Abstract
Purpose
Abnormalities in semen parameters are often associated with reduced fertility in males, and may, in part, be attributed to genetic variation. Aim of this study is to determine if genetic variants that were previously shown to be predictors of family size and birth rate in healthy men are also associated with sperm morphology in men recruited from an infertility laboratory.
Methods
Genetic associations with sperm morphology phenotypes in 126 ethnically diverse men from Chicago at 41 independent loci, previously shown to be predictors of family size and birth rate in healthy men, were tested.
Results
Two intronic SNPs, rs680730 (in DSCAML1) and rs10129954 (in DPF3), were associated with the percent of normal sperm morphology in Chicago men (P = 0.017 and 0.023, respectively). Furthermore, both loci were associated with increased occurrence of sperm head defects.
Conclusions
SNPs in two genes, both of which have roles in nervous system development, were associated with poor sperm morphology. These results may be helpful in identification of other novel genes and biological pathways whose proper functioning is crucial for sperm production and male reproductive processes.
Electronic supplementary material
The online version of this article (doi:10.1007/s10815-013-0140-9) contains supplementary material, which is available to authorized users.
Keywords: Male fertility, Sperm morphology, Association study, Genetics of infertility
Introduction
Approximately 1 in 8 couples in the western world is not able to conceive spontaneously within 1 year of unprotected intercourse. In nearly half of these couples, one or more semen parameters from the male are within WHO thresholds indicating subfertility [1–3]. Monogenic disorders (e.g., cystic fibrosis [CF], Kallman syndrome), cytogenetic abnormalities (e.g., Klinefelter syndrome [47,XXY]), and Y-chromosome deletions account for approximately 30 % of cases [4]. The etiology of the remaining male infertility (or subfertility) cases remains largely unknown. However, it is suggested that aberrations in many as yet unidentified genes may be responsible for large proportion of reproductive abnormalities, as the key steps in reproductive development, such as sperm production, require coordinated action of thousands of gene products. Supporting this hypothesis, previous studies in mouse models identified hundreds of genes, for which knock out animals display wide range of defects in reproductive processes, resulting in infertility or subfertility [5]. In contrast to studies in animal models, definitive identification of such genes in humans has proven challenging [1–5], and novel discovery strategies are required to fully characterize the genetic architecture of human fertility and infertility.
We previously performed a genome-wide association study (GWAS) of two reproductive phenotypes in 269 married men from a founder population of European descent, the Hutterites, who traditionally proscribes contraception and uniformly desires large families [6]. Forty-one independent genomic regions that were associated with either family size or birth rate in the Hutterite men were considered candidate loci and taken forward to a validation study of sperm count and/or motility in 123 ethnically diverse men from Chicago. We validated associations between nine loci and one or more of these measures. Here, we extend those validation studies to include the effects of these candidate single nucleotide polymorphisms (SNPs) on sperm morphology in the same sample of men from Chicago. We report associations between morphology and SNPs located in the introns of two genes, DPF3 and DSCAML1. Our observations further contribute to our understanding of the genetic basis of sperm development and male reproductive processes.
Materials and methods
Sample composition
The Chicago validation sample included 126 men who had undergone semen analyses at the University of Illinois at Chicago’s (UIC) Andrology Laboratory under IRB approval. All samples were de-identified, with only the subject’s age, ethnicity and results of semen analysis recorded. The ethnic composition of the sample was 57.1 % (n = 72) Hispanic, 26.2 % (n = 33) African American, 9.5 % (n = 12) Middle Eastern, 4.8 % (n = 6) individuals of European ancestry, and 2.4 % (n = 3) Asian as described previously [6]. The reasons for referral for semen analysis were not known, although it is likely that most were due to infertility.
Out of 126 men, 71 (56.4 %) were normozoospermic, whereas 24 (19.0 %) had asthenozoospermia, 30 (23.8 %) had oligoasthenozoospermia and 1 (0.8 %) had oligozoospermia. Four men (three Hispanic, one African American) were diagnosed with severe oligo-asthenozoospermia, with sperm concentration ranging between 0.6 and 3.2 million/ml, and sperm motility ranging between 0 and 27 %; and morphological evaluation could not be completed. Consequently, the final sample size included in these studies was 122.
Sperm morphology evaluation and parameters
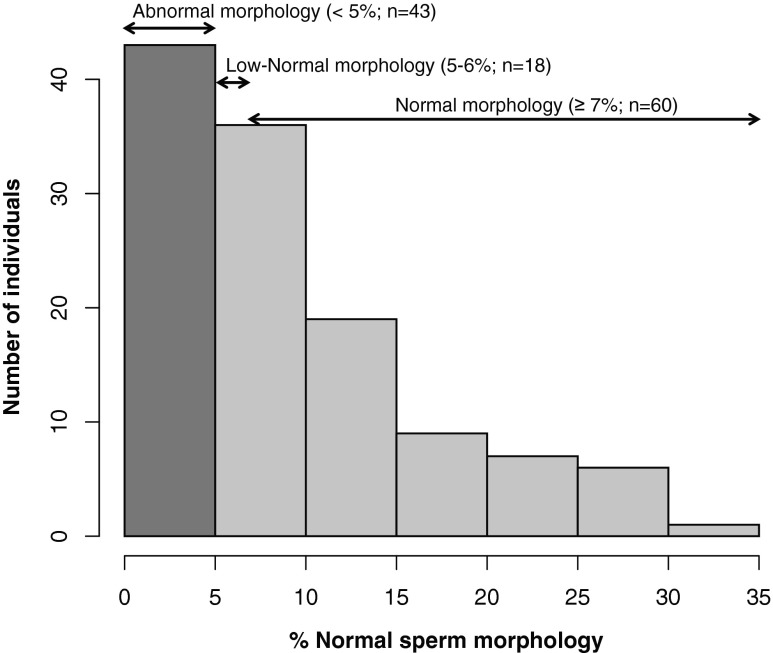
Semen samples, obtained at the laboratory site after 3–4 days of sexual abstinence, were allowed to liquefy at room temperature, and then analyzed within 1 h of sample collection. The same technologist performed all semen analyses with morphology results confirmed in a blinded manner by a second technologist. When discrepancies arose, the morphology slides were re-reviewed with the laboratory director (G.S.P.) and a mutual decision was reached. Samples were prepared by dehydration, fixation and PAP staining, and sperm morphology was evaluated according to Kruger strict criteria [7]. Results are reported as the percent of sperm displaying normal morphology, which we used as a continuous variable in our analyses. We also considered a categorical variable of sperm morphology, with men having sperm with ≤4 % normal morphology classified as “abnormal” (teratozoospermia, n = 43; 34.4 %), 5–6 % normal morphology as “low-normal” (n = 18; 14.4 %), and ≥7 % normal morphology as “normal” (n = 60; 48.0 %; Fig. 1). For analyses of those data, we included only the two extreme groups (“abnormal” and “normal”) and excluded the intermediate group as measures of sperm morphology are subjective and subject to technician bias [8].
Fig. 1.
Distribution of sperm morphology measures in the Chicago sample
In addition to WHO fifth edition morphology [9], specific morphologic abnormalities were noted as percentage of sperm displaying the given defect. These included (i) head defects (percent of acrosome deficient, amorphous, large, amorphous large, double, tapering, pyriform, and vacuolated head), (ii) neck and midpiece defects, (iii) cytoplasmic defects and (iv) tail defects (percent of coiled and multiple tail) [10]. The frequencies of these defects in our sample are shown in Supplementary Table 1. All these traits were analyzed as continuous variables.
Genotyping and statistical analyses
DNA was obtained from the sperm samples at the University of Chicago after semen analyses were completed at the UIC, following a standard DNA isolation procedure with tissue lysis and ethanol precipitation. Genotyping was performed with iPLEX MassARRAY platform (Sequenom, San Diego, CA) for 38 SNPs; the three remaining SNPs failed the assay design and were genotyped by TaqMan allelic discrimination assays (Applied Biosystems, Foster City, CA), following the manufacturers’ instructions, as previously described [6].
To ensure that the heterogeneous ethnic composition of this sample did not cause spurious associations, ethnicity of the patient was included as a covariate in linear regression models for each of the sperm morphology parameters. Ethnicity was not a significant predictor of any of the phenotypes. The effects of age of the patient on each phenotype was also tested; but it also did not differ between men with abnormal, low normal and normal morphology (mean age 35.7, 35.6 and 34.7, respectively). Therefore, neither ethnicity nor age was considered further in the analyses. In order to minimize the deviation of the continuous traits from the normal distribution, we used square-root or log-transformed values as appropriate.
We used a regression-based model to test for association between each of the 41 validation SNPs and morphology parameters, requiring that the model being tested (additive, recessive or dominant) and the direction of the effect for a given allele (increased or decreased fertility) were consistent with the initial association observed in the Hutterites [6]. For continuous variables, we used a one-sided linear regression test; for categorical classification, we used a one-sided logistic regression test. We assessed significance by permutation, in which the genotypes for 41 SNPs were considered together and randomly permuted 10,000 times between individuals, retaining the correlation between phenotypes. Empirical P-values were then calculated from this data as the proportion of permuted test statistics (t- or z-values, as appropriate) that were greater than the observed test statistic. This approach has the advantage of retaining correlations between both phenotypes and genotypes in our data, and also minimizes the effects of any skewing from the normal distribution on the P-values. All statistical analyses were conducted with R statistical software [11].
Results
SNPs at two of the 41 candidate loci that were previously associated with reduced fertility in Hutterite men [6] were associated with abnormalities in sperm morphology in the Chicago men at P < 0.05 (Table 1). The TT genotype at rs10129954 was previously associated with smaller family sizes in the Hutterite men (Fig. 2a, modified from Kosova et al. [6]); here the TT genotype was associated with reduced percentage of normal sperm morphology in the Chicago men. The mean percentage of morphologically normal sperm was 7.21 % with a standard error (SE) of 1.03 in TT men compared to 10.0 % (SE 0.94) in CT and 9.18 % (SE 2.16) in CC men (P = 0.023, Fig. 2b). The second association was with rs680730. In the Hutterite men, increasing copies of the C allele were previously associated in an additive manner with increasingly lower birth rates (Fig. 2e, modified from Kosova et al. [6]). A similar additive pattern was observed in the Chicago men. The mean percentage of normal sperm morphology increased from 7.77 % (SE 1.03) in CC men, to 8.80 % (SE 0.96) in CT men, and to 13.58 % (SE 2.06) in TT men (P = 0.017, Fig. 2f). The C allele at this SNP was also associated with abnormal sperm morphology when tested as a binary variable. 50 % of men with the CC genotype at this SNP were classified as teratozoospermic. This fraction decreased to 37 % in the CT heterozygotes, and to 25 % in the TT homozygotes (P = 0.036, Fig. 2g). The observed trend at both associated loci remained similar when the analyses were repeated in Hispanic men only, although associations were less significant due to the smaller sample size (n = 72). For example, the mean percentage of normal sperm morphology in Hispanic men with TT genotype at rs10129954 was 7.68 % (SE 1.27), compared to 8.20 % (SE 3.32) and 10.97 % (SE 1.36) in men with CC and CT genotype at the same locus, respectively. Likewise, mean percentage of normal sperm morphology increased with each additional T allele at rs680730 locus, changing from 7.78 % (SE 1.22) for CC genotype, to 10.16 % (SE 1.48) and 12.71 % (SE 2.81) for CT and TT genotypes, respectively. This indicates that our findings cannot be attributed to genetic stratification or ethnic heterogeneity.
Table 1.
Results of the association tests between 41 validation SNPs and two sperm morphology traits, % normal morphology and morphology classification
| SNP | Chr | Position | Closest gene(s) | Relationship to the closest gene | Dist (kb) | Chicago validation sample | Hutterite men | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Allele: genotype count (2/1/0 copies) | Freq | % normal morphology P-value | Morphology classification P-value | Assoc. trait | P-value | Model | ||||||
| rs2991396 | 13 | 66,787,387 | PCDH9 | upst. | 84.9 | G: 14/53/55 | 0.33 | 1.27E-01 | 1.25E-01 | FS | 4.43E-06 | additive |
| rs10966811 | 9 | 25,223,484 | TUSC1 | dwnst. | 445.4 | T: 23/49/47 | 0.40 | 9.70E-01 | 9.21E-01 | FS | 5.57E-06 | recessive |
| rs11144790 | 9 | 78,002,292 | PCSK5 | dwnst. | 4.1 | A: 1/11/106 | 0.06 | 9.85E-01 | 9.68E-01 | FS | 6.45E-06 | additive |
| rs2423942 | 20 | 15,271,282 | MACROD2 | intron | — | T: 12/50/54 | 0.32 | 7.97E-01 | 8.24E-01 | FS | 9.26E-06 | recessive |
| rs7867029 | 9 | 80,210,238 | PSAT1 | dwnst. | 75.4 | G: 9/38/65 | 0.25 | 3.53E-01 | 2.97E-01 | FS | 1.04E-05 | recessive |
| rs4753939 | 11 | 98,493,518 | CNTN5 | intron | — | T: 26/56/40 | 0.44 | 8.88E-01 | 9.37E-01 | FS | 1.05E-05 | recessive |
| rs12339229 | 9 | 28,083,232 | LINGO2 | intron | — | A: 1/18/102 | 0.08 | 8.45E-01 | 7.63E-01 | FS | 1.23E-05 | additive |
| rs9851460 | 3 | 156,384,950 | MME | dwnst. | 0.7 | A: 11/53/58 | 0.31 | 8.69E-01 | 8.81E-01 | FS | 1.24E-05 | recessive |
| rs1152948 | 12 | 68,678,852 | RAB3IP | dwnst. | 175.6 | A: 11/52/59 | 0.30 | 9.96E-01 | 9.94E-01 | FS | 1.24E-05 | recessive |
| rs1472272 | 7 | 97,950,910 | BAIAP2L1 | upst. | 82.6 | G: 6/41/72 | 0.22 | 4.03E-01 | 1.76E-01 | FS | 1.44E-05 | recessive |
| rs12870438 | 13 | 42,378,205 | EPSTI1 | intron | — | A: 5/32/85 | 0.17 | 3.20E-01 | 3.98E-01 | FS | 2.07E-05 | recessive |
| rs1507417 | 3 | 78,733,335 | ROBO1 | intron | — | C: 11/40/71 | 0.25 | 3.99E-01 | 5.94E-01 | FS | 2.41E-05 | additive |
| rs12088543 | 1 | 84,490,571 | PRKACB | dwnst. | 13.8 | G: 11/46/64 | 0.28 | 5.29E-01 | 6.32E-01 | FS | 3.31E-05 | additive |
| rs7174015 | 15 | 48,504,360 | USP8 | intron | — | C: 24/52/39 | 0.43 | 3.31E-01 | 1.85E-01 | FS | 3.57E-05 | recessive |
| rs11723705 | 4 | 55,445,828 | KIT, KDR | dwnst. | 144.2, 193.6 | T: 2/24/92 | 0.12 | 7.66E-01 | 8.91E-01 | FS | 3.63E-05 | additive |
| rs10129954 | 14 | 72,220,454 | DPF3 | intron | — | C: 12/60/50 | 0.34 | 2.33E-02 | 9.88E-02 | FS | 3.86E-05 | recessive |
| rs7106789 | 11 | 21,526,300 | NELL1 | intron | — | A: 18/67/35 | 0.43 | 6.17E-01 | 6.85E-01 | FS | 4.36E-05 | recessive |
| rs1514673 | 12 | 41,930,420 | ADAMTS20 | dwnst. | 103.9 | T: 7/40/73 | 0.23 | 9.81E-01 | 9.86E-01 | FS | 7.18E-05 | additive |
| rs1487933 | 21 | 20,593,873 | BC047405 | dwnst. | 442.9 | G: 20/61/39 | 0.42 | 3.28E-01 | 1.78E-01 | FS | 5.18E-05 | recessive |
| rs6471590 | 8 | 143,795,554 | LY6K, C8orf55 | dwnst. | 12.9, 10.1 | C: 7/41/73 | 0.23 | 8.42E-01 | 6.33E-01 | FS | 5.32E-05 | recessive |
| rs11956144 | 5 | 160,248,178 | ATP10B | upst. | 36.4 | G: 1/30/81 | 0.14 | 5.83E-01 | 4.46E-01 | FS | 5.96E-05 | additive |
| rs10893363 | 11 | 124,670,989 | PKNOX2 | intron | — | G: 0/17/104 | 0.07 | 1.46E-01 | 9.06E-02 | FS | 7.16E-05 | additive |
| rs17088625 | 8 | 22,703,795 | PEBP4 | intron | — | C: 15/56/51 | 0.35 | 9.14E-01 | 9.65E-01 | FS | 7.20E-05 | additive |
| rs2330929 | 22 | 23,549,930 | SGSM1 | intron | — | C: 29/51/39 | 0.46 | 8.84E-01 | 8.10E-01 | FS | 7.91E-05 | additive |
| rs16917898 | 9 | 32,106,520 | ACO1 | upst. | 268.1 | T: 0/16/107 | 0.07 | 5.80E-01 | 2.72E-01 | FS | 7.99E-05 | recessive |
| rs11794651 | 9 | 38,728,489 | ANKRD18A | upst. | 117.8 | G: 3/31/88 | 0.15 | 5.50E-01 | 9.06E-01 | FS | 9.48E-05 | additive |
| rs1745689 | 14 | 57,140,645 | SLC35F4 | upst. | 7.3 | A: 16/45/62 | 0.31 | 3.63E-01 | 2.30E-01 | FS | 9.66E-05 | additive |
| rs9422913 | 10 | 127,335,597 | C10orf22 | intron | — | G: 22/58/37 | 0.44 | 3.82E-01 | 4.71E-01 | FS | 9.89E-05 | additive |
| rs433406 | 11 | 130,876,229 | NTM | intron | — | G: 32/51/32 | 0.50 | 4.06E-01 | 6.78E-01 | BR | 3.36E-06 | additive |
| rs4936891 | 11 | 123,425,855 | OR10G7 | upst. | 10.9 | C: 11/38/59 | 0.28 | 2.26E-01 | 8.78E-02 | BR | 8.62E-06 | additive |
| rs680730 | 11 | 116,980,443 | DSCAML1 | intron | — | T: 12/58/50 | 0.34 | 1.68E-02 | 3.60E-02 | BR | 2.45E-05 | additive |
| rs4538394 | 3 | 21,774,713 | ZNF385D | intron | — | A: 14/40/58 | 0.30 | 3.45E-01 | 2.59E-01 | BR | 3.67E-05 | recessive |
| rs13265504 | 8 | 3,532,499 | CSMD1 | intron | — | C: 35/48/38 | 0.49 | 7.41E-01 | 9.11E-01 | BR | 4.43E-05 | additive |
| rs1484095 | 18 | 33,971,576 | BRUNOL4 | upst. | 571.8 | T: 10/46/65 | 0.27 | 1.25E-01 | 5.78E-02 | BR | 5.37E-05 | additive |
| rs11236909 | 11 | 76,116,716 | TSKU, LRRC32 | upst. | 55.2, 58.0 | C: 8/43/70 | 0.24 | 4.42E-01 | 3.99E-01 | BR | 6.11E-05 | additive |
| rs768343 | 18 | 37,686,676 | PIK3C3 | upst. | 102.5 | A: 15/58/44 | 0.38 | 7.53E-01 | 8.97E-01 | BR | 6.60E-05 | additive |
| rs2035804 | 10 | 133,884,369 | STK32C | intron | — | T: 3/31/88 | 0.15 | 4.74E-01 | 5.20E-01 | BR | 7.55E-05 | additive |
| rs4128691 | 3 | 72,432,553 | RYBP | dwnst. | 73.9 | T: 18/45/53 | 0.35 | 5.76E-01 | 6.49E-01 | BR | 8.40E-05 | additive |
| rs10488786 | 11 | 100,245,404 | ARHGAP42 | intron | — | T: 0/11/106 | 0.05 | 3.66E-01 | 6.01E-01 | BR | 8.70E-05 | additive |
| rs79110092 | 10 | 49,808,465 | LRRC18, WDFY4 | intron | — | T: 1/20/101 | 0.09 | 5.92E-01 | 8.23E-01 | BR | 9.49E-05 | additive |
| rs724078 | 6 | 29,597,027 | MAS1L, UBD | upst. | 33.7, 34.4 | T: 26/59/34 | 0.47 | 1.84E-01 | 7.12E-02 | BR | 9.99E-05 | recessive |
In the Chicago validation sample, P-values from 10,000 permutations are shown. Bolded P-values show the significant associations in the Chicago validation sample
Fig. 2.
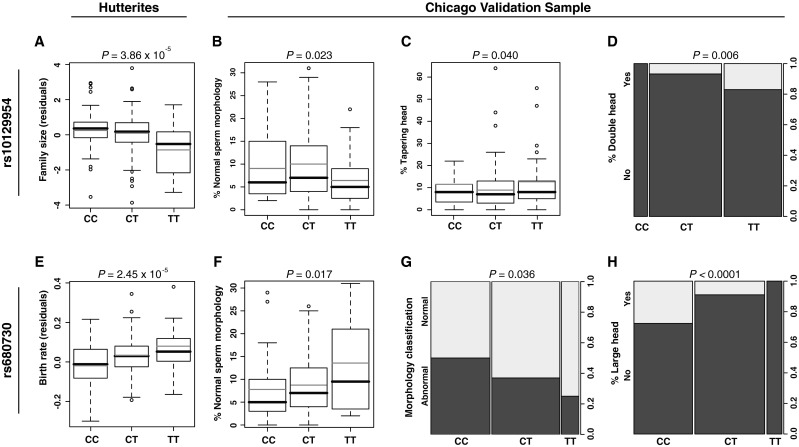
Effects of the associated SNPs, rs10129954 (a–d) and rs680730 (e–h), on fertility measures in Hutterite men (from Kosova et al. [6]) and sperm morphology in Chicago sample. The boxplots show the first and third quartiles; the whiskers extend to the minimum and maximum values, excluding the outliers. Black horizontal lines show the median, and grey horizontal lines show the mean value within each genotype group. Column plots in panels d, g and h show the presence or absence of each abnormality in the Chicago sample, with the proportion of men is shown on the right y-axis. P-values correspond to the single-locus SNP-specific GWAS P-value in the Hutterite men [6], and empirical P-values obtained after 10,000 permutations in the Chicago men. FS family size; BR birth rate
To gain insight into the specific types of defects associated with these two SNPs, we further tested for associations with additional morphological abnormalities (Table 2). The first SNP, rs10129954, was associated with both increased occurrence of tapering head (P = 0.040) and double head (P = 0.006) anomalies, whereas the second SNP, rs680370, was associated with a large head abnormality (P < 0.0001, Fig. 2c, d and h). These results suggest that both SNPs and the genes in which they reside may play a role in sperm head development.
Table 2.
P-values based on 10,000 permutations for the associations between the two sperm morphology-associated SNPs, rs10129954 and rs680730, and specific morphologic defects
| Morphology defect | rs10129954 | rs680730 |
|---|---|---|
| Head defects | ||
| % Acrosome deficient | 0.804 | 0.561 |
| % Amorphous head | 0.573 | 0.078 |
| % Large head | 0.661 | <0.0001 |
| % Amorphous large head | 0.213 | 0.248 |
| % Double head | 0.006 | 0.279 |
| % Tapering head | 0.040 | 0.550 |
| % Pyriform head | 0.372 | 0.961 |
| % Vacuolated head | 0.484 | 0.139 |
| % Neck/Midpiece defects | 0.933 | 0.950 |
| % Cytoplasmic defect | 0.816 | 0.840 |
| Tail defects | ||
| % Coiled tail | 0.080 | 0.566 |
| % Multiple tail | 0.069 | 0.083 |
Bold denotes most significant SNPs in analysis
Discussion
Our study demonstrates that genetic variants previously associated with reproductive success in healthy, fertile men are also with gross sperm morphology and specific sperm head defects in an unrelated sample of ethnically diverse men. In this study, we validate associations with two SNPs, rs10129954, located in an intronic region of the DPF3 gene on chromosome 14, and rs680730, located in an intronic region of the DSCAML1 gene on chromosome 11, and gross sperm morphology. The effects of SNP rs10129954 (DPF3) fit a recessive model, in which homozygosity for the T allele, associated previously with decreased fertility in Hutterite men [6], is now associated with reduced percentage of normal morphology in the ethnically diverse Chicago men (Fig. 2a–b). In contrast, the effects of SNP rs680730 (DSCAML1) best fit an additive model, with each additional copy of the C allele associated with reduced reproductive measures (Fig. 2e–f). The validation of associations that were initially observed in the Hutterites in ethnically diverse Chicago men suggests that our findings are robust to ethnic background, allele frequency differences or environment.
D4, zinc and double PHD finger 3 (DPF3) and down syndrome cell adhesion molecule-like 1 (DSCAML1) both could have a role in reproduction, although expression of neither gene has yet been examined in the testis. DPF3 was previously implicated as a key epigenetic factor in development that facilitates access of transcription factors to DNA through interaction with histones, specifically by binding acetylated and methylated H3 and H4 lysine residues [12]. Results of recent studies emphasize the key role of histone modification, such as that regulated through DPF3, in preparing mature sperm DNA for early embryogenesis [13, 14]. Evidence suggests that DSCAML1 is a key signaling factor balancing the intensity of adhesion forces necessary for normal neuronal cellular morphogenesis [15].
We previously reported associations between each of these two SNPs and total motile count (rs10129954), beat frequency (rs10129954), and linearity (both SNPs). Prior studies correlated mature human sperm morphology with metrics of motility [16, 17]. In our data, percentage of normal morphology and total motile count are correlated (r2 = 0.26). The association of these SNPs with head defects could result in abnormal sperm shapes that reduce motility (Table 2). Thus, the same underlying defects associated with these variants may result in both abnormal morphology and poor motility.
Finally, we note that the relatively small sample sizes in this study affect power, and as a result we may have missed other associations. For example, although other SNPs associated with reduced sperm count and motility parameters in our previous study [6], such as rs7174015 (USP8) and rs724078 (MAS1L, UBD), also show a trend with reduced percentage of normal morphology, these associations were not statistically significant after correcting for multiple testing (Table 1). Additional studies in larger cohorts are required to verify the effects of these variants on sperm parameters and function.
By conducting a large family-based study for gene discovery first, and validating findings in an independent sample of men undergoing semen analyses, we identified two candidate genes of sperm morphology for further investigation. This strategy addresses the limitations inherent in studying heterogenous conditions, such as male infertility, using a case–control design [3] and provides an unbiased method for the discovery of genes that influence male fertility.
Electronic supplementary material
(PDF 58 kb)
Acknowledgments
We would like to thank Dr. Rachel A. Myers for statistical consultation; Mathis Morrison and William Birch for technical assistance with semen analysis; and Mary Coppolillo for subject coordination at the UIC andrology clinic. This work is supported by National Institutes of Health (NIH) grant HD21244.
Footnotes
Capsule SNPs from two genes were found to be associated with abnormal sperm morphology.
Gülüm Kosova and James M. Hotaling would like to be considered similar in author order.
Contributor Information
James M. Hotaling, Email: jim.hotaling@hsc.utah.edu
Carole Ober, Email: c-ober@genetics.uchicago.edu.
References
- 1.de Kretser DM. Male infertility. Lancet. 1997;349(9054):787–790. doi: 10.1016/S0140-6736(96)08341-9. [DOI] [PubMed] [Google Scholar]
- 2.Massart A, Lissens W, Tournaye H, Stouffs K. Genetic causes of spermatogenic failure. Asian J Androl. 2012;14(1):40–48. doi: 10.1038/aja.2011.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Visser L, Repping S. Unravelling the genetics of spermatogenic failure. Reproduction. 2010;139(2):303–307. doi: 10.1530/REP-09-0229. [DOI] [PubMed] [Google Scholar]
- 4.Aston KI, Conrad DF. A review of genome-wide approaches to study the genetic basis for spermatogenic defects. Methods Mol Biol. 2013;927:397–410. doi: 10.1007/978-1-62703-038-0_34. [DOI] [PubMed] [Google Scholar]
- 5.Matzuk MM, Lamb DJ. The biology of infertility: research advances and clinical challenges. Nat Med. 2008;14(11):1197–1213. doi: 10.1038/nm.f.1895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kosova G, Scott NM, Niederberger C, Prins GS, Ober C. Genome-wide association study identifies candidate genes for male fertility traits in humans. Am J Hum Genet. 2012;90(6):950–961. doi: 10.1016/j.ajhg.2012.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cooper TG, Noonan E, von Eckardstein S, Auger J, Baker HW, Behre HM, et al. World Health Organization reference values for human semen characteristics. Hum Reprod Update. 2010;16(3):231–245. doi: 10.1093/humupd/dmp048. [DOI] [PubMed] [Google Scholar]
- 8.Morbeck DE, Leonard PH, Weaver AL, Shimek KM, Bouwsma EV, Coddington CC. Sperm morphology: classification drift over time and clinical implications. Fertil Steril. 2011;96(6):1350–1354. doi: 10.1016/j.fertnstert.2011.08.036. [DOI] [PubMed] [Google Scholar]
- 9.WHO. WHO laboratory manual for the Examination and processing of human semen. (Accessed at http://whqlibdoc.who.int/publications/2010/9789241547789_eng.pdf.). 2010 [PubMed]
- 10.Chemes EH, Rawe YV. Sperm pathology: a step beyond descriptive morphology. Origin, characterization and fertility potential of abnormal sperm phenotypes in infertile men. Hum Reprod Update. 2003;9(5):405–428. doi: 10.1093/humupd/dmg034. [DOI] [PubMed] [Google Scholar]
- 11.Team RC. R: A language and environment for statistical computing. 2013.
- 12.Lange M, Kaynak B, Forster UB, Tonjes M, Fischer JJ, Grimm C, et al. Regulation of muscle development by DPF3, a novel histone acetylation and methylation reader of the BAF chromatin remodeling complex. Genes Dev. 2008;22(17):2370–2384. doi: 10.1101/gad.471408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Carrell DT, Hammoud SS. The human sperm epigenome and its potential role in embryonic development. Mol Hum Reprod. 2010;16(1):37–47. doi: 10.1093/molehr/gap090. [DOI] [PubMed] [Google Scholar]
- 14.Hammoud SS, Nix DA, Zhang H, Purwar J, Carrell DT, Cairns BR. Distinctive chromatin in human sperm packages genes for embryo development. Nature. 2009;460(7254):473–478. doi: 10.1038/nature08162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Garrett AM, Tadenev AL, Burgess RW. DSCAMs: restoring balance to developmental forces. Front Mol Neurosci. 2012;5:86. doi: 10.3389/fnmol.2012.00086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bronson RA, Bronson SK, Oula LD. Ability of abnormally-shaped human spermatozoa to adhere to and penetrate zona-free hamster eggs: correlation with sperm morphology and postincubation motility. J Androl. 2007;28(5):698–705. doi: 10.2164/jandrol.106.001503. [DOI] [PubMed] [Google Scholar]
- 17.Hu J, Zhu W, Liu W, Fan L. Factors affecting fecundity among sperm donors: a multivariate analysis. Andrologia. 2011;43(3):155–162. doi: 10.1111/j.1439-0272.2009.01036.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(PDF 58 kb)