Abstract
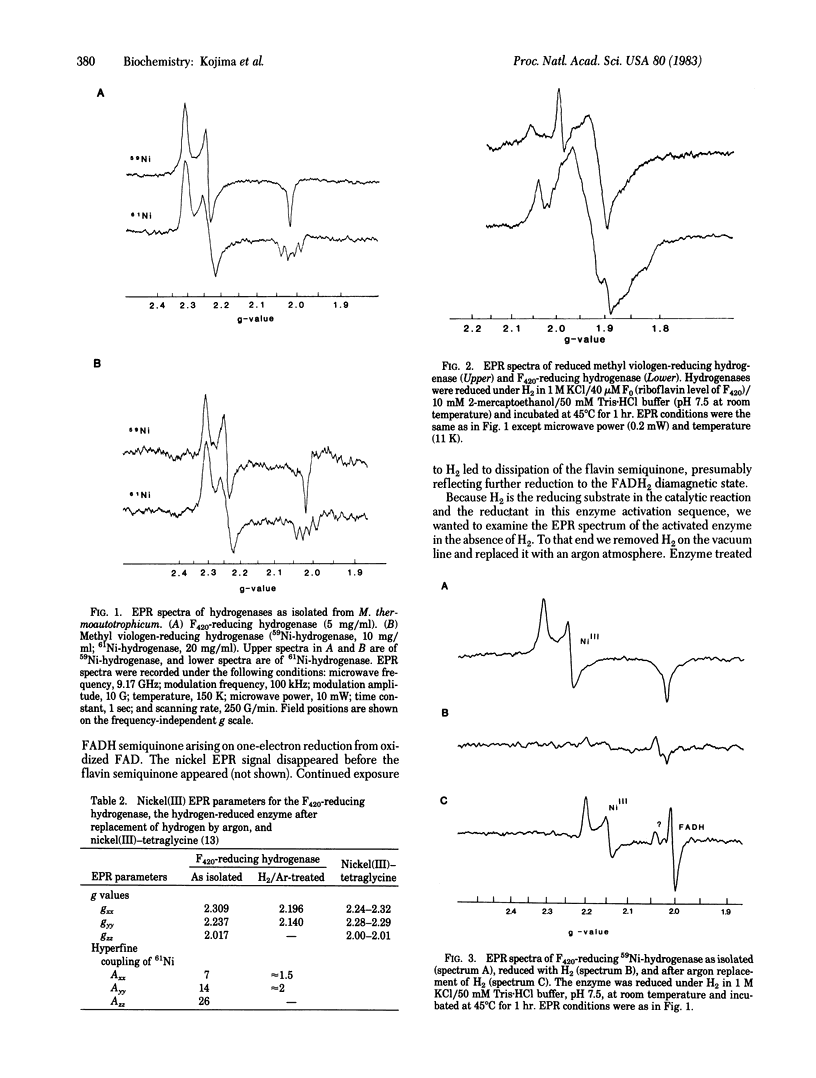
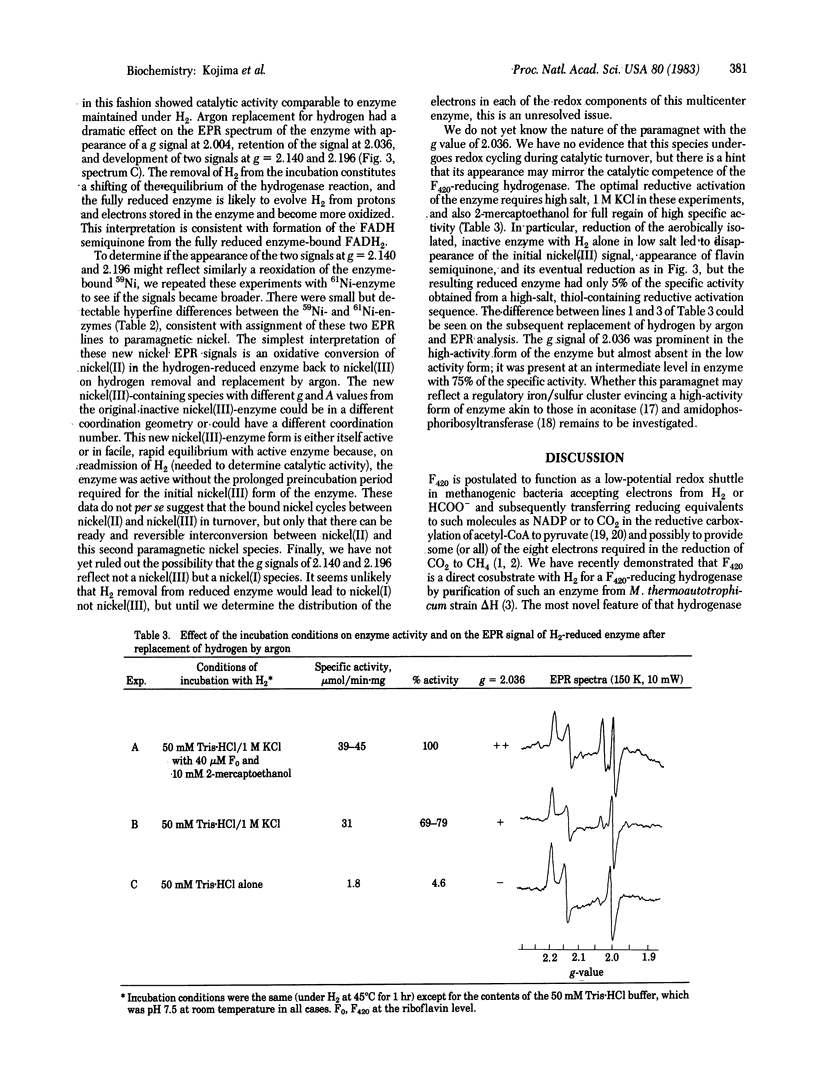
Two hydrogenases from the methanogenic bacterium Methanobacterium thermoautotrophicum strain ΔH have been purified and contain tightly bound nickel as well as the anticipated iron/sulfur atoms with a fixed ratio of 15-20 iron atoms per nickel. One hydrogenase reduces the 8-hydroxy-5-deazaflavin coenzyme factor 420 (F420), whereas the other has been purified as a methyl viologen-reducing hydrogenase. Both enzymes possess an EPR signal attributed to paramagnetic nickel as demonstrated by hyperfine coupling in 61Ni-containing hydrogenases. Comparison to model compounds suggests a nickel(III) oxidation state in the inactive forms of these aerobically purified enzymes. Loss of the nickel(III) signal accompanies reductive activation but is not kinetically correlated with regain of high specific activity. On replacement of H2 by argon in the gas phase over reduced, active, F420-reducing enzyme, several EPR signals appear, including a signal at g = 2.004 that is probably enzyme-bound FADH semiquinone, two signals at g = 2.140 and 2.196 that reflect a new form of paramagnetic nickel(III), and also a signal at g = 2.036 that may be an iron signal. The F420-reducing hydrogenase in the second paramagnetic nickel form is either itself active or in facile equilibrium with active enzyme. The size of the signal at g = 2.036 may correlate with the degree of activation of the enzyme. In contrast to the hydrogenase of Clostridium pasteurianum [Erbes, D. L., Burris, R. H. & Orme-Johnson, W. H. (1975) Proc. Natl. Acad. Sci. USA 72, 4795-4799], which appears to use only iron/sulfur prosthetic groups and which reacts with one-electron-transfer agents, this methanogen hydrogenase seems to utilize iron, nickel, and flavin redox sites and to reduce obligate one-electron (viologen) and two-electron (deazaflavin) oxidants.
Keywords: methanogenesis, archaebacteria, factor 420
Full text
PDF


Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Albracht S. P., Graf E. G., Thauer R. K. The EPR properties of nickel in hydrogenase from Methanobacterium. FEBS Lett. 1982 Apr 19;140(2):311–313. doi: 10.1016/0014-5793(82)80921-6. [DOI] [PubMed] [Google Scholar]
- Averill B. A., Dwivedi A., Debrunner P., Vollmer S. J., Wong J. Y., Switzer R. L. Evidence for a tetranuclear iron-sulfur center in glutamine phosphoribosylpyrophosphate amidotransferase from Bacillus subtilis. J Biol Chem. 1980 Jul 10;255(13):6007–6010. [PubMed] [Google Scholar]
- Diekert G., Klee B., Thauer R. K. Nickel, a component of factor F430 from Methanobacterium thermoautotrophicum. Arch Microbiol. 1980 Jan;124(1):103–106. doi: 10.1007/BF00407036. [DOI] [PubMed] [Google Scholar]
- Dixon N. E., Gazzola T. C., blakeley R. L., Zermer B. Letter: Jack bean urease (EC 3.5.1.5). A metalloenzyme. A simple biological role for nickel? J Am Chem Soc. 1975 Jul 9;97(14):4131–4133. doi: 10.1021/ja00847a045. [DOI] [PubMed] [Google Scholar]
- Drake H. L. Occurrence of nickel in carbon monoxide dehydrogenase from Clostridium pasteurianum and Clostridium thermoaceticum. J Bacteriol. 1982 Feb;149(2):561–566. doi: 10.1128/jb.149.2.561-566.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellefson W. L., Whitman W. B., Wolfe R. S. Nickel-containing factor F430: chromophore of the methylreductase of Methanobacterium. Proc Natl Acad Sci U S A. 1982 Jun;79(12):3707–3710. doi: 10.1073/pnas.79.12.3707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedrich C. G., Schneider K., Friedrich B. Nickel in the catalytically active hydrogenase of Alcaligenes eutrophus. J Bacteriol. 1982 Oct;152(1):42–48. doi: 10.1128/jb.152.1.42-48.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuchs G., Stupperich E., Thauer R. K. Acetate assimilation and the synthesis of alanine, aspartate and glutamate in Methanobacterium thermoautotrophicum. Arch Microbiol. 1978 Apr 27;117(1):61–66. doi: 10.1007/BF00689352. [DOI] [PubMed] [Google Scholar]
- Gunsalus R. P., Wolfe R. S. Methyl coenzyme M reductase from Methanobacterium thermoautotrophicum. Resolution and properties of the components. J Biol Chem. 1980 Mar 10;255(5):1891–1895. [PubMed] [Google Scholar]
- Jacobson F. S., Daniels L., Fox J. A., Walsh C. T., Orme-Johnson W. H. Purification and properties of an 8-hydroxy-5-deazaflavin-reducing hydrogenase from Methanobacterium thermoautotrophicum. J Biol Chem. 1982 Apr 10;257(7):3385–3388. [PubMed] [Google Scholar]
- Kent T. A., Dreyer J. L., Kennedy M. C., Huynh B. H., Emptage M. H., Beinert H., Münck E. Mössbauer studies of beef heart aconitase: evidence for facile interconversions of iron-sulfur clusters. Proc Natl Acad Sci U S A. 1982 Feb;79(4):1096–1100. doi: 10.1073/pnas.79.4.1096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lancaster J. R., Jr New biological paramagnetic center: octahedrally coordinated nickel(III) in the methanogenic bacteria. Science. 1982 Jun 18;216(4552):1324–1325. doi: 10.1126/science.216.4552.1324. [DOI] [PubMed] [Google Scholar]
- LeGall J., Ljungdahl P. O., Moura I., Peck H. D., Jr, Xavier A. V., Moura J. J., Teixera M., Huynh B. H., DerVartanian D. V. The presence of redox-sensitive nickel in the periplasmic hydrogenase from Desulfovibrio gigas. Biochem Biophys Res Commun. 1982 May 31;106(2):610–616. doi: 10.1016/0006-291x(82)91154-8. [DOI] [PubMed] [Google Scholar]
- Orme-Johnson W. H., Orme-Johnson N. R. Overview of iron--sulfur proteins. Methods Enzymol. 1978;53:259–268. doi: 10.1016/s0076-6879(78)53031-0. [DOI] [PubMed] [Google Scholar]
- Whitman W. B., Wolfe R. S. Presence of nickel in factor F430 from Methanobacterium bryantii. Biochem Biophys Res Commun. 1980 Feb 27;92(4):1196–1201. doi: 10.1016/0006-291x(80)90413-1. [DOI] [PubMed] [Google Scholar]
- Zeikus J. G., Fuchs G., Kenealy W., Thauer R. K. Oxidoreductases involved in cell carbon synthesis of Methanobacterium thermoautotrophicum. J Bacteriol. 1977 Nov;132(2):604–613. doi: 10.1128/jb.132.2.604-613.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]



