Abstract
Objective(s): Fascioliasis is a zoonotic parasitic disease caused by liver fluke species of Fasciola hepatica and Fasciola gigantica. Differentiation of these two species, based on their morphological characteristics, is difficult. The current study aimed to use PCR-RFLP assay to distinguish between F. hepatica and F. gigantica, based on profiles of RFLP, produced by effect of endonucleases on ITS2 of the ribosomal DNA genes from these two species.
Materials and Methods: Adult Fasciola spp. were isolated from bile duct of naturally infected animals. The species of Fasciola were confirmed by sequencing the 505 bp region of the ITS2 gene in the isolates. By running the sequences of the samples in NEBcutter, suitable restriction enzymes (MspI and KpnI) were selected. Eight F. gigantica and eighteen F. hepatica samples were evaluated.
Results: While RFLP pattern with MspI produced a profile by which it was difficult to differentiate these two species, KpnI along with MspI, produced a consistent pattern of a 231, 212 and 93 bp fragments in F. hepatica. This pattern was not seen in F. gigantica.
Conclusion: Findings of this study demonstrated that RFLP with KpnI and MspI produce a suitable pattern which simply differentiates F. hepatica from F. gigantica.
Key Words: Differentiation, Fasciola, Restriction enzymes, RFLP
Introduction
Fascioliasis is a zoonotic parasitic disease caused by the liver fluke species of the genus Fasciola. F. hepatica and F. gigantica are two species which infect human and animals. F. hepatica has a worldwide distribution and both species are present in the tropical and subtropical regions of Africa and Asia (1).
Differentiation of these two species, based on their morphological characteristics, is difficult. The differences in the intermediate hosts, control strategies, transmission patterns and also epidemiological characteristics which overlap in some area indicate that the proper differentiation of F. hepatica and F. gigantica infections in human or animals is crucial (2).
Due to the limitations of morphological methods, several molecular approaches, using different molecular targets, have been developed for the differentiation of F. hepatica and F. gigantica (3).
Several DNA-based approaches have been used for differentiation of Fasciola species (4-7). Among them, sequencing of the first and the second internal transcribed spacers (ITS-1 and ITS-2) of ribosomal DNA and mitochondrial DNA (mtDNA) provided reliable genetic markers for species-level identification of Fasciola species (3). ITS-2 sequence is located between the 5.8S and 28S coding regions of rDNA that are highly conserved and have few inter-specific nucleotides useful for genetic characterization and identification in both F. hepatica and F. gigantica (8-9).
Although sequencing of ribosomal or mitochondrial DNA which shows the differences between the nucleotides of the desired gene, offers reliable methods for differentiation of Fasciola species, RFLP-based approaches provide a relatively simple, cost-effective and appropriate method for differential diagnosis of F. hepatica and F. gigantica. In line with this, different restriction enzymes and target genes have been used (2). The current study was performed to examine the utility of a PCR-RFLP assay for differentiation of F. hepatica and F. gigantica, based on RFLP profiles, produced by the effect of endonucleases on ITS2 of the ribosomal DNA genes from these two species.
Materials and Methods
Parasite
Adult Fasciola spp. were isolated from bile duct of naturally infected sheep, goat and cattle at the slaughterhouses from various regions of Kohgiluyeh and Boyer-Ahmad province in Iran, where human cases of fasciolosis has been recently reported (10). Flukes were washed extensively in PBS (37°C) and subsequently fixed in 70% ethanol and maintained at 4°C for several weeks until used.
DNA extraction and polymerase chain reaction (PCR)
For genomic DNA extraction, a portion of the apical and lateral zone of adult flukes was removed and crushed. DNA from the crushed materials was extracted with phenol–chloroform method. Briefly, 500 μl of lysis buffer and 8 μl of proteinase K was added to the sample and incubated overnight at 37°C. Afterward, 100 μl of phenol–chloroform was added and centrifuged at 1000 g for 10 min at 25°C. Top aqueous phase was removed and absolute alcohol was used to precipitate the DNA. Extracted DNA was diluted in double distilled water and preserved at 4°C until used.
Amplification of the DNA was performed as described by Itagaki et al (2005) using a pair primer to amplify a 505 bp region of the ITS2 sequence (11). PCR reaction contained 3 μl of DNA solution, 0.25 μl of Taq DNA polymerase (Cinnagen, Iran), 2.5 μl of 10x PCR buffer, 1 μl of MgCl2, 1 μl of each primers (Forward: 5′-TGTGTCGATGAAGAGCGCAG-3′ and Reverse: 5′-TGGTTAGTTTCTTTTCCTCCGC-3′), 1 μl of dNTPs and 15.5 μl of DDW. The DNA product was sequenced and Fasciola species were identified.
Restriction fragment length polymorphism (RFLP)
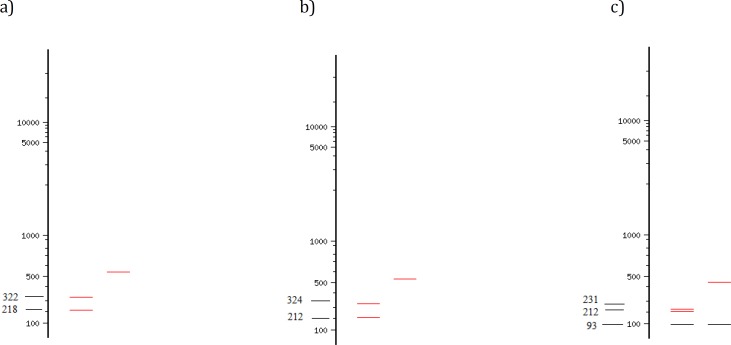
After sequencing, using NEBcutter V2.0 software (12), the cutting sites of commercially available restriction enzymes on ITS2 sequences of F. hepatica and F. gigantica were assessed (Figure 1). MspI and KpnI were selected as the enzymes which might produce the most informative profile. For RFLP, a total volume of 20 μl, including 10 μl of ITS2 PCR product was added with 1 μl of either MspI or KpnI, 4 μl of 10x Tango buffer (Fermentas, Lithuania) and 4 μl of DD-H2O. The tubes were incubated at 37°C for 12 hr, according to the manufacturer instruction to ensure full cutting of fragments. For analyzing the digestion products, 15 μl of each product in addition to 2 μl of loading buffer were run in 2% gel electrophoresis.
Figure 1.
Cutting sites of MspI restriction enzyme on a) Fasciola gigantica, b) F. hepatica, C) cutting sites of KpnI on F. hepatica. F. gigantica has no cutting sites for KpnI
Results
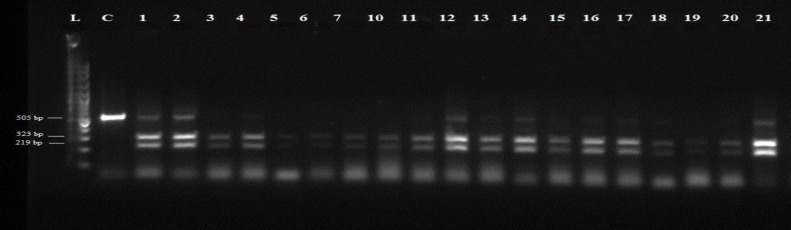
The species of Fasciola were confirmed by sequencing the 505 bp region of the ITS2 gene in the isolates. All amplified products of both F. hepatica and F. gigantica were digested with the MspI restriction endonuclease. Eight F. gigantica and eighteen F. hepatica samples were evaluated. RFLP pattern with MspI produced a 212 and 324 bp fragments for F. hepatica and 218 and 322 bp for F. gigantica (Figure 2). Based on these profiles, it was quite difficult to differentiate these two species. Using KpnI along with MspI (double digestion), a consistent pattern was found where KpnI cut ITS2 fragment of F. hepatica and produced a 231, 212 and 93 bp fragments (Figure 3). This pattern was not seen in F. gigantica (Figure 3).
Figure 2.
RFLP pattern of PCR products of F. hepatica and F. gigantica digested by MspI. L) 100-bp DNA ladder, C) undigested PCR product for control, lane 1, 2, 4, 10, 12, 14, 15 and 19 are F. gigantica; lane 3, 5-7, 11, 13, 16-18, 20 and 21 are F. hepatica
Figure 3.
RFLP pattern of PCR products of F. hepatica and F. gigantica digested by KpnI and MspI. L) 100-bp DNA ladder, C) undigested PCR product for control; lane 1, 2, 6, 7, 8, 11, 15 are F. gigantica and the rest are F. hepatica
Discussion
Differentiation of Fasciola species is crucial for proper implementation of any control measurement about both human and animal fascioliasis (1). Molecular markers based on DNA analysis have been employed for genetic characterization and identification of parasites especially helminthes (13, 14). In the present study, a rapid and simple method of RFLP assay, based on the partial rDNA of ITS2 gene, was utilized for the accurate differentiation and identification of Fasciola species.
Restriction enzymes are powerful and simple approaches toward the characterization of parasite species based on differences in their genomes. These methods have been used for differentiation among Fasciola species based on the profiles generated by the effects of endonucleases on ITS genes of these parasites (4, 5). In a study in China, effect of Hsp92II or RcaI on ITS2 region, Fasciola spp. were differentiated from one another by their unique restriction patterns (8). Isolates of Chinese Fasciola produced a mixture of patterns in two Fasciola species. Huang et al (2004) used the restriction endonucleases, Hsp92II and RcaI, on ITS2 region for molecular differentiation of Fasciola spp. and showed that Hsp92II produces different profiles in different isolates from different hosts. They reported that Hsp92II is better than RcaI for differentiating between these two species (8).
Fasciola species have been traditionally classified based on their morphological features, such as width and body length. Because of the size variations of these two species, the discrepancy of morphological features, and the presence of intermediate forms, it is difficult to distinguish the two species, solely based on these characteristics. RFLP is a powerful approach for discriminating these two species. RFLP pattern have been used for characterization of Fasciola species and also other organisms in Iran (15-18). Karimi (2008) showed that in 18S DNA region, BfrI restriction enzyme produce similar profile for both F. hepatica and F. gigantica whereas DraI generates different patterns for two species of Fasciola (15). Rokni et al (2010) used TasI restriction enzyme for ITS1 region which properly differentiated F. hepatica from F. gigantica (2). In another study, Saki et al (2011) showed that AvaII and DraII restriction enzymes in 28S DNA appropriately differentiate these two species (16). However, Ghavami et al (2009) showed that the pattern of restriction digestion in ITS2 sequence of Fasciola samples which was seen with BamHI and PagI restriction enzymes at the nucleotide positions of 230, 340 and 341 bp are specific to F. hepatica species and has no effect on F. gigantica (17).
Conclusion
In the current study, considering the sequences of ITS2 of F. hepatica and F. gigantica, two endonucleases, MspI and KpnI, were used to differentiate Fasciola spp. For MspI, the RFLP profiles of two species were very similar while for KpnI the profile was quite different and this enzyme was able to cut the ITS2 of two species of Fasciola at different sites which may be utilized for the differentiation of two species. The evaluation of the Fasciola spp. in the areas where two species of the fluke coexist is important and this simple RFLP can be used for discerning these two species.
Acknowledgment
This study was financially supported by the office of Vice-Chancellor for Research of Shiraz University of Medical Sciences, Shiraz, Iran. The results described in this paper were part of PhD student thesis (Reza Shafiei).
References
- 1.Mas-Coma S, Valero MA, Bargues MD. Chapter 2. Fasciola, lymnaeids and human fascioliasis, with a global overview on disease transmission, epidemiology, evolutionary genetics, molecular epidemiology and control. Adv Parasitol . 2009;69:41–146. doi: 10.1016/S0065-308X(09)69002-3. [DOI] [PubMed] [Google Scholar]
- 2.Rokni MB, Mirhendi H, Mizani A, Mohebali M, Sharbatkhori M, Kia EB, et al. Identification and differentiation of Fasciolahepatica and F. gigantica using a simple PCR-restriction enzyme method. Exp Parasitol . 2010;124:209–213. doi: 10.1016/j.exppara.2009.09.015. [DOI] [PubMed] [Google Scholar]
- 3.Ai L, Chen MX, Alasaad S, Elsheikha HM, Li J, Li HL, et al. Genetic characterization, species differentiation and detection of Fasciola spp. by molecular approaches. Parasit Vectors. 2011;4:101. doi: 10.1186/1756-3305-4-101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chaichanasak P, Ichikawa M, Sobhon P, Itagaki T. Identification of Fasciola flukes in Thailand based on their spermatogenesis and nuclear ribosomal DNA, and their intraspecific relationships based on mitochondrial DNA. Parasitol Int . 2012;61:545–459. doi: 10.1016/j.parint.2012.03.009. [DOI] [PubMed] [Google Scholar]
- 5.Dar Y, Amer S, Mercier A, Courtioux B, Dreyfuss G. Molecular identification of Fasciola spp. (Digenea: Fasciolidae) in Egypt. Parasite . 2012;19:177–182. doi: 10.1051/parasite/2012192177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ichikawa M, Itagaki T. Molecular analysis of aspermic Fasciola flukes from Korea on the basis of the nuclear ITS1 region and mitochondrial DNA markers and comparison with Japanese aspermic Fasciola flukes. J Vet Med Sci . 2012;74:899–904. doi: 10.1292/jvms.11-0523. [DOI] [PubMed] [Google Scholar]
- 7.Mahami-Oskouei M, Dalimi A, Forouzandeh-Moghadam M, Rokni M. Molecular identification and differentiation of Fasciola isolates using PCR- RFLP method based on internal transcribed spacer (ITS1, 5.8S rDNA, ITS2) Iran J Parasitol . 2011;6:35–42. [PMC free article] [PubMed] [Google Scholar]
- 8.Huang WY, He B, Wang CR, Zhu XQ. Characterisation of Fasciola species from Mainland China by ITS-2 ribosomal DNA sequence. Vet Parasitol . 2004;120:75–83. doi: 10.1016/j.vetpar.2003.12.006. [DOI] [PubMed] [Google Scholar]
- 9.Choe SE, Nguyen TT, Kang TG, Kweon CH, Kang SW. Genetic analysis of Fasciola isolates from cattle in Korea based on second internal transcribed spacer (ITS-2) sequence of nuclear ribosomal DNA. Parasitol Res . 2011;109:833–839. doi: 10.1007/s00436-011-2323-6. [DOI] [PubMed] [Google Scholar]
- 10.Sarkari B, Ghobakhloo N, Moshfea A, Eilami O. Seroprevalence of human fasciolosis in a new-emerging focus of fasciolosis in Yasuj district, southwest of Iran. Iran J Parasitol . 2012;7:15–20. [PMC free article] [PubMed] [Google Scholar]
- 11.Itagaki T, Kikawa M, Sakaguchi K, Shimo J, Terasaki K, Shibahara T, et al. Genetic characterization of parthenogenic Fasciola sp in Japan on the basis of the sequences of ribosomal and mitochondrial DNA. Parasitology . 2005;131:679–685. doi: 10.1017/S0031182005008292. [DOI] [PubMed] [Google Scholar]
- 12.New England Biolabs, authors. Nebcutter V2. Available at: http://tools.neb.com/NEBcutter2.
- 13.Hwang UW, Kim W. General properties and phylogenetic utilities of nuclear ribosomal DNA and mitochondrial DNA commonly used in molecular systematics. Korean J Parasitol . 1999;37:215–228. doi: 10.3347/kjp.1999.37.4.215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vilas R, Criscione CD, Blouin MS. A comparison between mitochondrial DNA and the ribosomal internal transcribed regions in prospecting for cryptic species of platyhelminth parasites. Parasitology. 2005;131:839–846. doi: 10.1017/S0031182005008437. [DOI] [PubMed] [Google Scholar]
- 15.Karimi A. Genetic diagnosis of Fasciola species based on 18S ribosomal DNA sequences. J Biol Sci . 2008;8:1166–1173. [Google Scholar]
- 16.Saki J, Khademvatan S, Yousefi E. Molecular identification of animal Fasciola isolates in Southwest of Iran. Aust J Basic Appl Sci. 2011;5:1878–1883. [Google Scholar]
- 17.Ghavami MB, Rahimi P, Haniloo A, Mosavinasab SN. Genotypic and phenotypic analysis of Fasciola Isolates. Iran J Parasitol . 2009;4:61–670. [Google Scholar]
- 18.Hossein M, Mirhendi SH, Brandão J, Mirdashti R, Rosado L. Comparison of enzymatic method rapid yeast plus system with RFLP-PCR for identification of isolated yeast from vulvovaginal candidiasis. Iran J Basic Med Sci . 2011;14:443–50. [PMC free article] [PubMed] [Google Scholar]