Abstract
Purpose
Several researchers have suggested that the rs4779584 (15q13.3) polymorphism is associated with an increased risk of developing colorectal cancer (CRC). However, past results remain inconclusive. We addressed this controversy by performing a meta-analysis of the relationship between rs4779584 of GREM1-SCG5 and colorectal cancer.
Methods
We selected 12 case-control studies involving 11,769 cases of CRC and 14,328 healthy controls. The association between the rs4779584 polymorphism and CRC was examined by the overall odds ratio (OR) with a 95% confidence interval (CI). We used different genetic model analyses, sensitivity analyses, and assessments of bias in our meta-analysis.
Results
GREM1-SCG5 rs4779584 polymorphisms were associated with CRC in all of the genetic models that were examined in this meta-analysis of 12 case-control studies.
Conclusion
GREM1-SCG5 rs4779584 polymorphisms may increase the risk of developing colorectal cancer.
Introduction
Colorectal cancer (CRC) is the third most common malignancy and the fourth most common cause of cancer-related death in the world [1]. The incidence of CRC is increasing each year [2], [3]. Although CRC is a heterogeneous disease, patients with advanced stages of disease generally have a poor prognosis [4]. The development of CRC is influenced by lifestyle and dietary factors. In addition, the prognosis of patients with CRC is largely affected by genetic components [5]–[7]. Current data suggests that the rs4779584 polymorphism within the 15q13.3 chromosomal region is associated with an increased risk of developing colorectal cancer [8]–[16]. Rs4779584 lies between the GREM1 and SCG5 genes. GREM1 encodes gremlin 1, which is a signaling molecule in the transforming growth factor-β (TGF-β) pathway. TGF-β signaling has been implicated in tumor invasion and metastasis [17]. SCG5 encodes secretogranin V, which is an important neuroendocrine signaling molecule that appears to influence cellular proliferation in the large bowel based on nutrient availability or systemic hormonal effects [18].
Several research groups have reported associations between this single-nucleotide polymorphism (SNP) and the risk of CRC. However, because single studies are often underpowered due to inadequate sample sizes, the results from past studies are inconclusive. Thus, we performed a meta-analysis to more precisely characterize the association between the rs4779584 polymorphism and colorectal cancer.
Materials and Methods
2.1 Search Strategy and Selection Criteria
We searched five electronic databases (PubMed, Medline, Web of Knowledge, CNKI, and Google Scholar) to identify eligible studies that were published before September 2013. Articles were sought with the following key words: “colorectal cancer”, “rs4779584”, “polymorphism”, and “SCG5 or GREM1”. We also supplemented this search by reviewing the reference lists of all of the retrieved publications and identifying additional relevant articles.
The following inclusion criteria were used to identify articles for our meta-analysis. (1) The study involved unrelated individuals. (2) Sufficient genotype data were presented to allow calculation of the odds ratios (ORs). (3) The study clearly described the diagnosis of CRC and the sources of the cases and controls. (4) The genotype distribution complied with the Hardy–Weinberg equilibrium (HWE). In addition, we excluded reviews and redundant studies. Finally, we selected 11 available studies, which involved 11,769 cases of CRC and 14,328 healthy controls.
2.2 Data Extraction
Two investigators used a standardized form to independently extract data to improve the reliability of our results. The following information was extracted from each study: first author, publication year, ethnicity (country), source of controls, number of cases and controls, and the genotype frequencies of the cases and controls.
2.3 Statistical Analyses
All analyses were performed with the STATA Version 11.0 software (Stata Corp, College Station, TX). All P values in this study were two-sided, and P = 0.05 was set as the threshold value for statistical significance. To evaluate associations between rs4779584 polymorphisms and risk of CRC, the pooled odds ratio (OR) and associated 95% confidence interval (CI) were calculated. We used the following models to calculate different ORs: the allele model (A vs. a), the additive genetic model (AA vs. aa), the dominant genetic model (AA+Aa vs. aa), and the recessive genetic model (AA vs. Aa+aa). If the P value was greater than 0.100 according to the Q-test, indicating a lack of heterogeneity among studies, the summary OR estimate of each study was calculated by a fixed-effects model (the Mantel–Haenszel method). Otherwise, the random-effects model (the DerSimonian-Laird method) was performed. Heterogeneity was estimated with the Cochran’s Q-statistic, and P<0.05 was considered to be an indication of statistically significant heterogeneity [19]. We also quantified the effect of heterogeneity with the I2 test [20]. As a guide, I2 values ranged from 0 to 100%, and values of 25%, 50%, and 75% were considered to represent low, moderate, and high levels of heterogeneity, respectively. The funnel plot was drawn to assess publication biases. The test suggested was used to test the funnel-plot symmetry. This test involved building a regression model, in which the standardized estimate of the size effect was the dependent variable, and the inverse of the standard error was the independent variable. If the intercept was significantly different from zero, the estimate of the effect was considered biased. The significance of the pooled OR was determined with the Z test. Each study was removed in turn for sensitivity analyses, and the remaining studies were reanalyzed to assess the stability of results.
Results
3.1 Characteristics of the Included Studies
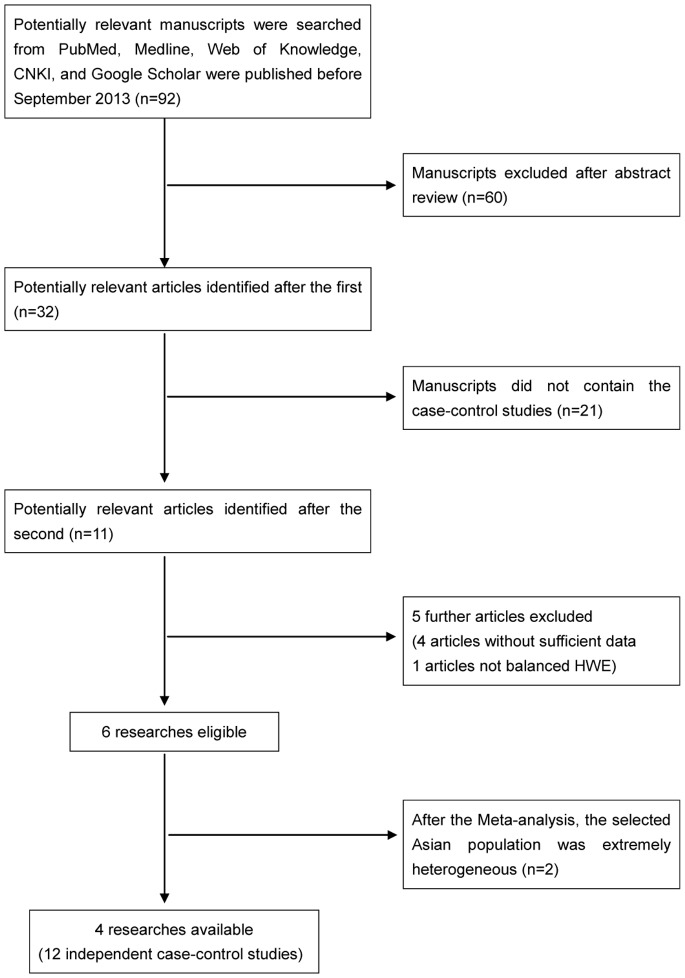
According to search, we identified 92 potentially relevant articles. On the basis of the abstract, 32 studies were reviewed in their entirety. During the extraction of data, 21 articles were removed, because of these articles did not contain the case-control studies. Among the 11 papers, four papers without sufficient data and one article that did not fit the HWE during calculations. Finally, we selected 6 researches eligible. After our meta-analysis found that the selected Asian population was extremely heterogeneous. Finally we selected 12 independent case-control studies from 4 articles involved 11,769 cases of CRC and 14,328 healthy controls in our meta-analysis. The characteristics of the studies are listed in Table 1. Flow diagram of study selection in Figure 1.
Table 1. Characteristics of studies included in the meta-analysis.
| ID | Study | year | Population | Source of controls | Ethnic group | Case/control | HWE p |
| 1 | Xiong et al. [16] | 2010 | Chinese | PB | Asian | 2124/2124 | 0.076 |
| 2 | Ho et al. [10] | 2011 | Hong Kong Chinese | HB | Asian | 716/714 | 0.763 |
| 3 | Jaeger et al. [7] | 2008 | British | PB | Caucasian | 730/960 | 0.726 |
| 4 | Houlston et al. a [29] | 2008 | British | HB | Caucasian | 922/929 | 0.577 |
| 5 | Houlston et al.b | 2008 | Scottish | HB | Caucasian | 922/930 | 0.209 |
| 6 | Houlston et al.c | 2008 | British | HB | Caucasian | 922/931 | 0.796 |
| 7 | Houlston et al.d | 2008 | Scottish | HB | Caucasian | 922/932 | 0.799 |
| 8 | Talseth-Palmer et al. [30] | 2010 | British | PB | Caucasian | 258/313 | 0.969 |
| 9 | Tomlinson et al.e [11] | 2011 | Australian | HB | Caucasian | 591/2353 | 0.877 |
| 10 | Tomlinson et al.f | 2011 | Spanish | HB | Caucasian | 1410/1410 | 0.266 |
| 11 | Tomlinson et al. g | 2011 | American, Canadian, Australian | HB | Caucasian | 1332/1084 | 0.333 |
| 12 | Tomlinson et al.h | 2011 | Finnish | HB | Caucasian | 988/864 | 0.671 |
| 13 | Tomlinson et al.i | 2011 | British | HB | Caucasian | 621/1121 | 0.324 |
| 14 | Tomlinson et al.j | 2011 | British | HB | Caucasian | 2151/2501 | 0.822 |
HB: hospital-based; PB: population-based.
Figure 1. Flow diagram of study selection.
3.2 Meta-analysis Databases
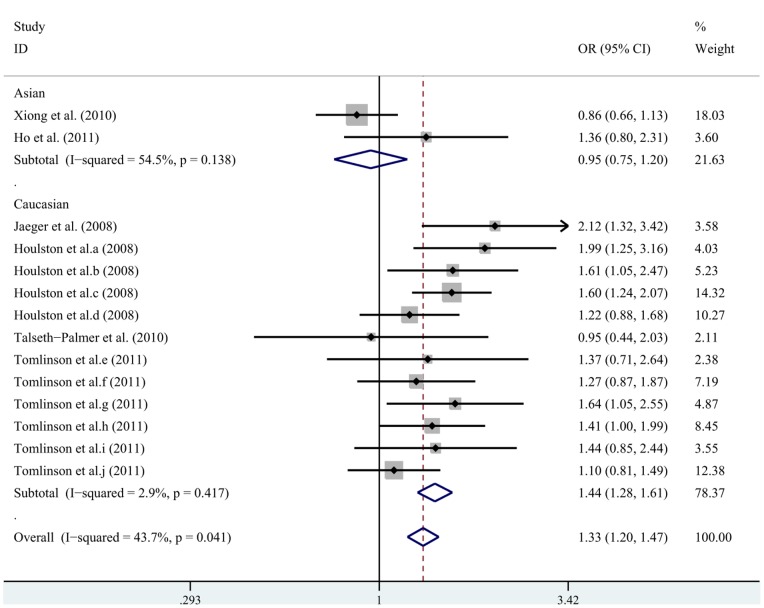
Different genetic models were used in our analysis (Table 2). After pooling all of the selected studies into the meta-analysis, we found that the rs4779584 polymorphism was significantly associated with an increased risk of CRC in Caucasian subjects. The following data were obtained: in the additive model (TT vs. CC; OR = 1.44, 95% CI: 1.28–1.61; TT vs. CT; OR = 1.29, 95% CI: 1.15–1.45); in the dominant model (TT/TC vs. CC; OR = 1.15, 95% CI: 1.07–1.24); in the recessive model (TT vs. CC/CT; OR = 1.38, 95% CI: 1.24–1.55). Forest plots for rs4779584 in the additive genetic models are shown in Figure 2.
Table 2. Meta-analysis of the rs4779584 polymorphism.
| geneticmodel | OR (95% CI) | Z | P | |||||||
| Caucasian | Chinese | Hong-KongChinese | Caucasian | Chinese | Hong-KongChinese | Caucasian | Chinese | Hong-KongChinese | ||
| additive | TT vs. CC | 1.44(1.28–1.61) | 0.86(0.66–1.13) | 1.36(0.80–2.31) | 6.30 | 1.07 | 1.12 | <0.001 | 0.283 | 0.262 |
| TT vs. CT | 1.29(1.15–1.45) | 1.10(0.97–1.26) | 1.32(1.05–1.66) | 4.29 | 1.47 | 2.36 | <0.001 | 0.142 | 0.018 | |
| dominant | TT/TC vs. CC | 1.15(1.07–1.24) | 0.84(0.64–1.09) | 1.25(0.73–2.11) | 3.94 | 1.33 | 0.81 | <0.001 | 0.184 | 0.416 |
| recessive | TT vs. CC/CT | 1.38(1.24–1.55) | 1.06(0.94–1.21) | 1.32(1.06–1.65) | 5.72 | 0.96 | 2.49 | <0.001 | 0.336 | 0.013 |
Figure 2. Forest plots for the rs4779584 polymorphism and risk of CRC in the additive genetic model.
3.3 Test of Heterogeneity
Significant heterogeneity existed in all of the genetic models (Table 3). The selected Asian population was extremely heterogeneous. Thus, we did not include the Asian population in our meta-analysis. However, we did compare the OR of the Asian population with that of the European population.
Table 3. Degree of heterogeneity in meta-analyses of the rs4779584 polymorphism.
| genetic model | Heterogeneity statistic | P | I- squared (%) | |||||||
| Asian | Caucasian | overall | Asian | Caucasian | overall | Asian | Caucasian | overall | ||
| additive | TT vs. CC | 2.20 | 11.32 | 23.09 | 0.138 | 0.417 | 0.041 | 54.5 | 2.9 | 43.7 |
| TT vs. CT | 1.72 | 5.36 | 8.84 | 0.190 | 0.912 | 0.785 | 41.9 | 0.0 | 0.0 | |
| dominant | TT/TC vs. CC | 1.74 | 21.50 | 27.01 | 0.187 | 0.029 | 0.012 | 42.7 | 48.8 | 51.9 |
| recessive | TT vs. CC/CT | 2.86 | 8.81 | 18.52 | 0.091 | 0.639 | 0.139 | 65.0 | 0.0 | 29.8 |
3.4 Bias Diagnostics
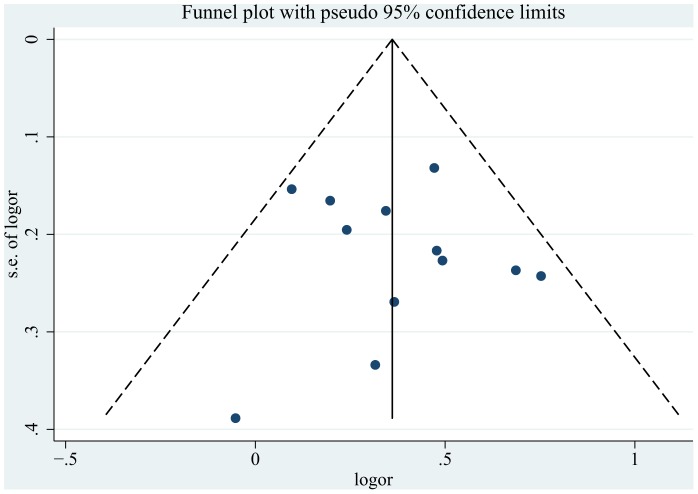
Begger’s funnel plot and Egger’s linear regression test were performed to assess the publication biases of the selected studies. The shape of the funnel plot for publication bias appeared to be symmetrical, although there was some uncertainty regarding the degree of symmetry (Figure 3). The estimate of the effect was considered to be biased. In the recessive model (TT vs. CC/CT), the results from the Begger’s test (P = 0.45) and Egger’s linear regression test (t = 0.42, P = 0.682) did not show evidence of publication bias. Furthermore, the 95% confidence interval (95% CI: −1.73–2.54) included zero, indicating a lack of publication bias. Other genetic models also suggested a lack of publication bias.
Figure 3. Funnel plot of the meta-analysis of CRC risk and the rs4779584 polymorphism.
3.5 Sensitivity Analyses
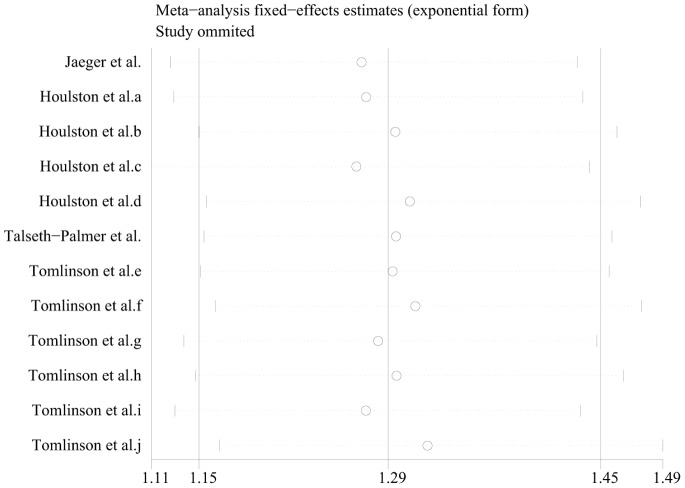
Sensitivity analyses were performed after sequentially removing each eligible study. This approach is regarded as an indispensable step for analyzing multiple criteria. The significance of the pooled ORs was not influenced by any single study in the recessive genetic model (Figure 4), indicating that our results were statistically robust.
Figure 4. Sensitivity analysis of the odds ratio coefficients among Caucasian subject.
Discussion
A growing number of studies suggest that the rs4779584 polymorphism is associated with an increased susceptibility to colorectal cancer. However, the results of past studies have been controversial, with some studies supporting a significant association, whereas others refute this association. In the present study, to confirm that the rs4779584 polymorphism plays a role in the development of CRC, we conducted a meta-analysis of 12 independent case-control studies, which included 11,769cases and 14,328 controls. The major finding of the present meta-analysis was that 15q13.3 rs4779584 was a risk factor for CRC in the Caucasian population. In comparison to previous meta-analyses, our analysis included a greater number of studies. Therefore, a larger sample size and increased statistical power were obtained. Moreover, the present meta-analysis included an acceptable quality evaluation system, minimizing the potential for bias.
A statistically significant level of heterogeneity was found in the Asian population but not in Caucasians. Thus, we eliminated the Asian population (Chinese and Hong-Kong Chinese) from our meta-analysis. In addition, data in HapMap (http://www.hapmap.org/) demonstrated that T was a minor allele in the Caucasian population, whereas it is a major allele in the Asian population. Different allelic frequencies in different ethnic groups may account for these discrepancies. Heterogeneity may be due to many factors, such as differences in the characteristics of controls, diverse genotyping methods, small sample size, and a mixed population from different geographic regions.
Rs4779584 is located between GREM1 and SCG5. Jaeger et al. [9] were the first to report that GREM1-SCG5 was strongly associated with an increased risk for CRC (for rs4779584, P = 4.44×10−14). Although the functional relevance of GREM1 rs4779584 is not completely understood, studies suggest that GREM1 maps to human chromosome 15q13-q15, specifically at 15q13.3 [21]. Gremlin1 (GREM1) is a bone morphogenetic protein (BMP) antagonist and putative angiogenesis-modulating protein. GREM1 is silenced by promoter hypermethylation in human malignancies [22]. In the colon, GREM1 is one of several BMP antagonists produced by sub-epithelial myofibroblasts. GREM1 binds to and inactivates the ligands BMP2 and BMP4, which are primarily produced by inter-cryptal stromal cells [13]. Gremlin 1 is a signaling component of the TGF-β pathway, which suppresses cellular proliferation and modulates cell invasion, immune regulation, and the tumor microenvironment [23]. It is generally accepted that excessive production and/or activation of TGF-β by tumor cells promotes cancer progression by mechanisms that include increased tumor neoangiogenesis, extracellular matrix production, upregulation of proteases, and inhibition of immune surveillance in the cancer host [24], [25]. The TGF-β/BMP pathway plays an important role in colorectal tumorigenesis [26], [27]. In particular, GREM1 initiates and maintains important developmental and disease-associated activities. Thus, GREM1 may increase tumor proliferation through stromal effects [28]. Although SCG5 is genetically and functionally a less critical candidate than GREM1, neuroendocrine signaling involving SCG5 may influence cellular proliferation in the large bowel based on nutrient availability or systemic hormonal effects [18].
There were some limitations in our meta-analysis. First, only published studies were included in the present meta-analysis. Second, our meta-analysis was based on unadjusted ORs estimates, because not all of the studies reported adjusted ORs. In addition, in cases in which adjusted ORs were presented, the ORs were not adjusted by the same potential confounders, such as ethnicity, age, gender, or geographic distribution. Finally, our meta-analysis focused on one population (the Caucasian population). We excluded the Asian population due to a relatively small sample size and significant heterogeneity.
Conclusion
In summary, this meta-analysis provides reliable evidence that the GREM1-SCG5 rs4779584 polymorphism may be a risk factor for CRC among Caucasian subjects. Moreover, future investigations into the combined effects of genes and the environment may improve current understanding of the associations between GREM1-SCG5 rs4779584 and the risk of developing colorectal cancer. Further work will help clarify the clinical and biological implications of these associations.
Supporting Information
PRISMA 2009 Checklist.
(DOC)
Funding Statement
This work was supported by the National 863 High-Technology Research and Development Program (No. 2012AA02A519). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Stewart BW, Kleihues P(2003) International Agency for Research on Cancer.World Cancer Report. IARC Press: Lyon. [Google Scholar]
- 2. Ferlay J, Parkin DM, Steliarova-Foucher E (2010) Estimates of cancer incidence and mortality in Europe in 2008. Eur J Cancer 46(4): 765–781. [DOI] [PubMed] [Google Scholar]
- 3. Center MM, Jemal A, Ward E (2009) International trends in colorectal cancer incidence rates. Cancer Epidemiol Biomarkers Prev 18(6): 1688–1694. [DOI] [PubMed] [Google Scholar]
- 4. Cui R, Okada Y, Jang SG, Ku JL, Park JG, et al. (2011) Common variant in 6q26-q27 is associated with distal colon cancer in an Asian population. Gut 2011 60(6): 799–805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Wei EK, Giovannucci E, Wu K, Rosner B, Fuchs CS, et al. (2004) Comparison of risk factors for colon and rectal cancer. International Journal of Cancer 108(3): 433–442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Hemminki K, Chen B (2004) Familial Risk for Colorectal Cancers Are Mainly Due to Heritable Causes. Cancer Epidemiol Biomarkers Prev 13: 1253. [PubMed] [Google Scholar]
- 7. Le Marchand L (2009) Genome-wide association studies and colorectal cancer. Surg Oncol Clin N Am 18(4): 663–668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. von Holst S, Picelli S, Edler D, Lenander C, Dalén J, et al. (2010) Association studies on 11 published colorectal cancer risk loci. British Journal of Cancer 103(4): 575–580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Jaeger E, Webb E, Howarth K, Carvajal-Carmona L, Rowan A, et al. (2007) Common genetic variants at the CRAC1 (HMPS) locus on chromosome 15q13.3 influence colorectal cancer risk. Nature Genetics 40(1): 26–28. [DOI] [PubMed] [Google Scholar]
- 10. Middeldorp A, Jagmohan-Changur S, van Eijk R, Tops C, Devilee P, et al. (2009) Enrichment of Low Penetrance Susceptibility Loci in a Dutch Familial Colorectal Cancer Cohort. Cancer Epidemiology Biomarkers & Prevention 18(11): 3062–3067. [DOI] [PubMed] [Google Scholar]
- 11. He J, Wilkens LR, Stram DO, Kolonel LN, Henderson BE, et al. (2010) Generalizability and Epidemiologic Characterization of Eleven Colorectal Cancer GWAS Hits in Multiple Populations. Cancer Epidemiology Biomarkers & Prevention 20(1): 70–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Ho JW, Choi Sc, Lee Yf, Hui TC, Cherny SS, et al. (2010) Replication study of SNP associations for colorectal cancer in Hong Kong Chinese. British Journal of Cancer 104(2): 369–375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tomlinson IPM(2011) Multiple Common Susceptibility Variants near BMP Pathway Loci GREM1, BMP4, and BMP2 Explain Part of the Missing Heritability of Colorectal Cancer. PLoS Genetics 7(6). [DOI] [PMC free article] [PubMed]
- 14. Dunlop MG, Tenesa A, Farrington SM, Ballereau S, Brewster DH, et al. (2013) Cumulative impact of common genetic variants and other risk factors on colorectal cancer risk in 42,103 individuals. Gut 62(6): 871–881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Jia WH, Zhang B, Matsuo K, Shin A, Xiang YB, et al. (2013) Genome-wide association analyses in East Asians identify new susceptibility loci for colorectal cancer. Nat Genet 45(2): 191–196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Xiong F, Wu C, Bi X, Yu D, Huang L, et al. (2010) Risk of Genome-Wide Association Study-Identified Genetic Variants for Colorectal Cancer in a Chinese Population. Cancer Epidemiology Biomarkers & Prevention 19(7): 1855–1861. [DOI] [PubMed] [Google Scholar]
- 17. Derynck R, Akhurst RJ, Balmain A (2001) TGF-beta signaling in tumor suppression and cancer progression. Nat Genet 29(2): 117–129. [DOI] [PubMed] [Google Scholar]
- 18. Seidah NG, Chretien M (1999) Proprotein and prohormone convertases: a family of subtilases generating diverse bioactive polypeptides. Brain Res 848(1–2): 45–62. [DOI] [PubMed] [Google Scholar]
- 19. Higgins JP, Thompson SG (2002) Quantifying heterogeneity in a meta-analysis. Stat Med 21(11): 1539–1558. [DOI] [PubMed] [Google Scholar]
- 20. Zintzaras E, Ioannidis JP (2005) Heterogeneity testing in meta-analysis of genome searches. Genet Epidemiol 28(2): 123–137. [DOI] [PubMed] [Google Scholar]
- 21.Topol LZ(2000) DRM/GREMLIN (CKTSF1B1) maps to human chromosome 15 and is highly expressed in adult and fetal brain. Cytogenet Cell Genet 89. [DOI] [PubMed]
- 22.Vlodrop IJHv(2010) Prognostic Significance of Gremlin1 (GREM1) Promoter CpG Island Hypermethylation in Clear Cell Renal Cell Carcinoma.The American Journal of Pathology 176(2). [DOI] [PMC free article] [PubMed]
- 23. Wordinger RJ, Zode G, Clark AF (2008) Focus on Molecules: Gremlin. Experimental Eye Research 87(2): 78–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Padua D, Massagué J (2009) Roles of TGFβ in metastasis. Cell Research 19(1): 89–102. [DOI] [PubMed] [Google Scholar]
- 25. Biswas S, Guix M, Rinehart C, Dugger TC, Chytil A, et al. (2007) Inhibition of TGF-beta with neutralizing antibodies prevents radiation-induced acceleration of metastatic cancer progression. J Clin Invest 117(5): 1305–1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Hahn SA (1995) Allelotype of Pancreatic Adenocarcinoma Using Xenograft Enrichment. Cancer Research 55: 6. [PubMed] [Google Scholar]
- 27. Levy L, Hill CS (2006) Alterations in components of the TGF-beta superfamily signaling pathways in human cancer. Cytokine Growth Factor Rev 17(1–2): 41–58. [DOI] [PubMed] [Google Scholar]
- 28. Sneddon JB, Zhen HH, Montgomery K, Rijn M, Tward AD, et al. (2006) Bone morphogenetic protein antagonist gremlin 1 is widely expressed by cancer-associated stromal cells and can promote tumor cell proliferation. PNAS103(40): 14842–14847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Houlston RS, Webb E, Broderick P, Pittman AM, Di Bernardo MC, et al. Meta-analysis of genome-wide association data identifies four new susceptibility loci for colorectal cancer. Nature Genetics 40(12): 1426–1435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Talseth-Palmer BA, Brenne IS, Ashton KA, Evans TJ, McPhillips M, et al. (2010) Colorectal cancer susceptibility loci on chromosome 8q23.3 and 11q23.1 as modifiers for disease expression in lynch syndrome. Journal of Medical Genetics 48(4): 279–284. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
PRISMA 2009 Checklist.
(DOC)