Abstract
Thymidylate synthase (TSase) is a 62 kDa homodimeric enzyme required for de novo synthesis of thymidine monophosphate (dTMP) in most organisms. This makes the enzyme an excellent target for anticancer and microbial antibiotic drugs. In addition, TSase has been shown to exhibit negative cooperativity and half-the-sites reactivity. For these collective reasons, TSase is widely studied, and much is known about its kinetics and structure as it progresses through a multi-step catalytic cycle. Recently, nuclear magnetic resonance (NMR) spin relaxation has been instrumental in demonstrating the critical role of dynamics in enzyme function in small model systems. These studies raise questions about how dynamics affect function in larger enzymes with more complex reaction coordinates. TSase is an ideal candidate given its size, oligomeric state, cooperativity, and status as a drug target. Here, as a pre-requisite to spin relaxation studies, we present the backbone and ILV methyl resonance assignments of TSase from Escherichia coli bound to a substrate analogue and cofactor.
Biological Context
NMR spin relaxation studies have been instrumental in bringing about the understanding that dynamics are critical for enzyme function. Most of the seminal NMR experiments establishing links between flexibility and substrate binding and release, or catalysis itself, were necessarily performed on small model systems (Eisenmesser et al. 2005; Boehr et al. 2006; Watt et al. 2007). It therefore remains a largely unanswered question as to whether insights gleaned from these small proteins apply to larger systems with added levels of complexity such as multimeric assembly and allostery. Further, it is reasonable to expect that by studying more complex systems, new principles governing enzyme catalysis will emerge. Recent improvements in hardware (higher magnetic field strengths and cryogenic probes), in concert with clever isotopic labeling schemes have substantially raised the size limit for detailed NMR spin relaxation studies to over 100 kDa. Thus, we are now poised to study by NMR spectroscopy, countless enzymes and enzyme assemblies that are of interest to human health.
TSase is such an enzyme in that it catalyzes the final chemical transformations in the pathway that yields the sole source of dTMP in most organisms. Given that cell proliferation cannot occur without DNA synthesis, inhibition of TSase is a common strategy in chemotherapy and anti-microbial treatments. The enzyme is large by NMR standards (62 kDa), is homodimeric, exhibits negative cooperativity and half-the-sites reactivity, and catalyzes a multistep reaction. Despite the fact that the kinetics of the reaction steps have been worked out (Spencer et al. 1997) and crystal structures are available for each intermediate in the reaction cycle (Stroud et al. 2003), the basis for communication between subunits is not understood. NMR spin relaxation, given its ability to monitor the flexibility of probes throughout the molecule on multiple timescales, is well suited to identify mechanisms of communication that are invisible to x-ray crystallography. In addition, identification of inhibitor bound enzyme conformations that are different than the biological cofactor-bound conformations highlights the importance of receptor plasticity in drug design (Fritz et al. 2001). Thus an understanding of the differences in flexibility between TSases from different species will be important in the design of novel inhibitors with improved specificity and resilience to resistance.
As a prerequisite to NMR spin relaxation studies, we present here the backbone and ILV methyl resonance assignments of TSase from Escherichia coli (ecTSase), bound to the biological cofactor, 5,10-Methylenetetrahydrofolate (CH2H4fol), and the inhibitor, 5-Fluoro-2′-deoxyuridine 5′-monophosphate (5F-dUMP). This state represents the only stable intermediate after methylene transfer and before hydride transfer. It is therefore the best state to interrogate the effects of flexibility on the rate limiting step of hydride transfer.
Methods and Experiments
Sample preparation
The gene for ecTSase was cloned into pET21a, which was then transformed into BL21 (DE3) cells lacking the rne131 gene (Invitrogen). To prepare U-[2H, 13C, 15N] ecTSase for backbone resonance assignments, cells were grown in 1 L of M9 (99.8% D2O, CIL) media supplemented with 1 g of 15NH4Cl2 (CIL), 2 g of U-[13C, 2H]-glucose (CIL) as the sole nitrogen and carbon sources, respectively. Cells were grown at 37 °C to an OD600 of 0.8, at which time 0.75 mM IPTG was added and protein was expressed for 24 hours at 18 °C. For ILV methyl resonance assignments, methyl protonated {I(δ1 only), L(13CH3, 12CD3), V(13CH3, 12CD3)} U-[2H, 13C, 15N] ecTSase was prepared as above with the only modification being that at OD600 = 0.8, 60 mg of 2-keto-3,3-d2-1,2,3,4-13C-butyrate (Isotec) and 100 mg of 2-keto-3-methyl-d3-3-d1-1,2,3,4-13C-butyrate (Isotec) were added to the culture, which was then grown for 1 hour at 37 °C prior to induction. To facilitate measurement of intra-methyl NOESYs, we prepared a methyl protonated {I(δ1 only), L(13CH , 13CH3), V(13CH3, 13CH3)} U-[2H, 15N] ecTSase sample (differs from the previous sample in that both methyl groups of LV are 13CH3), by adding the appropriate alpha-ketoacids (Isotec). Lastly for stereospecific assignments of the prochiral LV methyl groups, we prepared a 10% 13C fractionally labeled sample grown in M9 (H2O). Cells were harvested and ecTSase was purified as described (Changchien et al. 2000) with an added step of running the DEAE pool over a Superdex G75 size exclusion column equilibrated with NMR buffer (25 mM NaPO4, 150 mM NaCl, 1 mM EDTA, 10 mM DTT, 0.02% NaN3, pH 7.5).
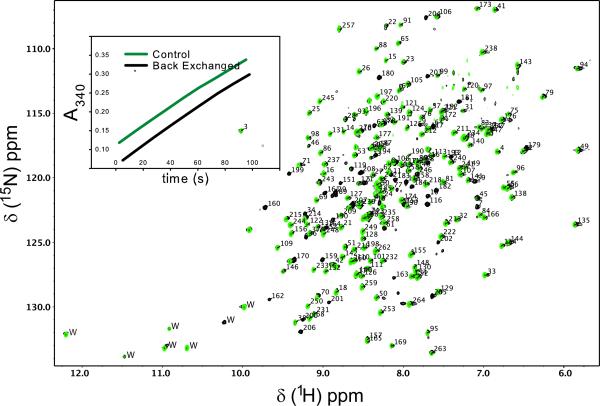
A comparison of 1H-15N TROSY-HSQC spectra of apo-ecTSase grown in H2O and D2O revealed that approximately 50 amides were resilient to back exchange after 1 week at room temperature. Back exchange efforts involving complete unfolding of the enzyme were unsuccessful, so we resorted to a more gentle approach; ecTSase (2 μM) was incubated at 4 °C for six days in NMR buffer supplemented with 2 M urea and 20 mM Tris, pH 9.0. Both urea and elevated pH were required for efficient back exchange and this treatment did not affect the specific activity of the enzyme (Figure 1 inset). For this study, samples used in backbone HN directed experiments were back exchanged and samples used in Hmethyl directed experiments were not.
Figure 1.
Backbone resonance assignments of ecTSase. 1H-15N TROSY HSQC spectra of ecTSase bound to 5F-dUMP and CH2H4fol. Spectra before and after 1H/2H back-exchange are shown in green and black respectively. Note that all green resonances have black peaks underneath and some resonances appear only after back-exchange. Inset shows that TS activity (Spencer et al. 1997) is not affected by back-exchange protocol. 233 out of 250 non-proline residues are assigned as indicated by residue numbers pointing to resonances. Trp side-chain resonances (labeled “W”) are not assigned. Data were acquired on U-[2H, 13C, 15N] enzyme at 25 °C at 700 MHz.
The covalent ecTSase-5F-dUMP-CH2H4fol complex studied in this work was prepared by reacting the enzyme with 4 molar equivalents of 5F-dUMP (Sigma), and 8 molar equivalents of CH2H4fol (Merck Eprova AG). Unincorporated ligands were removed via a G25 desalting column equilibrated with NMR buffer and the complex was concentrated using Amicon stirred cells and centrifugal devices equipped with 10k cutoff membranes.
NMR Experiments
For backbone resonance assignments a 0.5 mM (dimer) sample of U-[2H, 13C, 15N] ecTSase-5F-dUMP-CH2H4fol was prepared in NMR buffer supplemented with 5% D2O. Six TROSY triple resonance experiments were acquired on this sample: The HNCO, HN(CA)CO, HN(CO)CA, and HN(COCA)CB were acquired on a Varian INOVA 500 MHz spectrometer equipped with a 5mm triple resonance XYZ probe. The HNCA and HN(CA)CB were acquired on a Bruker AVANCE III 950 MHz spectrometer equipped with a TCI cryogenically cooled probe. For ILV methyl resonance assignments a 0.5 mM (dimer) sample of methyl protonated {I(δ1 only), L(13CH3, 12CD3), V(13CH3, 12CD3)} U-[2H, 13C, 15N] ecTSase-5F-dUMP-CH2H4fol was prepared in NMR buffer supplemented with 5% D2O. We acquired HMCM[CG]CBCA and Ile,Leu-HMCM(CGCBCA)CO spectra from this sample on a Varian INOVA 700 MHz spectrometer equipped with a cold probe. To aid in methyl assignments, a [13C-F1, 13C-F2]-edited NOESY (Zwahlen et al. 1998) (70 ms mixing time) was acquired on a 0.5 mM (dimer) sample of methyl protonated {I(δ1 only), L(13CH3, 13CH3), V(13CH3, 13CH3)} U-[2H, 15N] ecTSase-5F-dUMP-CH2H4fol in 99.8% D2O NMR buffer (700 MHz). Lastly, for stereospecific resonance assignments of prochiral LV methyl groups, we acquired a 1H-13C constant time HSQC (constant time period = 1/JCC = 28 ms) on 10% 13C fractionally labeled 0.5 mM (dimer) ecTSase-5F-dUMP-CH2H4fol in 99.8% D2O NMR buffer (700 MHz). All spectra were acquired at 25 °C. NMR data were processed using nmrPipe (Delaglio et al. 1995). The RunAbout module within NMRViewJ (Johnson 2004) was used to aid in the assignments of the backbone, and in-house NMRView scripts were used to facilitate methyl assignments.
Assignments and data deposition
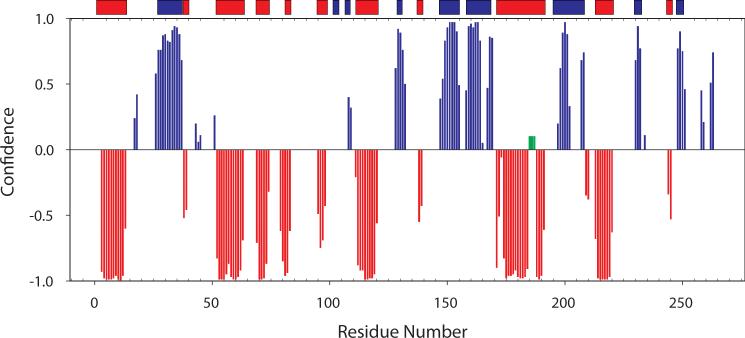
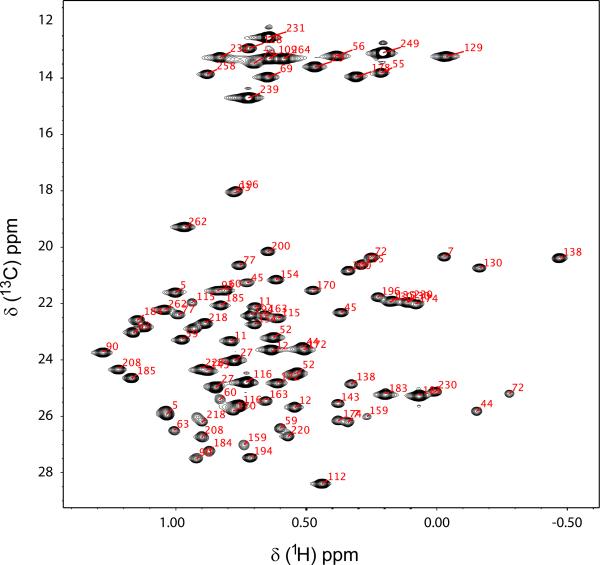
Figure 1 shows TROSY 1H-15N spectra of the U-[2H, 13C, 15N] ecTSase-5F-dUMP-CH2H4fol complex. First, given the reports in the literature that ecTSase is a half the sites-active enzyme (Maley et al. 1995), it is noteworthy that there is only a single set of resonances in spectra with nucleotide analogue and cofactor bound. Therefore, both active sites are competent to form the covalent complex with 5F-dUMP and CH2H4fol. Resonances are labeled with assignments for 93% (233/250) of non-proline 1H/15N amide pairs (Figure 1). In addition, based on the suite of six triple resonance experiments described above, we assigned 89% (455/510) of Cα + Cβ and 88% (232/264) of C′ resonances, respectively. The minor gaps in the sequential assignments are the result of four Pro-X-Pro motifs, several residues residing in solvent exposed loops, and chemical exchange. To validate the assignments, we used Talos+ (Shen et al. 2009) to predict the backbone dihedral angles of ecTSase in this complex and the predicted angles show excellent agreement with the secondary structure obtained from the high resolution x-ray model containing the same ligands used here (Figure 2). We also assigned ILV methyl resonances from this complex with an eye towards taking advantage of the favorable spectral properties of methyl groups in large proteins in future experiments. Methyl ILV assignments are shown on the 1H-13C CT-HSQC of methyl protonated {I(δ1 only), L(13CH3, 12CD3), V(13CH3, 12CD3)} U-[2H, 13C, 15N] ecTSase-5F-dUMP-CH2H4fol (Figure 3). We were able to assign 100% (42/42) of the Val+Ile (Cδ1 only) methyl resonances and 88% (49/56) of Leu methyl groups. It is noteworthy that complete assignment of the Ile+Val groups was possible using only the HMCM[CG]CBCA spectrum. We used the HMCM(CGCBCA)CO and the [13C-F1, 13C-F2]-edited NOESY to increase the completeness of Leu assignments and as independent check of the Val and Ile assignments. Backbone and side-chain chemical shift assignments have been deposited in the BioMagResBank (http://www.bmrb.wisc.edu/) under accession number 19082.
Figure 2.
Secondary structure from backbone chemical shifts agrees with the crystal structure. Talos+ (Shen et al. 2009) was used to predict backbone dihedral angles from the backbone resonance assignments. Beta-sheet probabilities are plotted in blue and alpha helical probabilities were multiplied by -1 and plotted in red. Bars on top of the graph represent the STRIDE algorithm (Heinig et al. 2004) predicted secondary structure from the x-ray model of ecTSase-5F-dUMP-CH2H4fol (PDBID 1TSN). Green bars highlight resonances that do not map to secondary structure elements because they are unassigned.
Figure 3.
ILV methyl assignments within the ecTSase-5F-dUMP-CH2H4fol complex. A 1H-13C CT-HSQC of a methyl protonated {I(δ1 only), L(13CH3, 12CD3), V(13CH3, 12CD3)} U-[2H, 13C, 15N] sample at 700 MHz is shown. Stereosepecific assignments for LV resonances are present in the BMRB deposition, but these designations are omitted from the labels above to improve clarity.
Acknowledgements
We thank Amnon Kohen for kindly providing a pBluescript vector containing the ecTSase coding sequence and Kevin Knagge at the David H. Murdoch Research Institute for help with data acquisition on the Bruker Avance 950 MHz spectrometer. 5,10-Methylenetetrahydrofolate was a kind gift from Merck & Cie (Schaffhausen, Switzerland).
References
- Boehr DD, McElheny D, Dyson HJ, Wright PE. The dynamic energy landscape of dihydrofolate reductase catalysis. Science. 2006;313(5793):1638–42. doi: 10.1126/science.1130258. [DOI] [PubMed] [Google Scholar]
- Changchien LM, Garibian A, Frasca V, Lobo A, Maley GF, Maley F. High-level expression of Escherichia coli and Bacillus subtilis thymidylate synthases. Protein Expr Purif. 2000;19(2):265–70. doi: 10.1006/prep.2000.1245. [DOI] [PubMed] [Google Scholar]
- Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J Biomol NMR. 1995;6(3):277–93. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- Eisenmesser EZ, Millet O, Labeikovsky W, Korzhnev DM, Wolf-Watz M, Bosco DA, Skalicky JJ, Kay LE, Kern D. Intrinsic dynamics of an enzyme underlies catalysis. Nature. 2005;438(7064):117–21. doi: 10.1038/nature04105. [DOI] [PubMed] [Google Scholar]
- Fritz TA, Tondi D, Finer-Moore JS, Costi MP, Stroud RM. Predicting and harnessing protein flexibility in the design of species-specific inhibitors of thymidylate synthase. Chem Biol. 2001;8(10):981–95. doi: 10.1016/s1074-5521(01)00067-9. [DOI] [PubMed] [Google Scholar]
- Heinig M, Frishman D. STRIDE: a web server for secondary structure assignment from known atomic coordinates of proteins. Nucleic Acids Res. 2004;32(Web Server issue):W500–2. doi: 10.1093/nar/gkh429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson BA. Using NMRView to visualize and analyze the NMR spectra of macromolecules. Methods Mol Biol. 2004;278:313–52. doi: 10.1385/1-59259-809-9:313. [DOI] [PubMed] [Google Scholar]
- Maley F, Pedersen-Lane J, Changchien L. Complete restoration of activity to inactive mutants of Escherichia coli thymidylate synthase: evidence that E. coli thymidylate synthase is a half-the-sites activity enzyme. Biochemistry. 1995;34(5):1469–74. doi: 10.1021/bi00005a001. [DOI] [PubMed] [Google Scholar]
- Shen Y, Delaglio F, Cornilescu G, Bax A. TALOS+: a hybrid method for predicting protein backbone torsion angles from NMR chemical shifts. J Biomol NMR. 2009;44(4):213–23. doi: 10.1007/s10858-009-9333-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spencer HT, Villafranca JE, Appleman JR. Kinetic scheme for thymidylate synthase from Escherichia coli: determination from measurements of ligand binding, primary and secondary isotope effects, and pre-steady-state catalysis. Biochemistry. 1997;36(14):4212–22. doi: 10.1021/bi961794q. [DOI] [PubMed] [Google Scholar]
- Stroud RM, Finer-Moore JS. Conformational dynamics along an enzymatic reaction pathway: thymidylate synthase, “the movie”. Biochemistry. 2003;42(2):239–47. doi: 10.1021/bi020598i. [DOI] [PubMed] [Google Scholar]
- Watt ED, Shimada H, Kovrigin EL, Loria JP. The mechanism of rate-limiting motions in enzyme function. Proc Natl Acad Sci U S A. 2007;104(29):11981–6. doi: 10.1073/pnas.0702551104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zwahlen C, Gardner KH, Sarma SP, Horita DA, Byrd RA, Kay LE. An NMR experiment for measuring methyl-methyl NOEs in C-13-labeled proteins with high resolution. Journal of the American Chemical Society. 1998;120(30):7617–7625. [Google Scholar]