Abstract
Exposure to maternal mood disorder in utero may program infant neurobehavior via DNA methylation of the glucocorticoid receptor (NR3C1) and 11β-hydroxysteroid dehydrogenase type 2 (11β-HSD-2), two placental genes that have been implicated in perturbations of the hypothalamic pituitary adrenocortical (HPA) axis. We tested the relations among prenatal exposure to maternal depression or anxiety, methylation of exon 1F of NR3C1 and 11β-HSD-2, and newborn neurobehavior. Controlling for relevant covariates, infants whose mothers reported depression during pregnancy and showed greater methylation of placental NR3C1 CpG2 had poorer self-regulation, more hypotonia, and more lethargy than infants whose mothers did not report depression. On the other hand, infants whose mothers reported anxiety during pregnancy and showed greater methylation of placental 11β-HSD-2 CpG4 were more hypotonic compared with infants of mothers who did not report anxiety during pregnancy. Our results support the fetal programming hypothesis and suggest that fetal adjustments to cues from the intrauterine environment, in this case an environment that could be characterized by increased exposure to maternal cortisol, may lead to poor neurodevelopmental outcomes.
Keywords: DNA methylation, maternal depression, maternal anxiety, newborn neurobehavior
As many as 22% of women experience clinical levels of depression or anxiety while pregnant.1 Fetal exposure to these maternal mood disorders is a serious public health problem as this exposure may lead to long-term emotional, behavioral, and social problems in offspring,2 though not for all newborns. Epigenetic processes involved in the physiologic and behavioral sequelae of exposure to maternal depression and anxiety in utero may help to explain why not all children exposed to maternal mood disorders develop psychosocial difficulties.
Adults with mood disorders often hypersecrete cortisol and exhibit prolonged elevations in cortisol.3 Infants whose mothers had a mood disorder during pregnancy had higher cortisol levels4 and depressive symptoms including irritability5 and altered newborn neurobehavior6 compared with infants of well mothers. Furthermore, infants whose mothers had a mood disorder in pregnancy and in infancy had higher baseline cortisol levels than children never exposed to a maternal mood disorder or exposed to a maternal mood disorder only in infancy.7 These studies provide evidence that the fetal environment has a strong influence on these problematic outcomes by altering programming of the developing brain including the infant neuroendocrine system.8 During early development, neural networks are formed and behavioral pathways are programmed making them vulnerable to environmental influences9 although the molecular mechanisms underlying these relationships are unclear.10
The fetus is exposed to physiological sequelae of the mother’s mood disorder in the intrauterine environment via placental transfer of altered hormones and neurotransmitters as well as restricted uterine blood flow.11 These changes may be due to changes in physiology, specifically, elevated circulating cortisol, elevated norepinephrine, and decreased serotonin.4,12 Eighty to 90 percent of maternal cortisol is metabolized while it passes through the placenta, although higher levels of maternal cortisol may overwhelm or alter this metabolism leading to higher levels of fetal exposure to cortisol, which can have harmful effects on the fetus and neonate.13 Urinary cortisol is elevated in neonates of mothers with depression, and baseline cortisol is highest among infants of mothers who were depressed during pregnancy and who exhibited less sensitive caregiving.14 Glucocorticoid exposure can affect synaptogenesis, neurotransmitter function, and glucocorticoid receptor expression in the offspring’s developing brain and impact stress responses systems including the development of the HPA axis, and the autonomic nervous system.
Fetal exposure to maternal cortisol disrupts the programming of fetal biological stress response systems. 11β-hydroxysteroid dehydrogenase type 2 (11β-HSD-2) and the glucocorticoid receptor (NR3C1) are placental genes that have been implicated in perturbations of the hypothalamic pituitary adrenocortical (HPA) axis. They protect the fetus from excess catecholamines and glucocorticoids, which have harmful effects on the fetus. 11β-HSD-2 in particular converts maternal cortisol to inert cortisone protecting the fetus from exposure to maternal cortisol. Rat pups born to mothers who did not express 11β-HSD-2 were exposed to higher levels of maternal glucocorticoids, had lower birth weights, and exhibited more anxiety compared with rat pups born to parents who did express 11β-HSD-215. Prenatal stress reduces 11β-HSD-2 activity in rodents, which is related to increased exposure to glucocorticoids.16 Increased exposure to glucocorticoids, or inhibition of 11β-HSD-2, resulted in decreased birth weight, increased hyperglycemia, hypertension, HPA axis reactivity and anxiety in rodent models.16 In humans, prenatal anxiety has been associated with reduced 11β-HSD-2 expression in the placenta.17 We found increased placental DNA methylation leading to decreased expression of 11β-HSD-2 in healthy term infants with the lowest birth weights. This increased methylation was associated with poorer quality of movement in those newborns18 based on a standardized neurobehavioral assessment.19
The NR3C1 gene encodes the glucocorticoid receptor, is activated by cortisol and also regulates 11β-HSD-2. Maternal depression during the third trimester of pregnancy is associated with increased methylation of NR3C11,20 and increased cortisol responses in infants at 3 mo postpartum20 at a region analogous to the site found to be hypermethylated in rat pups exposed to less maternal nurturing care.20 In an animal model of depression, male rodents exposed to chronic variable stress early in gestation spent more time immobile during a tail suspension test and a forced swim test, suggestive of learned helplessness, and they ingested significantly more of a sugar solution.21 They also exhibited increased NR3C1 methylation at the same site that had previously been linked to decreased expression of NR3C1,22 and increased HPA reactivity. We found increased methylation of NR3C1 in the placenta related to decrements in the newborn infant’s ability to track and follow animate and inanimate stimuli.23
The placenta, as a key regulator of the intrauterine environment, may play a critical role in mediating the neurodevelopment of the infant through epigenetic mechanisms such as methylation of 11β-HSD-2 and NR3C1. Here, we report findings suggesting that these same epigenetic changes and subsequent neurobehavioral effects are related to maternal depression and anxiety during pregnancy. We hypothesized that infants whose mothers reported experiencing depression or anxiety in utero would show increased methylation of NR3C1 and 11β-HSD-2 in the placenta, compared with infants of mothers who did not report depression or anxiety in utero. We expected that these increases in methylation would be related to more problematic newborn neurobehavior.
Results
Demographic and descriptive statistics of the sample of infants with completed NNNS assessments are presented in Table 1. All infants were born at term, with a mean gestational age of 39.0 weeks (range 37–41 weeks). The majority of participants were Caucasian. Approximately 13.7% of mothers were identified as having depression or anxiety during pregnancy, which is consistent with estimated prevalence rates in the general population.5 Complete NNNS data were available for 90.6% of the sample. There were no differences in the distribution of demographic variables including depression and anxiety in mothers whose children did or did not have complete NNNS exams (all P values > 0.07), with the exception of tobacco exposure. More infants with missing NNNS data were tobacco-exposed, compared with infants without missing NNNS data (χ2[2] = 6.11, P = 0.05).
Table 1. Demographics and study variables.
| Variable | Women with depression (n = 66) | Women with anxiety (n = 57) | Well women (n = 398) |
|---|---|---|---|
| Growth status, n* (%) | |||
| SGA | 14 (21.2) | 10 (17.5) | 74 (18.8) |
| AGA | 30 (45.5) | 30 (52.7) | 207 (52.5) |
| LGA | 22 (33.3) | 17 (29.8) | 113 (28.7) |
| Gestational age (weeks), n, M(SD) | 66, 38.9 (0.98) | 57, 38.7 (0.86) | 397, 39.0 (1.06) |
| Maternal age (years), n, M(SD) | 66, 29.2 (5.74) | 57, 30.3 (5.85) | 396, 29.2 (5.61) |
| Infant gender (n, %) | |||
|---|---|---|---|
| Female | 34 (51.5) | 31 (54.4) | 206 (51.8) |
| Male | 32 (48.5) | 26 (45.6) | 192 (48.2) |
| Maternal race (n, %) | |||
|---|---|---|---|
| Caucasian | 48 (72.7) | 49 (86.0) | 284 (71.4) |
| African American | 11 (16.7) | 6 (10.6) | 66 (16.6) |
| Other | 6 (9.1) | 1 (1.7) | 46 (11.5) |
| Unknown | 1 (1.5) | 1 (1.7) | 2 (0.5) |
| Maternal tobacco use during pregnancy (n, %) | |||
|---|---|---|---|
| No | 59 (89.4) | 53 (93.0) | 383 (96.2) |
| Yes | 7 (10.6) | 4 (7.0) | 15 (3.8) |
| Habituation, n, M(SD) | 37, 7.32 (0.99) | 34, 7.52 (1.02) | 213, 7.22 (1.33) |
| Attention, n, M(SD) | 60, 4.03 (1.35) | 52, 3.99 (1.50) | 357, 4.21 (1.26) |
| Stress abstinence, n, M(SD) | 66, 0.18 (0.07) | 57, 0.18 (0.07) | 398, 0.18 (0.07) |
| Quality of movement, n, M(SD) | 66, 4.04 (0.63) | 57, 4.20 (0.57) | 398, 4.08 (0.67) |
| Excitability, n, M(SD) | 66, 4.41 (2.81) | 57, 3.93 (2.87) | 398, 4.80 (2.92) |
| Handling, n, M(SD) | 60, 0.36 (0.21) | 52, 0.37 (0.22) | 377, 0.37 (0.23) |
| Self-regulation, n, M(SD) | 65, 4.87 (0.95) | 56, 5.01 (0.98) | 396, 4.74 (0.92) |
| Arousal, n, M(SD) | 66, 4.06 (0.83) | 57, 4.02 (0.83) | 398, 4.20 (0.81) |
| Hypertonicity, n, M(SD) | 66, 0.48 (0.81) | 57, 0.42 (0.78) | 398, 0.42 (0.80) |
| Hypotonicity, n, M(SD) | 66, 0.76 (1.15) | 57, 0.65 (1.11) | 398, 0.54 (0.72) |
| Asymmetrical reflexes, n, M(SD) | 66, 1.80 (1.42) | 57, 1.61 (1.33) | 398, 1.73 (1.35) |
| Lethargy, n, M(SD) | 66, 7.05 (2.84) | 57, 7.16 (2.93) | 398, 6.24 (2.39) |
| Non-optimal reflexes, n, M(SD) | 66, 6.11 (2.11) | 57, 6.54 (2.11) | 398, 6.00 (2.13) |
Four women had missing birth weight data
Analyses for NR3C1 methylation
We first ran Spearman rank correlations to identify associations between depression and anxiety, and our selected CpG sites for NR3C1. Depression (but not anxiety) was related to greater methylation of CpG2, (Spearmans’s ρ = 0.10, P = 0.03). We therefore only examined interactions between depression and methylation of NR3C1 CpG2 to limit the presence of Type 1 error. We next ran hierarchical linear regressions to predict NNNS scores from maternal prenatal depression, DNA methylation of NR3C1 CpG2, and the interaction between NR3C1 CpG2 and depression. Covariates in the models included maternal anxiety, infant sex, birth weight, and maternal age, as these variables have been found in the literature to be related to NR3C1 methylation.
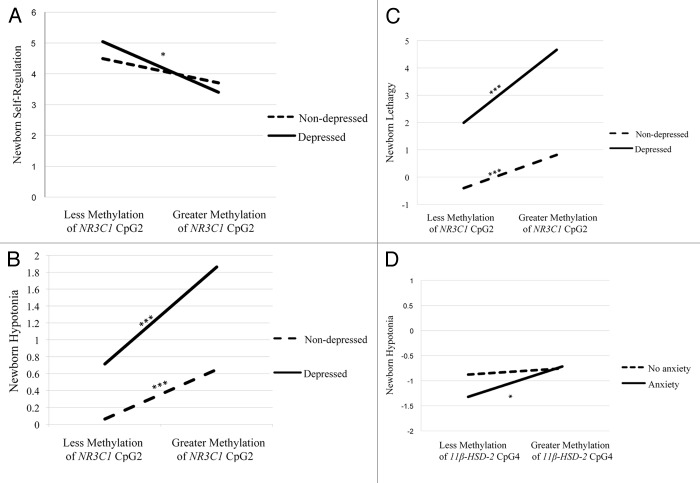
There was a statistically significant maternal depression x NR3C1 CpG2 methylation interaction for self-regulation (Table 2). There were no differences in self-regulation between infants of non-depressed mothers, regardless of the extent of methylation of NR3C1 CpG2. However, infants of depressed mothers with greater placental methylation of NR3C1 CpG2 showed poorer self-regulation (Fig. 1A).
Table 2. Hierarchical linear regression predicting newborn neurobehavior at birth.
| Predictors | β step 1 | β step 2 | R2 | F |
|---|---|---|---|---|
| Outcome: Self-regulation | ||||
| Infant sex | 0.07 | 0.05 | ||
| Maternal age | −0.04 | −0.06 | ||
| Birth weight | 0.05 | 0.06 | ||
| Maternal anxiety | 0.15 | 0.14 | ||
| Maternal depression | <0.001 | 0.05 | ||
| NR3C1 CpG2 methylation | 0.04 | 0.04 | ||
| 0.03 | 0.95 | |||
| Maternal depression x NR3C1 CpG2 methylation | - | −0.16* | ||
| ∆R2 = 0.02* | 0.06 | 10.46 | ||
| Outcome: Hypotonia | ||||
|---|---|---|---|---|
| Infant sex | 0.08 | 0.11 | ||
| Maternal age | −0.10 | −0.06 | ||
| Birth weight | −0.36‡ | −0.37‡ | ||
| Maternal anxiety | −0.04 | −0.02 | ||
| Maternal depression | 0.12 | 0.04 | ||
| NR3C1 CpG2 methylation | 0.03 | 0.03 | ||
| 0.16 | 50.55‡ | |||
| Maternal depression x NR3C1 CpG2 methylation | — | 0.25‡ | ||
| ∆R2 = 0.06‡ | 0.21 | 60.81‡ | ||
| Outcome: Lethargy | ||||
|---|---|---|---|---|
| Infant sex | 0.14 | 0.16* | ||
| Maternal age | −0.03 | 0.002 | ||
| Birth weight | −0.13 | −0.14 | ||
| Maternal anxiety | −0.05 | −0.03 | ||
| Maternal depression | 0.19* | 0.13 | ||
| NR3C1 CpG2 methylation | −0.09 | −0.09 | ||
| 0.06 | 20.04 | |||
| Maternal depression x NR3C1 CpG2 methylation | – | 0.19† | ||
| ∆R2 = 0.03† | 0.10 | 20.68† | ||
| Outcome: Hypotonia | ||||
|---|---|---|---|---|
| Infant sex | 0.08 | 0.09 | ||
| Maternal age | −0.06 | −0.05 | ||
| Birth weight | −0.27‡ | −0.27‡ | ||
| Maternal tobacco use during pregnancy | 0.13* | 0.13* | ||
| Maternal anxiety | −0.05 | −0.08 | ||
| Maternal depression | 0.15† | 0.16† | ||
| 11β-HSD-2 CpG4 methylation | 0.06 | 0.06 | ||
| 0.10 | 50.76‡ | |||
| Maternal anxiety x 11β-HSD-2 CpG4 methylation | — | 0.10* | ||
| ∆R2 = 0.01* | 0.11 | 30.68* | ||
P < 0.05, †P < 0.01, ‡P < 0.001
Figure 1. Interactions between maternal depression during pregnancy and NR3C1 CpG2 on newborn self-regulation (A), hypotonia (B), and lethargy (C), and the interaction between maternal anxiety during pregnancy and 11βHSD-2 CpG4 methylation on newborn hypotonia (D). Simple slopes were tested at ± 1 SD from the mean (A, D), or 1 SD and 2 SD above the mean (B, C). * P < 0.05; ** P < 0.01; *** P < 0.001.
There was also a significant NR3C1 CpG2 methylation x maternal depression interaction on hypotonia (Table 2). Infants of depressed mothers with greater placental methylation of NR3C1 CpG2 were more hypotonic and this effect was stronger when compared with infants of mothers who were not depressed (Fig. 1B). In addition, infants of mothers who were not depressed were more hypotonic with greater methylation of NR3C1 CpG2.
Lethargy also showed a statistically significant maternal depression x NR3C1 CpG2 methylation interaction (Table 2). Infants of depressed mothers with greater methylation placental of NR3C1 CpG2 were more lethargic (Fig. 1C) and this effect was stronger compared with infants of mothers who were not depressed. Infants of mothers who were not depressed also exhibited more lethargy with greater methylation of NR3C1 CpG2.
Analyses for 11β-HSD-2 methylation
We first ran Spearman rank correlations to determine associations between depression and anxiety, and our selected CpG sites for 11β-HSD-2. Controlling for maternal depression during pregnancy, maternal anxiety during pregnancy was related to greater methylation of 11β-HSD-2 CpG4, (Spearman’s ρ = 0.10, P = 0.04). We therefore only examined interactions between 11β-HSD-2 CpG4 and anxiety to limit the presence of Type 1 error. We tested associations between maternal anxiety during pregnancy, methylation of 11β-HSD-2, and the interaction between 11β-HSD-2 and anxiety, on our NNNS infant neurobehavioral outcomes. Covariates included maternal depression, infant sex, birth weight, maternal age, and maternal tobacco use, as these were related to methylation of 11β-HSD-2 CpG4.
There was a statistically significant maternal anxiety x 11β-HSD-2 CpG4 methylation interaction on hypotonia (Table 2). There were no differences in hypotonia among infants of mothers without anxiety during pregnancy, regardless of the extent of methylation of 11β-HSD-2 CpG4. However, infants of mothers who were anxious during pregnancy were more hypotonic with greater methylation of 11β-HSD-2 CpG4 (Fig. 1C).
Discussion
We found that the effects of maternal report of depression or anxiety during pregnancy on newborn neurobehavior depended upon increased DNA methylation of placental genes that regulate fetal exposure to cortisol. Interestingly, the association between self-reported depression was unique to NR3C1 CpG2, and self-reported anxiety to 11β-HSD-2 CpG4. Specifically, the group of infants whose mothers reported depression during pregnancy and showed greater methylation of placental NR3C1 CpG2 had poorer self-regulation than infants whose mothers were not depressed or anxious regardless of DNA methylation status of these placental genes. Both infants of depressed and non-depressed mothers who exhibited greater methylation of NR3C1 CpG2 exhibited more hypotonia and lethargy, though this effect was stronger for infants whose mothers reported depression. Infants whose mothers reported anxiety and who exhibited greater methylation of 11β-HSD-2 CpG4 had more hypotonia compared with well mothers. This work supports the fetal programming hypothesis and suggests that fetal adjustments to cues from the intrauterine environment, in this case an environment that could be characterized by increased exposure to maternal cortisol, may lead to poor neurodevelopmental outcomes.
Our findings support previous work20 relating maternal depression during pregnancy to increased methylation of NR3C1 CpG2 in cord blood mononuclear cells. We add to the literature by showing similar findings in placental genes, and by studying another gene that also regulates fetal exposure to maternal cortisol (11β-HSD-2). For the first time, we have also shown that epigenetic changes related to maternal-reported depression and anxiety have negative effects on newborn neurobehavior. These behaviors are related to risk of developing problem behavior later in life, particularly when the newborn is raised by a mother with a mood disorder.17,24 For instance, Field’s cumulative risk model purports that fetal exposure to an adverse intrauterine environment may result in neurobiological dysregulation which, when interacting with a poor caregiving environment, may result in risk for the development of psychopathology.25
Greater methylation of NR3C1 CpG2 appeared to confer risk for poor newborn neurobehavior, but only if the infant has been exposed to maternal depression during pregnancy. Adverse intrauterine experiences such as exposure to maternal stress lead to increases in the release of fetal catecholamines and glucocorticoids, possibly via methylation of genes such as NR3C1 and 11β-HSD-2. These catecholamines in turn may act on the fetus’ developing HPA axis, resulting in altered set points for physiologic, metabolic, and behavioral outcomes. Indeed there is evidence that maternal adrenocorticotropic hormone (ACTH) modulates the fetal HPA axis as increases in maternal ACTH resulted in increased offspring cortisol and ACTH levels.26 Increases in offspring cortisol, in turn, may be related to poorer self-regulation27 and motor behavior.28
An infant who experiences maternal depression during pregnancy may be exposed to greater levels of maternal cortisol in utero via increased methylation of 11β-HSD-216. 11β-HSD-2 functions to convert maternal cortisol to inert cortisone, thereby protecting the fetus from exposure to maternal cortisol. Maternal anxiety in utero is also related to greater fetal exposure to maternal cortisol.29 Therefore, greater maternal anxiety in utero may lead to increased methylation of 11β-HSD-2, which in turn could lead to alterations in HPA axis function or altered physiological responses to stress and associated neurobehavioral outcomes. For instance, in previous work using this sample, we18,23 found that increased methylation of 11β-HSD-2 and NR3C1 CpG6, 7, and 8 was related to poorer quality of movement in the newborn. We expand on this work by demonstrating that infants who experienced maternal-reported anxiety in utero and had greater methylation of 11β-HSD-2 CpG4 exhibited more hypotonia at birth. Other studies have found that greater cortisol exposure at specific times during pregnancy is related to poorer motor control.28 Deficits in motor control at birth may be a precursor to later neurodevelopmental problems. For instance, infants who exhibit delays in gross and fine motor skills are also at risk for later learning problems, as the ability to grasp, inspect, and cross the midline is essential for typical neurodevelopment.
We did not find associations between depression and DNA methylation of NR3C1 in CpG sites 1 or 3, as have others,20 or the other three CpG sites of 11β-HSD-2. As the literature on DNA methylation of NR3C1 and 11β-HSD-2 in infants is very small, it is unclear which CpG sites may be more likely to be associated with maternal mood or behavior outcomes. There may be specific sites within the regions sequenced that can bind to specific transcription factors, and so methylation at these different sites may represent different functional effects on downstream transcription. Therefore, in the case of NR3C1, we may have been less likely to find effects in other CpG sites because they are not as important in transcription. There is clearly a need for future mechanistic, in vitro studies to dissect the contribution of each of these sites within these regions on function, and the potential changes in transcription factor binding related to differential methylation.
Elsewhere, we have proposed a model of how risk for depression may be transmitted from mother to offspring via epigenetic processes.30 We argue that maternal depression during pregnancy leads to programming of the fetal HPA axis via epigenetic effects; specifically, exposure to maternal catecholamines and glucocorticoids may lead to alterations in the fetal HPA axis via methylation of genes that regulate fetal exposure to these hormones, including 11β-HSD-2 and NR3C1. Alterations of the fetal HPA axis could then lead to altered newborn neurobehavior because of HPA axis under or over-activity. We have found evidence for this hypothesized pathway as maternal-reported depression or anxiety was related to DNA methylation of 11β-HSD-2 or NR3C1 and methylation of these genes was in turn predictive of newborn neurobehavior. However, a key limitation of this study is that we cannot infer direction of effect with these data and mechanistic studies are needed to overcome this limitation. We have also argued that postnatal parenting practices and newborn exposure to other stressors may exacerbate risk for depression that was initiated prenatally, an important next step for future research.
While the literature is small, the majority of human behavioral epigenetic research thus far has examined associations between maternal mood and DNA methylation, without examining behavioral outcomes,1,20 or has focused on DNA methylation and newborn behavior, without considering the effects of exposure to maternal mood disorder in utero.23 This is the first study to integrate all three domains in order to articulate the processes involved in the development of newborn neurobehavior, individual differences in which may lay the foundation for later cognitive31 and behavioral functioning.32 Limitations of this study include our assessment of maternal depression and anxiety, as assessment was made based on chart review. Clinical interviews or symptom counts of depression and anxiety at specific points during gestation are needed to better understand how the timing of exposure to maternal mood disorders may result in epigenetic alterations in key genes associated with the infant response to stress. We are also unable to identify the specific mechanisms involved in the development of newborn neurobehavior, though our findings converge with others to suggest that epigenetic processes are likely involved. Nevertheless, this research adds to the growing body of work highlighting the importance of considering epigenetic effects when examining the development of behavioral outcomes, in order to identify novel targets for prevention and intervention.
Materials and Method
Participants
Mothers were recruited at birth from a local hospital following approval from the Women and Infants Hospital of Rhode Island and Dartmouth College IRB, and after informed consent. Participants were recruited based on birth weight and all infants had to be full-term (≥37 weeks GA) in order to meet criteria for entry into the study. Term infants born small for gestational age (SGA; lowest 10th percentile) or large for gestational age (LGA; highest 10th percentile) calculated from the Fenton growth chart were selected. Infants appropriate for gestational age (AGA) matched on gender, gestational age (±3 d), and maternal age (±2 y) were also enrolled. Only singleton, viable infants were included in the study. Other exclusion criteria were maternal age < 18 y or a life-threatening medical complication of the mother, and congenital or chromosomal abnormality of the infant. A structured chart review was used to collect information from the maternal inpatient medical record from delivery, and mothers were given a structured interview to obtain information on lifestyle, demographics, and exposure history of the infants. There were 27 mothers who reported depression only, 18 who reported anxiety only, 39 who reported both depression and anxiety, and 398 mothers who did not report depression or anxiety, resulting in 482 total participants.
Assessment of maternal depression and anxiety
A structured chart review was used to collect information about whether the mother endorsed depression or anxiety during pregnancy. All women were asked by a nurse or physician whether they had experienced depression or anxiety during pregnancy, and this information was recorded in their chart. Two trained abstractors obtained data on diagnoses of maternal depression and anxiety from the clinical records of the women in this sample. Underreporting of these undesirable behaviors was of concern. In this sample, however, the percentage of women who disclosed a diagnosis of depression and/or anxiety were similar to those reported in other studies.5,33,34 In addition, the validity of this method of assessment of depression and anxiety is bolstered by studies that found associations between chart-reviews of maternal-reported depression and HIV status,35 and high concordance between chart-review abstraction of maternal report of depression and clinical diagnoses of depression.36 It is unknown based on structured chart review at which trimester the depression or anxiety occurred. A dichotomous measure of the presence or absence of depression or anxiety was recorded. They were measured as separate predictors in our models.
Assessment of newborn neurobehavior
The NICU Network Neurobehavioral Scale (NNNS) was administered during the newborn inpatient stay, prior to discharge by certified psychometrists, who were blinded to study hypotheses.19 The NNNS is a comprehensive evaluation of the neurobehavioral performance of high-risk and preterm infants that includes neurological and behavioral measures and signs of stress. The NNNS was conducted 24 h prior to discharge. It was completed during the day between the hours of 9:00 am and 5:00 pm. There are strict criteria for assessing newborn neurobehavior using the NNNS that require all newborns to be asleep at the start of the NNNS examination. The infant also has to have been recently fed. The NNNS were conducted by certified NNNS examiners and as such they followed administration guidelines. As inclusion criteria included healthy women and infants with no significant medical complications, women spent an average of 1–4 d in the hospital; specifically, one day if they had a vaginal delivery and 3–4 d if they had a cesarean section.
Items for the NNNS were scored using previously established summary scores. Summary scores using the NNNS include: attention, handling, self-regulation, arousal, excitability, lethargy, hypertonicity, hypotonicity, non-optimal reflexes, asymmetrical reflexes, quality of movement, and stress abstinence signs. Psychometric properties of the exam have been established.19
Placenta sample collection, nucleic acid extraction, and bisulfite modification
For each participant within 2 h of delivery, 12 samples of placenta tissue, 3 from each of 4 quadrants (totaling approximately 1 g of tissue), 2 cm from the cord insertion site were excised. Samples of placental parenchyma were carefully dissected by trained research assistants to be free of maternal decidua, in order to assure that the samples from which DNA is extracted were of fetal origin. The samples were placed immediately in RNAlater and stored at 4 °C. At least 72 h later, placenta samples were removed from RNAlater, blotted dry, snap-frozen in liquid nitrogen, pulverized to homogeneity using a stainless steel cup and piston unit (Cell Crusher), and stored in sample tubes at −80 °C until needed for examination. DNA was extracted from the placenta samples using the QIAmp DNA Mini Kit (Qiagen, Inc.). Purified DNA was quantified using a ND-1000 spectrophotometer (Nanodrop), and DNA samples (1 µg) were bisulfite-modified using the EZ DNA Methylation Kit (Zymo Research) and stored at −20 °C.
In another study, we have genotyped infant cord blood, placenta, and maternal blood, and found that the infant’s genotype as assessed using the placental samples matched the infant cord blood genotype.37 We are therefore confident that the samples in this study are of fetal origin.
Bisulfite pyrosequencing DNA methylation analysis
NR3C1
We sought to interrogate the 13 CpG sites in the NR3C1 exon 1F promoter region. However, as justified below, our primary interest was on sites 1–3, which have previously showed variability in DNA methylation associated with maternal depression and cortisol response in infant cord blood20 and with infant growth status in placenta.38 DNA samples (1 μg) were bisulfite-modified using the EZ DNA methylation Kit (Zymo Research) following the manufacturer’s protocol. Pyrosequencing was performed on PCR product amplified from bisulfite modified DNA as described previously.38
The primers for amplification were Forward: 5′-TTTTTTTTTT GAAGTTTTTT TA-3′ and Reverse: 5′-Biotin-CCCCCAACTC CCCAAAAA-3′. The first sequencing primer was designed to sequence the first five CpG sites (5′-GAGTGGGTTT GGAGT-3′), and the second sequencing primer was designed to sequence the following eight CpG sites (5′-AGAAAAGAAT TGGAGAAATT-3′) for a total of 13 sites sequenced. Percent DNA methylation at each CpG site was quantified using the Pyro Q-CpG software, version 1.0.11 (Qiagen).
11β-HSD-2
Pyrosequencing was performed on PCR product amplified from bisulfite-modified DNA as described previously based on the region sequenced and displaying differential methylation in human placenta from Alikhani-Koopaei and colleagues.39 In brief, the Pyromark PCR Kit (Qiagen) and the following forward and biotinylated reverse primers were used for amplification: HSD11B2-F, 5′-GGAAGTGGGG TTGTGYGTTT TTAGGTTTAA GTT-3′ and HSD11B2-R, 5′-biotin-ATACCCTTTA CTAATCRCAC CACC-3′ (IDT Inc.). Cycling conditions were 94 °C for 15 min followed by 45 cycles of 94 °C for 30 s, 55°C for 1 min and 72 °C for 1 min with a final extension of 7 min at 72 °C. PCR products were sequenced using a PyroMark MD system and the following sequencing primer (IDT): HSD11B2-seq, 5′-GGGGTAGAGA TTTTAAGAA -3′. The sequencing primer was designed to sequence four CpG sites, and the dispensation orders for the assays were GTCGATGTCA GTCGTTAGTT CGTCA. The percent methylation at each CpG site was quantified using the Pyro Q-CpG software, version 1.0.11 (Qiagen). For both assays, bisulfite conversion controls were included on each sequencing read. In order for the sample’s methylation extent to be called, the bisulfite conversion rate must be >93%, and for all samples examined the conversion rate was >95%. All assays were performed in triplicate on the same bisulfite converted DNA template on all samples, and if any of the repeats differed by >10% those assays on that sample were repeated. To prevent batch effects from bisulfite treatments interfering with the analysis, samples were randomized across batches.
Covariates
Covariates were included if correlated (P < 0.05) with maternal depression, maternal anxiety, or DNA methylation of NR3C1 CpG2 or 11β-HSD-2 CpG4, or if a variable was found in the literature to be related to NR3C1 methylation. Variables that met these criteria were infant sex, birth weight, and maternal age for the NR3C1 CpG2 analyses and infant sex, birth weight, maternal age, and maternal tobacco use for 11β-HSD-2 CpG4 analyses. Ethnicity was also tested as a covariate but was not related to maternal depression, anxiety, or DNA methylation of NR3C1 CpG2 or 11β-HSD-2 CpG4 (all P > 0.07). Maternal depression was included as a covariate in all analyses involving maternal anxiety, and anxiety was included as a covariate in all analyses that involved maternal depression as a predictor, due to the high comorbidity between anxiety and depression.40
Data analysis plan
Data were examined for outliers and violations of normality. NR3C1 CpG2 was log transformed because of positive skewness. We did not log-transform 11β-HSD-2 CpG4 because it was normally distributed. Spearman rank correlations were first run to identify significant associations between methylation of specific CpG sites and maternal depression or anxiety. Based on the work of Oberlander and colleagues,20 who found that methylation of NR3C1 at CpG sites 1, 2, and 3 were related to prenatal maternal depressed mood and anxiety, we examined correlations between these three sites and maternal depression or anxiety. As we found no other studies examining 11β-HSD-2 and depression or anxiety in humans, we analyzed all 4 CpG sites in our correlations. In addition, we chose to look at the following five NNNS summary scores, as they have been shown in previous work to be most sensitive to the intrauterine environment32: Self-regulation, lethargy, hypotonia, attention, and quality of movement. Quality of movement and attention were not predicted by mood x DNA methylation interactions. We therefore do not report on results related to quality of movement or attention.
Based on results from the Spearman rank correlations between depression/anxiety and NR3CI or 11β-HSD-2 we only examined interactions between depression or anxiety and the specific site that emerged as significant in our correlations, to limit Type I error. Hierarchical regression models were used to predict NNNS summary scores from covariates, maternal mood variables (depression and anxiety), methylation of NR3C1 CpG 2 (in separate models, as this site significantly correlated with depression) and 11β-HSD-2 CpG 4 (also in separate models, as this site was significantly correlated with anxiety), and the interaction between maternal mood and methylation of specific CpG sites within these genes. We only report on significant interaction effects in our tables. Significant interactions were found for the sample as a whole. Following Aiken and West,41 interactions were probed at ± 1 or 2 SD from the mean of DNA methylation using the online computational tools provided by Preacher, Curran, and Bauer (2006; http://www.quantpsy.org/interact/mlr2.htm) to probe significant interactions. We also tested for mediation but found no effects. Effect sizes are reported for regression analyses in Table 2. Data were analyzed using SPSS version 20.
Acknowledgments
We would like to thank Gilda Ferro, Joyce Lee, Erica Oliveria, and Susan Capobianco for their hard work in recruitment of subjects and the support and staff of the Brown Center for the Study of Children at Risk.
Glossary
Abbreviations:
- NNNS
NICU network neurobehavioral scale
- HPA
hypothalamic pituitary adrenal axis
- ACTH
adrenocorticotropic hormone
- GA
gestational age
- SGA
small for gestational age
- AGA
appropriate for gestational age
- LGA
large for gestational age
Potential Conflicts of Interest
The authors report no conflict of interest.
Financial Disclosure
This study was supported by the National Institute of Mental Health R01MH094609 (to Marsit CJ) and a National Research Service Award from the National Institute on Drug Abuse F32DA032175 (to Conradt E). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institute of Mental Health, National Institute on Drug Abuse, or the National Institutes of Health.
Footnotes
Previously published online: www.landesbioscience.com/journals/epigenetics/article/26634
References
- 1.Hompes T, Izzi B, Gellens E, Morreels M, Fieuws S, Pexsters A, Schops G, Dom M, Van Bree R, Freson K, et al. Investigating the influence of maternal cortisol and emotional state during pregnancy on the DNA methylation status of the glucocorticoid receptor gene (NR3C1) promoter region in cord blood. J Psychiatr Res. 2013;47:880–91. doi: 10.1016/j.jpsychires.2013.03.009. [DOI] [PubMed] [Google Scholar]
- 2.Salisbury AL, Wisner KL, Pearlstein T, Battle CL, Stroud L, Lester BM. Newborn neurobehavioral patterns are differentially related to prenatal maternal major depressive disorder and serotonin reuptake inhibitor treatment. Depress Anxiety. 2011;28:1008–19. doi: 10.1002/da.20883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Parker KJ, Schatzberg AF, Lyons DM. Neuroendocrine aspects of hypercortisolism in major depression. Horm Behav. 2003;43:60–6. doi: 10.1016/S0018-506X(02)00016-8. [DOI] [PubMed] [Google Scholar]
- 4.Field T, Diego M, Dieter J, et al. Prenatal depression effects on the fetus and the newborn. Infant Behav Dev. 2004;27:216–29. doi: 10.1016/j.infbeh.2003.09.010. [DOI] [PubMed] [Google Scholar]
- 5.Gaynes BN, Gavin N, Meltzer-Brody S, et al. Perinatal Depression: Prevalence, Screening Accuracy, and Screening Outcomes. Summary (Indianap Ind) 2005 doi: 10.1037/e439372005-001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Monk C, Spicer J, Champagne FA. Linking prenatal maternal adversity to developmental outcomes in infants: the role of epigenetic pathways. Dev Psychopathol. 2012;24:1361–76. doi: 10.1017/S0954579412000764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Essex MJ, Klein MH, Cho E, Kalin NH. Maternal stress beginning in infancy may sensitize children to later stress exposure: effects on cortisol and behavior. Biol Psychiatry. 2002;52:776–84. doi: 10.1016/S0006-3223(02)01553-6. [DOI] [PubMed] [Google Scholar]
- 8.Sandman CA, Davis EP, Buss C, Glynn LM. Prenatal programming of human neurological function. Int J Pept. 2011;2011:837596. doi: 10.1155/2011/837596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Millan MJ. An epigenetic framework for neurodevelopmental disorders: From pathogenesis to potential therapy. Neuropharmacology. 2012;68:2–82. doi: 10.1016/j.neuropharm.2012.11.015. [DOI] [PubMed] [Google Scholar]
- 10.Sandman CA, Davis EP. Neurobehavioral risk is associated with gestational exposure to stress hormones. Expert Rev Endocrinol Metab. 2012;7:445–59. doi: 10.1586/eem.12.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gitau R, Fisk NM, Teixeira JM, Cameron A, Glover V. Fetal hypothalamic-pituitary-adrenal stress responses to invasive procedures are independent of maternal responses. J Clin Endocrinol Metab. 2001;86:104–9. doi: 10.1210/jc.86.1.104. [DOI] [PubMed] [Google Scholar]
- 12.Lundy BL, Jones NA, Field T, et al. Prenatal depression effects on neonates. Infant Behav Dev. 1999;22:119–29. doi: 10.1016/S0163-6383(99)80009-5. [DOI] [Google Scholar]
- 13.Meyer JS. Biochemical effects of corticosteroids on neural tissues. Physiol Rev. 1985;65:946–1020. doi: 10.1152/physrev.1985.65.4.946. [DOI] [PubMed] [Google Scholar]
- 14.Kaplan LA, Evans L, Monk C. Effects of mothers’ prenatal psychiatric status and postnatal caregiving on infant biobehavioral regulation: can prenatal programming be modified? Early Hum Dev. 2008;84:249–56. doi: 10.1016/j.earlhumdev.2007.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Holmes MC, Abrahamsen CT, French KL, Paterson JM, Mullins JJ, Seckl JR. The mother or the fetus? 11beta-hydroxysteroid dehydrogenase type 2 null mice provide evidence for direct fetal programming of behavior by endogenous glucocorticoids. J Neurosci. 2006;26:3840–4. doi: 10.1523/JNEUROSCI.4464-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Harris A, Seckl J. Glucocorticoids, prenatal stress and the programming of disease. Horm Behav. 2011;59:279–89. doi: 10.1016/j.yhbeh.2010.06.007. [DOI] [PubMed] [Google Scholar]
- 17.Cicchetti D, Toth SL. The development of depression in children and adolescents. Am Psychol. 1998;53:221–41. doi: 10.1037/0003-066X.53.2.221. [DOI] [PubMed] [Google Scholar]
- 18.Marsit CJ, Maccani MA, Padbury JF, Lester BM. Placental 11-beta hydroxysteroid dehydrogenase methylation is associated with newborn growth and a measure of neurobehavioral outcome. PLoS One. 2012;7:e33794. doi: 10.1371/journal.pone.0033794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lester BM, Tronick EZ. History and description of the Neonatal Intensive Care Unit Network Neurobehavioral Scale. Pediatrics. 2004;113:634–40. [PubMed] [Google Scholar]
- 20.Oberlander TF, Weinberg J, Papsdorf M, Grunau R, Misri S, Devlin AM. Prenatal exposure to maternal depression, neonatal methylation of human glucocorticoid receptor gene (NR3C1) and infant cortisol stress responses. Epigenetics. 2008;3:97–106. doi: 10.4161/epi.3.2.6034. [DOI] [PubMed] [Google Scholar]
- 21.Mueller BR, Bale TL. Sex-specific programming of offspring emotionality after stress early in pregnancy. J Neurosci. 2008;28:9055–65. doi: 10.1523/JNEUROSCI.1424-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Weaver IC, Cervoni N, Champagne FA, D’Alessio AC, Sharma S, Seckl JR, Dymov S, Szyf M, Meaney MJ. Epigenetic programming by maternal behavior. Nat Neurosci. 2004;7:847–54. doi: 10.1038/nn1276. [DOI] [PubMed] [Google Scholar]
- 23.Bromer C, Marsit CJ, Armstrong DA, Padbury JF, Lester B. Genetic and epigenetic variation of the glucocorticoid receptor (NR3C1) in placenta and infant neurobehavior. Dev Psychobiol. 2012 doi: 10.1002/dev.21061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Goodman SH, Gotlib IH. Risk for psychopathology in the children of depressed mothers: a developmental model for understanding mechanisms of transmission. Psychol Rev. 1999;106:458–90. doi: 10.1037/0033-295X.106.3.458. [DOI] [PubMed] [Google Scholar]
- 25.Field T. Infants of depressed mothers. Dev Psychopathol. 1992;4:49–66. doi: 10.1017/S0954579400005551. [DOI] [PubMed] [Google Scholar]
- 26.Slone-Wilcoxon J, Redei EE. Maternal-fetal glucocorticoid milieu programs hypothalamic-pituitary-thyroid function of adult offspring. Endocrinology. 2004;145:4068–72. doi: 10.1210/en.2004-0473. [DOI] [PubMed] [Google Scholar]
- 27.Rieger M, Pirke KM, Buske-Kirschbaum A, Wurmser H, Papousek M, Hellhammer DH. Influence of stress during pregnancy on HPA activity and neonatal behavior. Ann N Y Acad Sci. 2004;1032:228–30. doi: 10.1196/annals.1314.026. [DOI] [PubMed] [Google Scholar]
- 28.Ellman LM, Schetter CD, Hobel CJ, Chicz-Demet A, Glynn LM, Sandman CA. Timing of fetal exposure to stress hormones: effects on newborn physical and neuromuscular maturation. Dev Psychobiol. 2008;50:232–41. doi: 10.1002/dev.20293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Van den Bergh BR, Mulder EJ, Mennes M, Glover V. Antenatal maternal anxiety and stress and the neurobehavioural development of the fetus and child: links and possible mechanisms. A review. Neurosci Biobehav Rev. 2005;29:237–58. doi: 10.1016/j.neubiorev.2004.10.007. [DOI] [PubMed] [Google Scholar]
- 30.Lester BM, Conradt E, Marsit CJ. Epigenetic basis for the development of depression in children. Clin Obstet Gynecol. 2013;56:556–65. doi: 10.1097/GRF.0b013e318299d2a8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Feldman R, Eidelman AI. Neonatal state organization, neuromaturation, mother-infant interaction, and cognitive development in small-for-gestational-age premature infants. Pediatrics. 2006;118:e869–78. doi: 10.1542/peds.2005-2040. [DOI] [PubMed] [Google Scholar]
- 32.Liu J, Bann C, Lester B, Tronick E, Das A, Lagasse L, Bauer C, Shankaran S, Bada H. Neonatal neurobehavior predicts medical and behavioral outcome. Pediatrics. 2010;125:e90–8. doi: 10.1542/peds.2009-0204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Edge D. Ethnicity, psychosocial risk, and perinatal depression--a comparative study among inner-city women in the United Kingdom. J Psychosom Res. 2007;63:291–5. doi: 10.1016/j.jpsychores.2007.02.013. [DOI] [PubMed] [Google Scholar]
- 34.Leahy-Warren P, McCarthy G, Corcoran P. Postnatal depression in first-time mothers: prevalence and relationships between functional and structural social support at 6 and 12 weeks postpartum. Arch Psychiatr Nurs. 2011;25:174–84. doi: 10.1016/j.apnu.2010.08.005. [DOI] [PubMed] [Google Scholar]
- 35.Kadivar H, Garvie PA, Sinnock C, Heston JD, Flynn PM. Psychosocial profile of HIV-infected adolescents in a Southern US urban cohort. AIDS Care. 2006;18:544–9. doi: 10.1080/13548500500228763. [DOI] [PubMed] [Google Scholar]
- 36.Ponder KL, Salisbury A, McGonnigal B, Laliberte A, Lester B, Padbury JF. Maternal depression and anxiety are associated with altered gene expression in the human placenta without modification by antidepressant use: implications for fetal programming. Dev Psychobiol. 2011;53:711–23. doi: 10.1002/dev.20549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lesseur C, Armstrong DA, Paquette AG, Koestler DC, Padbury JF, Marsit CJ. Tissue-specific Leptin promoter DNA methylation is associated with maternal and infant perinatal factors. Mol Cell Endocrinol. 2013;381:160–7. doi: 10.1016/j.mce.2013.07.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Filiberto AC, Maccani MA, Koestler DC, Wilhelm-Benartzi C, Avissar-Whiting M, Banister CE, Gagne LA, Marsit CJ. Birthweight is associated with DNA promoter methylation of the glucocorticoid receptor in human placenta. Epigenetics. 2011;6:566–72. doi: 10.4161/epi.6.5.15236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Alikhani-Koopaei R, Fouladkou F, Frey FJ, Frey BM. Epigenetic regulation of 11 β-hydroxysteroid dehydrogenase type 2 expression. J Clin Invest. 2004;114:1146–57. doi: 10.1172/JCI21647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kessler RC, Chiu WT, Demler O, Merikangas KR, Walters EE. Prevalence, severity, and comorbidity of 12-month DSM-IV disorders in the National Comorbidity Survey Replication. Arch Gen Psychiatry. 2005;62:617–27. doi: 10.1001/archpsyc.62.6.617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Aiken LS, West SG. Multiple regression: Testing and interpreting interactions. Sage. 1991 [Google Scholar]