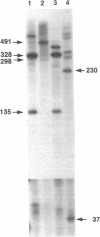
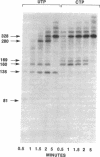
Abstract
The pyrBI operon of Escherichia coli K-12 encodes the subunits of the pyrimidine biosynthetic enzyme aspartate transcarbamylase (carbamoylphosphate:L-aspartate carbamoyltransferase, EC 2.1.3.2). Expression of this operon apparently is negatively regulated by the intracellular levels of UTP. To elucidate the regulatory mechanism in which UTP functions, the nucleotide sequence of the promoter-regulatory region of the pyrBI operon was determined and DNA fragments containing this region were transcribed in vitro. These experiments revealed a rho-independent transcriptional terminator (attenuator) located only 23 base pairs before the promoter-proximal end of the structural genes. Transcription initiated upstream at either of two potential pyrBI promoters was efficiently (approximately equal to 98%) terminated at this site, indicating that the regulation of pyrBI expression involves attenuation control. Additional features identified suggest a model for regulation in which the relative rates of UTP-dependent transcription within the pyrBI leader region and coupled translation of the leader transcript control transcriptional termination at the attenuator.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bachmann B. J., Low K. B. Linkage map of Escherichia coli K-12, edition 6. Microbiol Rev. 1980 Mar;44(1):1–56. doi: 10.1128/mr.44.1.1-56.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birnboim H. C., Doly J. A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 1979 Nov 24;7(6):1513–1523. doi: 10.1093/nar/7.6.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolivar F., Rodriguez R. L., Greene P. J., Betlach M. C., Heyneker H. L., Boyer H. W., Crosa J. H., Falkow S. Construction and characterization of new cloning vehicles. II. A multipurpose cloning system. Gene. 1977;2(2):95–113. [PubMed] [Google Scholar]
- Brutlag D. L., Clayton J., Friedland P., Kedes L. H. SEQ: a nucleotide sequence analysis and recombination system. Nucleic Acids Res. 1982 Jan 11;10(1):279–294. doi: 10.1093/nar/10.1.279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgess R. R., Jendrisak J. J. A procedure for the rapid, large-scall purification of Escherichia coli DNA-dependent RNA polymerase involving Polymin P precipitation and DNA-cellulose chromatography. Biochemistry. 1975 Oct 21;14(21):4634–4638. doi: 10.1021/bi00692a011. [DOI] [PubMed] [Google Scholar]
- Chamberlin M. J., Nierman W. C., Wiggs J., Neff N. A quantitative assay for bacterial RNA polymerases. J Biol Chem. 1979 Oct 25;254(20):10061–10069. [PubMed] [Google Scholar]
- Christie G. E., Farnham P. J., Platt T. Synthetic sites for transcription termination and a functional comparison with tryptophan operon termination sites in vitro. Proc Natl Acad Sci U S A. 1981 Jul;78(7):4180–4184. doi: 10.1073/pnas.78.7.4180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farnham P. J., Platt T. Rho-independent termination: dyad symmetry in DNA causes RNA polymerase to pause during transcription in vitro. Nucleic Acids Res. 1981 Feb 11;9(3):563–577. doi: 10.1093/nar/9.3.563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gigot D., Glansdorff N., Legrain C., Piérard A., Stalon V., Konigsberg W., Caplier I., Strosberg A. D., Hervé G. Comparison of the N-terminal sequences of aspartate and ornithine carbamoyltransferases of Escherichia coli. FEBS Lett. 1977 Sep 1;81(1):28–32. doi: 10.1016/0014-5793(77)80920-4. [DOI] [PubMed] [Google Scholar]
- Gold L., Pribnow D., Schneider T., Shinedling S., Singer B. S., Stormo G. Translational initiation in prokaryotes. Annu Rev Microbiol. 1981;35:365–403. doi: 10.1146/annurev.mi.35.100181.002053. [DOI] [PubMed] [Google Scholar]
- Gonzalez N., Wiggs J., Chamberlin M. J. A simple procedure for resolution of Escherichia coli RNA polymerase holoenzyme from core polymerase. Arch Biochem Biophys. 1977 Aug;182(2):404–408. doi: 10.1016/0003-9861(77)90521-5. [DOI] [PubMed] [Google Scholar]
- Jagusztyn-Krynicka E. K., Smorawinska M., Curtiss R., 3rd Expression of Streptococcus mutans aspartate-semialdehyde dehydrogenase gene cloned into plasmid pBR322. J Gen Microbiol. 1982 May;128(5):1135–1145. doi: 10.1099/00221287-128-5-1135. [DOI] [PubMed] [Google Scholar]
- Jensen K. F. Apparent involvement of purines in the control of expression of Salmonella typhimurium pyr genes: analysis of a leaky guaB mutant resistant to pyrimidine analogs. J Bacteriol. 1979 Jun;138(3):731–738. doi: 10.1128/jb.138.3.731-738.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen K. F., Neuhard J., Schack L. RNA polymerase involvement in the regulation of expression of Salmonella typhimurium pyr genes. Isolation and characterization of a fluorouracil-resistant mutant with high, constitutive expression of the pyrB and pyrE genes due to a mutation in rpoBC. EMBO J. 1982;1(1):69–74. doi: 10.1002/j.1460-2075.1982.tb01126.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston H. M., Barnes W. M., Chumley F. G., Bossi L., Roth J. R. Model for regulation of the histidine operon of Salmonella. Proc Natl Acad Sci U S A. 1980 Jan;77(1):508–512. doi: 10.1073/pnas.77.1.508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Justesen J., Neuhard J. pyrR identical to pyrH in Salmonella typhimurium: control of expression of the pyr genes. J Bacteriol. 1975 Sep;123(3):851–854. doi: 10.1128/jb.123.3.851-854.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kassavetis G. A., Chamberlin M. J. Pausing and termination of transcription within the early region of bacteriophage T7 DNA in vitro. J Biol Chem. 1981 Mar 25;256(6):2777–2786. [PubMed] [Google Scholar]
- Kikuchi A., Gorini L. Similarity of genes argF and argI. Nature. 1975 Aug 21;256(5519):621–624. doi: 10.1038/256621a0. [DOI] [PubMed] [Google Scholar]
- Lee F., Yanofsky C. Transcription termination at the trp operon attenuators of Escherichia coli and Salmonella typhimurium: RNA secondary structure and regulation of termination. Proc Natl Acad Sci U S A. 1977 Oct;74(10):4365–4369. doi: 10.1073/pnas.74.10.4365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maniatis T., Jeffrey A., van deSande H. Chain length determination of small double- and single-stranded DNA molecules by polyacrylamide gel electrophoresis. Biochemistry. 1975 Aug 26;14(17):3787–3794. doi: 10.1021/bi00688a010. [DOI] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- McDonell M. W., Simon M. N., Studier F. W. Analysis of restriction fragments of T7 DNA and determination of molecular weights by electrophoresis in neutral and alkaline gels. J Mol Biol. 1977 Feb 15;110(1):119–146. doi: 10.1016/s0022-2836(77)80102-2. [DOI] [PubMed] [Google Scholar]
- McKenney K., Shimatake H., Court D., Schmeissner U., Brady C., Rosenberg M. A system to study promoter and terminator signals recognized by Escherichia coli RNA polymerase. Gene Amplif Anal. 1981;2:383–415. [PubMed] [Google Scholar]
- Morrison D. A. Transformation and preservation of competent bacterial cells by freezing. Methods Enzymol. 1979;68:326–331. doi: 10.1016/0076-6879(79)68023-0. [DOI] [PubMed] [Google Scholar]
- O'Donovan G. A., Gerhart J. C. Isolation and partial characterization of regulatory mutants of the pyrimidine pathway in Salmonella typhimurium. J Bacteriol. 1972 Mar;109(3):1085–1096. doi: 10.1128/jb.109.3.1085-1096.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Donovan G. A., Neuhard J. Pyrimidine metabolism in microorganisms. Bacteriol Rev. 1970 Sep;34(3):278–343. doi: 10.1128/br.34.3.278-343.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pauza C. D., Karels M. J., Navre M., Schachman H. K. Genes encoding Escherichia coli aspartate transcarbamoylase: the pyrB-pyrI operon. Proc Natl Acad Sci U S A. 1982 Jul;79(13):4020–4024. doi: 10.1073/pnas.79.13.4020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piérard A., Glansdorff N., Gigot D., Crabeel M., Halleux P., Thiry L. Repression of Escherichia coli carbamoylphosphate synthase: relationships with enzyme synthesis in the arginine and pyrimidine pathways. J Bacteriol. 1976 Jul;127(1):291–301. doi: 10.1128/jb.127.1.291-301.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenberg M., Court D. Regulatory sequences involved in the promotion and termination of RNA transcription. Annu Rev Genet. 1979;13:319–353. doi: 10.1146/annurev.ge.13.120179.001535. [DOI] [PubMed] [Google Scholar]
- Salser W. Globin mRNA sequences: analysis of base pairing and evolutionary implications. Cold Spring Harb Symp Quant Biol. 1978;42(Pt 2):985–1002. doi: 10.1101/sqb.1978.042.01.099. [DOI] [PubMed] [Google Scholar]
- Schwartz M., Neuhard J. Control of expression of the pyr genes in Salmonella typhimurium: effects of variations in uridine and cytidine nucleotide pools. J Bacteriol. 1975 Mar;121(3):814–822. doi: 10.1128/jb.121.3.814-822.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Travers A. A. Promoter sequence for stringent control of bacterial ribonucleic acid synthesis. J Bacteriol. 1980 Feb;141(2):973–976. doi: 10.1128/jb.141.2.973-976.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanofsky C. Attenuation in the control of expression of bacterial operons. Nature. 1981 Feb 26;289(5800):751–758. doi: 10.1038/289751a0. [DOI] [PubMed] [Google Scholar]