Abstract
To understand the genetic makeup and impact on pharmacokinetics (PK) in the Taiwanese population, we analyzed the pharmacogenetic (PG) profile and demonstrated its effects on enzyme metabolism using indapamide as an example. A multiplex mass spectrometry method was used to examine the single nucleotide polymorphism (SNP) profile of eight major phases I and II metabolic enzymes in 1,038 Taiwanese subjects. A PG/PK study was conducted in 24 healthy subjects to investigate the possible effects of 28 SNPs on drug biotransformation. Among the genetic profile analyzed, eight SNPs from CYP2A6, CYP2C19, CYP2D6, CYP2E1, CYP3A5, and UGT2B7 showed higher variant frequencies than those previously reported in Caucasians or Africans. For instance, we observed 14.7% frequency of the SNP rs5031016 (I471T) from CYP2A6 in Taiwanese, whereas 0% variation was reported in Caucasians and Africans. The PG/PK study of indapamide demonstrated that the polymorphic SNPs CYP2C9 rs4918758 and CYP2C19 rs4244285 appeared to confer lowered enzyme activity, as indicated by increased Cmax (25% ∼ 64%), increased area under the plasma level-time curves (30∼76%), increased area under the time infinity (43% ∼ 80%), and lower apparent clearance values than PK for wild-type indapamide. Our results reinforce the biochemical support of CYP2C19 in indapamide metabolism and identify a possible new participating enzyme CYP2C9. The PG/PK approach contributed toward understanding the genetic makeup of different ethnic groups and associations of enzymes in drug metabolism. It could be used to identify two genetic markers that enable to differentiate subjects with varied PK outcomes of indapamide.
KEY WORDS: indapamide, metabolic enzymes, pharmacogenetics, pharmacokinetics, SNPs
INTRODUCTION
The ultimate goal of personalized medicine is to identify specific genetic features with the differential risk of human diseases or the efficacy of certain therapeutic interventions (1). Mounting evidence has linked the genetic make-up to a significant portion of drug-induced toxicity (2–5). The primary focus of pharmacogenetics (PG) is drug-metabolizing enzymes (DMEs).
The pharmacology of drugs subject to inherited variability in metabolism is often complex. Few have simple or single pathways of elimination (6). In addition, ethnicity may account for the observed differences in both pharmacokinetics (PK) and pharmacodynamics of drugs, thus resulting in variability in response to drug therapy (7). Furthermore, genetic polymorphism is one of the most important factors that may contribute to the ethnic sensitivity of a drug in its metabolic pathways (7). The E5 guidance, published by the US Food and Drug Administration, in 1999, provides a general framework for evaluating medicines in terms of their sensitivity to ethnic factors by considering the three major racial groups: Asian, Black, and Caucasian. Traditionally, drug-dosage determination has long been based on Caucasian data, but Caucasians represent <6% of the total population. By contrast, Asian people represent approximately 60% of the world’s population. It is important to understand the genetic make-up of Asians, specifically Chinese, which represent more than half of the Asian population. Taiwanese are the descendants of mainland China and also show genetic affinities to southern Asian populations (8), so Taiwanese people are representative ethnic group for investigating PG variabilities in Asians and Chinese.
Single nucleotide polymorphism (SNP) is the most frequently observed mutation in all organisms (9,10). The cost for mapping of the genetic variance among thousands of SNPs could be extremely high. Recent advances in whole-genome sequencing (WGS) have led to burst of bioinformation and a significant knowledge base for investigating the genetic architecture of drug metabolisms and treatment efficacies. However, the clinical use of whole-genome analysis is not yet achievable, presumably because of the relatively high cost and the scarcity of such high-end techniques (1,11). Moreover, serious limitations in WGS genotyping platforms are still present in the low coverage for different ethnic groups (12). In addition to WGS, various modern techniques are readily applied in large-scale SNP-genotyping association studies to enable robust assays in clinical study design, including mass detections (13).
Indapamide is a thiazide-type diuretic commonly prescribed to treat mild to moderate hypertension. As it has fewer side effects in inducing metabolic derangements than other thiazide diuretics, indapamide is well accepted to be used as initial therapy to treat hypertension in patients with previous stroke or older people (over 80) or as an add-on treatment (14). However, severe hyponatremia and hypokalemia have been reported (15). Therefore, limiting the dose to that necessary to achieve maximal blood pressure reduction and improved cardiovascular outcomes is relevant (16).
Although the major metabolites of indapamide have been identified, the pathways of cytochrome P450 (CYP450)-catalyzed indapamide biotransformation have not been fully elucidated (17–19). One recent report suggested that CYP2C19, CYP2C8, and CYP3A4 are involved in the metabolism (18). CYP3A4 has the highest activity, as indicated by specific substrate inhibition of DMEs with an in silico-based approach. We have little information on PG studies of indapamide. The effect of genetic variation on DME activity and the clinical impact on indapamide in particular are not fully understood.
To investigate the effect of genetic variation on drug metabolism among Taiwanese people, we conducted an extensive SNP PG analysis of 1,038 healthy Taiwanese subjects. We chose 28 SNPs in eight phase I and one phase II enzymes (CYP2A6, 2C9, 2C19, 2D6, 2E1, 3A4, 3A5, and UGT2B7) that showed varied distributions in Han Chinese and Japanese (http://www.ncbi.nlm.nih.gov/projects/SNP/). We focused on the SNP-oriented PK/PG study of indapamide. To further investigate the association between subjects carrying different enzyme genotypes, 24 healthy subjects were included in the PK/PG study.
MATERIALS AND METHODS
Volunteers for PG Survey and Sample Acquisition
A total of 1,038 normal healthy subjects were included in the PG survey during 2007 to 2012. All volunteers were selected from Taiwan residents, and informed consent was obtained from all subjects. Except for agreeing to donate 5 mL peripheral blood for the genetic analysis, no particular criteria were required for entry into this study. Genomic DNA was extracted from blood samples by use of a commercial extraction kit for whole blood from QIAGEN GmbH (Hilden, Germany).
Genomic DNA Isolation, Genotyping, and Sequencing
DNA isolation and SNP genotyping were performed in the National Genotyping Center at Academia Sinica (Taipei). Genomic DNA was isolated by use of the PURGENE DNA purification system (Gentra System, Minneapolis, MN, USA). Matrix-assisted laser desorption and ionization-time of flight (MALDI-TOF) mass spectrometry (SEQUENOM MassARRAY system, Sequenom, San Diego, CA, USA) was used to identify the SNPs (20). Primers and the extending probes were designed in multiplex format by using SpectroDESIGNER (Sequenom, San Diego, CA, USA). The primer specificity and the generated amplicons were checked by use of BLAST and UCSC In silico PCR, respectively. After amplification, the unincorporated dNTPs were dephosphorylated by use of shrimp alkaline phosphatase (Hoffman-LaRoche, Basel, Switzerland) followed by primer extension. The purified extension reactions were spotted onto a 384-element silicon chip (Sequenom), and analyzed by a Bruker Biflex III MALDI-TOF SpectroREADER mass spectrometer. The spectra were processed with SpectroTYPER (Sequenom).
SNPs that failed to perform in the high throughput MALDI-TOF were identified by direct sequencing. Primers were designed by use of Primer3 (http://primer3.wi.mit.edu/). The specificity and the generated amplicons were confirmed by BLAST and UCSC in silico PCR, respectively. All PCR products were sequenced by use of the BigDye Terminator Cycle Sequencing Kit v1.1/3.1 (Applied Biosystems, Foster City, CA, USA). Sequencing products were separated by use of the Applied Biosystems 3730 DNA Analyzer (Applied Biosystems). Raw data were analyzed by use of DNA Sequencing Analysis Software v3.7 (Applied Biosystems).
Healthy Volunteers for Indapamide PK Study
A total of 24 healthy volunteers participated in this study. Volunteers were randomly recruited among Taiwan residents and comprehensive informed consent was obtained. Volunteers were included if they (1) were healthy male adults 20 to 40 years old and (2) had ±20% the ideal body weight (kilogram), 62+ (height (centimeter) − 170) × 0.6 and acceptable physical condition and medical history, including complete blood counts, differential, platelets, urinalysis (including microscopic evaluation), electrocardiography (ECG), chest X-ray, electrolytes tests (sodium, potassium, chloride, and calcium), liver function tests (serum glutamyl oxaloacetic transaminase, serum glutamyl pyruvic transaminase, alkaline phosphatase, total and direct bilirubin, albumin, gamma glutamyl transferase, and total protein), and kidney function tests (blood urea nitrogen, creatinine, and uric acid). Other blood tests included fasting blood sugar, total cholesterol, triglycerides. In addition, healthy volunteers were excluded if they had a history of adverse or allergy reactions to indapamide or a related drug, significant drug or alcohol abuse, took any drug that may affect the result of the study within 14 days before to the start of the study, took any alcoholic drink, and grapefruit juice within 48 h before the start of the study, had an acute illness or surgery within 4 weeks before entering the study, had a history of a psychiatric disorder and under the care of a psychiatrist and/or on medications for treatment of a psychiatric disorder such as somatoform disorders, conversion disorders, current depression, or a history of schizophrenia or a bipolar disorder.
Before drug administration, 5 mL blood was drawn, then subjects took one 1.5-mg tablet of indapamide SR. All volunteers were given a once-daily dose of 1.5 mg indapamide SR for 9 days, and 8 mL blood was drawn at 24 h on days 7, 8, 9, and 10 after drug administration. The dosing regimen was to insure that indapamide reached a steady-state prior the last dosing. On the last dosing day, volunteers entered the clinical study place and were tested by sitting to determine if they exhibited normal vital signs of heart rate and blood pressure. During the half-hour before drug administration, volunteers had an indwelling catheter inserted in their forearm vein for blood samples. After administration, 8 mL blood was obtained at 0.25, 0.5, 1, 1.5, 2, 3, 4, 6, 8, 12 and 24 h for PK analysis. At the end of the study, the participants underwent the same physical and laboratory evaluations as those conducted at the start of the study, except the chest X-ray and ECG. Blood samples were collected into heparinizied vacuum tubes. Samples were centrifuged at 3,000 rpm for 15 min and the plasma was separated and stored in light-resistant plastic tubes at −20°C. The Tri-Service General Hospital Institutional Review Board approved the protocol, and written informed consent was obtained from all subjects. PK analysis of indapamide concentrations in plasma was performed using Winnonlin (version 5.2.1, PharSight CORP, CA, USA) with a noncompartment model.
Sample Preparation for PK study
For each sample, 2 mL of subject blood was used for analysis. Each of the following solutions were added with and vortexed: H2O (500 μL, 1 min followed by 10 min incubation at room temperature and sonicated for 5 min), flunitrazepam (50 μL, 30 s), and diethyl ether (4.0 mL and 3 min). The resulting solution was then centrifuged at 2,800 rpm for 5 min and frozen at −70°C freezer for 40 min. The organic layer was transferred to a new tube and evaporated to dryness under N2 gas. Once dried, the sample was reconstituted with 0.5 mL of 50% CH3CN followed by a 1-min vortex and transferred to sample vial for analysis.
Instruments and Analytical Conditions for PK study
HPLC Conditions
The HPLC system consisted of a Waters 2795 pump and an autosampler; the mass spectrometry model was the Waters Quattro Ultima Triple Quadrupole (Waters Instruments, Milford, MA, USA). The Phenomenex Luna C18 column (4.6 × 50 mm/5 μL) was maintained at room temperature for the duration of analysis. The mobile phase consisted of 50% CH3CN(aq) with 0.4 mM HCOOH and 0.2 mM CH3COONH4 CH3CN. The flow rate was set at 1.00 mL min−1 and the injection volume was 8 μL. The internal standard used in this experiment was flunitrazepam.
Mass Spectrometry Conditions
The Waters Quattro Ultima Triple Quadrupole mass spectrometer was used for analysis (Waters). The positive mode of ionization was selected for analysis. Capillary voltage and cone voltage were set at 3,500 and 20 V, respectively. The temperature of ion source was 85°C with ultra high-purity nitrogen as cone gas and nebulizer gas (450 L h−1). Desolvation gas was heated to 400°C and set at a flow rate of 450 L h−1. With argon as collision gas, multiple reaction monitoring was applied to detect indapamide and flunitrazepam by monitoring the ion transition of 366.10→132 and 314.10→1268.1 m/z, respectively, and collision energy was set at 13 eV. Data acquisition involved use of MassLynx 3.5 (Micromass, Manchester, UK).
Linkage Disequilibrium Analysis
The software Haploview was used to calculate the linkage disequilibrium (LD) between the SNPs (in terms of Lewontin’s D’) (21).
Statistical Analysis
Data are expressed as mean ± SD. The PK parameters were analyzed for statistical significance by one-way ANOVA or Wilcoxon signed rank test with SPSS 13.0 (SPSS Inc. Chicago, IL, USA). To account for multiple testing, we used the Bonferroni correction and considered significant only those SNPs for which p < 0.05/28 = 0.0018. The least-significant difference post hoc test of multiple comparisons was used to identify significant differences among groups (p < 0.05).
RESULTS
Minor Allele Frequency of Phases I and II Enzymes in Taiwanese
We prioritized our SNP selections based on the following criteria: SNP that changes amino acid coding (nonsynonymous SNP), may affect gene expression (5′ promotor region) or mRNA splicing (splice site) and has substantial allele frequency in Asians, particularly in Han Chinese and Japanese. Thus, we chose 28 SNPs from the SNP database (dbSNP) at NCBI (http://www.ncbi.nlm.nih.gov/projects/SNP/): CYP2A6 (rs28399468, rs1809810, rs5031017, and rs5031016), CYP2C9 (rs4918758 and rs1057910), CYP2C19 (rs11568732, rs4244285, rs3758581, and rs4986893), CYP2D6 (rs1058172, rs3915951, rs3892097, rs5030865, rs1065852, rs1135840, rs16947, rs28371735, and rs1135835), CYP2E1 (rs3813865, rs2031921, and rs2070676), CYP3A4 (rs28371759), CYP3A5 (rs776741 and rs776746), and UGT2B7 (rs7668282, rs12233719, and rs7439366) (Table I). Among them, 18 SNPs are nonsynonymous, 1 SNP is synonymous, 6 SNPs reside in the promotor region, 1 SNPs is located at the splice site, and 2 SNPs are found in introns. The minor allele frequency of the SNPs ranged from 0.2% (rs3915951) to 39.2% (rs4918758) in our 1,038 healthy Taiwanese. Five SNPs, rs3915951 (0.2%), rs3892097 (0.4%), rs28371735 (0.6%), rs1135835 (0.6%), and rs1058172 (0.7%), existed at a very low frequency in Taiwanese and their frequencies in other populations have yet to be determined.
Table I.
Summary of Allele Frequency of the 28 SNPs in 1,038 Taiwanese People and Comparison with Different Ethnic Groups
| Enzyme | RefSNP ID | SNP Ref/Var | Ancestral allelea | Position in locus | Amino acid change | Frequencies of the variant SNPb | ||||
|---|---|---|---|---|---|---|---|---|---|---|
| Taiwanese | Han Chinese | Japanese | Caucasian | African | ||||||
| CYP2A6 | rs1809810 | A/T | NA | Exon 8 | F392Y | 0.017 | NA | NA | 0 | 0 |
| rs5031016 | T/C | NA | Exon 9 | I471T | 0.147 | NA | NA | 0 | 0 | |
| rs5031017 | G/T | NA | Exon 9 | G479V | 0.018 | NA | NA | 0 | 0.125 | |
| rs28399468 | G/T | NA | Exon 9 | R485L | 0.052 | NA | 0.022 | NA | NA | |
| CYP2C9 | rs1057910 | A/C | A | Exon 7 | I359L | 0.031 | 0.044 | 0.033 | 0.058 | 0 |
| rs4918758 | T/C | C | 5′ promotor | NA | 0.392 | 0.333 | 0.400 | 0.383 | 0.275 | |
| CYP2C19 | rs4244285 | G/A | G | Exon 5 | P227P | 0.312 | 0.256 | 0.284 | 0.150 | 0.167 |
| rs3758581 | G/A | G | ±9-exon 7 | V331I | 0.033 | 0.067 | 0.102 | 0.051 | 0 | |
| rs11568732 | T/G | T | 5′ promotor | NA | 0.076 | 0.089 | 0.133 | 0.059 | 0.075 | |
| rs4986893 | G/A | G | Exon 4 | W212X | 0.058 | 0.033 | 0.045 | 0 | 0 | |
| CYP2D6 | rs1065852 | C/T | C | Exon 1 | P34S | 0.775 | 0.667 | 0.500 | 0.211 | 0.167 |
| rs3892097 | G/A | G | Intron 3 | Splice-site/acceptor | 0.004 | NA | NA | 0.183 | 0 | |
| rs28371735 | C/T | C | Exon 9 | H478Y | 0.006 | NA | NA | NA | 0.042 | |
| rs1135835 | A/G | A | Exon 9 | T470A | 0.006 | NA | NA | NA | 0.042 | |
| rs1135840 | C/G | C | Exon 9 | T486S | 0.281 | 0.211 | 0.407 | 0.456 | 0.389 | |
| rs3915951 | G/T | G | Exon 7 | G329V | 0.002 | NA | NA | 0.380d | NA | |
| rs1058172 | G/A | G | Exon 7 | R314H | 0.007 | NA | NA | 0.280 | NA | |
| rs5030865 | G/A | Cc | Exon 3 | G169C | 0.011 | 0 | NA | 0 | 0 | |
| rs16947 | A/G | G | Exon 6 | C296R | 0.126 | 0.122 | 0.116 | 0.333 | 0.444 | |
| CYP2E1 | rs2031921 | T/C | T | 5′ promotor | NA | 0.214 | 0.188 | NA | 0.022 | 0.119 |
| rs3813865 | G/C | G | 5′ promotor | NA | 0.202 | 0.222 | 0.239 | 0.025 | 0.167 | |
| rs2070676 | C/G | G | Intron 7 | NA | 0.167 | 0.156 | 0.156 | 0.100 | 0.692 | |
| CYP3A4 | rs28371759 | T/C | T | Exon 10 | L293P | 0.029 | 0.044 | 0.023 | 0 | 0 |
| CYP3A5 | rs776741 | T/C | C | 5′ promotor | NA | 0.259 | 0.322 | 0.227 | 0.017 | 0.517 |
| rs776746 | G/A | A | Intron 3 | NA | 0.277 | 0.333 | 0.250 | 0.058 | 0.850 | |
| UGT2B7 | rs7668282 | T/C | T | 5′ promotor | NA | 0.040 | 0.080 | 0.058 | 0.008 | 0.053 |
| rs12233719 | G/T | G | Exon 1 | A71S | 0.151 | 0.125 | 0.148 | 0 | 0 | |
| rs7439366 | C/T | T | Exon 2 | Y268H | 0.222 | 0.278 | 0.318 | 0.500 | 0.008 | |
NA data not available
aInformation for ancestral allele was obtained from the dbSNP database on NCBI (http://www.ncbi.nlm.nih.gov/snp)
bThe population diversity data (Han Chinese, Japanese, Caucasian, and African) were obtained from the dbSNP database based on HapMap project (HapMap) or SNP500CANCER submission
cVariants of A/G were only observed in our study, while the ancestral allele for rs5030865 was reported to be C in the dbSNP database
dData derived from CEPH pedigree, Caucasians of Northern and Western European descent
Population Diversity
We compared the minor allele frequency of the 28 SNPs in Taiwanese to current dbSNP data and published data for other populations, including Han Chinese, Japanese, Caucasians, and Africans (http://www.ncbi.nlm.nih.gov/projects/SNP/). Taiwanese, Han Chinese, and Japanese reside in geographic regions in close proximity, therefore genetic material can be interchanged more frequently as compared with populations that are in distant geographical regions. Thus, most of the minor allele frequencies are similar in Taiwanese, Han Chinese, and Japanese. In addition, we expected that the minor allele frequency among Taiwanese and Han Chinese would have greater similarities than in Japanese because a large proportion of the Taiwanese population emigrated from mainland China. However, one SNP (rs4918758) exhibited better similarity in Taiwanese and Japanese than in Han Chinese: 39.2%, 40%, and 33.2%, respectively (Table I). Three SNPs (rs4244285, rs2031921, and rs12233719) showed relatively higher allele frequencies in Taiwanese (31.2%, 21.4%, and 15.1%), Han Chinese (25.6%, 18.8%, and 12.5%), and Japanese (28.4%, NA, and 14.8%) than in Caucasians and Africans. Three SNPs (rs776741, 51.7%%; rs2070676, 69.2%; and rs776746, 85%) had much higher allele frequencies in Africans than the rest of the populations. SNP rs7439366 had a very low allele frequency in Africans (0.8%) but was found at a much higher frequency in Taiwanese, Han Chinese, Japanese, and Caucasians: 22.2%, 27.8%, 31.8%, and 50%, respectively.
Pharmcokinetics of Indapamide
PK parameters of indapamide in 24 recruited subjects are shown in Table II. For Cmax, the minimum, maximum and mean concentration were 37.60, 100.40, and 64.32 ng/mL, respectively, whereas the minimum, maximum, and mean of the area under the plasma level-time curves (AUCt) were 751, 1,995, and 1,236, respectively. We used the indapamide PK previously reported for the UK (22); demographic data were similar in gender, age, and body mass index between the two ethnicities, and also the Cmax and AUCt. The half-life of indapamide SR in Taiwanese was 31.0 ± 12.3 h, which was much longer than previously reported in Caucasians, 19.2 ± 9.7 h. Hence, we studied the PK profile after a consecutive 9 daily doses to ensure that the steady state was reached in the present study. Meanwhile, Tmax in Taiwanese was 5.9 ± 3.2 h after administration of the last dose, which was earlier than previously reported, 11 ± 7.0 h (22). However, considering the nature of sustained release formulation and no significant influence found on Cmax and AUCt, we assume that such differences in Tmax did not lead to a significant change of drug absorption between the two ethnic groups.
Table II.
Comparison of the Characteristics and Pharmacokinetics of Indapamide Between Taiwanese and Caucasians
| Taiwanese (n = 24) | Caucasian (n = 12) (22) | |
|---|---|---|
| Sex | Male only | Male only |
| Age (year) | 24.4 ± 5.5 | 23.5 ± 4.7 |
| Body mass index (kg/m2) | 22.3 ± 3.9 | 22.7 ± 2.7 |
| Starting time of the PK study (h) | 216 | 168 |
| C max (ng/mL) | 64 ± 17 | 58 ± 20 |
| AUCt (h ng mL−1) | 1,240 ± 316 | 1,090 ± 310 |
| AUCinf (h ng mL−1) | 3,120 ± 1,110 | – |
| T max (h) | 5.9 ± 3.2 | 11 ± 7.0 |
| Half-life (h) | 31.0 ± 12.3 | 19.2 ± 9.7 |
| CLss/F (mL/h) | 1,300 ± 328 | – |
Data are mean ± SD unless stated. PK data of Caucasian group were adopted from (22)
Significant SNP Association with Indapamide PK Parameters
Statistical analysis first focused on known enzymes involved in indapamide metabolism, CYP2C19 and CYP3A4, then expanded to other phases I and II enzymes. Of the 8 selected enzymes and 28 SNPs, 2 SNPs were associated with indapadmide PK parameters.
CYP2C9 rs4918758
SNP rs4918758 causes a transition mutation that changes the wild-type T allele to the mutant form C allele with unkown consequences. As shown in Fig. 1 and Table III, 2 of 24 subjects were genotyped as homozygous variant (CC), 12 and 10 were heterozygous (CT), and reference wild type (TT), respectively. The mutation significantly affects several indapamide PK parameters (Table III). Homozygous variants had the highest mean Cmax, AUCt, area under the time infinity (AUCinf), and the lowest clearance among the three genotypes. Post hoc analysis demonstrated that the differences were significant as compared with heterozygous or homozygous mutants with wild-type individuals. The mean Cmax of indapamide in homozygous and heterozygous variants were 89.4 ± 15.6 and 68.4 ± 14.2, significantly higher than that of the wild type, 54.5 ± 13.3 ng/mL (p = 0.004 and p = 0.029, respectively). Significant increase of AUCt and AUCinf in both variants (CT versus TT, p = 0.005 and p = 0.046; CC versus TT, p = 0.000 and p = 0.027, respectively) was also in line with the increased Cmax. CLss/F of indapamide in homozygous and heterozygous variants were both reduced as compared with the wild type (CT versus TT, p = 0.003; CC versus TT, p = 0.002). Although the mean oral half-life was higher with heterozygous and homozygous variants than the wild type, the three groups did not significantly differ. No significant differences in Tmax were found.
Fig. 1.
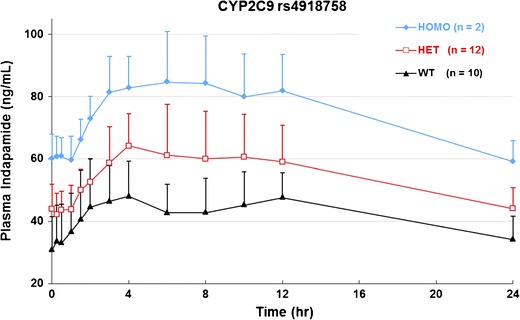
Steady-state mean plasma concentration-time profile according to genotypes for SNP rs4918758 after administration of the last dose of indapamide 1.5 mg at 216 h. HOMO homozygous, HET heterozygous, WT wild type, n sample size. Data are mean ± SD at specific times
Table III.
SNPs with Significant Effects on Indapamide Metabolism
| Parameter genotype | Number | C max (ng/mL) | T max (h) | Half-life (h) | AUCt (h ng mL−1) | AUCinf (h ng mL−1) | CLss/F (mL/h) |
|---|---|---|---|---|---|---|---|
| CYP2C9:rs4918758 | |||||||
| TT | 10 | 54.5 ± 13.3 a | 6.3 ± 4.0 a | 29.0 ± 10.9 a | 1,020 ± 200 a | 2,480 ± 950 a | 1,530 ± 310 a |
| CT | 12 | 68.4 ± 14.2 b | 5.5 ± 2.7 a | 33.0 ± 14.3 a | 1,330 ± 240 b | 3,550 ± 1,020 b | 1,170 ± 200 b |
| CC | 2 | 89.4 ± 15.6 c | 6.0 ± 2.8 a | 29.4 ± 6.7 a | 1,790 ± 290 c | 4,450 ± 1,290 b | 850 ± 140 c |
| P value | 0.007 | 0.935 | 0.757 | 0.000* | 0.036 | 0.001* | |
| CYP2C19:rs4244285 | |||||||
| GG | 9 | 56.3 ± 12.6 a | 5.8 ± 4.0 a | 28.2 ± 10.9 a | 1,050 ± 190 a | 2,500 ± 1,010 a | 1,480 ± 280 a |
| AG | 14 | 69.6 ± 18.0 b | 6.0 ± 2.8 a | 33.4 ± 12.5 a | 1,360 ± 330 b | 3,670 ± 1,070 a | 1,180 ± 320 b |
| P value | 0.049 | 0.630 | 0.144 | 0.009 | 0.016 | 0.013 | |
Data are mean ± SD; statistical significance was compared with the wild type using one-way ANOVA or independent-samples t test, except a Wilcoxon signed rank test was used for T max. LSD post hoc test was used for multiple comparisons of the CYP2C9 rs4918758 genotypes. Means with different characters differ significantly (p < 0.05). p values are those of one-way ANOVA or independent-samples t tests with Bonferroni adjustment for comparison among genotype groups on each parameter
*p < 0.0018
CYP2C19 rs4244285
SNP rs4344285 causes a transversion mutation that changes the wild-type G allele to the mutant A allele. The mutation is synonymous and does not affect amino acid coding (Pro227Pro). We did not identify any homozygous mutants for this SNP; thus, we can only compare the 14 heterozygous mutants with 9 of wild-type individuals. One was excluded because we were unable to determine the genotype. The AUCt and AUCinf of heterozygous variants were higher and the clearance was lower than that of the wild type, despite the level of significance was not qualified as significant when using a Bonferroni correction approach (p = 0.0018). The mean of Cmax and half-life were both higher in rs4344285 mutants but did not significantly differ, presumably because of relatively high variance among the groups (25.9% and 41% CV for heterozygous mutants in Cmax and half-life, respectively). Nevertheless, the mean plasma concentration plot over the observed time course showed separated curves between the variant and wild-type groups (Fig. 2).
Fig. 2.
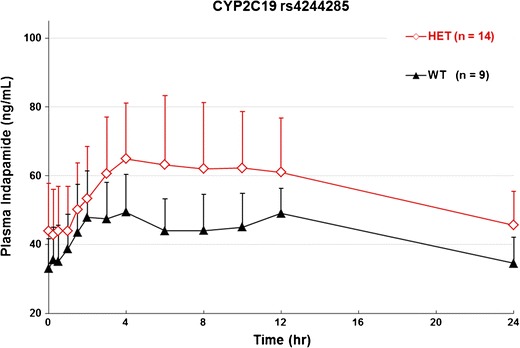
Steady-state mean plasma concentration-time profile according to genotypes for SNP rs4244285 after administration of the last dose of indapamide 1.5 mg at 216 h. HOMO homozygous, HET heterozygous, WT wild type, n sample size. Data are mean ± SD at specific times
Linkage Disequilibrium Analysis
The LD between the two analyzed SNPs was calculated based on Haploview method. The calculation shows strong LD between CYP2C9 rs4918758 and CYP2C19 rs4244285 (D′ = 0.88) with correlation coefficient r2 at 0.635.
DISCUSSION
Many PG studies have been performed in Japanese, East Indians, and Koreans, but little has been done to analyze the PG of Taiwanese or Chinese, the largest races in Asia (23–27). The current study provides an extensive survey of the genetic variances among eight selected major DMEs in Taiwanese, which is considered the closest population to Han Chinese. Most differences between Asians and Caucasians have been shown, especially in enzymatic activity of several phase I enzymes such as CYP2D6 and the CYP2C subfamily (28). However, most studies are allele oriented, in most cases analyzing a combination of mutations instead of single mutations, which is quite commonly observed in CYP enzymes. Thus, in allele-oriented analyses it is sometimes difficult to determine the contribution of a specific mutation in an affected metabolic pathway or a phenotype. To date, the human genome has more than 3.1 million SNPs and this number is likely to increase with additional DNA sources from diverse backgrounds of ethnicity (9,10). As compared with other types of genetic abnormalities, SNPs are more frequently observed in the general population. In addition, SNPs can be analyzed individually or in combination with adjacent SNPs (haplotype analysis) with the phenotype of interest to reveal genetic and phenotypic association (9,10).
The first objective of this study was to understand the genetic makeup of most phases I and II metabolic enzymes in Taiwanese. In comparing with the current data at NCBI, Taiwanese, Han Chinese, and Japanese exhibited greater similarity in genetic makeup than Caucasians and Africans. The result is not surprising because Taiwan, Mainland China, and Japan are in close geographic proximity. Similarities in population structure can arise due to ethnicity, human migration, mating, marriage, or founder effects, which allow for frequent exchange of genetic material and equilibrates the mutation selection. However, for some SNPs, the allele frequency we found between Taiwanese and the other two populations was quite different. For instance, CYP2C19 rs37558581 had an allele frequency of 3.3% in Taiwanese but 6.7% and 10.2% in Han Chinese and Japanese, respectively. The difference between Taiwanese and Han Chinese is twofold, while the difference with Japanese is threefold. This may be attributed to the cross-breeding in the micro-environment of Taiwan by Chinese immigrants and indigenous tribes during the past hundreds of years (29).
Simultaneously, we performed PG and PK studies that can reveal the relationship between drug absorption, disposition, and genetic polymorphisms. Variation in genotypes for DMEs, drug receptors, and drug transporters is associated with interindividual and interethnic variation in drug response (28). The most common situations causing these variabilities are differences in drug metabolism (30) (31). Indapamide undergoes extensive hepatic metabolism, and only 5–7% of a dose is excreted in the urine as unchanged drug (32,33). However, only one report recently postulated that CYP2C19, CYP2C8 and CYP3A4 may be involved in the metabolism of indapamide (18), although it was based on in silico predictions. Our extensive association study on the effects of SNPs on indapamide metabolism explored 8 enzymes and 28 SNPs with the hope to identify additional players in indapamide metabolism.
CYP2C9 rs4918758 causes a transversion mutation that changes the wild-type C allele to the mutant T allele. The mutation is located near the 5′ promotor region and may affect the expression of CYP2C9. From Table III and Fig. 1, we concluded that variants of SNP rs4918758 may lower the metabolism of CYP2C9 that results in lowered CL/F, increased concentration of indapamide in plasma and, consequently, increased overall exposure to indapamide by elevated AUC. Furthermore, the plasma concentration of indapamide is allele-dosage dependent, with homozygous mutants having the highest mean indapamide plasma concentration followed by heterozygous mutants and wild-type individuals. This presents a classic case of allele-dosage-dependent drug metabolism.
CYP2C19 rs4244285 causes a transition mutation that changes the wild-type G allele to the mutant A allele. The mutation is synonymous in that it does not change the coding of amino acid proline at position 227 of CYP2C19. Although a synonymous mutation does not change the coding of the amino acid, it may substitute the wild-type codon with a rare codon and result in a less efficient protein translation. A less-efficient codon can thus lower the concentration of the enzymes and lead to slower drug metabolism. In fact, we observed this with this SNP. By having one copy of the mutant allele, the mean indapamide plasma concentration was relatively higher as compared with wild-type individuals (Table III and Fig. 2). Unfortunately, we did not identify any homozygous mutants, so we were not able to determine whether the effect of rs4244285 on indapamide was allele dependent. The SNP rs4244285 suggests that synonymous mutations may not be without functional consequences for certain drugs.
Despite the two SNPs belonged to different enzymes, strong LD by LD analysis must be recognized (D′ = 0.88). The correlation between genetic variations of CYP2C19 and other CYP2C genes, including CYP2C8 and CYP2C9, has been discussed earlier (34). However, the existence of these two variants on the same haplotypes is still a theoretical possibility that inferred by statistical models. The clinical outcome is clear showing that either CYP2C9 rs4918758 or CYP2C19 rs4244285 independently conferred lower enzyme activity against indapamide. More studies are needed to distinguish the contribution of different enzymes participating in the indapamide metabolism.
Although the Cmax and AUC of indapamide were significantly increased among variant genotype groups in rs4918758 and rs4244285, the half-life of indapamide did not change significantly, whereas lowered CL/F was observed in carriers of rs4918758 and rs4244285. Therefore, we assume that metabolic transformation of indapamide is decreased with the variant genotypes, thus leading to incomplete bioavailability (F). This result is in accordance with observations of previous studies in which the presence of CYP2C19*2 allele had a more pronounced effect on the metabolism of other CYP2C19 substrates, such as leflunomide (35)
All above hypotheses offer possible explanations to the observed phenotypes. To firmly cement the claims, in vitro and/or in vivo data are necessary to validate the hypotheses. In this study, we could not conclude firmly on the effect of gene dosage because only a few homozygous healthy volunteers were included. Therefore, to obtain a clearer picture of gene dosage effect on indapamide metabolism, more homozygous volunteers are needed. Our efforts to understand the genetic makeup of Taiwanese and its effect on drug metabolisms will be useful for physicians to prescribe individualized drug dosage to achieve the maximum therapeutic effect and the minimum unwanted side effects. Furthermore, the proper usage of PG information will help to reduce the waste of national medical insurance in drug expenditure.
CONCLUSIONS
The ultimate goal of PG is to achieve the maximized therapeutic effect and minimized drug-induced toxicity for each individual patient. We created a PG database of 1,038 healthy Taiwanese subjects and investigated 28 SNPs for 8 major metabolic enzymes. Among them, eight SNPs from CYP2A6, 2C19, 2D6, 2E1, 3A5, and UGT2B7 showed remarkably increased variant frequencies than those previously reported in Caucasians. The genetic impacts on indapamide metabolism were further manifested by CYP2C9 SNP rs4918758 that showed significant correlation with indapamide PK parameters in healthy subjects. The PG/PK approach could be used to analyze the genetic makeup and its effects on drug metabolism. Further comprehensive PG analysis will be useful for obtaining valuable information to maximize therapeutic effect for individualized therapy.
ACKNOWLEDGMENTS
The authors are grateful to Hung-Lun Chiang and Yie-Wen Liao at the Institute of Biomedical Sciences, Academia Sinica, for genotyping analysis. The statistical advices from Mr. Chin Lin at the School of Public Health, National Defense Medical Center, were also appreciated.
Conflict of Interest
The authors declare no conflict of interest.
REFERENCES
- 1.Lee C, Morton CC. Structural genomic variation and personalized medicine. N Engl J Med. 2008;358(7):740–741. doi: 10.1056/NEJMcibr0708452. [DOI] [PubMed] [Google Scholar]
- 2.Ma MK, Woo MH, McLeod HL. Genetic basis of drug metabolism. Am J Health Syst Pharm. 2002;59(21):2061–2069. doi: 10.1093/ajhp/59.21.2061. [DOI] [PubMed] [Google Scholar]
- 3.Wang KS, Zahn LE, Favor J, Huang KM, Stambolian D. Genetic and phenotypic analysis of Tcm, a mutation affecting early eye development. Mamm Genome. 2005;16(5):332–343. doi: 10.1007/s00335-004-2444-7. [DOI] [PubMed] [Google Scholar]
- 4.Sakurai Y, Hirayama M, Hashimoto M, Tanaka T, Hasegawa S, Irie S, et al. Population pharmacokinetics and proton pump inhibitory effects of intravenous lansoprazole in healthy Japanese males. Biol Pharm Bull. 2007;30(12):2238–2243. doi: 10.1248/bpb.30.2238. [DOI] [PubMed] [Google Scholar]
- 5.Tomalik-Scharte D, Lazar A, Fuhr U, Kirchheiner J. The clinical role of genetic polymorphisms in drug-metabolizing enzymes. Pharmacogenomics J. 2008;8(1):4–15. doi: 10.1038/sj.tpj.6500462. [DOI] [PubMed] [Google Scholar]
- 6.Gardiner SJ, Begg EJ. Pharmacogenetics, drug-metabolizing enzymes, and clinical practice. Pharmacol Rev. 2006;58(3):521–590. doi: 10.1124/pr.58.3.6. [DOI] [PubMed] [Google Scholar]
- 7.Yasuda SU, Zhang L, Huang SM. The role of ethnicity in variability in response to drugs: focus on clinical pharmacology studies. Clin Pharmacol Ther. 2008;84(3):417–423. doi: 10.1038/clpt.2008.141. [DOI] [PubMed] [Google Scholar]
- 8.Lin M, Chu CC, Chang SL, Lee HL, Loo JH, Akaza T, et al. The origin of Minnan and Hakka, the so-called “Taiwanese”, inferred by HLA study. Tissue Antigens. 2001;57(3):192–199. doi: 10.1034/j.1399-0039.2001.057003192.x. [DOI] [PubMed] [Google Scholar]
- 9.Pattaro C, Ruczinski I, Fallin DM, Parmigiani G. Haplotype block partitioning as a tool for dimensionality reduction in SNP association studies. BMC Genomics. 2008;9(1):405. doi: 10.1186/1471-2164-9-405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sethupathy P, Giang H, Plotkin JB, Hannenhalli S. Genome-wide analysis of natural selection on human cis-elements. PLoS One. 2008;3(9):e3137. doi: 10.1371/journal.pone.0003137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hunter DJ, Khoury MJ, Drazen JM. Letting the genome out of the bottle—will we get our wish? N Engl J Med. 2008;358(2):105–107. doi: 10.1056/NEJMp0708162. [DOI] [PubMed] [Google Scholar]
- 12.Ding C, Jin S. High-throughput methods for SNP genotyping. Methods Mol Biol. 2009;578:245–254. doi: 10.1007/978-1-60327-411-1_16. [DOI] [PubMed] [Google Scholar]
- 13.Ragoussis J. Genotyping technologies for genetic research. Annu Rev Genomics Hum Genet. 2009;10(1):117–133. doi: 10.1146/annurev-genom-082908-150116. [DOI] [PubMed] [Google Scholar]
- 14.Reilly RF, Peixoto AJ, Desir GV. The evidence-based use of thiazide diuretics in hypertension and nephrolithiasis. Clin J Am Soc Nephrol. 2010;5(10):1893–1903. doi: 10.2215/CJN.04670510. [DOI] [PubMed] [Google Scholar]
- 15.Chan TY. Indapamide-induced severe hyponatremia and hypokalemia. The Annals of Pharmacotherapy. 1995;29(11):1124–1128. doi: 10.1177/106002809502901111. [DOI] [PubMed] [Google Scholar]
- 16.Psaty BM, Smith NL, Siscovick DS, Koepsell TD, Weiss NS, Heckbert SR, et al. Health outcomes associated with antihypertensive therapies used as first-line agents. A systematic review and meta-analysis. JAMA J Am Med Assoc. 1997;277(9):739–745. doi: 10.1001/jama.1997.03540330061036. [DOI] [PubMed] [Google Scholar]
- 17.Campbell DB, Phillips EM. Short term effects and urinary excretion of the new diuretic, indapamide, in normal subjects. Eur J Clin Pharmacol. 1974;7(6):407–414. doi: 10.1007/BF00560352. [DOI] [PubMed] [Google Scholar]
- 18.Sun H, Moore C, Dansette PM, Kumar S, Halpert JR, Yost GS. Dehydrogenation of the indoline-containing drug 4-chloro-N-(2-methyl-1-indolinyl)-3-sulfamoylbenzamide (indapamide) by CYP3A4: correlation with in silico predictions. Drug Metab Dispos. 2009;37(3):672–684. doi: 10.1124/dmd.108.022707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Van Hee W, Thomas J, Brems H. Indapamide in the treatment of essential arterial hypertension in the elderly. Postgrad Med J. 1981;57(Suppl 2):29–33. [PubMed] [Google Scholar]
- 20.Jurinke C, van den Boom D, Cantor C, Köster H, et al. The use of MassARRAY technology for high throughput genotyping chip technology. In: Hoheisel J, Brazma A, Büssow K, Cantor C, Christians F, Chui G, et al., editors. Advances in biochemical engineering/biotechnology. Advances in biochemical engineering/biotechnology, 77. Heidelberg: Springer; 2002. pp. 57–74. [DOI] [PubMed] [Google Scholar]
- 21.Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21(2):263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- 22.Schiavi P, Jochemsen R, Guez D. Pharmacokinetics of sustained and immediate release formulations of indapamide after single and repeated oral administration in healthy volunteers. Fundam Clin Pharmacol. 2000;14(2):139–146. doi: 10.1111/j.1472-8206.2000.tb00402.x. [DOI] [PubMed] [Google Scholar]
- 23.Nakashima K, Narukawa M, Kanazu Y, Takeuchi M. Differences between Japan and the United States in dosages of drugs recently approved in Japan. J Clin Pharmacol. 2011;51(4):549–560. doi: 10.1177/0091270010375958. [DOI] [PubMed] [Google Scholar]
- 24.Arnold FL, Kusama M, Ono S. Exploring differences in drug doses between Japan and Western countries. Clin Pharmacol Ther. 2010;87(6):714–720. doi: 10.1038/clpt.2010.31. [DOI] [PubMed] [Google Scholar]
- 25.Kim DH, Sriharsha L, Xu W, Kamel-Reid S, Liu X, Siminovitch K, et al. Clinical relevance of a pharmacogenetic approach using multiple candidate genes to predict response and resistance to imatinib therapy in chronic myeloid leukemia. Clin Cancer Res. 2009;15(14):4750–4758. doi: 10.1158/1078-0432.CCR-09-0145. [DOI] [PubMed] [Google Scholar]
- 26.Chowbay B, Zhou S, Lee EJ. An interethnic comparison of polymorphisms of the genes encoding drug-metabolizing enzymes and drug transporters: experience in Singapore. Drug Metab Rev. 2005;37(2):327–378. doi: 10.1081/DMR-200028805. [DOI] [PubMed] [Google Scholar]
- 27.Zhou Q, Yu XM, Lin HB, Wang L, Yun QZ, Hu SN, et al. Genetic polymorphism, linkage disequilibrium, haplotype structure and novel allele analysis of CYP2C19 and CYP2D6 in Han Chinese. Pharmacogenomics J. 2009;9(6):380–394. doi: 10.1038/tpj.2009.31. [DOI] [PubMed] [Google Scholar]
- 28.Kim K, Johnson JA, Derendorf H. Differences in drug pharmacokinetics between East Asians and Caucasians and the role of genetic polymorphisms. J Clin Pharmacol. 2004;44(10):1083–1105. doi: 10.1177/0091270004268128. [DOI] [PubMed] [Google Scholar]
- 29.Lin M, Chu C, Chang S, Lee H, Loo J, Akaza T, et al. The origin of Minnan and Hakka, the so-called “Taiwanese”, inferred by HLA study. Tissue Antigens. 2001;57:192–199. doi: 10.1034/j.1399-0039.2001.057003192.x. [DOI] [PubMed] [Google Scholar]
- 30.Evans WE, McLeod HL. Pharmacogenomics—drug disposition, drug targets, and side effects. N Engl J Med. 2003;348(6):538–549. doi: 10.1056/NEJMra020526. [DOI] [PubMed] [Google Scholar]
- 31.Ingelman-Sundberg M, Oscarson M, McLellan RA. Polymorphic human cytochrome P450 enzymes: an opportunity for individualized drug treatment. Trends Pharmacol Sci. 1999;20(8):342–349. doi: 10.1016/S0165-6147(99)01363-2. [DOI] [PubMed] [Google Scholar]
- 32.Caruso FS, Szabadi RR, Vukovich RA. Pharmacokinetics and clinical pharmacology of indapamide. Am Heart J. 1983;106(1 Pt 2):212–220. doi: 10.1016/0002-8703(83)90119-9. [DOI] [PubMed] [Google Scholar]
- 33.Robinson DM, Wellington K. Indapamide sustained release: a review of its use in the treatment of hypertension. Drugs. 2006;66(2):257–271. doi: 10.2165/00003495-200666020-00011. [DOI] [PubMed] [Google Scholar]
- 34.Pedersen RS, Brasch-Andersen C, Sim SC, Bergmann TK, Halling J, Petersen MS, et al. Linkage disequilibrium between the CYP2C19*17 allele and wildtype CYP2C8 and CYP2C9 alleles: identification of CYP2C haplotypes in healthy Nordic populations. Eur J Clin Pharmacol. 2010;66(12):1199–1205. doi: 10.1007/s00228-010-0864-8. [DOI] [PubMed] [Google Scholar]
- 35.Bohanec Grabar P, Grabnar I, Rozman B, Logar D, Tomsic M, Suput D, et al. Investigation of the influence of CYP1A2 and CYP2C19 genetic polymorphism on 2-cyano-3-hydroxy-N-[4-(trifluoromethyl)phenyl]-2-butenamide (A77 1726) pharmacokinetics in leflunomide-treated patients with rheumatoid arthritis. Drug Metab Dispos. 2009;37(10):2061–2068. doi: 10.1124/dmd.109.027482. [DOI] [PubMed] [Google Scholar]


