Abstract
Objective(s): The major antibiotic efflux pump of Esherichia coli is AcrAB-TolC. The first part of the pump, AcrAB, is encoded by acrAB operon. The expression of this operon can be kept elevated by overexpression of an activator, MarA following inactivation of MarR and AcrR repressors due to mutation in encoding genes, marR and acrR, respectively. The aims of this research were to use E. coli mutants with or without mutation in marR to search for the presence of possible mutation in acrR and to quantify the expression of acrAB.
Materials and Methods: The DNA binding region of acrR gene in these mutants were amplified by PCR and sequenced. The relative expression of acrA and acrB were determined by real time PCR.
Results: Results showed that W26 and C14 had the same mutation in acrR, but none of the mutants overexpressed acrA and acrB in comparison with wild type strain.
Conclusions: The effect of marR or acrR mutation on acrAB overexpression is dependent on levels of resistance to tetracycline and ciprofloxacin.
Key Words: acrAB operon, acrR, marR, Multiple antibiotic resistance
Introduction
Generation of multiple drug resistant phenotypes of pathogenic bacteria, like Esherichia coli is a worldwide clinical concern. These phenotypes are associated with increase in the activity of membrane transporters mainly AcrAB-TolC, which belongs to the resistance-nodulation-division (RND) family of transporters (1). This transporter or efflux pump consists of three ingredients, including AcrA, a periplasmic membrane-fusion protein; AcrB, the inner membrane protein; and TolC, an outer membrane channel. These ingredients are encoded by acrA, acrB and tolC. The first two genes are located in the same operon, while tolC is placed on different site of bacterial chromosome (2, 3).
AcrR, the repressor of acrAB operon is encoded by acrR gene (4). Its location is upstream of acrAB operon and transcribed divergently from the same promoter (Figure 1). Attachment of AcrR through its DNA-binding helix-turn-helix (HTH) motif to operator site of acrAB operon causes operon repression (6). On the other hand, this operon is under the positive regulation of MarA, a transcriptional activator (7). Its binding site is shown in Figure 1.
Figure 1.
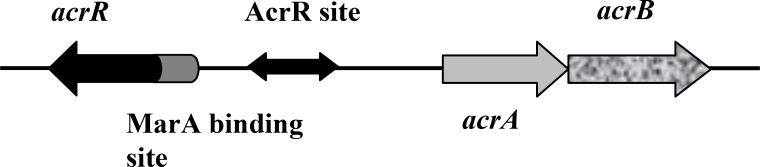
acrAB operon, acrR and the regulatory region between them. Modified and adapted from Dzwokai et al (12)
Expression of MarA happens following dissociation of MarR repressor from operator site of marRAB operon (8). Both AcrR and MarR repressors possess DNA binding and drug binding sites (6, 9). In separate studies, it was shown that mutation in their encoding genes, marR and acrR, can maintain the overactivity of the AcrAB-TolC pump (10, 11). It was found that two clones C14 (without a mutation in marR) and C16 (with a mutation in marR) slightly overexpressed marA (12, 13). This in turn may promote overexpression of acrAB operon. The aims of this research were first, to study the possible presence of mutations in acrR gene and then to quantify the expression of acrA and acrB in these clones.
Materials and Methods
Antimicrobial agent and media
The stock of 4 mg/ml tetracycline hydrochloride (Tc) (Sigma) was used in this research. LB broth (Merck, Germany) and LBA containing 1.5% agar (Merck, Germany) were used for cultivation of control strain and mutants.
Bacterial strain and mutants
Bacterial strain and mutants are listed and described in Table 1. MG1655 was the wild type strain. W26 and W49 are mutants isolated from cultivation of wild type strain on LBA plus 40 ng/ml ciprofloxacin (14). Clones C14 and C16 were generated during the previous work (15). They were derived from mutants W26 and W49 following cultivation on LBA agar containing up to 20 µg/ml Tc (15). Based on the previous data, these clones and mutants show low to medium levels of resistance to ciprofloxacin and tetracycline (16, 17).
Table 1.
Bacterial strain and mutants
| Strain/Mutant/Clone | Relevant properties | MIC |
Source/Reference | |
|---|---|---|---|---|
| Ciprofloxacin (ng/ml) | Tetracycline (µg/ml) | |||
| MG1655 | Wild type | 35 | 3 | A gift from Prof. R. G. Lloyd |
| W26 | Wild type; gyrA (Ser83→Leu) | 75 | 4 | Pourahmad & Mohiti, 2010 |
| W49 | Wild type; gyrA and marOR (20 bp duplication in operator) | 625 | 4 | Pourahmad & Mohiti, 2010 |
| C14 | W26; gyrA (Ser83→Leu) | 1000 | 30 | Pourahmad & Ebadi, 2013 |
| C16 | W49; gyrA and marOR (20 bp duplication in operator) | 1000 | 30 | Pourahmad & Ebadi, 2013 |
PCR amplification and DNA sequencing of acrR
PCR was used to amplify the 5′ end of acrR gene in wild type and mutants (14). A single colony from each mutant and clone on LB agar was suspended in 100 µl of sterile water and after boiling at 95˚C for 3 min; it was cooled on ice and used as a PCR template for acrR amplification. Forward and reverse primers for amplification were 5΄-CACGAACATATGGCACG-3΄ and 5΄-GCCTGATACTCAAGCTC-3΄, respectively. The amplified PCR products were 240 bp. The sequence of these products was compared with that of MG1655 obtained from NCBI (NC_000913.2) following DNA sequencing.
acrA and acrB expression analysis by real time PCR
A fresh culture of bacteria was prepared in LB broth plus 3 µg/ml Tc (except for the wild type) and incubated at 37°C with shaking at 150 rpm and grown to mid-logarithmic phase. Each culture was used for extraction of RNA using an RNeasy Mini Kit (Qiagen, Germany) following stabilization in RNA protect bacterial reagent (Qiagen. Germany). RNase-free DNase I was used to eliminate contaminating genomic DNA according to the manufacturer's instruction (Fermentas, Life science research) and the absence of DNA was confirmed by amplification of RNA samples plus a DNA sample as a positive control. The concentration of total RNA was estimated at OD260 using spectrophotometer (Ultrospec 1100, Amersham Pharmacia Biothech).
Purified total RNA (2 µg) was used as a template in RT-PCR using a RevertAid Reverse Transcriptase kit (Fermentas, Life science research). The cDNAs obtained from reverse transcription were used to quantify the level of acrA, acrB and gapA, as an endogenous reference gene by real time PCR in a Rotor Gene 6000 thermocycler (Corbett Research, Australia) using a SYBR Green kit (Takara, Japan). The specific primers used for real time PCR are listed in Table 2. Thermal cycling conditions were described previously (3). Relative gene expression was calculated using the efficiency method pfaffl (ratio of target gene expression, acrA and acrB, to gapA expression) (18). All data on gene expression are the mean of triplicate analyses. The data were presented as mean±SD. Statistical analysis of relative expression was done by SPSS version 16. T-test was used for comparison of relative gene expression data.
Table 2.
List of real time PCR primers
| Gene | Primer sequence (5′-3′) | Length of amplicon (bp) | Reference |
|---|---|---|---|
| acrA | F: TTGAAATTACGCTTCAGGAT | 189 | Viveiros et al, 2007 |
| R: CTTAGCCCTAACAGGATGTG | |||
| acrB | F: CGTACACAGAAAGTGCTCAA | 183 | Viveiros et al, 2007 |
| R: CGCTTCAACTTTGTTTTCTT | |||
| gapA | F: ACTTACGAGCAGATCAAAGC | 170 | Viveiros et al, 2007 |
| R: AGTTTCACGAAGTTGTCGTT |
Results
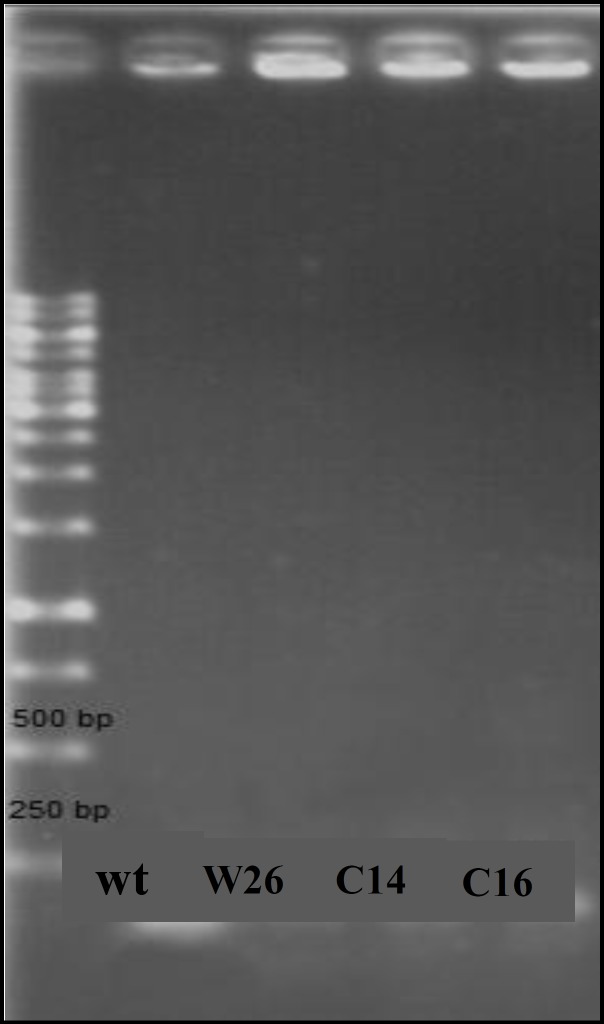
Mutants were used to be evaluated for the presence of possible mutation in 5΄ end of acrR gene corresponding to HTH motif of encoded protein and for quantification of acrAB expression. Figure 2 shows the result of gel electrophoresis of the acrR PCR product of MG1655 and mutants. The comparison of nucleotide sequence of PCR products following DNA sequencing with published sequence of acrR in MG1655 showed that W26 and C14 had the same changes in acrR. Figure 3 shows the comparison of nucleotide sequence of C14 PCR product with that of the wild type. However, other mutants and clones were the same as the wild type. Thus, all mutants and clones had just a single mutation either in marR or acrR.
Figure 2.
PCR products of acrR gene in wild type (wt) and mutants. First lane shows the 1 Kb ladder and other lanes show PCR products
Figure 3.
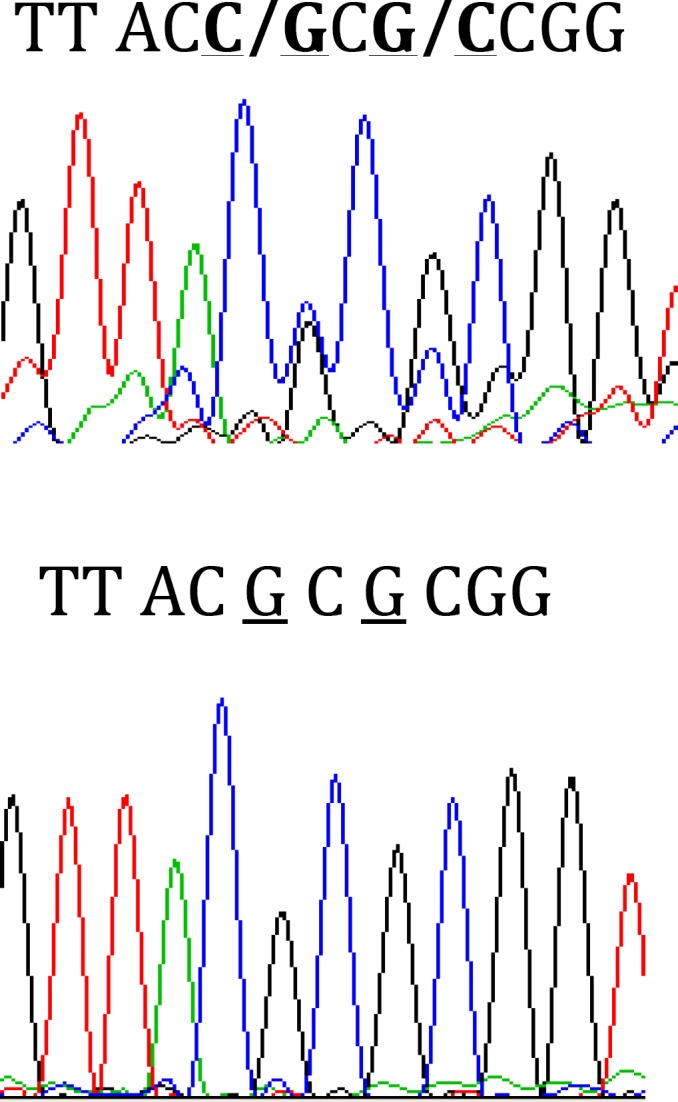
Sequence output from acrR PCR product of C14 mutant (first part) and wild type (second part) using forward and reverse primers. Underlined nucleotides show the differences between nucleotide sequences of two parts
A G/C heterozygote genotype at nucleotide position 131 in coding region of acrR in W26 and C14 would not change amino acid Thr at codon 44 and a G/C heterozygote genotype at position 133 could change Arg (CGC) to Pro (CCC) at codon 45. Substitution of Arg-45 with Cys, but not with Pro was reported previously (10).
Real time PCR results reveal that the efficiency of acrA, acrB and gapA were 1.96, 1.99 and 2.1, respectively. The melting curve of two genes showed just one major peak which indicates the purity of samples. The melting point of three genes was 86-88oC. Table 3 shows the acrA and acrB relative expression in wild type and others. The t-test analysis showed no significant difference among the expression of these genes in the wild type and in mutants and clones (P<0.05). This shows the low induction of acrAB promoter in these bacteria.
Table 3.
Relative expression of acrA and acrB in wild type (MG1655) and mutants as determined by real time PCR
| Strain/mutant | Relative expression of acrA | Relative expression of acrB |
|---|---|---|
| Wild type (MG1655) | 1±0 | 1±0 |
| W26 | 1.16±0.021 | 1.13±0.012 |
| W49 | 1.62±0.01 | 1.45±0.015 |
| C14 | 1.28±0.013 | 1.17±0.011 |
| C16 | 1.4±0.013 | 1.31±0.02 |
Furthermore, as acrA and acrB are in the same operon, it was expected that both of them show almost the same result for the level of expression.
Discussion
The increased level of resistance to fluroroquinolones, such as ciprofloxacin and other structurally unrelated antibiotics, like tetracycline which causes multiple resistance phenotypes is attributed to over activation of multidrug efflux pumps, mainly AcrAB-TolC pump in E. coli (1-3). Overactivity of this pump was seen even in other bacteria following the induction with increasing amounts of tetracycline or acquiring high levels of resistance (19).
Generally, fluroroquinolone resistance has been attributed to point mutations in the quinolone resistance-determining regions of the target genes, such as gyrA (20). However, higher levels of resistance can be achieved following overactivation of AcrAB-TolC pump (21). This happens following the overexpression of marA and thereby overactivation of acrAB and tolC genes. The expression of these genes has been determined in clinical isolates by real time PCR (22-24). Therefore, it was decided to quantify the expression of acrA and acrB genes in mutants and clones with or without a mutation in marR after evaluation of the possibility of acquiring mutation in acrR.
It was found that none of the mutants overexpresses acrA and acrB following the addition of 3 µg/ml Tc. This may be due to the levels of resistance to antibiotics in mutants and clones. This is also possible that the level of marA overexpression in clones was not enough to overexpress acrAB. This possibility arises following the suggestion that the level of marA overexpression is important for activation of acrAB operon (25).
It was shown that Arg at position 45 of AcrR is highly conserved and its alteration to cystein enhances the expression of acrB in mutants with high levels of resistance to ciprofloxacin (10). In the present work, it was found that W26 and its derived clone C14 had alteration at position 45. However, this alteration did not promote overexpression of acrAB. This may imply that mutation at this location is not the only cause of acrAB overexpression. This is consistent with the previous findings indicating that in stress conditions, expression of acrAB enhances independent of AcrR activity. However, after overexpression of acrAB, the presence of active AcrR is important to regulate the levels of acrAB expression (26).
Moreover, the importance of the repressor binding site on DNA, and repressor DNA binding motif for MarR and AcrR repressor were mentioned previously as mutations in these locations along with overactivity of MarA promote overexpression of acrAB operon (10, 11). The finding that mutants and clones harboring mutations in either of these locations in marR or acrR could not promote overexpression of acrA and acrB, again reveals that the level of resistance is important for overexpression of acrAB operon.
In addition, it was shown that tolerance of organic solvants, such as cyclohexane happens following overexpression of acrAB-tolC (27). It was previously found that none of these mutants and clones could tolerate cyclohexane (13, 15). Thus, the findings of this study reconfirm the relation between organicsolvent tolerance and the level of acrAB expression.
Conclusion
Upregulation of acrAB operon occurs after acquisition of high levels of resistance to Tc and ciprofloxacin. Thus, the effect of marR or acrR mutation on acrAB overexpression is dependent on the levels of resistance to these antibiotics. At high levels of resistance, evaluation of the synthesis of AcrAB-TolC pump ingredients along with mRNA quantification by real time PCR would reconfirm AcrAB-TolC overactivity.
Acknowledgment
This work was financially supported by the University of Shahrekord, Iran. We thank Prof R G Lloyd for his kind gift of MG1655.
References
- 1.Piddock LJV. Clinically relevant chromosomally encoded multidrug resistance efflux pumps in bacteria. Clinic Microbiol Rev . 2006;19:382–402. doi: 10.1128/CMR.19.2.382-402.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Grkovic S, Brown MH, Skurray RA. Regulation of bacterial drug export systems. Microbiol Mol Biol Rev. 2002;66:671–701. doi: 10.1128/MMBR.66.4.671-701.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Viveiros M, Dupont M, Rodrigues L, Couto I, Davin-Regli A, Martins M, et al. Antibiotic stress, genetic response and altered permeability of Escherichia coli. PLoS . 2007;4:e365 . doi: 10.1371/journal.pone.0000365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Su CC, Rutherford DJ, Yu EW. Characterization of the multidrug efflux regulator AcrR from Escherichia coli. Biochem. Biophys Res Commun . 2007;361:85–90. doi: 10.1016/j.bbrc.2007.06.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dzwokai M, Alberti M, Lynch C, Nikaido H, Hearst JE. The local repressor AcrR plays a modulating role in the regulation of acrAB genes of Escherichia coli by global stress signals. Mol Microbiol . 1996;19:101–112. doi: 10.1046/j.1365-2958.1996.357881.x. [DOI] [PubMed] [Google Scholar]
- 6.Gu R, Li M, Su CC, Long F, Routh MD, Yang F, et al. Conformational change of the AcrR regulator reveals a possible mechanism of induction. Acta Cryst . 2008;64:584–588. doi: 10.1107/S1744309108016035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rhee S, Martin RG, Rosner JL, Davies DR. A novel DNA-binding motif in MarA: the first structure for an AraC family transcriptional activator. Proc Natl Acad Sci USA . 1998;95:10413–10418. doi: 10.1073/pnas.95.18.10413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Martin RG, Rosner JL. Binding of purified multiple antibiotic resistance repressor protein (MarR) to mar operator sequences. Proc Natl Acad Sci USA. 1995;92:5456–5460. doi: 10.1073/pnas.92.12.5456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Perera IC, Grove A. Molecular mechanisms of ligand-mediated attenuation of DNA binding by MarR family transcriptional regulators. J Mol Cell Biol . 2010;2:243–254. doi: 10.1093/jmcb/mjq021. [DOI] [PubMed] [Google Scholar]
- 10.Webber MA, Talukder A, Piddock LJV. Contribution of mutation at amino acid 45 of AcrR to acrB expression and ciprofloxacin resistance in clinical and veterinary Escherichia coli isolates. Antimicrobial Agents Chemother . 2005;49:4390–4392. doi: 10.1128/AAC.49.10.4390-4392.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Maneewannakul K, Levy SB. Identification of mar mutants among quinolone resistant clinical isolates of Escherichia coli. Antimicrobial Agents Chemother . 1996;40:1695–1698. doi: 10.1128/aac.40.7.1695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pourahmad Jaktaji R, Ebadi R. Expression of marA gene in ciprofloxacin and tetracycline resistant mutants of Esherichia coli. Iran J Pharm Res . 2013 in press. [PMC free article] [PubMed] [Google Scholar]
- 13.Pourahmad Jaktaji R, Ebadi R, Karimi M. Study of organic solvent tolerance and increased antibiotic resistance properties in Escherichia coli gyrA mutants. Iran J Pharm Res . 2011;11:595–600. [PMC free article] [PubMed] [Google Scholar]
- 14.Pourahmad Jaktaji R, Mohiti E. Study of mutations in the DNA gyrase gyrA gene of Escherichia coli. Iran J Pharm Res. 2010;9:43–45. [PMC free article] [PubMed] [Google Scholar]
- 15.Pourahmad Jaktaji R, Ebadi R. Generation of clones with higher resistance to tetracycline and chloramphenicol from ciprofluoxacin resistant Escherichia coli mutants. Iran J Antibiotics . 2013;4:3063–3067. [Google Scholar]
- 16.George AM, Levy SB. Amplifiable resistance to tetracycline, chloramphenicol and other antibiotics in Escherichia coli: involvement of a non-plasmid determined efflux of tetracycline. J Bacteriol . 1983;155:531–540. doi: 10.1128/jb.155.2.531-540.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kishii R, Takei M. Relationship between the expression of OmpF and quinolone resitance in Escherichia coli. J Infect Chemother . 2009;15:361–366. doi: 10.1007/s10156-009-0716-6. [DOI] [PubMed] [Google Scholar]
- 18.Pfaffl MW, Horgan GW, Dempfle L. Relative expression software tool (REST ©) for group wise comparison and statistical analysis of relative expression results in real time PCR. Nucleic Acids Res . 2002;30:1–10. doi: 10.1093/nar/30.9.e36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Piddock LJV, White DG, Gensberg K, Pumbwe L, Griggs DJ. Evidence for an efflux pump mediating multiple antibiotic resistance in Salmonella enteric serovar typhimurium. Antimicrobial Agents Chemother. 2000;44:3118–3121. doi: 10.1128/aac.44.11.3118-3121.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ruiz J. Mechanisms of resistance to quinolones: target alteration, decreased accumulation and DNA gyrase protection. J Antimicrobial Chemother. 2003;51:1109–1117. doi: 10.1093/jac/dkg222. [DOI] [PubMed] [Google Scholar]
- 21.Yasufuku T, Shigemura K, Shirakawa T, Matsumoto M, Nakano Y, Tanaka K, et al. Correlation of overexpression of efflux pump genes with antibiotic resistance in Escherichia coli strains clinically isolated from urinary tract infection patients. J Clin Microbiol . 2011;49:189–194. doi: 10.1128/JCM.00827-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mazzariol A, Tokue Y, Kanegawa TM, Cornaglia G, Nikaido H. High level fluoroquinolone resistant clinical isolates of Escherichia coli overproduce multidrug efflux protein AcrA. Antimicrobial Agents Chemother . 2000;44:3441–3443. doi: 10.1128/aac.44.12.3441-3443.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rand JD, Danby SG, Greenway DL, England RR. Increased expression of the multidrug efflux genes acrAB occurs during slow growth of Escherichia coli. FEMS Microbiol Lett . 2002;207:91–95. doi: 10.1111/j.1574-6968.2002.tb11034.x. [DOI] [PubMed] [Google Scholar]
- 24.Swick MC, Morgan-Linnell SK, Carlson KM, Zechiedrich L. Expression of multidrug efflux pump genes acrAB-tolC, mdfA and norE in Escherichia coli clinical isolates as a function of fluoroquinolone and multidrug resistance. Antimicrobial Agents Chemother . 2011;55:921–924. doi: 10.1128/AAC.00996-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Martin RG, Bartlett ES, Rosner JL, Wall ME. Activation of the E. colimarA/soxS/rob regulon in response to transcriptional activator concentration. J Mol Biol . 2008;380:278–284. doi: 10.1016/j.jmb.2008.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ma D, Alberti M, Lynch C, Nikaido H, Hearst JE. The local repressor AcrR plays a modulating role in the regulation of acrAB genes of Escherichia coli by global stress signals. Mol Microbiol . 1996;19:101–112. doi: 10.1046/j.1365-2958.1996.357881.x. [DOI] [PubMed] [Google Scholar]
- 27.Aono R, Tsukagoshi N, Yamamoto M. Involvement of outer membrane protein TolC a possible member of the mar-sox regulon in maintenance and improvement of organic solvent tolerance of Escherichia coli K-12. J Bacteriol. 1998;180:938–944. doi: 10.1128/jb.180.4.938-944.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]