed and Cbx7 decreases. As PcG proteins are evicted from the Kiss1 promoter, association of histone marks involved in gene activation (H3K9,14ac, and H3K4me3) is enhanced and Kiss1 expression increases. When the pubertal decline in hypothalamic EED and CBX7 expression was prevented by overexpressing these proteins in the ARC of early juvenile rats, the number of immunoreactive KNDy neurons in the ARC was reduced, puberty was delayed, estrous cyclicity was disrupted, and importantly, fertility was severely compromised (Lomniczi et al., 2012b). These results suggest that similar or complementary mechanisms of epigenetic regulation may coordinate and dynamically integrate signal propagation within gene networks involved in the neuroendocrine control of puberty (Fig. 4). PcG, POK and ZNF proteins illustrate this feature; while PcG proteins repress gene transcription by imposing histone modifications associated with gene silencing (Sawarkar and Paro, 2010; Simon et al., 2009), POK proteins exert their repressive effect by recruiting co-repressors and histone deacetylases (Costoya, 2007; Koh and others, 2009; Lemercier and others, 2002). Similarly, ZNFs impose gene silencing by recruiting co-repressors, which in turn recruit histone methyltransferases needed for the synthesis of repressive histone marks (Frietze and others, 2010).
Conclusions and Perspectives
The aforementioned observations are consistent with the view that the onset of puberty depends on the contribution of more than one gene. Puberty also appears to be controlled at the transcriptional/post-transcriptional level by discrete groups of genes. We envision the participation of “activators” that move the process along by promoting key developmental events, and “repressors” that prevent the untimely activation of activating genes (Fig. 3 and 4). These elements may be organized in at least three functionally connected sub-networks, one centered about LIN28b, another composed of TRGs and a third one containing ZNFs (Fig. 4). It would be reasonable to assume that – even though of different compositions – these regulatory systems operate within both the neuronal and glial populations involved in controlling the onset of puberty. The results thus far obtained suggest that a two-tiered hierarchy of repressive nodes provides “hard-wiring” to the networks. One of the repressive layers, composed of transcriptional and post-transcriptional “repressors” (such as PcG genes and Lin28b) may directly prevent the premature activation of puberty-inducing genes (such as Kiss1, Tac3, Ttf1, etc). Another layer, located at a higher hierarchical level in the network is formed by repressors such as POK/ZNF genes. Because these genes may modulate the expression of repressors (Fig. 4), they can be considered as “repressors of repressors”. Both layers may directly regulate subordinate genes of the network involved in the stimulatory (Kisspeptin, glutamatergic, glial) and inhibitory (GABA, opioid, RFRP3) control of GnRH secretion. In our view, an essential feature of this model is the presence of a dynamic flow of epigenetic information (Lomniczi et al., 2010; Ojeda and Lomniczi, 2010), which may provide a granular level of coordination and transcriptional plasticity to the sub-networks. We hope that the concepts proposed here provide a framework for future investigation in the area of mammalian puberty.
Acknowledgments
Funding Source
This work was supported by the US National Science Foundation (NSF: IOS1121691), and by the US National Institute of Health (NIH: HD025123-ARRA, and P51-OD 011092-53 supporting the operation of the Oregon National Primate Research Center).
References
- 1.Alexa A, Rahnenfuhrer J, Lengauer T. Improved scoring of functional groups from gene expression data by decorrelating GO graph structure. Bioinformatics. 2006;22:1600–1607. doi: 10.1093/bioinformatics/btl140. [DOI] [PubMed] [Google Scholar]
- 2.Artenstein AW, Opal SM. Proprotein convertases in health and disease. N Engl J Med. 2011;365:2507–2518. doi: 10.1056/NEJMra1106700. [DOI] [PubMed] [Google Scholar]
- 3.Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, Harris MA, Hill DP, Issel-Tarver L, Kasarskis A, Lewis S, Matese JC, Richardson JE, Ringwald M, Rubin GM, Sherlock G. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ball MP, Li JB, Gao Y, Lee JH, LeProust EM, Park IH, Xie B, Daley GQ, Church GM. Targeted and genome-scale strategies reveal gene-body methylation signatures in human cells. Nat Biotechnol. 2009;27:361–368. doi: 10.1038/nbt.1533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Basso K, Margolin AA, Stolovitzky G, Klein U, Dalla-Favera R, Califano A. Reverse engineering of regulatory networks in human B cells. Nat Genet. 2005;37:382–390. doi: 10.1038/ng1532. [DOI] [PubMed] [Google Scholar]
- 6.Bedecarrats GY, Kaiser UB. Mutations in the human gonadotropin-releasing hormone receptor: insights into receptor biology and function. Semin Reprod Med. 2007;25:368–378. doi: 10.1055/s-2007-984743. [DOI] [PubMed] [Google Scholar]
- 7.Burdach J, O’Connell MR, Mackay JP, Crossley M. Two-timing zinc finger transcription factors liaising with RNA. Trends Biochem Sci. 2012;37:199–205. doi: 10.1016/j.tibs.2012.02.001. [DOI] [PubMed] [Google Scholar]
- 8.Carro MS, Lim WK, Alvarez MJ, Bollo RJ, Zhao X, Snyder EY, Sulman EP, Anne SL, Doetsch F, Colman H, Lasorella A, Aldape K, Califano A, Iavarone A. The transcriptional network for mesenchymal transformation of brain tumours. Nature. 2010;463:318–325. doi: 10.1038/nature08712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chang TC, Zeitels LR, Hwang HW, Chivukula RR, Wentzel EA, Dews M, Jung J, Gao P, Dang CV, Beer MA, Thomas-Tikhonenko A, Mendell JT. Lin-28B transactivation is necessary for Myc-mediated let-7 repression and proliferation. Proc Natl Acad Sci U S A. 2009;106:3384–3389. doi: 10.1073/pnas.0808300106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Choi J, Ha CM, Choi EJ, Jeong CS, Park JW, Baik JH, Park JY, Costa ME, Ojeda SR, Lee BJ. Kinesin superfamily-associated protein 3 is preferentially expressed in glutamatergic neurons and contributes to the excitatory control of female puberty. Endocrinology. 2008;149:6146–6156. doi: 10.1210/en.2008-0432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Clarkson J, d’Anglemont dTX, Colledge WH, Caraty A, Herbison AE. Distribution of kisspeptin neurones in the adult female mouse brain. J Neuroendocrinol. 2009;21:673–682. doi: 10.1111/j.1365-2826.2009.01892.x. [DOI] [PubMed] [Google Scholar]
- 12.Clasadonte J, Poulain P, Hanchate NK, Corfas G, Ojeda SR, Prevot V. Prostaglandin E2 release from astrocytes triggers gonadotropin-releasing hormone (GnRH) neuron firing via EP2 receptor activation. Proc Natl Acad Sci U S A. 2011;108:16104–16109. doi: 10.1073/pnas.1107533108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Classen AK, Bunker BD, Harvey KF, Vaccari T, Bilder D. A tumor suppressor activity of Drosophila Polycomb genes mediated by JAK-STAT signaling. Nat Genet. 2009;41:1150–1155. doi: 10.1038/ng.445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Costoya JA. Functional analysis of the role of POK transcriptional repressors. Brief Funct Genomic Proteomic. 2007;6:8–18. doi: 10.1093/bfgp/elm002. [DOI] [PubMed] [Google Scholar]
- 15.d’Anglemont dTX, Colledge WH. The role of kisspeptin signaling in reproduction. Physiology (Bethesda) 2010;25:207–217. doi: 10.1152/physiol.00009.2010. [DOI] [PubMed] [Google Scholar]
- 16.Davidson EH, Rast JP, Oliveri P, Ransick A, Calestani C, Yuh CH, Minokawa T, Amore G, Hinman V, Arenas-mena C, Otim O, Brown CT, Livi CB, Lee PY, Revilla R, Rust AG, Pan ZJ, Schilstra MJ, Clarke PJC, Arnone MI, Rowen L, Cameron RA, McClay DR, Hood L, Bolouri H. A genomic regulatory network for development. Science. 2002;295:1669–1678. doi: 10.1126/science.1069883. [DOI] [PubMed] [Google Scholar]
- 17.de Roux N, Genin E, Carel JC, Matsuda F, Chaussain JL, Milgrom E. Hypogonadotropic hypogonadism due to loss of function of the KiSS1-derived peptide receptor GPR54. Proc Natl Acad Sci U S A. 2003;100:10972–10976. doi: 10.1073/pnas.1834399100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.DeFazio RA, Heger S, Ojeda SR, Moenter SM. Activation of A-type g-aminobutyric acid receptors excites gonadotropin-releasing hormone neurons. Mol Endocrinol. 2002;16:2872–2891. doi: 10.1210/me.2002-0163. [DOI] [PubMed] [Google Scholar]
- 19.Dissen GA, Lomniczi A, Heger S, Neff TL, Ojeda SR. Hypothalamic enhanced at puberty 1 (EAP1) is required for menstrual cyclicity in non-human primates. Endocrinology. 2012;153:350–361. doi: 10.1210/en.2011-1541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ducret E, Anderson GM, Herbison AE. RFamide-related peptide-3, a mammalian gonadotropin-inhibitory hormone ortholog, regulates gonadotropin-releasing hormone neuron firing in the mouse. Endocrinology. 2009;150:2799–2804. doi: 10.1210/en.2008-1623. [DOI] [PubMed] [Google Scholar]
- 21.Dudas B, Merchenthaler I. Three-dimensional representation of the neurotransmitter systems of the human hypothalamus: inputs of the gonadotrophin hormone-releasing hormone neuronal system. J Neuroendocrinol. 2006;18:79–95. doi: 10.1111/j.1365-2826.2005.01398.x. [DOI] [PubMed] [Google Scholar]
- 22.Eaves L, Silberg J, Foley D, Bulik C, Maes H, Erkanli A, Angold A, Costello EJ, Worthman C. Genetic and environmental influences on the relative timing of pubertal change. Twin Res. 2004;7:471–481. doi: 10.1375/1369052042335278. [DOI] [PubMed] [Google Scholar]
- 23.Ebling FJ, Luckman SM. RFAmide-related peptide: another sexy peptide? Endocrinology. 2008;149:899–901. doi: 10.1210/en.2007-1765. [DOI] [PubMed] [Google Scholar]
- 24.Elks CE, Perry JR, Sulem P, Chasman DI, Franceschini N, He C, Lunetta KL, Visser JA, Byrne EM, Cousminer DL, Gudbjartsson DF, Esko T, Feenstra B, Hottenga JJ, Koller DL, Kutalik Z, Lin P, Mangino M, Marongiu M, McArdle PF, Smith AV, Stolk L, van Wingerden SH, Zhao JH, Albrecht E, Corre T, Ingelsson E, Hayward C, Magnusson PK, Smith EN, Ulivi S, Warrington NM, Zgaga L, Alavere H, Amin N, Aspelund T, Bandinelli S, Barroso I, Berenson GS, Bergmann S, Blackburn H, Boerwinkle E, Buring JE, Busonero F, Campbell H, Chanock SJ, Chen W, Cornelis MC, Couper D, Coviello AD, d’Adamo P, de FU, de Geus EJ, Deloukas P, Doring A, Smith GD, Easton DF, Eiriksdottir G, Emilsson V, Eriksson J, Ferrucci L, Folsom AR, Foroud T, Garcia M, Gasparini P, Geller F, Gieger C, Gudnason V, Hall P, Hankinson SE, Ferreli L, Heath AC, Hernandez DG, Hofman A, Hu FB, Illig T, Jarvelin MR, Johnson AD, Karasik D, Khaw KT, Kiel DP, Kilpelainen TO, Kolcic I, Kraft P, Launer LJ, Laven JS, Li S, Liu J, Levy D, Martin NG, McArdle WL, Melbye M, Mooser V, Murray JC, Murray SS, Nalls MA, Navarro P, Nelis M, Ness AR, Northstone K, Oostra BA, Peacock M, Palmer LJ, Palotie A, Pare G, Parker AN, Pedersen NL, Peltonen L, Pennell CE, Pharoah P, Polasek O, Plump AS, Pouta A, Porcu E, Rafnar T, Rice JP, Ring SM, Rivadeneira F, Rudan I, Sala C, Salomaa V, Sanna S, Schlessinger D, Schork NJ, Scuteri A, Segre AV, Shuldiner AR, Soranzo N, Sovio U, Srinivasan SR, Strachan DP, Tammesoo ML, Tikkanen E, Toniolo D, Tsui K, Tryggvadottir L, Tyrer J, Uda M, van Dam RM, van Meurs JB, Vollenweider P, Waeber G, Wareham NJ, Waterworth DM, Weedon MN, Wichmann HE, Willemsen G, Wilson JF, Wright AF, Young L, Zhai G, Zhuang WV, Bierut LJ, Boomsma DI, Boyd HA, Crisponi L, Demerath EW, van Duijn CM, Econs MJ, Harris TB, Hunter DJ, Loos RJ, Metspalu A, Montgomery GW, Ridker PM, Spector TD, Streeten EA, Stefansson K, Thorsteinsdottir U, Uitterlinden AG, Widen E, Murabito JM, Ong KK, Murray A. Thirty new loci for age at menarche identified by a meta-analysis of genome-wide association studies. Nat Genet. 2010;42:1077–1085. doi: 10.1038/ng.714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Falcon S, Gentleman R. Using GOstats to test gene lists for GO term association. Bioinformatics. 2007;23:257–258. doi: 10.1093/bioinformatics/btl567. [DOI] [PubMed] [Google Scholar]
- 26.Filion GJ, Zhenilo S, Salozhin S, Yamada D, Prokhortchouk E, Defossez PA. A family of human zinc finger proteins that bind methylated DNA and repress transcription. Mol Cell Biol. 2006;26:169–181. doi: 10.1128/MCB.26.1.169-181.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Finney A, Hucka M. Systems biology markup language: Level 2 and beyond. Biochem Soc Trans. 2003;31:1472–1473. doi: 10.1042/bst0311472. [DOI] [PubMed] [Google Scholar]
- 28.Frietze S, O’Geen H, Blahnik KR, Jin VX, Farnham PJ. ZNF274 recruits the histone methyltransferase SETDB1 to the 3′ ends of ZNF genes. PLoS ONE. 2010;5:e15082. doi: 10.1371/journal.pone.0015082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fukusumi S, Fujii R, Hinuma S. Recent advances in mammalian RFamide peptides: the discovery and functional analyses of PrRP, RFRPs and QRFP. Peptides. 2006;27:1073–1086. doi: 10.1016/j.peptides.2005.06.031. [DOI] [PubMed] [Google Scholar]
- 30.Gajdos ZK, Butler JL, Henderson KD, He C, Supelak PJ, Egyud M, Price A, Reich D, Clayton PE, Le ML, Hunter DJ, Henderson BE, Palmert MR, Hirschhorn JN. Association studies of common variants in 10 hypogonadotropic hypogonadism genes with age at menarche. J Clin Endocrinol Metab. 2008;93:4290–4298. doi: 10.1210/jc.2008-0981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Garcia-Rudaz C, Deng V, Matagne V, Ronnekleiv O, Bosch M, Han V, Percy AK, Ojeda SR. FXYD1, a modulator of Na(+),K(+)-ATPase activity, facilitates female sexual development by maintaining GnRH neuronal excitability. J Neuroendocrinol. 2008;21:108–122. doi: 10.1111/j.1365-2826.2008.01812.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gibson EM, Humber SA, Jain S, Williams WP, III, Zhao S, Bentley GE, Tsutsui K, Kriegsfeld LJ. Alterations in RFamide-related peptide expression are coordinated with the preovulatory luteinizing hormone surge. Endocrinology. 2008;149:4958–4969. doi: 10.1210/en.2008-0316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gottsch ML, Cunningham MJ, Smith JT, Popa SM, Acohido BV, Crowley WF, Seminara S, Clifton DK, Steiner RA. A role for kisspeptins in the regulation of gonadotropin secretion in the mouse. Endocrinology. 2004;145:4073–4077. doi: 10.1210/en.2004-0431. [DOI] [PubMed] [Google Scholar]
- 34.Guenther MG, Levine SS, Boyer LA, Jaenisch R, Young RA. A chromatin landmark and transcription initiation at most promoters in human cells. Cell. 2007;130:77–88. doi: 10.1016/j.cell.2007.05.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ha CM, Choi J, Choi EJ, Costa ME, Lee BJ, Ojeda SR. NELL2, a neuron-specific EGF-like protein, is selectively expressed in glutamatergic neurons and contributes to the glutamatergic control of GnRH neurons at puberty. Neuroendocrinology. 2008;88:199–211. doi: 10.1159/000139579. [DOI] [PubMed] [Google Scholar]
- 36.Hagan JP, Piskounova E, Gregory RI. Lin28 recruits the TUTase Zcchc11 to inhibit let-7 maturation in mouse embryonic stem cells. Nat Struct Mol Biol. 2009;16:1021–1025. doi: 10.1038/nsmb.1676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.He C, Kraft P, Chen C, Buring JE, Pare G, Hankinson SE, Chanock SJ, Ridker PM, Hunter DJ, Chasman DI. Genome-wide association studies identify loci associated with age at menarche and age at natural menopause. Nat Genet. 2009;41:724–728. doi: 10.1038/ng.385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Heger S, Mastronardi C, Dissen GA, Lomniczi A, Cabrera R, Roth CL, Jung H, Galimi F, Sippell W, Ojeda SR. Enhanced at puberty 1 (EAP1) is a new transcriptional regulator of the female neuroendocrine reproductive axis. J Clin Invest. 2007;117:2145–2154. doi: 10.1172/JCI31752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Heo I, Joo C, Kim YK, Ha M, Yoon MJ, Cho J, Yeom KH, Han J, Kim VN. TUT4 in concert with Lin28 suppresses microRNA biogenesis through pre-microRNA uridylation. Cell. 2009;138:696–708. doi: 10.1016/j.cell.2009.08.002. [DOI] [PubMed] [Google Scholar]
- 40.Hinuma S, Shintani Y, Fukusumi S, Iijima N, Matsumoto Y, Hosoya M, Fujii R, Watanabe T, Kikuchi K, Terao Y, Yano T, Yamamoto T, Kawamata Y, Habata Y, Asada M, Kitada C, Kurokawa T, Onda H, Nishimura O, Tanaka M, Ibata Y, !Lost Data. New neuropeptides containing carboxy-terminal RFamide and their receptor in mammals. Nat Cell Biol. 2000;2:703–708. doi: 10.1038/35036326. [DOI] [PubMed] [Google Scholar]
- 41.Iancu OD, Darakjian P, Walter NA, Malmanger B, Oberbeck D, Belknap J, McWeeney S, Hitzemann R. Genetic diversity and striatal gene networks: focus on the heterogeneous stock-collaborative cross (HS-CC) mouse. BMC Genomics. 2010;11:585. doi: 10.1186/1471-2164-11-585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ideker T, Thorsson V, Ranish JA, Christmas R, Buhler J, Eng JK, Bumgarner R, Goodlett DR, Aebersold R, Hood L. Integrated genomic and proteomic analyses of a systematically perturbed metabolic network. Science. 2001;292:929–934. doi: 10.1126/science.292.5518.929. [DOI] [PubMed] [Google Scholar]
- 43.Kauffman AS, Clifton DK, Steiner RA. Emerging ideas about kisspeptin- GPR54 signaling in the neuroendocrine regulation of reproduction. Trends Neurosci. 2007;30:504–511. doi: 10.1016/j.tins.2007.08.001. [DOI] [PubMed] [Google Scholar]
- 44.Kelly KF, Daniel JM. POZ for effect--POZ-ZF transcription factors in cancer and development. Trends Cell Biol. 2006;16:578–587. doi: 10.1016/j.tcb.2006.09.003. [DOI] [PubMed] [Google Scholar]
- 45.Keshava Prasad TS, Goel R, Kandasamy K, Keerthikumar S, Kumar S, Mathivanan S, Telikicherla D, Raju R, Shafreen B, Venugopal A, Balakrishnan L, Marimuthu A, Banerjee S, Somanathan DS, Sebastian A, Rani S, Ray S, Harrys Kishore CJ, Kanth S, Ahmed M, Kashyap MK, Mohmood R, Ramachandra YL, Krishna V, Rahiman BA, Mohan S, Ranganathan P, Ramabadran S, Chaerkady R, Pandey A. Human Protein Reference Database--2009 update. Nucleic Acids Res. 2009;37:D767–D772. doi: 10.1093/nar/gkn892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Khan AR, Kauffman AS. The Role of Kisspeptin and RFRP-3 Neurons in the Circadian-Timed Preovulatory Luteinizing Hormone Surge. J Neuroendocrinol. 2011;24:131–143. doi: 10.1111/j.1365-2826.2011.02162.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kinoshita F, Nakai Y, Katakami H, Imura H. Suppressive effect of dynorphin-(1-13) on luteinizing hormone release in conscious castrated rats. Life Sci. 1982;30:1915–1919. doi: 10.1016/0024-3205(82)90472-6. [DOI] [PubMed] [Google Scholar]
- 48.Kinsey-Jones JS, Grachev P, Li XF, Lin YS, Milligan SR, Lightman SL, O’Byrne KT. The inhibitory effects of neurokinin B on GnRH pulse generator frequency in the female rat. Endocrinology. 2012;153:307–315. doi: 10.1210/en.2011-1641. [DOI] [PubMed] [Google Scholar]
- 49.Klipp E, Herwig R, Kowald A, Wierling C, Lehrach H. Systems Biology in Practice. Wiley-VCH; Weinheim, Germany: 2005. [Google Scholar]
- 50.Koch CM, Andrews RM, Flicek P, Dillon SC, Karaoz U, Clelland GK, Wilcox S, Beare DM, Fowler JC, Couttet P, James KD, Lefebvre GC, Bruce AW, Dovey OM, Ellis PD, Dhami P, Langford CF, Weng Z, Birney E, Carter NP, Vetrie D, Dunham I. The landscape of histone modifications across 1% of the human genome in five human cell lines. Genome Res. 2007;17:691–707. doi: 10.1101/gr.5704207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Koh DI, Choi WI, Jeon BN, Lee CE, Yun CO, Hur MW. A novel POK family transcription factor, ZBTB5, represses transcription of p21CIP1 gene. J Biol Chem. 2009;284:19856–19866. doi: 10.1074/jbc.M109.025817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kohler C, Villar CB. Programming of gene expression by Polycomb group proteins. Trends Cell Biol. 2008;18:236–243. doi: 10.1016/j.tcb.2008.02.005. [DOI] [PubMed] [Google Scholar]
- 53.Kordon C, Drouva SV, Martínez de la Escalera G, Weiner RI. Role of classic and peptide neuromediators in the neuroendocrine regulation of luteinizing hormone and prolactin. In: Knobil E, Neill JD, editors. The Physiology of Reproduction. 2. Vol. 1. Raven Press; New York: 1994. pp. 1621–1681. [Google Scholar]
- 54.Kotani M, Detheux M, Vandenbogaerde A, Communi D, Vanderwinden JM, Le Poul E, Brezillon S, Tyldesley R, Suarez-Huerta N, Vandeput F, Blanpain C, Schiffmann SN, Vassart G, Parmentier M. The metastasis suppressor gene KiSS-1 encodes kisspeptins, the natural ligands of the orphan G protein-coupled receptor GPR54. J Biol Chem. 2001;276:34631–34636. doi: 10.1074/jbc.M104847200. [DOI] [PubMed] [Google Scholar]
- 55.Krewson TD, Supelak PJ, Hill AE, Singer JB, Lander ES, Nadeau JH, Palmert MR. Chromosomes 6 and 13 harbor genes that regulate pubertal timing in mouse chromosome substitution strains. Endocrinology. 2004;145:4447–4451. doi: 10.1210/en.2004-0543. [DOI] [PubMed] [Google Scholar]
- 56.Kuramochi M, Fukuhara H, Nobukuni T, Kanbe T, Maruyama T, Ghosh HP, Pletcher M, Isomura M, Onizuka M, Kitamura T, Sekiya T, Reeves RH, Murakami Y. TSLC1 is a tumor-suppressor gene in human non-small-cell lung cancer. Nat Genet. 2001;27:427–430. doi: 10.1038/86934. [DOI] [PubMed] [Google Scholar]
- 57.Kurian JR, Keen KL, Terasawa E. Epigenetic changes coincide with in vitro primate GnRH neuronal maturation. Endocrinology. 2010;151:5359–5368. doi: 10.1210/en.2010-0555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Langfelder P, Horvath S. WGCNA: an R package for weighted correlation network analysis. BMC Bioinformatics. 2008;9:559. doi: 10.1186/1471-2105-9-559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Langfelder P, Zhang B, Horvath S. Defining clusters from a hierarchical cluster tree: the Dynamic Tree Cut package for R. Bioinformatics. 2008;24:719–720. doi: 10.1093/bioinformatics/btm563. [DOI] [PubMed] [Google Scholar]
- 60.Lapatto R, Pallais JC, Zhang D, Chan YM, Mahan A, Cerrato F, Le WW, Hoffman GE, Seminara SB. Kiss1−/− mice exhibit more variable hypogonadism than Gpr54−/− mice. Endocrinology. 2007;148:4927–4936. doi: 10.1210/en.2007-0078. [DOI] [PubMed] [Google Scholar]
- 61.Lee BJ, Cho GJ, Norgren R, Junier MP, Hill DF, Tapia V, Costa ME, Ojeda SR. TTF-1, a homeodomain gene required for diencephalic morphogenesis, is postnatally expressed in the neuroendocrine brain in a developmentally regulated and cell-specific fashion. Mol Cell Neurosci. 2001;17:107–126. doi: 10.1006/mcne.2000.0933. [DOI] [PubMed] [Google Scholar]
- 62.Lehman MN, Coolen LM, Goodman RL. Minireview: kisspeptin/neurokinin B/dynorphin (KNDy) cells of the arcuate nucleus: a central node in the control of gonadotropin-releasing hormone secretion. Endocrinology. 2010;151:3479–3489. doi: 10.1210/en.2010-0022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lehrbach NJ, Armisen J, Lightfoot HL, Murfitt KJ, Bugaut A, Balasubramanian S, Miska EA. LIN-28 and the poly(U) polymerase PUP-2 regulate let-7 microRNA processing in Caenorhabditis elegans. Nat Struct Mol Biol. 2009;16:1016–1020. doi: 10.1038/nsmb.1675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lemercier C, Brocard MP, Puvion-Dutilleul F, Kao HY, Albagli O, Khochbin S. Class II histone deacetylases are directly recruited by BCL6 transcriptional repressor. J Biol Chem. 2002;277:22045–22052. doi: 10.1074/jbc.M201736200. [DOI] [PubMed] [Google Scholar]
- 65.Lomniczi A, Cornea A, Costa ME, Ojeda SR. Hypothalamic tumor necrosis factor-a converting enzyme (TACE) mediates excitatory amino acid-dependent neuron-to-glia signaling in the neuroendocrine brain. J Neurosci. 2006;26:51–62. doi: 10.1523/JNEUROSCI.2939-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lomniczi A, Garcia-Rudaz C, Ramakrishnan R, Wilmont B, Khouangsathiene S, Ferguson B, Dissen GA, Ojeda SR. A single nucleotide polymorphism in the EAP1 gene is associated with amenorrhea/oligomenorrhea in nonhuman primates. Endocrinology. 2012a;153:339–349. doi: 10.1210/en.2011-1540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Lomniczi A, Loche A, Castellano JM, Ronnekleiv O, Bosh M, Kaidar G, Knoll G, Pfeifer GP, Ojeda SR. Epigenetic control of female puberty. 2012b. Submitted. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lomniczi A, Loche A, Ojeda SR. Epigenetic regulation of female puberty. 40th Ann Mtg Society for Neuroscience; San Diego, CA. November 13–17. 2010. [Google Scholar]
- 69.Lomniczi A, Ojeda SR. A role for glial cells of the neuroendocrine brain in the central control of female sexual development. In: Parpura V, Haydon P, editors. Astrocytes in (Patho)Physiology of the Nervous System. Springer; NY: 2009. pp. 487–511. [Google Scholar]
- 70.Longabaugh WJ, Davidson EH, Bolouri H. Computational representation of developmental genetic regulatory networks. Dev Biol. 2005;283:1–16. doi: 10.1016/j.ydbio.2005.04.023. [DOI] [PubMed] [Google Scholar]
- 71.Luscombe NM, Babu MM, Yu H, Snyder M, Teichmann SA, Gerstein M. Genomic analysis of regulatory network dynamics reveals large topological changes. Nature. 2004;431:308–312. doi: 10.1038/nature02782. [DOI] [PubMed] [Google Scholar]
- 72.Mahesh VB, Dhandapani KM, Brann DW. Role of astrocytes in reproduction and neuroprotection. Mol Cell Endocrinol. 2006;246:1–9. doi: 10.1016/j.mce.2005.11.017. [DOI] [PubMed] [Google Scholar]
- 73.Martinez AM, Schuettengruber B, Sakr S, Janic A, Gonzalez C, Cavalli G. Polyhomeotic has a tumor suppressor activity mediated by repression of Notch signaling. Nat Genet. 2009;41:1076–1082. doi: 10.1038/ng.414. [DOI] [PubMed] [Google Scholar]
- 74.Mastronardi C, Smiley GG, Raber J, Kusakabe T, Kawaguchi A, Matagne V, Dietzel A, Heger S, Mungenast AE, Cabrera R, Kimura S, Ojeda SR. Deletion of the Ttf1 gene in differentiated neurons disrupts female reproduction without impairing basal ganglia function. J Neurosci. 2006;26:13167–13179. doi: 10.1523/JNEUROSCI.4238-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Matagne V, Ramaswamy S, Lomniczi A, Plant TM, Ojeda SR. Program No.703.9, 2009 Neuroscience Meeting Planner. Chicago, IL: Society for Neuroscience; 2009. Hypothalamic expression of a gene cluster encoding transcriptional repressors and mapping to chromosome 19 is developmentally regulated and linked to sexual maturation in the rhesus monkey. [Google Scholar]
- 76.Mayer C, Boehm U. Female reproductive maturation in the absence of kisspeptin/GPR54 signaling. Nat Neurosci. 2011;14:704–710. doi: 10.1038/nn.2818. [DOI] [PubMed] [Google Scholar]
- 77.Miao F, Natarajan R. Mapping global histone methylation patterns in the coding regions of human genes. Mol Cell Biol. 2005;25:4650–4661. doi: 10.1128/MCB.25.11.4650-4661.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Mikkelsen TS, Ku M, Jaffe DB, Issac B, Lieberman E, Giannoukos G, Alvarez P, Brockman W, Kim TK, Koche RP, Lee W, Mendenhall E, O’Donovan A, Presser A, Russ C, Xie X, Meissner A, Wernig M, Jaenisch R, Nusbaum C, Lander ES, Bernstein BE. Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature. 2007;448:553–560. doi: 10.1038/nature06008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Moenter SM, DeFazio RA. Endogenous gamma-aminobutyric acid can excite gonadotropin-releasing hormone neurons. Endocrinology. 2005;146:5374–5379. doi: 10.1210/en.2005-0788. [DOI] [PubMed] [Google Scholar]
- 80.Mueller JK, Dietzel A, Lomniczi A, Loche A, Tefs K, Kiess W, Danne T, Ojeda SR, Heger S. Transcriptional regulation of the human KiSS1 gene. Mol Cell Endocrinol. 2011;342:8–19. doi: 10.1016/j.mce.2011.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Mueller JK, Koch I, Lomniczi A, Loche A, Rulfs T, Castellano JM, Kiess W, Ojeda S, Heger S. Transcription of the human EAP1 gene is regulated by upstream components of a puberty-controlling Tumor Suppressor Gene network. Mol Cell Endocrinol. 2012;351:184–198. doi: 10.1016/j.mce.2011.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Navarro VM, Gottsch ML, Chavkin C, Okamura H, Clifton DK, Steiner RA. Regulation of gonadotropin-releasing hormone secretion by kisspeptin/dynorphin/neurokinin B neurons in the arcuate nucleus of the mouse. J Neurosci. 2009;29:11859–11866. doi: 10.1523/JNEUROSCI.1569-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Navarro VM, Gottsch ML, Wu M, Garcia-Galiano D, Hobbs SJ, Bosch MA, Pinilla L, Clifton DK, Dearth A, Ronnekleiv OK, Braun RE, Palmiter RD, Tena-Sempere M, Alreja M, Steiner RA. Regulation of NKB Pathways and Their Roles in the Control of Kiss1 Neurons in the Arcuate Nucleus of the Male Mouse. Endocrinology. 2011;152:4265–4275. doi: 10.1210/en.2011-1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Oakley AE, Clifton DK, Steiner RA. Kisspeptin signaling in the brain. Endocr Rev. 2009;30:713–743. doi: 10.1210/er.2009-0005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Ojeda SR, Dubay C, Lomniczi A, Kaidar G, Matagne V, Sandau US, Dissen GA. Gene networks and the neuroendocrine regulation of puberty. Mol Cell Endocrinol. 2010;324:3–11. doi: 10.1016/j.mce.2009.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Ojeda SR, Hill J, Hill DF, Costa ME, Tapia V, Cornea A, Ma YJ. The Oct-2 POU-domain gene in the neuroendocrine brain: A transcriptional regulator of mammalian puberty. Endocrinology. 1999;140:3774–3789. doi: 10.1210/endo.140.8.6941. [DOI] [PubMed] [Google Scholar]
- 87.Ojeda SR, Lomniczi A. The Epigenetics of Mammalian Puberty. 14th International Congress of Endocrinology; Kyoto, Japan. March 26–30. 2010. [Google Scholar]
- 88.Ojeda SR, Lomniczi A, Mastronardi C, Heger S, Roth C, Parent AS, Matagne V, Mungenast AE. Minireview: The neuroendocrine regulation of puberty: Is the time ripe for a systems biology approach? Endocrinology. 2006;147:1166–1174. doi: 10.1210/en.2005-1136. [DOI] [PubMed] [Google Scholar]
- 89.Ojeda SR, Lomniczi A, Sandau U. Glial-GnRH neuron interactions in the median eminence and the control of GnRH secretion. J Neuroendocrinol. 2008a;20:732–742. doi: 10.1111/j.1365-2826.2008.01712.x. [DOI] [PubMed] [Google Scholar]
- 90.Ojeda SR, Lomniczi A, Sandau U. Contribution of glial-neuronal interactions to the neuroendocrine control of female puberty. Eur J Neurosci. 2010;32:2003–2010. doi: 10.1111/j.1460-9568.2010.07515.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Ojeda SR, Lomniczi A, Sandau US. Glial-gonadotrophin hormone (GnRH) neurone interactions in the median eminence and the control of GnRH secretion. J Neuroendocrinol. 2008b;20:732–742. doi: 10.1111/j.1365-2826.2008.01712.x. [DOI] [PubMed] [Google Scholar]
- 92.Ojeda SR, Skinner MK. Puberty in the rat. In: Neill JD, editor. The Physiology of Reproduction. 3. Academic Press/Elsevier; San Diego: 2006. pp. 2061–2126. [Google Scholar]
- 93.Ojeda SR, Terasawa E. Neuroendocrine regulation of puberty. In: Pfaff D, Arnold A, Etgen A, Fahrbach S, Moss R, Rubin R, editors. Hormones, Brain and Behavior. Vol. 4. Elsevier; New York: 2002. pp. 589–659. [Google Scholar]
- 94.Ojeda SR, Urbanski HF. Puberty in the rat. In: Knobil E, Neill JD, editors. The Physiology of Reproduction. 2. Vol. 2. Raven Press; New York: 1994. pp. 363–409. [Google Scholar]
- 95.Ong KK, Elks CE, Li S, Zhao JH, Luan J, Andersen LB, Bingham SA, Brage S, Smith GD, Ekelund U, Gillson CJ, Glaser B, Golding J, Hardy R, Khaw KT, Kuh D, Luben R, Marcus M, McGeehin MA, Ness AR, Northstone K, Ring SM, Rubin C, Sims MA, Song K, Strachan DP, Vollenweider P, Waeber G, Waterworth DM, Wong A, Deloukas P, Barroso I, Mooser V, Loos RJ, Wareham NJ. Genetic variation in LIN28B is associated with the timing of puberty. Nat Genet. 2009;41:729–733. doi: 10.1038/ng.382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Parent AS, Matagne V, Westphal M, Heger S, Ojeda S, Jung H. Gene expression profiling of hypothalamic hamartomas: a search for genes associated with central precocious puberty. Horm Res. 2008;69:114–123. doi: 10.1159/000111815. [DOI] [PubMed] [Google Scholar]
- 97.Parent AS, Mungenast AE, Lomniczi A, Sandau US, Peles E, Bosch MA, Ronnekleiv OK, Ojeda SR. A contactin-receptor-like protein tyrosine phosphatase beta complex mediates adhesive communication between astroglial cells and gonadotrophin-releasing hormone neurones. J Neuroendocrinol. 2007;19:847–859. doi: 10.1111/j.1365-2826.2007.01597.x. [DOI] [PubMed] [Google Scholar]
- 98.Parkash J, Kaur G. Neuronal-glial plasticity in gonadotropin-releasing hormone release in adult female rats: role of the polysialylated form of the neural cell adhesion molecule. J Endocrinol. 2005;186:397–409. doi: 10.1677/joe.1.06156. [DOI] [PubMed] [Google Scholar]
- 99.Perera AD, Lagenaur CF, Plant TM. Postnatal expression of polysialic acid-neural cell adhesion molecule in the hypothalamus of the male rhesus monkey (Macaca mulatta) Endocrinology. 1993;133:2729–2735. doi: 10.1210/endo.133.6.7694845. [DOI] [PubMed] [Google Scholar]
- 100.Peri S, Navarro JD, Amanchy R, Kristiansen TZ, Jonnalagadda CK, Surendranath V, Niranjan V, Muthusamy B, Gandhi TK, Gronborg M, Ibarrola N, Deshpande N, Shanker K, Shivashankar HN, Rashmi BP, Ramya MA, Zhao Z, Chandrika KN, Padma N, Harsha HC, Yatish AJ, Kavitha MP, Menezes M, Choudhury DR, Suresh S, Ghosh N, Saravana R, Chandran S, Krishna S, Joy M, Anand SK, Madavan V, Joseph A, Wong GW, Schiemann WP, Constantinescu SN, Huang L, Khosravi-Far R, Steen H, Tewari M, Ghaffari S, Blobe GC, Dang CV, Garcia JG, Pevsner J, Jensen ON, Roepstorff P, Deshpande KS, Chinnaiyan AM, Hamosh A, Chakravarti A, Pandey A. Development of human protein reference database as an initial platform for approaching systems biology in humans. Genome Res. 2003;13:2363–2371. doi: 10.1101/gr.1680803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Perry JR, Stolk L, Franceschini N, Lunetta KL, Zhai G, McArdle PF, Smith AV, Aspelund T, Bandinelli S, Boerwinkle E, Cherkas L, Eiriksdottir G, Estrada K, Ferrucci L, Folsom AR, Garcia M, Gudnason V, Hofman A, Karasik D, Kiel DP, Launer LJ, van MJ, Nalls MA, Rivadeneira F, Shuldiner AR, Singleton A, Soranzo N, Tanaka T, Visser JA, Weedon MN, Wilson SG, Zhuang V, Streeten EA, Harris TB, Murray A, Spector TD, Demerath EW, Uitterlinden AG, Murabito JM. Meta-analysis of genome-wide association data identifies two loci influencing age at menarche. Nat Genet. 2009;41:648–650. doi: 10.1038/ng.386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Plant TM, Witchel SF. Puberty in nonhuman primates and humans. In: Neill JD, editor. The Physiology of Reproduction. 3. Academic Press/Elsevier; San Diego: 2006. pp. 2177–2230. [Google Scholar]
- 103.Poling MC, Kim J, Dhamija S, Kauffman AS. Development, Sex Steroid Regulation, and Phenotypic Characterization of RFamide-Related Peptide (Rfrp) Gene Expression and RFamide Receptors in the Mouse Hypothalamus. Endocrinology. 2012;153:1827–1840. doi: 10.1210/en.2011-2049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Ponomarev I, Wang S, Zhang L, Harris RA, Mayfield RD. Gene coexpression networks in human brain identify epigenetic modifications in alcohol dependence. J Neurosci. 2012;32:1884–1897. doi: 10.1523/JNEUROSCI.3136-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Prevot V. Glial-neuronal-endothelial interactions are involved in the control of GnRH secretion. J Neuroendocrinol. 2002;14:247–255. doi: 10.1046/j.0007-1331.2001.00772.x. [DOI] [PubMed] [Google Scholar]
- 106.Przulj N. Biological network comparison using graphlet degree distribution. Bioinformatics. 2007;23:e177–e183. doi: 10.1093/bioinformatics/btl301. [DOI] [PubMed] [Google Scholar]
- 107.Rampazzo A, Pivotto F, Occhi G, Tiso N, Bortoluzzi S, Rowen L, Hood L, Nava A, Danieli GA. Characterization of C14orf4, a novel intronless human gene containing a polyglutamine repeat, mapped to the ARVD1 critical region. Biochem Biophys Res Commun. 2000;278:766–774. doi: 10.1006/bbrc.2000.3883. [DOI] [PubMed] [Google Scholar]
- 108.Ravasz E, Somera AL, Mongru DA, Oltvai ZN, Barabasi AL. Hierarchical organization of modularity in metabolic networks. Science. 2002;297:1551–1555. doi: 10.1126/science.1073374. [DOI] [PubMed] [Google Scholar]
- 109.Reshef DN, Reshef YA, Finucane HK, Grossman SR, McVean G, Turnbaugh PJ, Lander ES, Mitzenmacher M, Sabeti PC. Detecting novel associations in large data sets. Science. 2011;334:1518–1524. doi: 10.1126/science.1205438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Roth CL, Mastronardi C, Lomniczi A, Wright H, Cabrera R, Mungenast AE, Heger S, Jung H, Dubay C, Ojeda SR. Expression of a tumor-related gene network increases in the mammalian hypothalamus at the time of female puberty. Endocrinology. 2007;148:5147–5161. doi: 10.1210/en.2007-0634. [DOI] [PubMed] [Google Scholar]
- 111.Roth CL, McCormack AL, Lomniczi A, Mungenast AE, Ojeda SR. Quantitative proteomics identifies a major change in glial glutamate metabolism at the time of female puberty. Mol Cell Endocrinol. 2006;254–255:51–59. doi: 10.1016/j.mce.2006.04.017. [DOI] [PubMed] [Google Scholar]
- 112.Sandau US, Mungenast AE, Alderman Z, Sardi SP, Fogel AI, Taylor B, Parent AS, Biederer T, Corfas G, Ojeda SR. SynCAM1, a Synaptic Adhesion Molecule, is Expressed in Astrocytes and Contributes to erbB4 Receptor-Mediated Control of Female Sexual Development. Endocrinology. 2011a;152:2364–2376. doi: 10.1210/en.2010-1435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Sandau US, Mungenast AE, McCarthy J, Biederer T, Corfas G, Ojeda SR. The Synaptic Cell Adhesion Molecule, SynCAM1, Mediates Astrocyte-to-Astrocyte and Astrocyte-to-GnRH Neuron Adhesiveness in the Mouse Hypothalamus. Endocrinology. 2011b;152:2353–2363. doi: 10.1210/en.2010-1434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Sauro HM, Hucka M, Finney A, Wellock C, Bolouri H, Doyle J, Kitano H. Next generation simulation tools: the Systems Biology Workbench and BioSPICE integration. OMICS. 2003;7:355–372. doi: 10.1089/153623103322637670. [DOI] [PubMed] [Google Scholar]
- 115.Sawarkar R, Paro R. Interpretation of developmental signaling at chromatin: the Polycomb perspective. Dev Cell. 2010;19:651–661. doi: 10.1016/j.devcel.2010.10.012. [DOI] [PubMed] [Google Scholar]
- 116.Schulz R, Wilhelm A, Pirke KM, Gramsch C, Herz A. Beta-endorphin and dynorphin control serum luteinizing hormone level in immature female rats. Nature. 1981;294:757–759. doi: 10.1038/294757a0. [DOI] [PubMed] [Google Scholar]
- 117.Schwartz YB, Pirrotta V. Polycomb silencing mechanisms and the management of genomic programmes. Nat Rev Genet. 2007;8:9–22. doi: 10.1038/nrg1981. [DOI] [PubMed] [Google Scholar]
- 118.Semaan SJ, Dhamija S, Kim J, Ku EC, Kauffman AS. Assessment of epigenetic contributions to sexually-dimorphic kiss1 expression in the anteroventral periventricular nucleus of mice. Endocrinology. 2012;153:1875–1886. doi: 10.1210/en.2011-1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Seminara SB, Crowley WF., Jr Perspective: the importance of genetic defects in humans in elucidating the complexities of the hypothalamic-pituitary-gonadal axis. Endocrinology. 2001;142:2173–2177. doi: 10.1210/endo.142.6.8261. [DOI] [PubMed] [Google Scholar]
- 120.Seminara SB, Messager S, Chatzidaki EE, Thresher RR, Acierno JS, Jr, Shagoury JK, Bo-Abbas Y, Kuohung W, Schwinof KM, Hendrick AG, Zahn D, Dixon J, Kaiser UB, Slaugenhaupt SA, Gusella JF, O’Rahilly S, Carlton MB, Crowley WF, Jr, Aparicio SA, Colledge WH. The GPR54 gene as a regulator of puberty. N Engl J Med. 2003;349:1614–1627. doi: 10.1056/NEJMoa035322. [DOI] [PubMed] [Google Scholar]
- 121.Shahab M, Mastronardi C, Seminara SB, Crowley WF, Ojeda SR, Plant TM. Increased hypothalamic GPR54 signaling: A potential mechanism for initiation of puberty in primates. Proc Natl Acad Sci U S A. 2005;102:2129–2134. doi: 10.1073/pnas.0409822102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Shannon M, Ashworth LK, Mucenski ML, Lamerdin JE, Branscomb E, Stubbs L. Comparative analysis of a conserved zinc finger gene cluster on human chromosome 19q and mouse chromosome 7. Genomics. 1996;33:112–120. doi: 10.1006/geno.1996.0166. [DOI] [PubMed] [Google Scholar]
- 123.Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Simon JA, Kingston RE. Mechanisms of polycomb gene silencing: knowns and unknowns. Nat Rev Mol Cell Biol. 2009;10:697–708. doi: 10.1038/nrm2763. [DOI] [PubMed] [Google Scholar]
- 125.Smith JT, Popa SM, Clifton DK, Hoffman GE, Steiner RA. Kiss1 neurons in the forebrain as central processors for generating the preovulatory luteinizing hormone surge. J Neurosci. 2006;26:6687–6694. doi: 10.1523/JNEUROSCI.1618-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Steeg PS, Ouatas T, Halverson D, Palmieri D, Salerno M. Metastasis suppressor genes: Basic biology and potential clinical use. Clin Breast Cancer. 2003;4:51–62. doi: 10.3816/cbc.2003.n.012. [DOI] [PubMed] [Google Scholar]
- 127.Storey JR. The positive false discovery rate: A bayesian interpretation and the q-value. The Annals of Statistics. 2003;31:2013–2035. [Google Scholar]
- 128.Sulem P, Gudbjartsson DF, Rafnar T, Holm H, Olafsdottir EJ, Olafsdottir GH, Jonsson T, Alexandersen P, Feenstra B, Boyd HA, Aben KK, Verbeek AL, Roeleveld N, Jonasdottir A, Styrkarsdottir U, Steinthorsdottir V, Karason A, Stacey SN, Gudmundsson J, Jakobsdottir M, Thorleifsson G, Hardarson G, Gulcher J, Kong A, Kiemeney LA, Melbye M, Christiansen C, Tryggvadottir L, Thorsteinsdottir U, Stefansson K. Genome-wide association study identifies sequence variants on 6q21 associated with age at menarche. Nat Genet. 2009;41:734–738. doi: 10.1038/ng.383. [DOI] [PubMed] [Google Scholar]
- 129.Tena-Sempere M. KiSS-1 and reproduction: focus on its role in the metabolic regulation of fertility. Neuroendocrinology. 2006;83:275–281. doi: 10.1159/000095549. [DOI] [PubMed] [Google Scholar]
- 130.Terasawa E, Fernandez DL. Neurobiological mechanisms of the onset of puberty in primates. Endocr Rev. 2001;22:111–151. doi: 10.1210/edrv.22.1.0418. [DOI] [PubMed] [Google Scholar]
- 131.Topaloglu AK, Reimann F, Guclu M, Yalin AS, Kotan LD, Porter KM, Serin A, Mungan NO, Cook JR, Ozbek MN, Imamoglu S, Akalin NS, Yuksel B, O’Rahilly S, Semple RK. TAC3 and TACR3 mutations in familial hypogonadotropic hypogonadism reveal a key role for Neurokinin B in the central control of reproduction. Nat Genet. 2008;41:354–358. doi: 10.1038/ng.306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Topaloglu AK, Tello JA, Kotan LD, Ozbek MN, Yilmaz MB, Erdogan S, Gurbuz F, Temiz F, Millar RP, Yuksel B. Inactivating KISS1 mutation and hypogonadotropic hypogonadism. N Engl J Med. 2012;366:629–635. doi: 10.1056/NEJMoa1111184. [DOI] [PubMed] [Google Scholar]
- 133.Tsutsui K, Bentley GE, Bedecarrats G, Osugi T, Ubuka T, Kriegsfeld LJ. Gonadotropin-inhibitory hormone (GnIH) and its control of central and peripheral reproductive function. Front Neuroendocrinol. 2010;31:284–295. doi: 10.1016/j.yfrne.2010.03.001. [DOI] [PubMed] [Google Scholar]
- 134.Ueda HR, Hayashi S, Chen W, Sano M, Machida M, Shigeyoshi Y, Iino M, Hashimoto S. System-level identification of transcriptional circuits underlying mammalian circadian clocks. Nat Genet. 2005;37:187–192. doi: 10.1038/ng1504. [DOI] [PubMed] [Google Scholar]
- 135.Urbanski HF, Ojeda SR. The juvenile-peripubertal transition period in the female rat: Establishment of a diurnal pattern of pulsatile luteinizing hormone secretion. Endocrinology. 1985;117:644–649. doi: 10.1210/endo-117-2-644. [DOI] [PubMed] [Google Scholar]
- 136.Urrutia R. KRAB-containing zinc-finger repressor proteins. Genome Biol. 2003;4:231. doi: 10.1186/gb-2003-4-10-231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Vogel MJ, Guelen L, de WE, Peric-Hupkes D, Loden M, Talhout W, Feenstra M, Abbas B, Classen AK, van SB. Human heterochromatin proteins form large domains containing KRAB-ZNF genes. Genome Res. 2006;16:1493–1504. doi: 10.1101/gr.5391806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Wakabayashi Y, Nakada T, Murata K, Ohkura S, Mogi K, Navarro VM, Clifton DK, Mori Y, Tsukamura H, Maeda K, Steiner RA, Okamura H. Neurokinin B and dynorphin A in kisspeptin neurons of the arcuate nucleus participate in generation of periodic oscillation of neural activity driving pulsatile gonadotropin-releasing hormone secretion in the goat. J Neurosci. 2010;30:3124–3132. doi: 10.1523/JNEUROSCI.5848-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Warde-Farley D, Donaldson SL, Comes O, Zuberi K, Badrawi R, Chao P, Franz M, Grouios C, Kazi F, Lopes CT, Maitland A, Mostafavi S, Montojo J, Shao Q, Wright G, Bader GD, Morris Q. The GeneMANIA prediction server: biological network integration for gene prioritization and predicting gene function. Nucleic Acids Res. 2010;38:W214–W220. doi: 10.1093/nar/gkq537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Watabe K, Ito A, Koma YI, Kitamura Y. IGSF4: a new intercellular adhesion molecule that is called by three names, TSLC1, SgIGSF and SynCAM, by vurtue of its diverse function. Histol Histopathol. 2003;18:1321–1329. doi: 10.14670/HH-18.1321. [DOI] [PubMed] [Google Scholar]
- 141.Weber M, Davies JJ, Wittig D, Oakeley EJ, Haase M, Lam WL, Schubeler D. Chromosome-wide and promoter-specific analyses identify sites of differential DNA methylation in normal and transformed human cells. Nat Genet. 2005;37:853–862. doi: 10.1038/ng1598. [DOI] [PubMed] [Google Scholar]
- 142.Weber M, Hellmann I, Stadler MB, Ramos L, Paabo S, Rebhan M, Schubeler D. Distribution, silencing potential and evolutionary impact of promoter DNA methylation in the human genome. Nat Genet. 2007;39:457–466. doi: 10.1038/ng1990. [DOI] [PubMed] [Google Scholar]
- 143.Whelan C, Sonmez K. Computing graphlet signatures of network nodes and motifs in Cytoscape with GraphletCounter. Bioinformatics. 2012;28:290–291. doi: 10.1093/bioinformatics/btr637. [DOI] [PubMed] [Google Scholar]
- 144.Winslow MM, Dayton TL, Verhaak RG, Kim-Kiselak C, Snyder EL, Feldser DM, Hubbard DD, DuPage MJ, Whittaker CA, Hoersch S, Yoon S, Crowley D, Bronson RT, Chiang DY, Meyerson M, Jacks T. Suppression of lung adenocarcinoma progression by Nkx2-1. Nature. 2011;473:101–104. doi: 10.1038/nature09881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Wright H, Cohen A, Sonmez K, Yochum G, McWeeney S. Occupancy classification of position weight matrix-inferred transcription factor binding sites. PLoS ONE. 2011;6:e26160. doi: 10.1371/journal.pone.0026160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Yang JJ, Caligioni CS, Chan YM, Seminara SB. Uncovering novel reproductive defects in neurokinin B receptor null mice: closing the gap between mice and men. Endocrinology. 2012;153:1498–1508. doi: 10.1210/en.2011-1949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Yeung KT, Das S, Zhang J, Lomniczi A, Ojeda S, Xu CF, Neubert TA, Samuels HH. A novel transcription complex that selectively modulates apoptosis of breast cancer cells through regulation of FASTKD2. Mol Cell Biol. 2011;31:2287–2298. doi: 10.1128/MCB.01381-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Zhang B, Horvath S. A general framework for weighted gene co-expression network analysis. Stat Appl Genet Mol Biol. 2005;4 doi: 10.2202/1544-6115.1128. [DOI] [PubMed] [Google Scholar]